Abstract
c-Jun NH2-terminal Kinases (JNKs) play a central role in the cellular response to a wide variety of stress signals. After their activation, JNKs induce phosphorylation of substrates, which control proliferation, migration, survival, and differentiation. Recent studies suggest that JNKs may also play a role in cell cycle control, although the underlying mechanisms are largely unexplored. Here we show that JNK directly phosphorylates Cdc25C at serine 168 during G2 phase of the cell cycle. Cdc25C phosphorylation by JNK negatively regulates its phosphatase activity and thereby Cdk1 activation, enabling a timely control of mitosis onset. Unrestrained phosphorylation by JNK, as obtained by a cell cycle-stabilized form of JNK or as seen in some human tumors, results in aberrant cell cycle progression. Additionally, UV irradiation-induced G2/M checkpoint requires inactivation of Cdc25C by JNK phosphorylation. JNK phosphorylation of Cdc25C as well as Cdc25A establishes a novel link between stress signaling and unperturbed cell cycle and checkpoint pathways.
Keywords: CDK (Cyclin-dependent Kinase), Cell Cycle, Checkpoint Control, DNA Damage, JNK, Cdc25C, DNA Damage Checkpoint
Introduction
Mitogen-activated protein kinases (MAPKs)3 are implicated in proliferation, apoptosis, and differentiation. MAPKs are commonly divided into three subgroups: extracellular-regulated kinases (ERKs), p38s, and JNKs. Notably, JNKs phosphorylate key regulatory proteins that control the cellular response to cytokine stimulation, stress factors, and cytotoxic and genotoxic agents that ultimately dictate cell fate -survival or death. Three major JNK isoforms have been characterized in mammals (JNK1, JNK2, and JNK3) that might have distinct physiological functions (1).
The ability of a cell to divide, which translates into its capacity to undergo a regulated cell cycle, is mostly controlled by cyclin-dependent kinases (CDKs) (2). In particular, Cdk1 has emerged as the master regulator of progression through mitosis. Cdk1 activation is a highly regulated event that involves binding to the regulatory/activatory subunit cyclin B together with the removal of inhibitory phosphorylation events occurring in the activation loop of Cdk1 (threonine 14 and tyrosine 15) that are mediated by the Wee1-Myt1 family of kinases. Thus, dephosphorylation of Cdk1 is essential for its activation, and it is accomplished by the family of Cdc25 phosphatases composed of three members: Cdc25A, Cdc25B, and Cdc25C (3, 4).
Growing evidence points to fine-tuning of cell cycle by MAPKs (5). For instance, ERK1/2 were recently found to regulate the activation of Cdc25C during the G2/M transition both in mammals and in Xenopus laevis (6). Moreover, activation of p38α was recently reported during the G2/M transition of the unperturbed cell cycle when it negatively targets Cdc25B (7). In addition, JNKs (and in particular JNK2) have been found to be essential for the proliferation and survival of several cell lines obtained from gastrointestinal and prostate tumors, leukemia, melanoma, and glioblastoma (8–16). Finally, initial studies in murine embryonic fibroblasts (MEFs) from JNK knock-out mice suggested that JNK1 is a negative and JNK2 a positive cell cycle regulator (17, 18), although more recent data generated using “chemical genetic” approaches suggest that both JNK1 and JNK2 are required for cell survival and proliferation (19, 20).
Members of the Cdc25 family also appear to be exquisite targets of the G2/M checkpoint that delays entry into mitosis until DNA is faithfully and completely replicated. First, UV-induced G2/M checkpoint activation induces the phosphorylation and subsequent degradation of Cdc25A (21, 22). Moreover, Chk1 and Chk2 phosphorylates Cdc25B/C and negatively regulates their activity via cytoplasmic sequestration instigated by 14-3-3 proteins (23, 24). Finally, other kinases, such as p38s and MAPKAPK2, are also known to regulate Cdc25B/C localization and action by regulating, via phosphorylation, their interaction with 14-3-3s (25–27).
We have initially reported that stress-induced JNK activation (by anisomycin, sorbitol, or UV) causes phosphorylation of Cdc25C at serine 168 (Ser-168), inhibiting its phosphatase activity (28). However, the functional role of this event remained unclear. Here, we show that phosphorylation of Cdc25C at Ser-168 occurs during both unperturbed cell cycle progression and induction of the G2/M DNA damage checkpoint. Moreover, our data reveal that this phosphorylation is required for proper entry into mitosis and efficient checkpoint arrest after UV irradiation.
EXPERIMENTAL PROCEDURES
Cells and Transfection
Human embryonic kidney 293T (HEK-293T) and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% bovine serum without antibiotics. HFF-1 (diploid human normal skin fibroblasts) cells were purchased from the ATCC and cultured following the manufacturer's recommendations. Freshly isolated primary MEFs, human malignant melanoma A375, and human glioblastoma/astrocytoma U87-MG cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin at 37 °C. Transfections of HeLa and HEK-293T cells were performed with either Lipofectamine with the addition of Plus reagent (Invitrogen), Lipofectamine 2000 (Invitrogen), or FuGENE (Roche Diagnostics) following the manufacturer protocols. HFF-1 cells were nucleofected with the help of an Amaxa Nucleofector following the manufacturer protocols and indications. Efficiency of transfection was estimated by using a green fluorescent protein-encoding vector to be between 80 and 100% in all cases. All experiments performed with MEFs were done with cultures between passages 2 and 5. Cell viability assays were regularly performed in cell lines kept in culture by staining with crystal violet. All cell lines used in this study tested negative for mycoplasma.
Cell Cycle Synchronization
For double-thymidine-block, either HeLa or HFF-1 cells were grown in the presence of thymidine (2 mm) for 18 h and then released into thymidine-free media for 6–8 h and finally grown again for 12 h with thymidine (2 mm) before their final release from G1/S-induced arrest. MEFs were synchronized by serum starvation (0.1% fetal bovine serum) for 48 h. For Fig. 1B, adherent HeLa cells were grown in the presence of thymidine (2 mm) for 18 h and then released into thymidine-free media for 3 h. The cells were then grown in 330 nm nocodazole for 12 h before release into nocodazole-free media. For Fig. 2B, mitotic shake-off fractions of HeLa cells were prepared by mechanical detachment of nocodazole-arrested cells. These were washed in PBS and used to make extracts. In all synchronization experiments, cell cycle position was determined by flow cytometric measurement of nuclear DNA content (using propidium iodide staining) and/or by analysis of common biochemical markers by either immunoblotting or immunokinase assays as indicated.
FIGURE 1.
