Abstract
Bone growth and remodeling depend upon the opposing rates of bone formation and resorption. These functions are regulated by intrinsic seven transmembrane-spanning receptors, the parathyroid hormone receptor (PTH1R) and frizzled (FZD), through their respective ligands, parathyroid hormone (PTH) and Wnt. FZD activation of canonical β-catenin signaling requires the adapter protein Dishevelled (Dvl). We identified a Dvl-binding motif in the PTH1R. Here, we report that the PTH1R activates the β-catenin pathway by directly recruiting Dvl, independent of Wnt or LRP5/6. PTH1R coimmunoprecipitated with Dvl. Deleting the carboxyl-terminal PTH1R PDZ-recognition domain did not abrogate PTH1R-Dvl interactions; nor did truncating the receptor at position 480. However, further deletion eliminating the putative Dvl recognition domain abolished PTH1R interactions with Dvl. PTH activated β-catenin in a time- and concentration-dependent manner and translocated β-catenin to the nucleus. β-Catenin activation was inhibited by Dvl2 dominant negatives and by short hairpin RNA sequences targeted against Dvl2. PTH-induced osteoclastogenesis was also inhibited by Dvl2 dominant negative mutants. These findings demonstrate that G protein-coupled receptors other than FZD directly activate β-catenin signaling, thereby mimicking many of the functions of the canonical Wnt-FZD pathway. The distinct modes whereby FZD and PTH1R activate β-catenin control convergent or divergent effects on osteoblast differentiation, and osteoclastogenesis may arise from PTH1R-induced second messenger phosphorylation.
Keywords: Beta-catenin, Bone, G Protein-coupled Receptors (GPCR), Glycogen Synthase Kinase 3, Lipoprotein-like Receptor (LRP), Trafficking, Dishevelled, Parathyroid Hormone, Parathyroid Hormone Receptor
Introduction
Wnts are secreted lipid-modified glycoproteins that act as ligands to stimulate signal transduction pathways through FZD (frizzled) receptors and LRP5/6 (lipoprotein receptor-related protein 5/6) co-receptors. Canonical and non-canonical Wnt signaling pathways have been described (1). In the canonical pathway, in the absence of Wnt ligands, β-catenin is targeted to a destruction complex with APC (adenomatous polyposis coli), CK1 (casein kinase 1), GSK3β (glycogen synthase kinase 3β), and axin. Amino-terminal phosphorylation by CK1 and GSK3β followed by subsequent ubiquitination targets β-catenin for proteasomal degradation (2). Wnt binding to cognate FZD receptors and LRP5/6 causes recruitment of the PDZ (PSD-95, Discs-large, and ZO-1) protein Dvl (Dishevelled) to the plasma membrane by direct interaction with FZD receptors. The recruitment of Dvl to the plasma membrane results in the formation of Dvl oligomers, which interact with axin. Parallel phosphorylation of the co-receptors LRP5 and LRP6 induces binding to axin, resulting in disruption of the destruction complex. As a consequence, β-catenin escapes proteasomal degradation and translocates to the nucleus, where it regulates the activity of the transcription factors TCF (T cell factor) and Lef (lymphocyte enhancer-binding factor).
Ten FZD receptors constitute a distinct family of seven transmembrane-spanning receptors (3). Whether or not they couple to G proteins remains controversial (4). Notably, they bear the closest phylogenetic relation to Family B1 GPCRs including the type 1 parathyroid hormone receptor (PTH1R) (5). Four of the 10 frizzled receptors (FZD1, -2, -4, and -7) contain a canonical carboxyl-terminal PDZ recognition domain, (D/E)(S/T)XΦ, where Φ represents a hydrophobic residue, generally L/I/V but sometimes M, as in the PTH1R (6, 7). This motif mediates interactions with PDZ proteins, such as NHERF1 (Na/H exchange regulatory factor-1) (8).
