Abstract
Mechanisms controlling human multipotent mesenchymal (stromal) stem cell (hMSC) differentiation into osteoblasts or adipocytes are poorly understood. We have previously demonstrated that Wnt signaling in hMSC enhanced osteoblast differentiation and inhibited adipogenesis by comparing two hMSC cell lines overexpressing mutated forms of the Wnt co-receptor LRP5: T253I (hMSC-LRP5T253) and T244M (hMSC-LRP5T244) conducting high and low level of Wnt signaling, respectively. To explore the underlying molecular mechanisms, we compared gene expression profiles of hMSC-LRP5T253 and hMSC-LRP5T244 treated with Wnt3a using whole genome expression microarrays and found that TNFRSF19 is differentially up-regulated between the two cells lines. Bioinformatic analysis and dual luciferase assay of its promoter revealed that TNFRSF19 transcript 2 (TNFRSF19.2) is a target of canonical Wnt signaling. Knocking down TNFRSF19 in hMSC-LRP5T253 cells decreased Wnt3a-induced osteoblast differentiation marker alkaline phosphate activity and its overexpression in hMSC-LRP5T244 cells increased alkaline phosphate activity. In addition, TNFRSF19 was negatively regulated by adipogenic transcription factor CCAAT/enhancer-binding proteins (C/EBP). Knocking down TNFRSF19 in hMSC-LRP5T253 cells or its overexpression in hMSC-LRP5T244 cells significantly increased or decreased adipogenesis, respectively. In conclusion, we revealed a novel function of TNFRSF19 as a factor mediating differentiation signals that determine the hMSC differentiating fate into osteoblasts or adipocytes.
Keywords: C/EBP Transcription Factor, Cell Differentiation, Gene Regulation, Stem Cell, Wnt Pathway, Adipogenesis, Human Mesenchymal Stem Cell, Osteogenesis
Introduction
Wnt signaling pathway plays important roles in a variety of cellular activities, including cell fate determination, proliferation, migration, polarity, and gene expression (1). Wnt/β-catenin or the canonical Wnt signaling pathway is initiated by binding of Wnt ligands to their cognate membrane receptors including Frizzled receptor and low-density lipoprotein receptor-related proteins 5 and 6 (LRP5/6)3 leading to formation of a stabilized cytosolic β-catenin, which translocates to the nucleus and binds to T-cell factor/lymphoid enhancer binding factor (TCF/LEF) binding sites in promoters of target genes (2). Identification of both activated and inactivated mutations in LRP5 as the cause of high bone mass phenotype (also known as osteopetrosis type I) or low bone mass (known as osteoporosis pseudoglioma syndrome), respectively (3–7), suggested that Wnt signaling in bone cells is responsible for such altered bone mass phenotypes (8).
A large body of evidence has demonstrated that canonical Wnt signaling is important for bone cell biology. Evidence from animal models with modification of different Wnt components has demonstrated a regulatory function of Wnt signaling in bone development and in postnatal bone homeostasis (9). Wnt signaling is thought to be not only important in osteoblastic cell proliferation, differentiation, synthesis of bone matrix, and osteoclast differentiation, but also in osteocytes transmitting signals of mechanical loading to cells on the bone surface (10, 11). The role of LRP5 as a Wnt co-receptor (12, 13), its involvement in transduction of canonical Wnt signaling (3, 14, 15), the resistance of activated LRP5 mutations to Wnt antagonists SOST or DKK1 inhibition (16–18) or disrupted interaction with LRP5/6 chaperone Mesd (19), as well as the impaired canonical Wnt signaling in inactivated mutations of LRP5 (4), have provided strong evidence for the involvement of LRP5 as a Wnt co-receptor in osteoblastic bone biology.
We have previously reported that patients with activated mutation T253I of LRP5 was responsible for the high bone mass phenotype in a large family in Denmark. Iliac bone biopsies from these patients exhibited increased bone mass and decreased adipose tissue mass in bone marrow suggesting enhanced osteoblastogenesis and inhibition of adipogenesis of human mesenchymal (multipotent stromal) stem cells (hMSC) in bone marrow (15). To establish a model for this human disease, we have established an hMSC cell line overexpressing the LRP5 T253I mutation (named hMSC-LRP5T253) as well as another hMSC cell line overexpressing an inactivated mutation of LRP5 known to cause osteoporosis pseudoglioma syndrome, T244M (named hMSC-LRP5T244). We have also demonstrated that hMSC-LRP5T253 and hMSC-LRP5T244 transmit high and low Wnt signaling, respectively. In addition, in vitro and in vivo studies have demonstrated that hMSC-LRP5T253 cells exhibit enhanced osteogenic differentiation and impaired adipogenic differentiation. The hMSC-LRP5T244 cells exhibited the opposite phenotype (15). These experiments suggested that canonical Wnt signaling mediated through LRP5 targets MSC and determines their differentiation fate into osteoblasts or adipocytes. This notion has been supported by other studies employing Wnt10b as a canonical Wnt ligand (20, 21).
To further identify downstream molecules and/or signaling pathways responsible for cell fate determination of hMSC in this model, we compared global gene expression patterns of hMSC-LRP5T253 and hMSC-LRP5T244 cells treated with Wnt3a. We found that tumor necrosis factor receptor superfamily member 19 (TNFRSF19) is regulated by both canonical Wnt signaling and C/EBP. Modulation of TNFRSF19 expression resulted in significant changes in hMSC differentiation ability to the osteoblastic and adipocytic lineage suggesting an important role as a mediator in regulation of differentiation fate of hMSC.
EXPERIMENTAL PROCEDURES
Cell Culture, Condition Medium, and Compound
hMSC cells (hMSC-LRP5T253 and hMSC-LRP5T244) were cultured in minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum (PAA Laboratories) and 1% penicillin/streptomycin (Invitrogen). The phoenix A amphotropic packaging cells and 293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Control condition medium (CM) and Wnt3a condition medium (Wnt3a-CM) were prepared from L control cells (CRL-2648, ATCC) and L-Wnt3a cells (CRL-2647, ATCC) as described (15).
Protein synthesis inhibitor cycloheximide (CHX, Fluka) was dissolved in dimethyl sulfoxide and stored at −80 °C. To inhibit protein synthesis, 1 μg/ml of CHX was added into culture medium for 8 h.
Microarray Analysis
hMSC-LRP5T253 and hMSC-LRP5T244 cells were treated with 50% control CM or Wnt3a-CM in triplicates for 1 or 24 h and total RNA was prepared by RNeasy mini kit (Qiagen). Synthesis of biotinylated cRNA was performed with 500 ng of total RNA of each sample using the Illumina TotalPrep RNA Amplification Kit (Ambion) according to the manufacturer's instructions. The quantification and quality analysis of total RNA and amplified cRNA were performed on a ND-1000 (NanoDrop Technologies, Wilmington, DE). Size distribution of the extracted total RNA and the amplified cRNA were checked with the Agilent RNA 6000 Nano LabChip kit and 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). HumanRef-8 v2 BeadChips from Illumina (Illumina, San Diego, CA) were used to generate expression profiles of more than 22,000 well annotated RefSeq transcripts following the manufacturer's recommended protocols.
Bioinformatic Analysis
The processing of the microarray expression profiles and identification of differentially expressed genes were performed by Beadstudio software provided by Illumina. We used DAVID (the Data base for Annotation, Visualization and Integrated Discovery) to cluster differentially expressed genes into relevant signaling pathways.
Promoter sequences of selected genes including all of transcripts were retrieved from the Transcriptional Regulatory Element data base (TRED) and DataBase of Transcriptional Start Sites (DBTSS). Promoter analysis was performed using online software TFsitescan and TFSEARCH. Besides, the consensus sequence of the TCF/LEF binding element (TBE, CTTTGWW) was also screened manually.
Construction of Plasmids
To construct human TNFRSF19 promoter-firefly luciferase reporter plasmids, the promoter regions containing 1328 bp for TNFRSF19 transcript 1 (TNFRSF19.1, NM_018647) and 1318 bp for transcript 2 (TNFRSF19.2, NM_148957) were amplified by PCR using PfuUltra II fusion HS DNA polymerase (Stratagene) according to the manufacturer's instructions. The added MluI and NheI restriction site primers used for amplification were: 5′-ATTCACGCGTAACCTCTATCAAGGTGTGACATCG-3′ and 5′-ATTCGCTAGCAGTACTCCGCCCGTGA-3′ for the promoter of TNFRSF19.1 (19.1p), and 5′-ATTCACGCGTCGGAAGAAACAGCCCTAAAAG-3′ and 5′-ATTCGCTAGCTGCGAAAAATGCAGTGAAAGC-3′ for TNFRSF19.2 (19.2p). The obtained PCR products were digested with MluI/NheI (Promega), purified with the SV gel and PCR clean-up system (Promega) and cloned into MluI/NheI-digested promoterless firefly luciferase reporter vector pGL3-Basic (Promega). To construct TNFRSF19 expressing vector, the full-length open reading frame of TNFRSF19.2 was PCR amplified using forward, 5′-AGGAGAAACTAAGTTGCTGAACG-3′, and reverse primers containing the SalI restriction site, 5′-ATTCGTCGACTCAGTCCATAAGCCTAACAAAGG-3′, digested with SalI, purified as above, and cloned into SnaBI/SalI-digested retrovirus vector pBABEpuro.