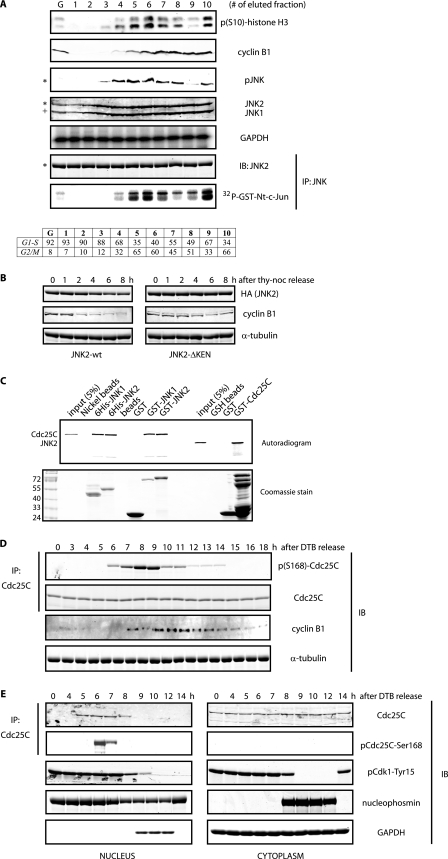
Cell cycle-dependent phosphorylation of Cdc25C by JNK. A, HeLa cells were fractionated by elutriation, and extracts prepared from elutriated cells were analyzed either by immunoblotting (IB) with the indicated antibodies or by immunokinase assays for JNK. G refers to exponentially growing cells, and fractions 1–4 correspond to cells in interphase, whereas fractions 5–10 correspond to cells in mitotic stages, as indicated by the FACS analysis (bottom table). The asterisk refers to the 54-kDa JNK2 isoform, and + refers to the 46-kDa JNK1 isoform present in HeLa cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IP, immunoprecipitation. B, HeLa cells were transfected with hemagglutinin (HA)-tagged either JNK2 wild type or KEN-deleted mutant and arrested in prometaphase by means of a thymidine-nocodazole (noc) protocol and then released for the indicated time points. Levels of hemagglutinin-tagged JNK2 and cyclin B1 were monitored by immunoblotting. Tubulin was used as a loading control. C, in vitro binding assay between JNKs and Cdc25C using bacterially purified recombinant proteins and reticulocyte extracts programmed to produce radiolabeled proteins as indicated. D, HeLa cells were synchronized along the cell cycle by a double-thymidine block (DTB) in G1/S and then released and analyzed by immunoblotting using the indicated antibodies during a 18 h kinetic. E, shown is a similar analysis as in D, but cells were fractionated into nuclear and cytosolic extracts. B and E, images originate from the same gel/membrane.
FIGURE 2.
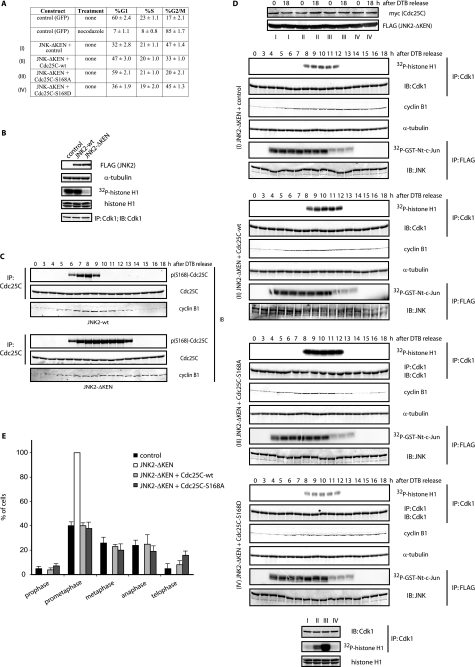
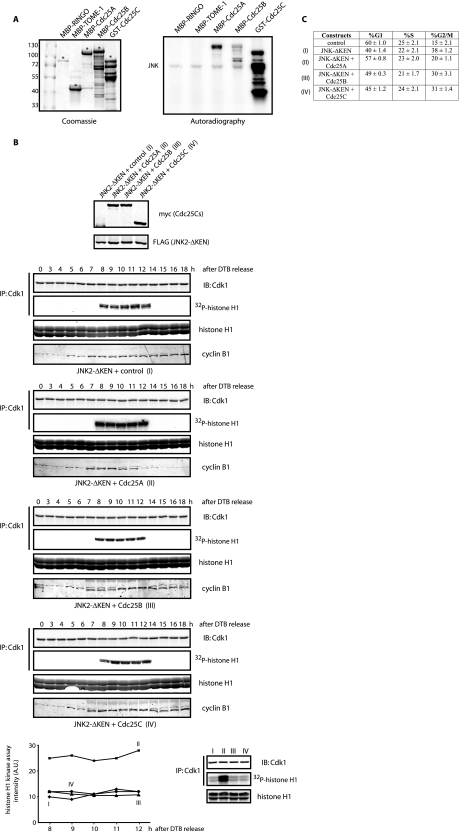
JNK-mediated Cdc25C phosphorylation regulates cell cycle. A, FACS analysis of HFF-1 cells expressing the indicated plasmids for 48 h is shown. The percentage (%) of cells in G1, S, and G2/M is indicated as the mean ± S.D. for three independent experiments. B, HeLa cells were transfected with FLAG-tagged either JNK2-wild type (wt) or KEN-deleted mutant for 24 h. Cell extracts from mitotic cells (obtained by mitotic shake-off) were prepared and analyzed by immunoblotting (IB) using the indicated antibodies and Cdk1 immunokinase assays. IP, immunoprecipitation. C, HeLa cells were transfected with either JNK2 wild type (wt) or KEN-deleted mutant, synchronized by a double-thymidine block (DTB), and released into the cell cycle and analyzed by immunoblotting for cyclin B1, Cdc25C, and phosphorylated Cdc25C at serine 168 levels. D, HFF-1 cells were cell cycle-synchronized by a double-thymidine block and nucleofected with empty plasmid (control), the JNK2-ΔKEN mutant alone or together with wild-type Cdc25C, non-phosphorylatable mutant (S168A), or phosphomimetic mutant (S168D) of Cdc25C, as indicated (conditions I–IV), and analyzed by immunoblotting or immunokinase assays for either Cdk1 (using histone H1 as substrate) or JNK (using a glutathione S-transferase (GST)-tagged N terminal fragment of c-Jun (amino acids 1–89) as substrate). The bottom panels show Cdk1 activity in the four different conditions (I–IV) as assessed in a same gel for comparison. E, HFF-1 cells were nucleofected with the indicated plasmids for 48 h, fixed, and stained for tubulin and DNA, and the percentage of cells in the different mitotic stages was quantified as indicated under “Experimental Procedures.”