Site-directed Mutagenesis
Site-directed mutagenesis of TBE by deleting CTTTG was performed by the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The mutations were confirmed by sequencing.
Transfection and Luciferase Assay
293T cells were plated in 24-well plates and transfected with 50 ng of TNFRSF19 promoter-firefly luciferase reporter vector and 5 ng of pRL-TK Renilla luciferase vector as internal control (Promega) by FuGENE 6 (Roche) when approximately 50–70% confluent. In addition, stabilized β-catenin (pCI-neo β-catenin S33Y), wild type TCF4 (pcDNA/Myc TCF4), dominant-negative TCF4 (pcDNA/Myc DeltaN TCF4, dnTCF4) (provided by Dr. Bert Vogelstein, Addgene plasmids 16519, 16512, and 16513), C/EBPα expressing vector CMV-rC/EBP-42 (obtained from Dr. M. Daniel Lane), pcDNA3-C/EBPβ (obtained from Prof. Karsten Kristiansen), or pCMV TAG 3B-C/EBPδ (obtained from Dr. Ez-Zoubir Amri) were also cotransfected for promoter analysis. Topflash firefly reporter vector containing TCF/LEF-binding elements (Upstate) and pGL3-basic vector were used as positive and negative controls, respectively. Luciferase assay was performed 24 h after transfection using the dual luciferase reporter assay system (Promega) following the manufacturer's protocol.
Transfection, Lentivirus Production, and Knockdown of TNFRSF19
To knockdown TNFRSF19, we used the MISSIONTM TRC shRNA target set (TRCN0000058884 or sh19-2, 0000058886 or sh19-4, and TRCN0000058887 or sh19-5) and non-target shRNA control (SHC002 or shCtrl) from Sigma to manufacture lentiviruses for infection. Briefly, 293T cells were seeded to 6-well plates and transfected by FuGENE 6 with 800 ng of lentiviral vector together with 1.6 μg of packaging plasmid psPAX2 (Addgene plasmid 12260) and 800 ng of envelope plasmid pMD2.G (Addgene plasmid 12259). The medium were changed after overnight incubation and collected 24 and 48 h later. For infection, hMSC cells were seeded 2 × 104/cm2 (for Wnt3a-induced osteogenesis) or 4 × 104/cm2 (for adipogenic differentiation) in 12- (for RNA isolation), 24- (for cytochemical staining), or 96-well (for ALP quantitation) plates and infected with lentivirus through a 0.45-μm filter.
In addition, we designed siRNA targeting specifically to TNFRSF19.1 (target sequence 5′-AACACAGCACTGACTTACAGT-3′) and TNFRSF19.2 (target sequence 5′-AACCAAACTGACGGCATTTGA-3′) using siRNAtarget finder. siRNA were transfected by Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions.
Transfection, Retrovirus Production, and Overexpressing of TNFRSF19
For transient overexpressing of TNFRSF19, 3 μg of retrovirus vector pBABEpuro or pBABE-TNFRSF19.2 were transfected into the Phoenix A packaging cell line and retrovirus containing medium were collected for infection as above.
Osteogenic and Adipogenic Differentiation
For osteogenic differentiation, cells were treated with 50% Wnt3a-CM or control CM for 7 days. For adipogenic differentiation, cells were incubated with adipocyte induction medium (AIM) containing 10% fetal bovine serum, 10% horse serum, 10 nm dexamethasone, 450 μm 1-methyl-3-isobutylxanthine (Sigma), 1 μm rosiglitazone (BRL49653, kindly provided by Novo Nordisk, Bagsvaerd, Denmark), and 3 μg/ml of human recombinant insulin (Sigma) for 7 days. The medium was renewed every 3 days.
Real-time RT-PCR
Real-time RT-PCR analysis of osteogenic marker alkaline phosphatase (ALP) and adipogenic markers including peroxisome proliferators-activator receptor γ (PPARγ2), adipocyte-specific fatty acid-binding protein (aP2), adiponectin (APM1), and lipoprotein lipase were performed using fast SYBR® Green master mix (Applied Biosystem) on a StepOnePlusTM system (Applied Biosystem) according to the manufacturer's protocol. RNA isolation, cDNA synthesis, primers for ALP, adipogenic markers, and GAPDH and data analysis were as described previously (15). Special primers used in this study are: TNFRSF19.1, 5′-CCAGGTGGCTGGGAAGA-3′ and 5′-CACATTCCTTAGACAACTCCATGC-3′; endogenous TNFRSF19.2, 5′-CATTTCATCTCCCTGCTCG-3′ and 5′-GCCACATTCCTTAGACAACTCC-3′; C/EBPα, 5′-CACGAAGCACGATCAGTCC-3′ and 5′-CATTGCACAAGGCACTGC-3′; C/EBPβ, 5′-CAAACCAACCGCACATGC-3′ and 5′-AACCGATTGCATCAACTTCG-3′; C/EBPδ, 5′-GGAGAGACTCAGCAACGACC-3′ and 5′-CAGTTTAGTGGTGGTAAGTCCAGG-3′. For detecting expression of ectopic TNFRSF19.2, the forward primer for TNFRSF19.2 cloning and reverse primer for endogenous TNFRSF19.2 amplification were used.
ALP Quantitation
ALP activity was measured by using p-nitrophenyl phosphate (Fluka) as substrate and normalized against the cell number. Briefly, cell number was quantitated by adding CellTiter-Blue reagent (Promega) to culture medium, incubating at 37 °C for 1 h, and measuring fluorescent intensity (560EX/590EM). Cells were then washed with phosphate-buffered saline and Tris-buffered saline, fixed in 3.7% formaldehyde, 90% ethanol for 30 s at room temperature, incubated with substrate (1 mg/ml of p-nitrophenyl phosphate in 50 mm NaHCO3, pH 9.6, and 1 mm MgCl2) at 37 °C for 20 min, and the absorbance measured at 405 nm (22).
Cytochemical Staining
Oil-red O staining were performed 7 days after differentiation as described (15).
Statistical Analysis
Statistical testing was determined by Student's t test and p < 0.05 was considered significant.
RESULTS
TNFRSF19 Is Up-regulated by Canonical Wnt Signaling
To explore the mechanisms underlying cellular phenotypes of hMSC-LRP5T253 and hMSC-LRP5T244 cells, we treated the cells with control CM or Wnt3a-CM for 1 or 24 h and compared their global gene expression profiles by Illumina® microarray system. Upon Wnt3a stimulation for 1 h, the gene expression profiles of hMSC-LRP5T253 and hMSC-LRP5T244 cells did not exhibit significant changes (data not shown). However, upon Wnt3a stimulation of hMSC-LRP5T244 cells for 24 h, 131 Illumina target IDs representing 124 non-redundant genes were changed at least 2-fold, 73 of were up-regulated and 51 were down-regulated (supplemental Table S1). In hMSC-LRP5T253, 176 Illumina target IDs representing 167 non-redundant genes were changed 2-fold including 94 up-regulated and 73 down-regulated non-redundant genes (supplemental Table S2). By clustering significantly regulated genes into KEGG signaling pathways using the online DAVID program, we found similar signaling pathways affected upon Wnt3a stimulation in both cell lines including cytokine-cytokine receptor interaction, transforming growth factor-β signaling, and the Wnt signaling pathway (supplemental Tables S1 and S2). The microarray data reported here was deposited in Gene Expression Omnibus (GEO) with accession number GSE16186.