Cellular Fractionation and Proliferation Assays
Nuclear/cytosolic separation was performed using HeLa cells and the fractionation kit from BioVision (#K266-25). Briefly, cells were detached from plates with 0.05% trypsin-EDTA and washed with cold PBS. Cells were first lysed with a hypotonic buffer. Centrifugation yielded the cytosolic fraction as the supernatant and the nucleus fraction as the pellet. The pellet was further lysed to yield the nuclear extract. Cell viability/proliferation was determined using the cell proliferation kit I (MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)-based assay), according to the manufacturer's protocols (Roche Applied Science).
Plasmids and Chemicals
The vectors pEF-FLAG-JNK2α2, pEF-FLAG-JNK2α2-ΔKEN, pCDNA3.1-JNK2α2-APF, pX-myc-Cdc25C (and Ser-168 mutants) and their respective empty plasmids (used as controls) were employed for cell transfections. The vectors pET15b-JNK1α1, pET15b-JNK2α2, pGEX4T3-JNK1α1, pGEX-4T3-JNK2α2, pMalc2E-RINGO-ls26_5, pMalc2E-TOME-1, pMalc2E-Cdc25A, pMalc2E-Cdc25B, and pGEX-KG-Cdc25C were used to purify recombinant proteins from Escherichia coli BL21-DE3-pLysS bacteria using standard techniques. All mutations described in this paper were generated by site-directed mutagenesis (QuikChange; Stratagene) and confirmed by sequencing. All chemicals, unless otherwise indicated, were purchased from Sigma. The cell-permeable peptide-based TAT-JIP1 (also known as JNK inhibitor VII) was purchased from Calbiochem (#420134) and used at 20 μm.
Antibodies
The following antibodies were used in this study: cyclin B1 (Santa Cruz, GNS1 clone), Cdc2/Cdk1 (Santa Cruz: sc-54, sc-954, and POH-1 clone), pan-JNK (Santa Cruz, sc-571 and Upstate 06-748), JNK1 (Pharmingen, 554268), JNK2 (Cell Signaling, 4672), phospho-JNK (Cell Signaling, 9255 and 9251; Promega, V793A), glyceraldehyde-3-phosphate dehydrogenase (Abcam, ab9484), phospho-Ser10-histone H3 (Cell Signaling, 9701), phospho-Ser-168-Cdc25C (homemade, see below), Cdc25C (Santa Cruz: sc-6950, sc-5620, and sc-327), proliferating cell nuclear antigen (Abcam, ab15497), phospho-Tyr-15-Cdk1 (Cell Signaling, 9111), nucleophosmin (Genway, NA24 clone), c-Myc (Santa Cruz, 9E10 and C-19 clones), FLAG (Sigma, M2 clone), hemagglutinin (home-made monoclonal antibody), hemagglutinin (Santa Cruz, polyclonal), β-actin (Sigma, monoclonal), α-tubulin (Sigma, monoclonal), Wee1 (Santa Cruz, C-20 clone), Chk1 (Cell Signaling, 2345), and phospho-Ser139-H2A.X (Cell Signaling, 2577).
Generation of Ser-168 Phospho-specific Antibody
The generation of this antibody has been previously reported (28). Briefly, rabbits were immunized with a peptide (YLGpSPITTC) that spans phosphorylated Ser-168 of human Cdc25C coupled to keyhole limpet hemocyanin maleimide through the terminal cysteine residue. Immune antisera were preincubated with unphosphorylated peptide and then purified on a column of phosphorylated peptide. Eluted antibody was used for immunoblotting as indicated after immunoprecipitation of Cdc25C with monoclonal antibodies of mouse origin.
In Vitro Binding Assays
Bacterially expressed and -purified recombinant proteins, either glutathione S-transferase or His6-tagged (1–5 μg), were prebound to 10 μl of glutathione-Sepharose beads (Amersham Biosciences) or nickel-nitrilotriacetic acid-agarose beads (Qiagen), respectively, by incubation for 1 h at 4 °C on a rotating wheel followed by 3 washes with IP buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5% Nonidet P-40, 5 mm EGTA, 5 mm EDTA, 20 mm NaF, 2 mm MgCl2, 100 μm sodium orthovanadate, 2 mm β-glycerophosphate, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 100 μm leupeptin, 4 μg/ml aprotinin). The beads were then mixed with 5–10 μl of reticulocyte lysate (that was programmed to express the protein of interest, labeled with [35S]methionine) in a total volume of around 500 μl of IP buffer. After incubation for 2 h at 4 °C on a rotating wheel, the beads were gently centrifuged, washed 3 times with IP buffer, and resuspended in 25 μl of 2× sample buffer. The beads were finally boiled for 5 min and subjected to SDS-PAGE followed by Coomassie staining of the gels (to verify equal amounts of recombinant proteins in every sample), dried, and analyzed with the help of an FLA-5100 imaging system (FujiFilm). The input lanes of the pulldown experiments typically correspond to 5% of the total radiolabeled sample used in the reaction mixture.
Immunoprecipitation
Cell extracts were prepared in IP buffer. Equal amounts of protein extracts (calculated by the Bradford method using Coomassie Plus protein assay reagent (Thermo)) were incubated with either control IgGs or affinity-purified antibodies (0.5–1 μg) covalently bound to protein A-Sepharose beads. The mixture was incubated in a rotating wheel for 1–2 h at 4 °C. The beads were gently centrifuged, washed three times with IP buffer, and resuspended in 30 μl of 2× sample buffer. The beads were finally boiled for 5 min and subjected to SDS-PAGE and immunoblotting.
Immunoblotting
For Western blots of total cell lysates, cells were detached from plates with 0.05% trypsin-EDTA, washed with cold PBS, and lysed on ice for 20 min after resuspension in IP buffer. After 20 min of centrifugation (13,200 rpm, 4 °C), supernatants were saved, and protein concentration was quantified. Depending on the experiment, either Laemmli SDS- or Anderson-PAGE (mostly in the study of phosphorylated proteins) was utilized. Gels were transferred into nitrocellulose membranes by liquid Western blot transfer performed overnight (8–12 h) at 4 °C. Detection was performed by classical methods using a LiCOR instrument and the Odyssey software.
In Vitro Kinase Assays
For immune-complex kinase assays, whole cell extracts were lysed in 20 mm HEPES (pH 7.5) containing 150 mm NaCl, 5 mm MgCl2, 250 μm EDTA, 1 mm dithiothreitol, 50 μm sodium orthovanadate, 10 mm NaF, 2 μg/ml leupeptin, aprotinin, pepstatin, 1 mm phenylmethylsulfonyl fluoride, and 0.01% Triton X-100. Supernatants from these lysates were mixed with 0.5 μg of anti-JNK, anti-FLAG (for JNK), or anti-Cdk1antibodies and protein-A-Sepharose beads and rotated at 4 °C for 3 h in Eppendorf tubes. The immunoprecipitates were washed twice in wash buffer (20 mm HEPES (pH 7.5), 100 mm NaCl, 5 mm MgCl2, 1 mm EDTA, 0.5 mm dithiothreitol, 0.05% Triton X-100, supplemented with phosphatase and protease inhibitors) and twice with histone H1 kinase buffer (15 mm MgCl2, 20 mm EGTA, 80 mm β-glycerophosphate (pH 7.4), 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride) (29). Washed immune complexes on beads were resuspended in 30 μl of histone H1 kinase buffer together with 10 μCi of [γ-32P]ATP, 40 μm cold ATP (final concentration), and 2 μg of histone H1 protein (for Cdk1 assays) or glutathione S-transferase-N terminus-c-Jun (amino acids 1–89) protein (for JNK assays). After 20 min at 30 °C, the reaction was terminated with the addition of 12 μl of 4× sample buffer and resolved by 12% Laemmli SDS-PAGE, Coomassie-stained, dried, and autoradiographed or analyzed with an FLA-5100 imaging system (FujiFilm). For JNK in vitro kinase assays, 50 ng of bacterially purified recombinant active human JNK2α2 protein (30) were used in the presence of the indicated substrates (2 μg), 10 μCi of [γ-32P]ATP, 40 μm cold ATP, and histone H1 kinase buffer. In Figs. 4E and 5A, JNK kinase activities were measured using total extracts, prepared the same way described above for immunokinase reactions, in the presence of histone H1 kinase buffer and glutathione S-transferase-tagged N terminus c-Jun protein as specific substrate.
FIGURE 4.
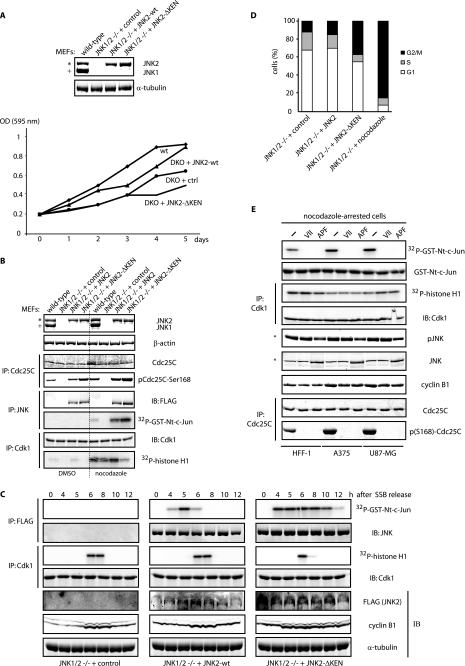
JNK is required for cell division. A, MEFs isolated from double JNK1 and JNK2 knock-out (DKO) mice were transiently reconstituted with either JNK2 wild-type (wt) or JNK2-ΔKEN mutant. Western blotting was performed 96 h after nucleofection. Growth curves for the cultures indicated were performed as described under “Experimental Procedures.” ctrl, control. IP, immunoprecipitate; GST, glutathione S-transferase; OD, optical density. B, shown are Western blot and kinase activation analyses of the cultures described in A in the absence or presence of a 16 h nocodazole treatment. C, MEFs as in A were cell cycle-arrested in G1 by serum starvation blockade (SSB) and released into the cell cycle by the addition of fresh medium and biochemically analyzed during the indicated kinetic by immunoblotting (IB) and immunokinase reactions as shown. Images for the three panels depicted were generated from a same gel/membrane. D, shown is a FACS analysis performed 48 h after nucleofection of the indicated MEFs. E, shown is comparison of cell cycle markers in extracts prepared from three different cell lines (HFF-1, A375, and U87-MG) arrested in G2/M by nocodazole treatment (330 nm nocodazole for 18 h) in the absence or presence of JNK activation. JNK inhibition was accomplished by either treatment with a peptidic cell permeable JNK inhibitor (VII) for 8 h or by previous transfection (24 h) of an untagged JNK2 version mutated at its phosphorylation-dependent activation loop (Thr-183—Pro—Tyr-185 to Ala-Pro-Phe) (APF mutant). In all panels, the asterisk refers to the 54-kDa JNK2 isoform, and + refers to the 46-kDa JNK1 isoform present in these cells.
FIGURE 5.
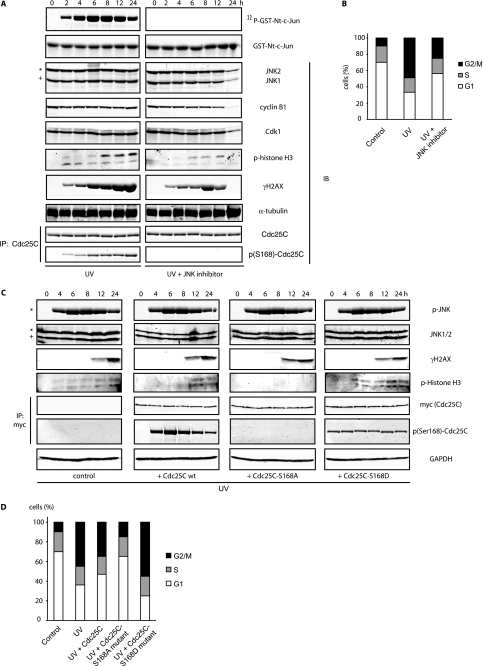
The JNK-Cdc25C axis is a component of the G2/M DNA damage checkpoint pathway. A, HeLa cells treated with UV (40 J/m2) were incubated and analyzed during a 24-h time-course by immunoblotting and JNK kinase reactions (in total extracts) in the absence or presence of a peptide-base JNK inhibitor. IP, immunoprecipitate; GST, glutathione S-transferase; IB, immunoblot. B, FACS profiles were performed and quantified for each condition at 24 h. C, shown is a similar analysis as in A in HeLa cells overexpressing Cdc25C wild type (wt) or Cdc25C mutated forms in serine 168 after 60 J/m2 UV irradiation. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. D, FACS profiles for the different conditions were performed at 24 h. A and C, images from different panels for a particular immunoblot or kinase assay originate from a same gel/membrane. In all panels the asterisk refers to the 54-kDa JNK2 isoform, and + refers to the 46-kDa JNK1 isoform present in HeLa cells.