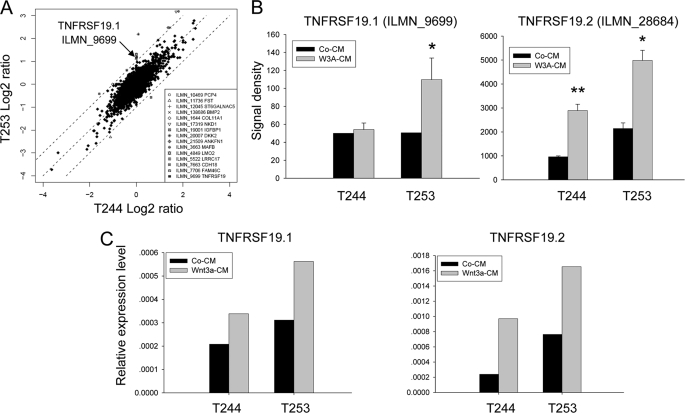
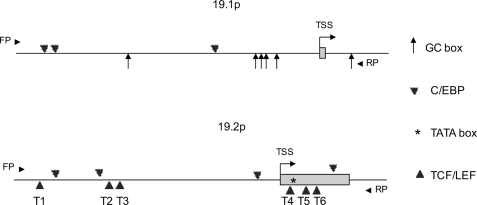
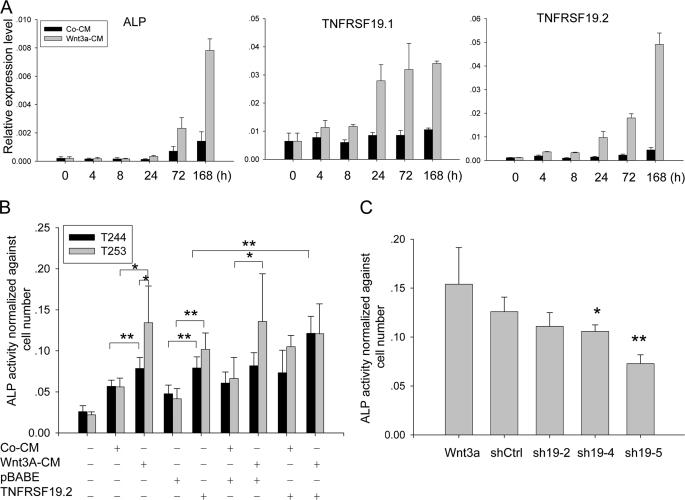
To detect Wnt target genes, we identified genes differentially changed upon Wnt3a treatment in hMSC-LRP5T253 and hMSC-LRP5T244. As shown in Fig. 1A, the x and y axis represent fold-induction (log2 ratio) of each Illumina target ID by Wnt3a in hMSC-LRP5T244 and hMSC-LRP5T253, respectively. Most of genes were changed in a similar direction, except there were 15 genes including BMP2, DKK2, NKD1, and TNFRSF19 (here it is TNFRSF19.1) at least 2-fold differentially up- or down-regulated between the two cell lines (Fig. 1A, inset). To further identify novel canonical Wnt target genes, we retrieved the promoter sequences from all of transcript variants of these 15 genes as well as from 21 genes up-regulated at least 3-fold by Wnt3a in hMSC-LRP5T253 (supplemental Table S2) and searched for TBE (CTTTGWW) within 1 kb upstream and 200 bp downstream of transcription start site defined by the reference mRNA sequence in GenBankTM. One of the genes identified was TNFRSF19 that has two different transcripts: TNFRSF19.1 and TNFRSF19.2. We found that the promoter of TNFRSF19.2 (19.2p) but not the promoter of TNFRSF19.1 (19.1p) has six canonical TBE within the defined promoter region (Fig. 2). The microarray data demonstrated that the TNFRSF19.2 expression level was higher in hMSC-LRP5T253 cells than in hMSC-LRP5T244 under basal conditions (signal density 2141 versus 960) and after Wnt3a stimulation (signal density 4985 versus 2891) (Fig. 1B). Interestingly, TNFRSF19.1 was differentially up-regulated by Wnt3a in hMSC-LRP5T253 (2.27-fold) and exhibited no change in hMSC-LRP5T244 cells (Fig. 1, A and B) but its promoter (19.1p) does not contain any TBE (Fig. 2). The expression patterns of TNFRSF19.1 and TNFRSF19.2 were examined by real-time RT-PCR analysis (Fig. 1C). TNFRSF19.2 was up-regulated by Wnt3a in both hMSC-LRP5T244 and hMSC-LRP5T253 cells (2.5- versus 2.3-fold, respectively). However, the expression level in hMSC-LRP5T253 cells was higher than hMSC-LRP5T244 as observed in microarray data. The up-regulation of TNFRSF19.1 by Wnt3a was also confirmed by real-time RT-PCR (Fig. 1C).
FIGURE 1.
TNFRSF19 is differentially up-regulated by Wnt3a between hMSC-LRP5T244 (T244) and hMSC-LRP5T253 (T253) cells. T244 and T253 cells were treated with 50% control condition medium (Co-CM) or Wnt3a condition medium (Wnt3a-CM) for 24 h and their gene expression profiles were compared by the Illumina microarray system and confirmed by real-time RT-PCR. A, global gene expression differences between T244 and T253 cells. x and y axes represent fold-induction (log2 ratio) after Wnt3a treatment compared with control CM in T244 and T253 cells, respectively. Genes differentially changed at least 2-fold between T244 and T253 were listed and the arrow indicates that TNFRSF19 transcript 1 (TNFRSF19.1, Illumina target ID ILMN_9699) is up-regulated 2-fold higher in T253 cells than in T244 cells. B, microarray signal density of TNFRSF19.1 and TNFRSF19 transcript 2 (TNFRSF19.2, Illumina target ID ILMN_28684) in T244 and T253 cells. Results shown are mean ± S.D. of three replicates. *, p < 0.05; **, p < 0.01. C, up-regulation of TNFRSF19 transcripts by Wnt3a was confirmed by real-time RT-PCR and normalized to GAPDH.
FIGURE 2.
Schematic representation of TNFRSF19 promoters for both transcripts. Promoter sequences were retrieved from the Transcriptional Regulatory Element Data base and analyzed by TFsitescan and TFSEARCH. FP and RP represent forward primer and reverse primer employed for promoter cloning. TSS represents the transcription start site defined by RefSeq and the first exon was indicated as box. The promoter of TNFRSF19 transcript 1 (19.1p) contains six GC box and three C/EBP binding sites within the −1149 to +178 bp region (upper). The promoter of TNFRSF19 transcript 2 (19.2p) contains a TATA box in the first exon, six canonical TCF/LEF binding sites (CTTTGWW, T1 to T6), and four C/EBP binding sites within the −975 to 342 bp region (bottom).
Promoters of TNFRSF19 Transcripts Have Different Compositions
In depth bioinformatic analysis of 19.1p and 19.2p promoters revealed that both promoters have different cis-regulatory elements. As shown in Fig. 2, 19.1p does not contain any TBE but have three C/EBP binding sites (−1048 to −1035 bp, −1010 to −998 bp, and −408 to −395 bp), which bind key adipogenic transcription factors C/EBPs. Besides, 19.1p is a TATA-less promoter with higher G + C content (64%, but 42% in 19.2p) and six GC boxes (−731 to −725 bp, −254 to −248 bp, −237 to −231 bp, −207 to −201 bp, −164 to −158 bp, and +153 to +159 bp), which are typical features of a housekeeping gene promoter (23). The 19.2p contains six TBE (−916 to −910 bp, −656 to −650 bp, −612 to −606 bp, +33 to +39 bp, +70 to 76 bp, and +134 to 140 bp) including three TBE located in the first exon, four C/EBP binding sites (−840 to −827 bp, −687 to −675 bp, −68 to −55 bp, and +202 to 215 bp), and one TATA box located in the first exon (+51 to +57 bp) (Fig. 2). Existence of TBE and C/EBP binding sites in 19.1p and 19.2p suggested TNFRSF19 may be involved in transmitting different signals to cell differentiation to osteoblasts or adipocytes. Based on these observations, we further examined the role of TNFRSF19 in hMSC differentiation.
TNFRSF19.2 but Not TNFRSF19.1 Is a Canonical Wnt Target Transcript
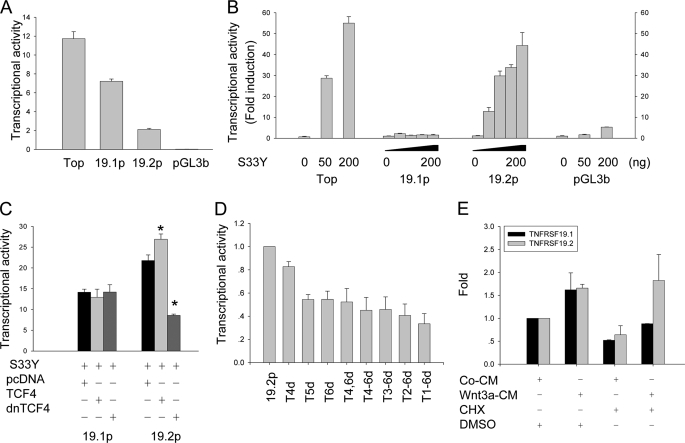
To further confirm that TNFRSF19 is regulated directly by Wnt signaling, we cloned 19.1p (1328 bp, −1149 to +178 bp) and 19.2p (1318 bp, −975 to +342 bp) into the promoterless firefly luciferase reporter vector pGL3-basic and the activities of the promoters were determined by dual luciferase assay. Without Wnt activation, 19.1p has a much higher basal activity than 19.2p (Fig. 3A) suggesting that TNFRSF19.1 is constitutively expressed by hMSC. We examined responsiveness of promoters to Wnt activation by co-transfection of 293T cells with a stabilized form of β-catenin (S33Y) (24). S33Y increased promoter activities of 19.2p to a maximal of 12–44-fold in a dose-dependent manner but the 19.1p promoter was not responsive (Fig. 3B). Furthermore, β-catenin-mediated transactivation of 19.2p could be further enhanced by co-transfection with wild type TCF4 and blocked by co-transfection with dnTCF4, which lacks of β-catenin binding domain (25) (Fig. 3C). The 19.1p did not respond to either wild type TCF4 or dnTCF4 (Fig. 3C). To determine whether TBE sites are essential for Wnt activation, we deleted each of the TBE sites in 19.2p. The single deletion of the distal three TBE sites (Fig. 2, T1, T2, or T3) did not affect the responsiveness of 19.2p to Wnt activation. On the other hand, single deletion of the proximal three TBE sites (Fig. 2, T4, T5, or T6) decreased the responsiveness of 19.2p to Wnt activation by 18 to 45%. Multiple TBE deletions further decreased 19.2p responsiveness to Wnt activation by 67% (Fig. 3D).
FIGURE 3.
TNFRSF19.2 is a direct target of canonical Wnt signaling. The promoter of TNFRSF19 transcript 1 (19.1p) and transcript 2 (19.2p) were cloned into the promoterless firefly luciferase reporter vector pGL3-basic (pGL3b). 293T cells were transfected with 50 ng of promoter firefly luciferase vector and 5 ng of pRL-TK Renilla luciferase vector as internal control by FuGENE 6. Topflash (Top) and pGL3b firefly vectors were used as positive and negative controls, respectively. A, basal transcriptional activity of TNFRSF19 promoters. B, dose-dependent activation of 19.2p by ectopic expression of a stable β-catenin S33Y (S33Y). 293T cells were co-transfected with 0, 25, 50, or 100 ng of S33Y for 19.1p and 19.2p analysis, and 0, 50, or 200 ng for controls. The data are presented as fold-induction compared with cells transfected without S33Y. C, dominant-negative TCF4 (dnTCF4) abolishes the activation of 19.2p by S33Y. 293T cells were co-transfected with 100 ng of S33Y and 200 ng of pcDNA, wild type TCF4, or dnTCF4 together with reporter vectors. D, proximal TBE are essential for Wnt activation. 19.2p harboring the single or multiple deletions of TBE were constructed and co-transfected with 50 ng of S33Y. The transcription activity of 19.2p was set to 1. The x axis indicates deletion of TBE in 19.2p. E, Wnt3a activates TNFRSF19.2 expression in the absence of protein synthesis. T253 cells were treated with 50% control condition medium (CM) or Wnt3a condition medium (Wnt3a-CM) with either dimethyl sulfoxide (DMSO) or 1 μg/ml of the protein synthesis inhibitor CHX for 8 h. Real-time RT-PCR was performed to detect TNFRSF19 expression, which was normalized against GAPDH. The expression level of TNFRSF19 in T253 cells treated with control CM and dimethyl sulfoxide was set to 1. All results are mean ± S.D. of three replicates. *, p < 0.01.
We further evaluated whether Wnt3a up-regulated TNFRSF19 expression requires protein synthesis. hMSC-LRP5T253 cells were treated with control CM or Wnt3a-CM in the presence of either dimethyl sulfoxide as control or the protein synthesis inhibitor CHX (1 μg/ml) for 8 h. Real-time RT-PCR analysis indicated that both TNFRSF19.1 and -19.2 were up-regulated by Wnt3a in the absence of CHX. In the presence of Wnt3a and CHX, expression of TNFRSF19.2 but not TNFRSF19.1 was stimulated (Fig. 3E), suggesting that TNFRSF19.2 but not TNFRSF19.1 is transcriptionally regulated by canonical Wnt signaling.
Expression of TNFRSF19 Is Inhibited by C/EBP
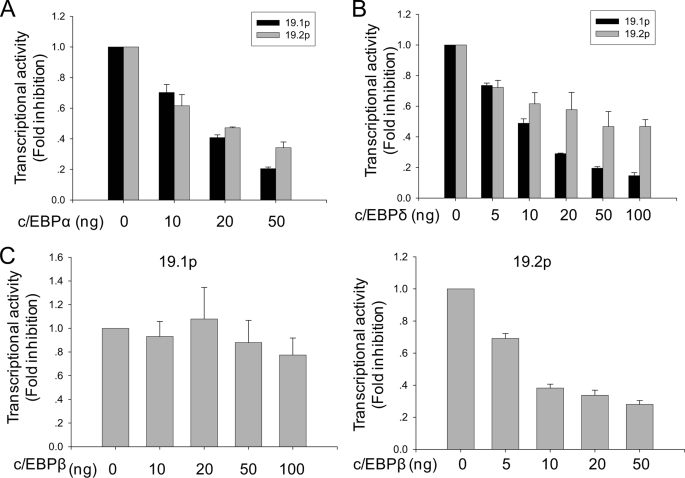
Because TNFRSF19.1 and TNFRSF19.2 promoters contain several C/EBP binding sites and expression of the transcription factor C/EBP has been shown to initiate adipogenesis by transactivating adipocyte-specific genes (26), we examined the regulation of TNFRSF19 expression by C/EBP. Co-transfection of 293T cells with C/EBPα and C/EBPδ significantly decreased 19.1p and 19.2 activities in a dose-dependent manner (Fig. 4, A and B) and the inhibitory effects were more pronounced for 19.1p than 19.2p. On the other hand, C/EBPβ did not inhibit 19.1p activity but only inhibited 19.2p (Fig. 4C). To further confirm that these C/EBP binding sites are functional elements, we performed site mutagenesis of these sites. Unfortunately, C/EBP mutant constructs displayed extremely low promoter activity that is 10% of the wild type promoter activity, which resulted in non-reproducible results (not shown).
FIGURE 4.
The promoter activity of TNFRSF19 is inhibited by C/EBP. 293T cells were transfected with 50 ng of promoter-firefly luciferase vector (19.1p or 19.2p) and 5 ng of pRL-TK Renilla luciferase vector as internal control by FuGENE 6. C/EBPα (A) and C/EBPδ (B) inhibit the activities of 19.1p and 19.2p in dose-dependent manner. C, C/EBPβ inhibits 19.2p activity but not 19.1p. Results are mean ± S.D. of three replicates.
Our data suggested that TNFRSF19 is repressed by C/EBP when adipogenesis is initiated. Therefore, up-regulation of TNFRSF19.2 by canonical Wnt signaling may be required for osteogenesis but suppression basal expression of TNFRSF19 via C/EBP may be important for adipogenesis.
TNFRSF19.2 Functions Downstream of Canonical Wnt Signaling to Regulate ALP Activity
We have previously demonstrated that Wnt activation in hMSC-LRP5T253 but not in hMSC-LRP5T244 induced osteoblast differentiation as demonstrated by up-regulation of the osteoblast differentiation marker ALP (15). To examine whether up-regulation of TNFRSF19 by Wnt3a is related to increased ALP activity, we first examined the time course expression of ALP and TNFRSF19 upon Wnt3a treatment. Compared with control CM, Wnt3a-induced ALP expression after 24 h (2.6-fold) and ALP levels continued to increase after 72 (3.3-fold) and 168 h (5.5-fold) (Fig. 5A). Wnt3a-induced up-regulation of TNFRSF19.2 was earlier than ALP and first observed at 8 h (3.4-fold) and increased continuously during Wnt3a treatment (6.7-, 7.9-, and 11-fold at 24, 72, and 168 h, respectively). TNFRSF19.1 was also up-regulated by Wnt3a treatment but its expression reached a maximal level at 24 h and was maintained until 168 h.
FIGURE 5.