Phosphatase Assay
Cdc25C phosphatase activity was assayed in a reaction mixture (0.1 ml) containing 10 mm p-nitrophenyl phosphate), 100 mm sodium chloride (pH 7.4), at 37 °C. Assays were initiated with the addition of Cdc25C that was immunoprecipitated from HEK-293T cells using Myc-conjugated agarose beads (9E10 clone from Santa Cruz) and terminated by the addition of 0.4 ml of 0.2 m NaOH. Nonenzymatic hydrolysis of p-nitrophenyl phosphate was corrected by including control assays including Myc beads incubated with extracts from cells without Cdc25C overexpression. The amount of product, p-nitrophenol, produced was calculated from the increase of absorbance at 410 nm using the molar extinction coefficient of 18,000 m−1 cm−1, which was determined with p-nitrophenol standards.
Flow Cytometry (FACS)
Cells were trypsinized and fixed in 70% ice-cold methanol for 20 min, washed in phosphate-buffered saline, and incubated for 30 min at 25 °C in propidium iodide (PI) buffer (10 mm Tris-HCl (pH 7.4), 5 mm MgCl2, 50 μg of PI per ml, and 10 μg of RNase A/ml). The stained cells were acquired by the FACSort flow cytometer (BD Biosciences), and the DNA content was analyzed using FlowJo software.
Immunofluorescence Microscopy
Cells were grown on coverslips and fixed with formaldehyde in PBS for 15 min at 25 °C. After fixation, samples were permeabilized with 0.5% Triton X-100 in PBS for 15 min and thereafter blocked with 10% fetal calf serum in PBS containing 0.01% Triton X-100. Coverslips were incubated for 1 h each at 25 °C with primary and secondary antibodies and mounted with Vectashield medium containing 4′,6-diamidino-2-phenylindole (DNA dye). The mouse monoclonal anti-α-tubulin (clone B-5-1-2, diluted 1:1000; Sigma) was used to detect microtubules dynamics. Cells were manually scored into different mitotic stages by virtue of the relative status of DNA condensation and spindle morphology (by microtubules staining). More than 100 mitotic cells in three individual experiments were photographed and counted in the different stages of mitosis (pro-, prometa-, meta-, ana-, and telo-phase).
RESULTS
JNK Regulates Cdc25C Activation during Entry into Mitosis
Earlier studies from several laboratories have revealed that JNK is activated during unperturbed cell cycle progression (31–34). However, most of these data rely on synchronization protocols based in the use of drugs (such as thymidine or nocodazole) that could indirectly activate JNK. To rule out this possibility, we subjected a large population of HeLa cells to elutriation procedures that allow separation of interphase and mitotic cell populations based on their size and density, which change during cell division (35). Biochemical analyses of elutriated cells using either phospho-specific JNK antibodies (that selectively detect active JNK when phosphorylated on its activation loop) or JNK immunokinase assays confirmed JNK activation in the collected fractions enriched on mitotic cells, as evidenced by the presence of phosphorylated histone H3 at serine 10 and high levels of cyclin B1 (Fig. 1A).
Activation of JNK during unperturbed cell cycle progression suggests that JNK itself might be regulated by the cell cycle machinery. Remarkably, we noticed the presence of a KEN motif in all JNK isoforms so-far identified in humans. KEN motifs are usually present in proteins in which cellular levels are regulated during the cell cycle by proteolytic events instigated by the anaphase-promoting complex or cyclosome (APC/C) ubiquitin ligase, especially when associated with its adaptor/activator/substrate recognition protein Cdh1/fzr (36). In accordance with a role for APC/CCdh1 in controlling JNK protein stability, we found that (i) JNK levels are reduced in cells that were released from a nocodazole-induced arrest in a KEN motif-dependent manner (Fig. 1B), and (ii) endogenous total JNK levels are reduced in cells that are in non-mitotic cell cycle phases (Fig. 1A). Interestingly, expression of a cell cycle-stabilized form of human JNK2α2 (JNK-ΔKEN mutant) in cells negatively affected cell cycle progression and Cdk1 activation during entry into mitosis (see below), indicating that Cdk1 activity was somehow controlled by JNK levels and activity in vivo.
We have reported that Cdc25C, a major direct phosphatase responsible for Cdk1 activation (37), is a bona fide substrate for JNK in cells treated with anisomycin and mapped Ser-168 as a JNK phospho-acceptor site (28). In vitro interaction assays and kinase reactions using both bacterially purified recombinant JNK and Cdc25C revealed JNK interaction with Cdc25C (Fig. 1C) and confirmed that JNK phosphorylates Cdc25C at Ser-168 (supplemental Fig. S1), which inhibits its phosphatase activity (supplemental Fig. S2). Notably, we found that in vivo phosphorylation of endogenous Cdc25C at Ser-168 transiently occurs before mitosis and early mitosis, as evidenced by noticeable accumulation of cyclin B1 protein during unperturbed cell cycle progression (Fig. 1D and supplemental Fig. S3). Interestingly, in vivo phosphorylation of Cdc25C at Ser-168 during G2/M transition (the time when JNK is also activated) was confirmed by analysis of elutriated cells along the cell cycle (supplemental Fig. S4). Consistent with these findings, we found an enrichment of the Ser-168-phosphorylated Cdc25C in the nucleus before Cdk1 activation, as evidenced by Cdc25-mediated dephosphorylation of Cdk1 at tyrosine 15 (Fig. 1E). Altogether, these results suggest that JNK-mediated Cdc25C inhibition through phosphorylation at Ser-168 could avoid premature activation of the phosphatase before mitosis onset and/or timely regulate the degree of activation of Cdc25C during entry into mitosis.