TNFRSF19 activates osteogenesis by increasing ALP activity. A, TNFRSF19 is up-regulated upon Wnt3a treatment. hMSC-LRP5T253 (T253) cells were treated with 50% control condition medium (Co-CM) or Wnt3a conditioned medium (Wnt3a-CM) for the indicated hours. The expression of osteogenic marker ALP and TNFRSF19 transcripts 1 and 2 (TNFRSF19.1 and TNFRSF19.2) were analyzed by real-time RT-PCR and normalized against GAPDH. Results are mean ± S.D. of three replicates. B, overexpression of TNFRSF19.2 restores ALP activity in hMSC-LRP5T244 (T244) cells. T244 and T253 cells were infected with pBABE empty vector or TNFRSF19.2 expression vector followed by culturing in normal medium, control CM, or Wnt3a-CM for 7 days. ALP activity was measured by using p-nitrophenyl phosphate as substrate and normalized against cell number. Results are mean ± S.D. of five replicates. *, p < 0.05; **, p < 0.01. C, knocking down TNFRSF19 in T253 cells decreased Wnt3a-induced ALP activity. T253 cells were infected with control shRNA (shCtrl) or three shRNAs targeting TNFRSF19 (sh19-2, sh19-4, or sh19-5). Results are mean ± S.D. of five replicates. *, p < 0.05; **, p < 0.01 compared with shCtrl.
Next, we overexpressed TNFRSF19.2 in hMSC-LRP5T244 and hMSC-LRP5T253 cells and measured ALP activity. As shown in Fig. 5B, ALP activity was increased significantly by Wnt3a compared with control CM in both hMSC-LRP5T253 and hMSC-LRP5T244 cells (p < 0.05), but hMSC-LRP5T253 had higher ALP activity than hMSC-LRP5T244 cells upon Wnt3a stimulation (p < 0.05), which corroborated our previous data (15). In the absence of Wnt3a, overexpression of TNFRSF19.2 in hMSC-LRP5T253 and hMSC-LRP5T244 cells significantly increased ALP activity compared with cells infected with the empty vector pBABE (p < 0.01). More importantly, TNFRSF19.2 overexpression in hMSC-LRP5T253 and hMSC-LRP5T244 cells increased ALP activity to the equivalent level induced by Wnt3a (p > 0.05) (Fig. 5B). Overexpression of TNFRSF19.2 in hMSC-LRP5T244 cells followed by Wnt3a treatment further increased ALP activity compared with hMSC-LRP5T244 cell overexpression of TNFRSF19.2 alone (p < 0.01). With or without Wnt3a, overexpression of TNFRSF19.2 in hMSC-LRP5T244 cells increased ALP activity to an equivalent level in hMSC-LRP5T253 cells, suggesting that TNFRSF19.2 can restore the osteoblastic differentiation capacity of hMSC-LRP5T244. Combined overexpression of TNFRSF19.2 and Wnt3a treatment in hMSC-LRP5T253 cells did not further increase the ALP activity possibly due to saturation effects.
Finally, we knocked down TNFRSF19 in hMSC-LRP5T253 cells using lentivirus-based shRNA. hMSC-LRP5T244 cells were not included in this experiment due to their very low ALP expression upon Wnt3a stimulation. Knockdown efficiency was first determined using the MISSION shTNFRSF19 set (TRCN0000058883-TRCN0000058887, Sigma), which targets both TNFRSF19.1 and TNFRSF19.2. We found all of shTNFRSF19 inhibited at least 70% Wnt3a-induced up-regulation of TNFRSF19 transcripts at day 3 (data now shown). We chose TRCN0000058884 (sh19-2), which had the lowest knock down efficiency and TRCN0000058886 (sh19-4) and TRCN0000058887 (sh19-5), which had the highest knockdown efficiency for further experiments. Wnt3a-induced ALP activity was blocked by sh19-4 and sh19-5 (p < 0.05) but not affected by sh19-2 compared with scramble shRNA (shCtrl) (Fig. 5C). Combined with promoter analysis results, TNFRSF19.2 seems to be a transcript regulated by canonical Wnt signaling to mediate Wnt3a-induced ALP activity.
TNFRSF19.2 Is a Negative Regulator of Adipocyte Differentiation
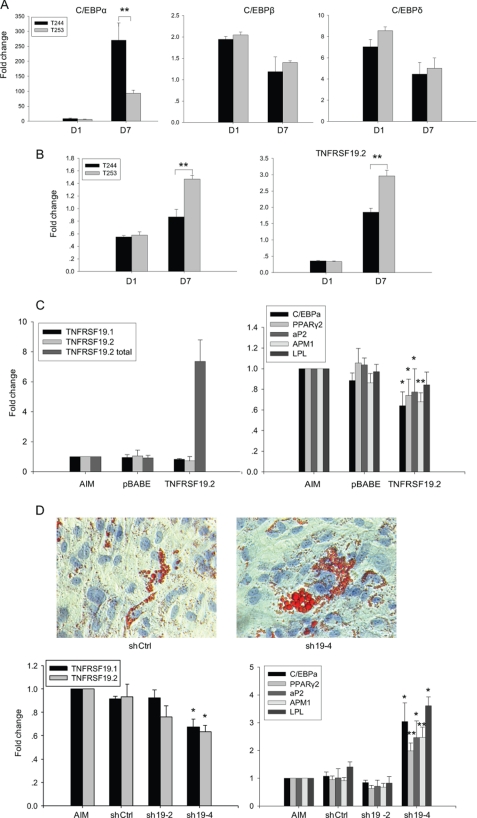
Because C/EBP proteins are transcription factors inducing adipocytic differentiation, their inhibitory effects on TNFRSF19 expression suggested that TNFRSF19 may function as a regulator of adipogenesis. We first examined the time course expression of C/EBP and TNFRSF19 during in vitro adipogenesis. hMSC-LRP5T253 and hMSC-LRP5T244 cells were treated with AIM for 1 or 7 days. The expression levels of C/EBP and TNFRSF19 in AIM was normalized against the expression levels in normal medium at the corresponding day. C/EBPα was increased 8.5-fold in hMSC-LRP5T244 cells and 6.2-fold in hMSC-LRP5T253 cells at day 1. At day 7, C/EBPα expression levels were severalfold higher in hMSC-LRP5T244 cells (270-fold) compared with hMSC-LRP5T253 cells (93-fold) (Fig. 6A) corroborating our previous findings of enhanced adipogenesis in hMSC-LRP5T244 cells (15). Similarly C/EBPβ and C/EBPδ were up-regulated in both hMSC-LRP5T253 and hMSC-LRP5T244 cells with higher levels at day 1 compared with day 7 (Fig. 6A). In contrast to up-regulation of C/EBP, TNFRSF19.1 was down-regulated more than 40% and TNFRSF19.2 was decreased about 60% in both hMSC-LRP5T244 and hMSC-LRP5T253 cells at day 1 (Fig. 6B). At day 7, both transcripts were expressed higher in hMSC-LRP5T253 cells supporting the reverse relationship between TNFRSF19 expression and adipocyte differentiation capacity. In addition, expression of TNFRSF19.1 and TNFRSF19.2 in hMSC-LRP5T244 and hMSC-LRP5T253 cells increased close to or above the expression levels in control cells (Fig. 6B) despite the continuous up-regulation of C/EBPα but accompanied by reduced up-regulation of C/EBPβ and C/EBPδ at day 7.
FIGURE 6.
TNFRSF19 inhibits adipogenic differentiation. hMSC-LRP5T244 (T244) and hMSC-LRP5T253 (T253) cells were treated without or with AIM for 1 or 7 days. The expression of C/EBP (A) and TNFRSF19 transcripts 1 and 2 (TNFRSF19.1 and TNFRSF19.2) (B) were determined by real-time RT-PCR and normalized against GAPDH. Data are represented as fold-change of cells treated with AIM relative to those treated with normal medium. Results are mean ± S.D. of three replicates. **, p < 0.01. C, overexpression of TNFRSF19.2 inhibits adipogenesis. T244 cells were infected with pBABE empty vector or TNFRSF19.2 expression vector followed by AIM treatment for 7 days. The expression of TNFRSF19.1, endogenous TNFRSF19.2, TNFRSF19.2 total (endogenous and ectopic TNFRSF19.2), and adipogenic markers including C/EBPα, PPARγ2, aP2, APM1, and lipoprotein lipase were determined by real-time RT-PCR and normalized against GAPDH. The expression levels in cells treated with AIM only was set to 1. Results are mean ± S.D. of three replicates. *, p < 0.05; **, p < 0.01. D, knocking down TNFRSF19 promotes adipogenesis. T253 cells were infected with control shRNA (shCtrl) or two shRNA targeting TNFRSF19 (sh19-2 and sh19-4) followed by AIM treatment for 7 days. Adipocytes containing lipid droplets were stained by Oil-red O staining (×200) (upper). The expression of TNFRSF19.1, TNFRSF19.2, and adipogenic markers were determined by real-time RT-PCR and normalized against GAPDH. The expression level in cells treated with AIM only was set to 1. Results shown are mean ± S.D. of three replicates. *, p < 0.05; **, p < 0.01 compared with shCtrl.
We then examined the effects of either overexpression of TNFRSF19.2 in hMSC-LRP5T244 or knocking down TNFRSF19 in hMSC-LRP5T253 on adipogenic differentiation, respectively. As shown in Fig. 6C, ectopic TNFRSF19.2 was expressed in hMSC-LRP5T244 cells infected with pBABE-TNFRSF19.2 retrovirus compared with non-infected cells and cells infected with empty vector pBABE virus. Correspondingly, the expression level of adipogenic markers including C/EBPα, PPARγ2, aP2, and APM1 were significantly reduced (p < 0.05) (Fig. 6C). On the other hand, knocking down TNFRSF19 in hMSC-LRP5T253 cells significantly increased its adipogenic differentiation evidenced by enhanced formation of a higher number of lipid-filled mature adipocytes (Fig. 6D). Real-time RT-PCR confirmed that sh19-4 has significantly higher knocking down efficiency compared with shCtrl (p < 0.05) up to 7–8 days post-infection and resulted in significant up-regulation of all examined adipogenic markers (p < 0.05) (Fig. 6D).
TNFRSF19.1 and TNFRSF19.2 Have Different Functions in hMSC Differentiation
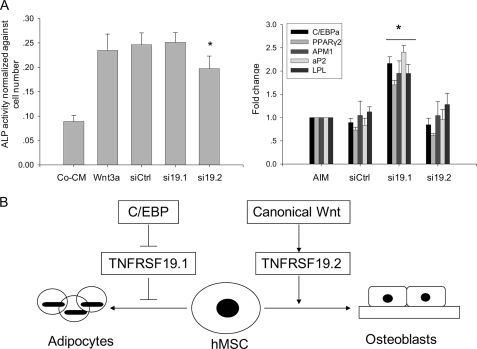
Due to high similarities between coding sequences of both transcripts, we designed siRNA targeting transcript-specific 3′-regions to distinguish the function of the two transcripts in hMSC differentiation. Previous studies have demonstrated that siRNAs targeting the 3′-untranslated region induce gene knock-down (27). As shown in Fig. 7A, si19.2 targeting TNFRSF19.2 decreased ALP activity but had no effect on adipogenesis. In contrast, si19.1 targeting TNFRSF19.1 significantly enhanced expression of adipogenic markers but did not affect ALP activity. si19.1 and si19.2 has about 50% knocking down efficiency for each target transcript (data not shown).
FIGURE 7.
TNFRSF19 transcripts 1 and 2 (TNFRSF19.1 and TNFRSF19.2) have different functions during hMSC differentiation. A, hMSC-LRP5T253 cells were transfected with siCtrl (scramble siRNA), si19.1 (TNFRSF19.1 siRNA), or si19.2 (TNFRSF19.2 siRNA) followed by osteoblastic differentiation (treated with control CM (Co-CM) or Wnt3a) or adipocyte differentiation (treated with AIM) for 7 days. ALP activity was quantitated and normalized against the cell number (left). The expression of adipogenic markers were normalized against GAPDH and the expression level in cells treated with AIM alone was set to 1 (right). Results are mean ± S.D. of 8 replicates (ALP) or 4 replicates (adipogenic markers). *, p < 0.01 comparing with siCtrl. B, proposed function of TNFRSF19 in determination of the differentiation fate of hMSC. Canonical Wnt signaling activates TNFRSF19.2 expression to drive hMSC to osteoblastic lineage. C/EBP proteins inhibit basal expression of TNFRSF19.1 to drive hMSC to adipocytic lineage.
DISCUSSION
TNFRSF proteins are type I transmembrane glycoproteins with a cysteine-rich extracellular ligand binding domain, a single transmembrane region, and a diverse cytoplasmic tail. TNFRSF play an important role in regulating diverse biological activities including key aspects of immune modulation (28). More than 22 TNFRSF have been identified and subgrouped according to their specific cytoplasmic domains that act as docking sites for signaling molecules, e.g. members containing cell death domain (e.g. TNFRSF1A, TNFRSF6, and TNFRSF12) mediate apoptosis and those containing TNFR-associated factor (TRAF) binding motif (e.g. TNFRSF1B and TNFRSF5) promote cell survival (28–30). Some members of TNFRSF, e.g. osteoprotegerin (TNFRSF11B) and RANK (TNFRSF11A), are the main regulators of osteoclast differentiation and function (31, 32) and thus TNFRSF is known to play an important role in bone biology.
Tnfrsf19 also known as toxicity and JNK inducer (Taj) or Troy is a newly identified member of TNFRSF (33–35). Gene expression studies revealed that Tnfrsf19 is highly expressed in brain and epithelium tissues during embryonic development (35, 36). In the postnatal organism, it is expressed in the brain and hair follicle (33, 35). In accordance with its expression profiles, Tnfrsf19 has been reported to be involved in brain and hair follicle development. During axon regeneration, the inhibitory molecules in central nervous system myelin signals through the NgR1-LINGO-p75 complex to inhibit successful axon regeneration. Observations that Tnfrsf19 were broadly expressed in postnatal and adult neurons compared with limited expression of p75 to certain types of neurons and its temporal expression during development, as well as Tnfrsf19, can replace p75 to form the NgR1-LINGO-Tnfrsf19 complex transducing signals from myelin offered a novel mechanisms of axon regeneration failure and a possible clinical application (37, 38). In addition, Tnfrsf19 exhibits sequence homology and an overlapped expression pattern with Edar, which is another TNFRSF member important in development of ectodermal organs. Studies in Tnfrsf19-deficient mice have suggested a potentially redundant role between Tnfrsf19 and Edar in hair follicle development (39, 40). Tnfrsf19 is absent in the spleen and thymus, which are major lymphoid tissues, suggesting that it does not play a role in the immune system (33, 35). TNFRSF19 was also reported to interact with TRAF family members to activate JNK or NF-κB pathways (34, 35) and overexpressing of Tnfrsf19 induce cell death (34, 41), although it does not contain cell death domain in its cytoplasmic region.
Tnfrsf19 is also expressed in mesenchymal tissues (39) but its role in mesodermal development is poorly studied. Here, we reported that human TNFRSF19 functions downstream of canonical Wnt signaling and C/EBP proteins to regulate differentiation fate of hMSC. The human TNFRSF19 gene has two transcripts: TNFRSF19.1 is 1485 bp in length and encodes a 423-amino acid polypeptide and TNFRSF19.2 is 4283 bp in length and encodes a 417-amino acid polypeptide. There is a structural homology between TNFRSF19.1 and TNFRSF19.2 except at the end of cytoplasmic tail where TNFRSF19.2 contains a major TRAF2-binding consensus sequence, (P/S/A/T)X(Q/E)E (SLQE at amino acid 413–416), which is absent in TNFRSF19.1. Mouse Tnfrsf19 has only one transcript encoding 416 amino acids with a major TRAF2-binding sequence, TLQE, in the cytoplasmic tail at position 276–279. The TRAF binding sequence is important for activating downstream JNK or NF-κB pathways (29) and its absence in TNFRSF19.1 suggests that both transcripts may have different biological functions. The position of the TLQE site is not conserved between mouse Tnfrsf19 (position 276–279) and human TNFRSF19.2 (position 413–416) and thus its differential function in the two species as a binding site of TRAF family members has yet to be determined.
The differences between human and mouse TNFRSF19 gene structure were further revealed by promoter analysis. Compared with human TNFRSF19 promoters, mouse Tnfrsf19 promoter (−1000 to 200 bp according to Refseq NM_013869) contains two TBE but no C/EBP binding sites, GC box, and TATA box, suggesting that the human gene has acquired novel functions during evolution. We observed that TNFRSF19.1 was up-regulated differentially by Wnt3a as evidenced by microarray data in the absence of TBE in the promoter. We conducted further analysis for the presence of the CTTTGWW sequence within 3 kb upstream of transcription start sites of 19.1p and 19.2p, and still did not identify TBE in 19.1p but several more in 19.2p (not shown). Up-regulation of TNFRSF19.1 may be mediated through an indirect effect of Wnt3a. It is also plausible that some sequences like CTTGWW or CTTTGXX may also respond to Wnt3a activation but need further confirmation.