Stability and Activity of JNK Regulate Cell Cycle through Cdc25s
To better characterize the link between JNK and Cdc25C in the regulation of cell cycle progression, we employed the JNK-ΔKEN mutant, whose stability is not regulated during the cell cycle (as shown in Fig. 1B). Interestingly, expression of JNK-ΔKEN caused impaired cell cycle progression in normal diploid fibroblasts, characterized by an accumulation of cells in the G2 and M populations (Fig. 2A) with reduced Cdk1 activation during mitosis (Fig. 2B). We hypothesized that the stabilized JNK-ΔKEN mutant would induce exacerbated phosphorylation of Cdc25C at Ser-168, impairing Cdc25C activation and, therefore, affecting Cdk1 activation. We indeed found enhanced phosphorylation of Cdc25C at Ser-168 in synchronized cells expressing the JNK-ΔKEN mutant when compared with JNK wild type (Fig. 2C). Of note, in cells expressing the JNK-ΔKEN mutant, cyclin B1 degradation was impaired (Fig. 2C). Furthermore, expression of Cdc25C wild type but more so of the Cdc25C-S168A mutant was able to biochemically rescue the gain-of-function phenotype induced by JNK-ΔKEN expression, restoring Cdk1 activation and cyclin B1 fluctuations (Figs. 2D and supplemental Fig. S5A). In agreement, a phosphomimic mutant of Cdc25C (S168D) failed to cause significant rescue of the gain-of-function phenotype seen in JNK-ΔKEN-expressing cells (Figs. 2D and supplemental Fig. S5A). In addition, in similar experiments we found that Cdc25C wild type but more so the Cdc25C-S168A mutant was able to attenuate the degree of G2/M arrest detected by FACS upon expression of JNK-ΔKEN (Fig. 2A and supplemental Fig. S5B). Finally, we also found that expression of Cdc25C counteracted the prometaphase-like arrest induced by JNK-ΔKEN, as detected by immunofluorescence microscopy of cells stained with 4′,6-diamidino-2-phenylindole and tubulin to detect chromosomal and spindle cell cycle dynamics, respectively (Figs. 2E and supplemental Figs. S5C and S6). Interestingly, the fact that Cdc25 wild type only produced a mild rescue of the latter phenotype in HeLa cells suggests that JNK-ΔKEN also affects other targets that directly regulate spindle and/or chromosomal dynamics during progression through mitosis in these cells. Our findings linking JNK to Cdc25C control establish a direct molecular and functional link between JNK and Cdc25C activities during cell cycle progression.
We next tested whether JNK could affect other members of the Cdc25 family of phosphatases, namely Cdc25A and Cdc25B. Significantly, Cdc25A and to a lesser degree Cdc25B, were phosphorylated by JNK in vitro (Fig. 3A). More importantly, expression of wild-type Cdc25A (but not of Cdc25B) in cells rescued normal Cdk1 activation and the G2/M transition arrest induced by JNK-ΔKEN expression (Figs. 3, B and C). Such a rescue, which resembles the effect of the Cdc25C-S168A mutant, provides strong support for the role of JNK in the regulation of these phosphatases and, consequently, the G2/M transition of the cell cycle.
FIGURE 3.
JNK regulates members of the Cdc25 family. A, shown are in vitro phosphorylation assays using active recombinant JNK and the indicated proteins as potential substrates. The asterisk refers to the full-length protein obtained from bacterial purification. B, shown is analysis of cyclin B1 fluctuations and Cdk1 activation (see “Experimental Procedures”) along the cell cycle using synchronized HeLa cells by means of a double-thymidine block (DTB) and release under the expression of the indicated constructs (conditions I–IV). The graph shows quantification of the Cdk1 activity during 8–12 h after release from the DTB. Panels at the bottom show Cdk1 activity in the four conditions (I–IV) as assessed in a same gel. IB, immunoblot; IP, immunoprecipitation. C, shown is FACS analysis of HFF-1 cells expressing the indicated constructs after 24 h. GST, glutathione S-transferase; AU, arbitrary units.
Availability and Activity of JNK Affect Cell Cycle Progression and Cell Survival
Genetic mouse models for JNK have been generated and studied. Some evidence points to change in proliferation among MEFs from JNK1 and JNK2 knock-out mice (38). Using MEFs derived from mice lacking both JNK1 and JNK2 (double-knock-out (DKO)), we have assessed changes in proliferation and phosphorylation of Cdc25C at Ser-168. As shown in Fig. 4A, DKO MEFs displayed defective proliferation compared with their wild-type counterparts. Significantly, such defective proliferation can be partially rescued upon ectopic expression of wild-type JNK but not by the JNK-ΔKEN mutant (Fig. 4A), indicating that a stabilized form of JNK does not properly fulfills the role(s) of JNK in cell cycle progression.
Consistent with the role of JNK in the phosphorylation of Cdc25C described in this study, we found that re-expression of JNK (wild-type or non-degradable mutant) in DKO MEFs is required to ensure phosphorylation of Cdc25C at Ser-168 in exponentially growing cells (Fig. 4B, left lanes). Furthermore, treatment of cells with nocodazole (which drastically enriches the fraction of cells in prometaphase) reveals a strong correlation between the degree of JNK activation (displayed by either wild-type or stabilized JNK), Cdc25C phosphorylation at Ser-168, and repression of Cdk1 activity (Figs. 4B, right lanes, and C). Additionally, in DKO MEFs reconstituted with either wild-type or stabilized JNK and cell cycle-synchronized, we found that JNK-ΔKEN expression (i) significantly impaired cyclin B1 degradation at the exit of mitosis (Fig. 4C) and (ii) induced a robust G2/M transition arrest (Fig. 4D).
To further validate the existence of the JNK-Cdc25C axis, we have assessed changes in cell cycle profiles of tumor cells that harbor constitutive activation of JNK. Earlier reports documented the presence of constitutively active JNK in glioblastoma and melanoma cells (11, 14, 16). Analysis of representative cultures from these tumors confirmed the presence of high levels of JNK activity under non-stressed conditions (data not shown). Remarkably, biochemical analysis of a prometaphase-arrested population of cells from these cultures revealed again a strong correlation between the activity of JNK, the degree of Cdc25C phosphorylation at Ser-168, and the decreased activation of Cdk1 (Fig. 4E). This observation suggests that JNK inhibition could be used as a strategy to correct the rewired cell division cycle displayed by these tumors. However, in glioblastoma cells, JNK inhibition severely affected their viability as well as their particular cell cycle profile (supplemental Fig. S7), indicating that JNK inhibition might have also affected some other substrates.
Role of JNK in the G2 DNA Damage Checkpoint
Given that JNK plays a key role in response to genotoxic stress (1, 39), we tested the possible contribution of JNK to cell cycle control after UV irradiation-induced DNA damage. To this end, changes in several cell cycle regulators were assessed after UV irradiation in the presence or absence of a specific peptidic JNK inhibitor. Notably, inhibition of JNK activity in UV-treated cells led to marked reduction of Cdc25C Ser-168 phosphorylation without affecting overall levels of Cdc25C (Fig. 5A). In addition, we detected decreased levels of cyclin B1 in cells co-treated with UV and the JNK inhibitor, which coincided with impaired mitotic arrest, as reflected by diminished phosphorylation of histone H3 at serine 10 (Fig. 5A) and diminished cell population arrested in the G2/M transition as evidenced by FACS (Fig. 5B).