In hMSC, canonical Wnt signaling enhance osteoblast differentiation as evidenced by increased ALP production (15). However, late osteoblast differentiation markers, e.g. osteocalcin, did not respond to Wnt3a treatment in our cell model possibly due to inhibition of the osteocalcin promoter (42), and down-regulation of Wnt signaling was reported to be required for late stage osteoblast differentiation (43). This may explain why previous studies employed ALP induction by canonical Wnt signaling as an indicator of osteogenesis (44, 45). In this study, we observed that this effect is mediated by up-regulation of TNFRSF19.2 evidenced by overexpression of TNFRSF19.2 alone or combined with Wnt3a treatment-rescued ALP activity in hMSC-LRP5T244 to the equivalent level of hMSC-LRP5T253. On the other hand, canonical Wnt signaling inhibited adipogenesis, suggesting that this signaling pathway may maintain an inactivated state so that TNFRSF19 is impossible to be up-regulated by canonical Wnt signaling during adipogenesis. We observed that C/EBP proteins inhibited TNFRSF19 expression suggesting that the basal expression of TNFRSF19 was required to be repressed at early stages of adipogenesis. In support of this notion, TNFRSF19 expression was reversely correlated with the expression of C/EBP. In our study, we chose different cell lines for overexpressing and knockdown experiments based on differences in the differentiation potential of hMSC-LRP5T244 and hMSC-LRP5T253 as reported previously (15), as well as the expression profile of TNFRSF19 transcripts in the two cell lines and the promoter analysis data. Unfortunately, we were not able to obtain an open reading frame of TNFRSF19.1 by PCR due to the very short untranslated region and biased nucleotide distribution. Besides, it was not possible to obtain specific shRNA targeting TNFRSF19.1 or TNFRSF19.2 due to the nearly identical coding sequence of the two transcripts. Thus, the detailed functional role of TNFRSF19.1 in MSC proliferation and in transferring C/EBP signals to adipocyte differentiation need to be further investigated.
The microarray data revealed significant down-regulation of other members of TNFRSF in hMSC by Wnt signaling (supplemental Tables S1 and S2). TNFRSF11B, also called osteoprotegerin, is an osteoblast-secreted decoy receptor that functions as a negative regulator of bone resorption (46). TNFRSF1B also called p75 was down-regulated only in hMSC-LRP5T253 cells. Genetic polymorphisms in TNFRSF1B have been reported to be possible determinants of bone mass (47–50) possibly through epistatic interaction with the collagen type 1, α1 (COL1A1) gene (51). Due to the biological functions of TNFRSF19 reported here, it is interesting to examine the role of TNFRSF19 in osteoblast-osteoclast differentiation.
A reciprocal relationship between bone mass and adipose tissue mass in the bone marrow exists in a number of physiological (e.g. aging) and pathophysiological conditions (e.g. osteoporosis, glucocorticoid, or glitazone therapy) (52). Because osteoblasts and adipocytes arise from MSC in the bone marrow microenvironment, understanding the ”switching mechanisms“ between osteoblast versus adipocyte differentiation can provide novel therapeutic targets for enhancing bone formation (52, 53). Previous studies have identified some factors that play a role in the switching mechanism. TAZ functions as a transcriptional modulator to stimulate MSC osteoblast cell differentiation and simultaneously block the differentiation to adipocytes through direct interaction with transcription factors Runx2 and PPARγ (54). On the other hand, PPARγ enhances adipocyte and inhibits osteoblast differentiation in vitro and in vivo (55). We have previously demonstrated that Wnt signaling in hMSC enhances osteoblast and inhibits adipocyte differentiation. In support of this, Wnt10b have been demonstrated to promote bone formation and inhibit adipogenesis (20, 21). Interestingly, TAZ has been reported to be regulated by canonical Wnt signaling (54). We propose here that TNFRSF19 is a possible novel factor mediating the ”switch“ of osteoblast versus adipocyte differentiation of MSC. The different cis-regulatory elements found in promoters of human TNFRSF19 transcripts, presence or absence of TRAF binding sequence in the two transcripts of TNFRSF19, expression profiles of TNFRSF19 transcripts at basal level and during MSC differentiation, as well as overexpression and knockdown experiments suggest that the major function of TNFRSF19.1 and TNFRSF19.2 is to mediate different differentiation signals driving hMSC to osteoblastic or adipocytic lineages (Fig. 7). The relative importance of these two transcripts and their signaling mechanisms in mediating the biological effects on MSC biology remain to be determined.
Supplementary Material
Acknowledgment
We thank Dr. Qin Hao for preparing C/EBP expression vectors.
This work was supported in part by grants from the Danish Medical Research Council, the Danish Stem Cell Center, the Novo Nordisk Foundation, and a grant from the local government of the region of Southern Denmark.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- LRP
- lipoprotein receptor-related protein
- TCF
- T-cell factor
- LEF
- lymphoid enhancer binding factor
- hMSC
- human mesenchymal stem cell
- TNFRSF
- tumor necrosis receptor superfamily
- AIM
- adipocyte induction medium
- ALP
- alkaline phosphatase
- PPARγ
- peroxisome proliferator activator receptor γ
- C/EBP
- CCAAT/enhancer-binding protein
- CM
- conditioned medium
- CHX
- cycloheximide
- TBE
- TCF/LEF binding element
- dnTCF4
- dominant-negative TCF4
- shRNA
- short hairpin RNA
- siRNA
- small interfering RNA
- RT
- reverse transcription
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- shCtrl
- short hairpin RNA control
- JNK
- c-Jun N-terminal kinase.
REFERENCES
- 1.Moon R. T., Bowerman B., Boutros M., Perrimon N. (2002) Science 296, 1644–1646 [DOI] [PubMed] [Google Scholar]
- 2.Westendorf J. J., Kahler R. A., Schroeder T. M. (2004) Gene 341, 19–39 [DOI] [PubMed] [Google Scholar]
- 3.Gong Y., Slee R. B., Fukai N., Rawadi G., Roman-Roman S., Reginato A. M., Wang H., Cundy T., Glorieux F. H., Lev D., Zacharin M., Oexle K., Marcelino J., Suwairi W., Heeger S., Sabatakos G., Apte S., Adkins W. N., Allgrove J., Arslan-Kirchner M., Batch J. A., Beighton P., Black G. C., Boles R. G., Boon L. M., Borrone C., Brunner H. G., Carle G. F., Dallapiccola B., De Paepe A., Floege B., Halfhide M. L., Hall B., Hennekam R. C., Hirose T., Jans A., Jüppner H., Kim C. A., Keppler-Noreuil K., Kohlschuetter A., LaCombe D., Lambert M., Lemyre E., Letteboer T., Peltonen L., Ramesar R. S., Romanengo M., Somer H., Steichen-Gersdorf E., Steinmann B., Sullivan B., Superti-Furga A., Swoboda W., van den Boogaard M. J., Van Hul W., Vikkula M., Votruba M., Zabel B., Garcia T., Baron R., Olsen B. R., Warman M. L.(2001) Cell 107, 513–523 [DOI] [PubMed] [Google Scholar]
- 4.Ai M., Heeger S., Bartels C. F., Schelling D. K. (2005) Am. J. Hum. Genet. 77, 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyden L. M., Mao J., Belsky J., Mitzner L., Farhi A., Mitnick M. A., Wu D., Insogna K., Lifton R. P. (2002) N. Engl. J. Med. 346, 1513–1521 [DOI] [PubMed] [Google Scholar]
- 6.Little R. D., Carulli J. P., Del Mastro R. G., Dupuis J., Osborne M., Folz C., Manning S. P., Swain P. M., Zhao S. C., Eustace B., Lappe M. M., Spitzer L., Zweier S., Braunschweiger K., Benchekroun Y., Hu X., Adair R., Chee L., FitzGerald M. G., Tulig C., Caruso A., Tzellas N., Bawa A., Franklin B., McGuire S., Nogues X., Gong G., Allen K. M., Anisowicz A., Morales A. J., Lomedico P. T., Recker S. M., Van Eerdewegh P., Recker R. R., Johnson M. L. (2002) Am. J. Hum. Genet. 70, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Wesenbeeck L., Cleiren E., Gram J., Beals R. K., Bénichou O., Scopelliti D., Key L., Renton T., Bartels C., Gong Y., Warman M. L., De Vernejoul M. C., Bollerslev J., Van Hul W. (2003) Am. J. Hum. Genet. 72, 763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baron R. (2009) IBMS BoneKEy 6, 86–93 [Google Scholar]
- 9.Glass D. A., 2nd, Karsenty G. (2007) Endocrinology 148, 2630–2634 [DOI] [PubMed] [Google Scholar]
- 10.Bonewald L. F., Johnson M. L. (2008) Bone 42, 606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass D. A., 2nd, Bialek P., Ahn J. D., Starbuck M., Patel M. S., Clevers H., Taketo M. M., Long F., McMahon A. P., Lang R. A., Karsenty G. (2005) Dev. Cell 8, 751–764 [DOI] [PubMed] [Google Scholar]
- 12.Tamai K., Semenov M., Kato Y., Spokony R., Liu C., Katsuyama Y., Hess F., Saint-Jeannet J. P., He X. (2000) Nature 407, 530–535 [DOI] [PubMed] [Google Scholar]
- 13.Wehrli M., Dougan S. T., Caldwell K., O'Keefe L., Schwartz S., Vaizel-Ohayon D., Schejter E., Tomlinson A., DiNardo S. (2000) Nature 407, 527–530 [DOI] [PubMed] [Google Scholar]
- 14.Kato M., Patel M. S., Levasseur R., Lobov I., Chang B. H., Glass D. A., 2nd, Hartmann C., Li L., Hwang T. H., Brayton C. F., Lang R. A., Karsenty G., Chan L. (2002) J. Cell Biol. 157, 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu W., Andersen T. E., Bollerslev J., Mandrup S., Abdallah B. M., Kassem M. (2007) J. Bone Miner. Res. 22, 1720–1731 [DOI] [PubMed] [Google Scholar]
- 16.Semenov M. V., He X. (2006) J. Biol. Chem. 281, 38276–38284 [DOI] [PubMed] [Google Scholar]
- 17.Ellies D. L., Viviano B., McCarthy J., Rey J. P., Itasaki N., Saunders S., Krumlauf R. (2006) J. Bone Miner. Res. 21, 1738–1749 [DOI] [PubMed] [Google Scholar]
- 18.Ai M., Holmen S. L., Van Hul W., Williams B. O., Warman M. L. (2005) Mol. Cell. Biol. 25, 4946–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Wang Y., Li X., Zhang J., Mao J., Li Z., Zheng J., Li L., Harris S., Wu D. (2004) Mol. Cell. Biol. 24, 4677–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross S. E., Hemati N., Longo K. A., Bennett C. N., Lucas P. C., Erickson R. L., MacDougald O. A. (2000) Science 289, 950–953 [DOI] [PubMed] [Google Scholar]
- 21.Bennett C. N., Longo K. A., Wright W. S., Suva L. J., Lane T. F., Hankenson K. D., MacDougald O. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3324–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodríguez J. P., González M., Ríos S., Cambiazo V. (2004) J. Cell Biochem. 93, 721–731 [DOI] [PubMed] [Google Scholar]
- 23.Dynan W. S. (1986) Trends Genet. 2, 196–197 [Google Scholar]
- 24.Morin P. J., Sparks A. B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K. W. (1997) Science 275, 1787–1790 [DOI] [PubMed] [Google Scholar]
- 25.Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 26.Gregoire F. M., Smas C. M., Sul H. S. (1998) Physiol. Rev. 78, 783–809 [DOI] [PubMed] [Google Scholar]
- 27.McManus M. T., Petersen C. P., Haines B. B., Chen J., Sharp P. A. (2002) RNA 8, 842–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locksley R. M., Killeen N., Lenardo M. J. (2001) Cell 104, 487–501 [DOI] [PubMed] [Google Scholar]
- 29.Arch R. H., Gedrich R. W., Thompson C. B. (1998) Genes Dev. 12, 2821–2830 [DOI] [PubMed] [Google Scholar]
- 30.Ashkenazi A., Dixit V. M. (1998) Science 281, 1305–1308 [DOI] [PubMed] [Google Scholar]
- 31.Anderson D. M., Maraskovsky E., Billingsley W. L., Dougall W. C., Tometsko M. E., Roux E. R., Teepe M. C., DuBose R. F., Cosman D., Galibert L. (1997) Nature 390, 175–179 [DOI] [PubMed] [Google Scholar]
- 32.Kong Y. Y., Yoshida H., Sarosi I., Tan H. L., Timms E., Capparelli C., Morony S., Oliveira-dos-Santos A. J., Van G., Itie A., Khoo W., Wakeham A., Dunstan C. R., Lacey D. L., Mak T. W., Boyle W. J., Penninger J. M. (1999) Nature 397, 315–323 [DOI] [PubMed] [Google Scholar]
- 33.Hu S., Tamada K., Ni J., Vincenz C., Chen L. (1999) Genomics 62, 103–107 [DOI] [PubMed] [Google Scholar]
- 34.Eby M. T., Jasmin A., Kumar A., Sharma K., Chaudhary P. M. (2000) J. Biol. Chem. 275, 15336–15342 [DOI] [PubMed] [Google Scholar]
- 35.Kojima T., Morikawa Y., Copeland N. G., Gilbert D. J., Jenkins N. A., Senba E., Kitamura T. (2000) J. Biol. Chem. 275, 20742–20747 [DOI] [PubMed] [Google Scholar]
- 36.Hisaoka T., Morikawa Y., Kitamura T., Senba E. (2003) Brain Res. Dev. Brain Res. 143, 105–109 [DOI] [PubMed] [Google Scholar]
- 37.Park J. B., Yiu G., Kaneko S., Wang J., Chang J., He X. L., Garcia K. C., He Z. (2005) Neuron. 45, 345–351 [DOI] [PubMed] [Google Scholar]
- 38.Shao Z., Browning J. L., Lee X., Scott M. L., Shulga-Morskaya S., Allaire N., Thill G., Levesque M., Sah D., McCoy J. M., Murray B., Jung V., Pepinsky R. B., Mi S. (2005) Neuron 45, 353–359 [DOI] [PubMed] [Google Scholar]
- 39.Pispa J., Mikkola M. L., Mustonen T., Thesleff I. (2003) Gene Expr. Patterns 3, 675–679 [DOI] [PubMed] [Google Scholar]
- 40.Pispa J., Pummila M., Barker P. A., Thesleff I., Mikkola M. L. (2008) Hum. Mol. Genet 17, 3380–3391 [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., Li X., Wang L., Ding P., Zhang Y., Han W., Ma D. (2004) J. Cell Sci. 117, 1525–1532 [DOI] [PubMed] [Google Scholar]
- 42.Kahler R. A., Westendorf J. J. (2003) J. Biol. Chem. 278, 11937–11944 [DOI] [PubMed] [Google Scholar]
- 43.van der Horst G., van der Werf S. M., Farih-Sips H., van Bezooijen R. L., Löwik C. W., Karperien M. (2005) J. Bone Miner. Res. 20, 1867–1877 [DOI] [PubMed] [Google Scholar]
- 44.Winkler D. G., Sutherland M. S., Ojala E., Turcott E., Geoghegan J. C., Shpektor D., Skonier J. E., Yu C., Latham J. A. (2005) J. Biol. Chem. 280, 2498–2502 [DOI] [PubMed] [Google Scholar]
- 45.Si W., Kang Q., Luu H. H., Park J. K., Luo Q., Song W. X., Jiang W., Luo X., Li X., Yin H., Montag A. G., Haydon R. C., He T. C. (2006) Mol. Cell. Biol. 26, 2955–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. (1998) Cell 93, 165–176 [DOI] [PubMed] [Google Scholar]
- 47.Albagha O. M., Tasker P. N., McGuigan F. E., Reid D. M., Ralston S. H. (2002) Hum. Mol. Genet. 11, 2289–2295 [DOI] [PubMed] [Google Scholar]
- 48.Spotila L. D., Rodriguez H., Koch M., Tenenhouse H. S., Tenenhouse A., Li H., Devoto M. (2003) Calcif. Tissue Int. 73, 140–146 [DOI] [PubMed] [Google Scholar]
- 49.Tasker P. N., Albagha O. M., Masson C. B., Reid D. M., Ralston S. H. (2004) Osteoporos. Int. 15, 903–908 [DOI] [PubMed] [Google Scholar]
- 50.Mullin B. H., Prince R. L., Dick I. M., Islam F. M., Hart D. J., Spector T. D., Devine A., Dudbridge F., Wilson S. G. (2008) Osteoporos. Int. 19, 961–968 [DOI] [PubMed] [Google Scholar]
- 51.Yang T. L., Shen H., Xiong D. H., Xiao P., Guo Y., Guo Y. F., Liu Y. Z., Recker R. R., Deng H. W. (2007) Ann. Hum. Genet. 71, 152–159 [DOI] [PubMed] [Google Scholar]
- 52.Gimble J. M., Zvonic S., Floyd Z. E., Kassem M., Nuttall M. E. (2006) J. Cell. Biochem. 98, 251–266 [DOI] [PubMed] [Google Scholar]
- 53.Justesen J., Stenderup K., Ebbesen E. N., Mosekilde L., Steiniche T., Kassem M. (2001) Biogerontology 2, 165–171 [DOI] [PubMed] [Google Scholar]
- 54.Hong J. H., Yaffe M. B. (2006) Cell Cycle 5, 176–179 [DOI] [PubMed] [Google Scholar]
- 55.Marie P. J., Kaabeche K. (2006) PPAR Res. 2006, 64807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.