Remarkably, overexpression of Cdc25C wild type and more so, S168A mutant (but not S168D mutant), rescued the UV-induced G2/M arrest (Figs. 5, C and D). These findings indicate that G2/M arrest (delay in mitosis progression), which is central in the cellular response to DNA damage, is mediated in part by JNK-dependent regulation of Cdc25C activity. These observations further establish that JNK plays a central function in cell cycle control; although important for proper G2/M transition under non-stressed growth conditions, it is required for the G2/M checkpoint after DNA damage. Our results are also consistent with recent observations indicating that inhibition of JNK also reduced phosphorylation of histone H3 (at serine 10) and H2AX (at serine 139) (Fig. 5A and Ref. 32, 40, and 41).
DISCUSSION
Mounting evidence links cell cycle regulation with JNK activity. The present study offers a mechanistic insight into important cell cycle components that are regulated by JNK. Here we demonstrate that JNK phosphorylation of Cdc25C at Ser-168 during G2 phase and early mitosis negatively regulates its phosphatase activity and thereby Cdk1 activation, enabling a timely control of mitosis onset.
Our findings were uncovered in part through the use of the JNK-ΔKEN mutant, a cell cycle-stabilized form of JNK that elicits a gain-of-function phenotype through accumulation of elevated levels of activatable JNK. We show that JNK-ΔKEN preferentially targets a larger population of Cdc25C (and presumably other Cdc25 family members) than the endogenous JNK. This increased regulation might also be explained by the localization of the JNK-ΔKEN mutant that seems to selectively accumulate in the nucleus,4 where activated Cdc25C has been reported to be localized during the G2/M transition (37). Regulation of other members of the Cdc25 family by JNK and other stress-related kinases clearly will become an area for future investigation (42).
The unrestrained phosphorylation by JNK, as obtained by its cell cycle-stabilized form, mimics a constitutive activation of JNK, as often seen in human tumors. Consistent with this idea are the findings that glioblastoma and melanoma tumor cell lines harboring high levels of JNK activity also display increased phosphorylation of Cdc25C at Ser-168. However, JNK hyperactivation is also required for the viability and the (abnormal) proliferation of these tumor cells. These findings suggest that constitutively active JNK, as observed in these tumors, is likely to elicit a gain-of-function phenotype that participates in the cancer-associated aberrant cell cycle and genomic instability features. Therefore, identification of relevant targets for JNK related to cell cycle control will be of exquisite interest for cancer research.
Finally, our data reveal that induction of the G2/M checkpoint after DNA damage, such as under UV irradiation, requires inactivation of Cdc25C by JNK phosphorylation. Consistent with the notion that JNK activity is important for cell cycle progression are the findings that inhibition of JNK activity, either by pharmacological inhibitors, RNAi, or genetic deletions, impairs the G2/M transition and cell cycle progression (12, 43–47). Of note, histone H3 and aurora B were recently suggested as cell cycle regulatory proteins that could be affected by either JNK or the JNK-c-Jun axis, respectively, during the cell cycle (31, 32), raising the possibility that JNK may also affect substrates other than members of the Cdc25 family of phosphatases. Overall, the finding that JNK phosphorylation of Cdc25C is required for this important checkpoint offers new insight into the underlying direct mechanisms linking stress signals to cell cycle control.
Supplementary Material
Acknowledgments
We are grateful to Drs. H. Piwnica-Worms, B. Ducommun, and V. Baldin for providing the Cdc25 plasmids used in this study. We are indebted to M. Chen, E. Lau, and Y. Altman for help with FACS analyses and P. Russell for help with elutriation experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant CA78419 (NCI; to Z. A. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
G. J. Gutierrez and Z. A. Ronai, unpublished results.
- MAPK
- mitogen-activated protein kinase
- JNK
- c-Jun N-terminal kinase
- ERK
- extracellular-regulated kinase
- CDK
- cyclin-dependent kinase
- HEK
- human embryonic kidney
- MEF
- murine embryonic fibroblast
- DKO
- double-knock-out
- PBS
- phosphate-buffered saline
- FACS
- fluorescence-activated cell sorter.
REFERENCES
- 1.Weston C. R., Davis R. J. (2007) Curr. Opin. Cell Biol. 19, 142–149 [DOI] [PubMed] [Google Scholar]
- 2.Malumbres M., Barbacid M. (2005) Trends Biochem. Sci. 30, 630–641 [DOI] [PubMed] [Google Scholar]
- 3.Boutros R., Dozier C., Ducommun B. (2006) Curr. Opin. Cell Biol. 18, 185–191 [DOI] [PubMed] [Google Scholar]
- 4.Boutros R., Lobjois V., Ducommun B. (2007) Nat. Rev. Cancer 7, 495–507 [DOI] [PubMed] [Google Scholar]
- 5.MacCorkle R. A., Tan T. H. (2005) Cell Biochem. Biophys. 43, 451–461 [DOI] [PubMed] [Google Scholar]
- 6.Wang R., He G., Nelman-Gonzalez M., Ashorn C. L., Gallick G. E., Stukenberg P. T., Kirschner M. W., Kuang J. (2007) Cell 128, 1119–1132 [DOI] [PubMed] [Google Scholar]
- 7.Cha H., Wang X., Li H., Fornace A. J., Jr. (2007) J. Biol. Chem. 282, 22984–22992 [DOI] [PubMed] [Google Scholar]
- 8.Xia H. H., He H., De Wang J., Gu Q., Lin M. C., Zou B., Yu L. F., Sun Y. W., Chan A. O., Kung H. F., Wong B. C. (2006) Cancer Lett. 241, 268–274 [DOI] [PubMed] [Google Scholar]
- 9.Yang Y. M., Bost F., Charbono W., Dean N., McKay R., Rhim J. S., Depatie C., Mercola D. (2003) Clin. Cancer Res. 9, 391–401 [PubMed] [Google Scholar]
- 10.Yang S. H., Chien C. M., Chang L. S., Lin S. R. (2007) Toxicon 49, 966–974 [DOI] [PubMed] [Google Scholar]
- 11.Alexaki V. I., Javelaud D., Mauviel A. (2008) Pigment Cell Melanoma Res. 21, 429–438 [DOI] [PubMed] [Google Scholar]
- 12.Potapova O., Gorospe M., Bost F., Dean N. M., Gaarde W. A., Mercola D., Holbrook N. J. (2000) J. Biol. Chem. 275, 24767–24775 [DOI] [PubMed] [Google Scholar]
- 13.Antonyak M. A., Kenyon L. C., Godwin A. K., James D. C., Emlet D. R., Okamoto I., Tnani M., Holgado-Madruga M., Moscatello D. K., Wong A. J. (2002) Oncogene 21, 5038–5046 [DOI] [PubMed] [Google Scholar]
- 14.Cui J., Han S. Y., Wang C., Su W., Harshyne L., Holgado-Madruga M., Wong A. J. (2006) Cancer Res. 66, 10024–10031 [DOI] [PubMed] [Google Scholar]
- 15.Tsuiki H., Tnani M., Okamoto I., Kenyon L. C., Emlet D. R., Holgado-Madruga M., Lanham I. S., Joynes C. J., Vo K. T., Wong A. J. (2003) Cancer Res. 63, 250–255 [PubMed] [Google Scholar]
- 16.Lopez-Bergami P., Huang C., Goydos J. S., Yip D., Bar-Eli M., Herlyn M., Smalley K. S., Mahale A., Eroshkin A., Aaronson S., Ronai Z. (2007) Cancer Cell 11, 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tournier C., Hess P., Yang D. D., Xu J., Turner T. K., Nimnual A., Bar-Sagi D., Jones S. N., Flavell R. A., Davis R. J. (2000) Science 288, 870–874 [DOI] [PubMed] [Google Scholar]
- 18.Sabapathy K., Hochedlinger K., Nam S. Y., Bauer A., Karin M., Wagner E. F. (2004) Mol. Cell 15, 713–725 [DOI] [PubMed] [Google Scholar]
- 19.Jaeschke A., Karasarides M., Ventura J. J., Ehrhardt A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. (2006) Mol. Cell 23, 899–911 [DOI] [PubMed] [Google Scholar]
- 20.Ventura J. J., Hübner A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. (2006) Mol. Cell 21, 701–710 [DOI] [PubMed] [Google Scholar]
- 21.Busino L., Donzelli M., Chiesa M., Guardavaccaro D., Ganoth D., Dorrello N. V., Hershko A., Pagano M., Draetta G. F. (2003) Nature 426, 87–91 [DOI] [PubMed] [Google Scholar]
- 22.Gutierrez G. J., Ronai Z. (2006) Trends Biochem. Sci. 31, 324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng C. Y., Graves P. R., Thoma R. S., Wu Z., Shaw A. S., Piwnica-Worms H. (1997) Science 277, 1501–1505 [DOI] [PubMed] [Google Scholar]
- 24.Sanchez Y., Wong C., Thoma R. S., Richman R., Wu Z., Piwnica-Worms H., Elledge S. J. (1997) Science 277, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 25.Bulavin D. V., Higashimoto Y., Popoff I. J., Gaarde W. A., Basrur V., Potapova O., Appella E., Fornace A. J., Jr. (2001) Nature 411, 102–107 [DOI] [PubMed] [Google Scholar]
- 26.Manke I. A., Nguyen A., Lim D., Stewart M. Q., Elia A. E., Yaffe M. B. (2005) Mol. Cell 17, 37–48 [DOI] [PubMed] [Google Scholar]
- 27.Reinhardt H. C., Yaffe M. B. (2009) Curr. Opin. Cell Biol. 21, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goss V. L., Cross J. V., Ma K., Qian Y., Mola P. W., Templeton D. J. (2003) Cell. Signal. 15, 709–718 [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez G. J., Vögtlin A., Castro A., Ferby I., Salvagiotto G., Ronai Z., Lorca T., Nebreda A. R. (2006) Nat. Cell Biol. 8, 1084–1094 [DOI] [PubMed] [Google Scholar]
- 30.Pimienta G., Ficarro S. B., Gutierrez G. J., Bhoumik A., Peters E. C., Ronai Z., Pascual J. (2007) Cell Cycle 6, 1762–1771 [DOI] [PubMed] [Google Scholar]
- 31.Lee K., Song K. (2008) Cell Cycle 7, 216–221 [DOI] [PubMed] [Google Scholar]
- 32.Oktay K., Buyuk E., Oktem O., Oktay M., Giancotti F. G. (2008) Cell Cycle 7, 533–541 [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez M. E., Makarova O., Peterson E. A., Privette L. M., Petty E. M. (2009) Cell. Signal. 21, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chuang J. Y., Wang Y. T., Yeh S. H., Liu Y. W., Chang W. C., Hung J. J. (2008) Mol. Biol. Cell 19, 1139–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis P. K., Ho A., Dowdy S. F. (2001) BioTechniques 30, 1322–1326, 1328,, 1330–1331 [DOI] [PubMed] [Google Scholar]
- 36.Peters J. M. (2006) Nat. Rev. Mol. Cell Biol. 7, 644–656 [DOI] [PubMed] [Google Scholar]
- 37.Perry J. A., Kornbluth S. (2007) Cell Div. 2, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronai Z. (2004) Mol. Cell 15, 843–844 [DOI] [PubMed] [Google Scholar]
- 39.Karin M., Gallagher E. (2005) IUBMB Life 57, 283–295 [DOI] [PubMed] [Google Scholar]
- 40.Sluss H. K., Davis R. J. (2006) Mol. Cell 23, 152–153 [DOI] [PubMed] [Google Scholar]
- 41.Lu C., Zhu F., Cho Y. Y., Tang F., Zykova T., Ma W. Y., Bode A. M., Dong Z. (2006) Mol. Cell 23, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uchida S., Yoshioka K., Kizu R., Nakagama H., Matsunaga T., Ishizaka Y., Poon R. Y., Yamashita K. (2009) Cancer Res. 69, 6438–6444 [DOI] [PubMed] [Google Scholar]
- 43.MacCorkle R. A., Tan T. H. (2004) J. Biol. Chem. 279, 40112–40121 [DOI] [PubMed] [Google Scholar]
- 44.Moon D. O., Kim M. O., Kang C. H., Lee J. D., Choi Y. H., Kim G. Y. (2009) Exp. Mol. Med. 41, 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mingo-Sion A. M., Marietta P. M., Koller E., Wolf D. M., Van Den Berg C. L. (2004) Oncogene 23, 596–604 [DOI] [PubMed] [Google Scholar]
- 46.Potapova O., Anisimov S. V., Gorospe M., Dougherty R. H., Gaarde W. A., Boheler K. R., Holbrook N. J. (2002) Cancer Res. 62, 3257–3263 [PubMed] [Google Scholar]
- 47.Potapova O., Gorospe M., Dougherty R. H., Dean N. M., Gaarde W. A., Holbrook N. J. (2000) Mol. Cell. Biol. 20, 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.