Bone growth and remodeling are regulated by parallel signaling pathways involving the PTH1R and FZD (9). Considerable evidence now strongly implicates Wnt/FZD signaling in regulating bone formation (9–11). β-Catenin signaling is required for suppression of chondrocyte differentiation and induction of osteoblastogenesis. Multiple lines of genetic evidence establish the critical participation of canonical β-catenin activity for early osteoblast differentiation (12). Acting on mature osteoblasts, stimulation of the PTH1R produces RANKL (receptor activator of NFκB ligand), which binds the RANK (receptor activator of NFκB) receptor on osteoclast precursors and induces formation of osteoclasts by signaling through NFκB2 and JNK. Homologous recombination of PTH1R, RANKL, or RANK results in mice with profound bone phenotypes (13–15). Gene knock-out of elements of FZD and the canonical β-catenin signaling pathway likewise produce profound disruption of normal bone formation or turnover (9–11). Recent work shows that PTH promotes β-catenin activation (16–18). The interaction between PTH1R and FZD signaling pathways is largely unexplored. Nissenson and co-workers (19) observed that PTH increased FZD1/2 mRNA levels in UMR cells. Other findings establish that PTH increases β-catenin levels in UMR, MC3T3-E1, and SAOS cells (16, 17, 20) and that ablation of the Wnt antagonist, secreted frizzled-related protein, blunts the anabolic action of PTH (21). More recently, Cao and co-workers (18) showed that PTH1R signals through LRP6. Further evidence for the interaction of PTH and β-catenin pathways in regulating bone turnover comes from studies showing that overexpression of sFRP1 attenuates PTH-dependent bone anabolism (22). Together, these and other studies imply that the actions of PTH may be partially mediated through β-catenin signaling. The mechanisms underlying PTH1R and FZD cross-talk are unknown. We now describe multiple lines of cross-talk between the two pathways and show that PTH activates β-catenin in an LRP- and Wnt-independent manner.
EXPERIMENTAL PROCEDURES
Cell Culture
CHO cells (Invitrogen) were cultured in Ham's F-12 medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10 μg/ml blasticidin. UMR-106 and UAMS-32P cells were cultured in Dulbecco's modified Eagle's medium/F-12 medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. MC4 cells were obtained from Dr. G. Xiao (University of Pittsburgh) and cultured in α-modified minimum essential medium with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2, 95% air.
Short Hairpin RNA Treatments
The expression of Dvl1, Dvl2, and Dvl3 in UMR cells was determined by quantitative RT-PCR. Dvl2 mRNA accounted for over 90% of the Disheveled mRNA expressed in these cells (not shown). Constructs coding for specific shRNA targeted against rat Dvl2 were purchased from SA Biosciences (Frederick, MD). Four different constructs were screened for their ability to knock down Dvl2 expression in UMR cells. The two most efficient constructs were selected for further use. The sequences targeted by these constructs (shDvl2-1 and shDvl2-2, respectively) are GCCTACCTTCTCCTACCAATACC and TTCAACTTGGTGCTCTTCTTAGT. UMR-106 cells were co-transfected with 1 μg of TOP-Flash luciferase reporter and 2 μg of either a scrambled shRNA or shDvl2 plasmids. Two days following transfection, the cells were treated with either vehicle, 100 ng/ml recombinant Wnt3a, 100 nm PTH(1–34), or 5 mm LiCl for 24 h. Cell lysates were assayed for luciferase expression using a commercial luciferase assay system (Promega, Madison, WI) as per the manufacturer's instructions.
Coimmunoprecipitation and Immunoblot Analysis
Interactions of PTH1R or FZD with Dvl were analyzed as described (23). Briefly, 6-well plates of the indicated cells were transiently transfected with HA-PTH1R, HA-PTH1R(ETVA), HA-PTH1R(480-stop), HA-PTH1R(470-stop), HA-FZD, Myc-Dvl, or the respective empty vector. 48 h later, the cells were lysed with Nonidet P-40 (50 mm Tris, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40) supplemented with protease inhibitor mixture I and incubated for 15 min on ice. Solubilized materials were incubated overnight at 4 °C with HA.11 monoclonal affinity matrix. Total lysates and immunoprecipitated protein, eluted by the addition of SDS sample buffer, were analyzed by SDS-polyacrylamide gels and transferred to Immobilon-P membranes (Millipore) using the semidry method (Bio-Rad). Membranes were blocked overnight at 4 °C with 5% nonfat dried milk in Tris-buffered saline plus Tween 20 (TBST) and incubated with the indicated antibodies (HA (Covance, catalog no. MMS-101R), Myc (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), catalog no. SC-40), active β-catenin (Millipore, catalog no. 05-665), or β-catenin (Millipore, catalog no. 06-734)) for 2 h at room temperature. The membranes were then washed and incubated with goat anti-rabbit IgG or sheep anti-mouse IgG conjugated to horseradish peroxidase at a 1:5000 dilution for 1 h at room temperature. Protein bands were visualized with a luminol-based enhanced chemiluminescence substrate.
β-Catenin
MC4 cells were passaged onto 6-well plates and 24 h later were transfected with 1 μg/well TopFlash (Super 8× TopFlash, Addgene) and 1 μg/well Xdd1, a mutant of Xenopus Xdsh1 harboring a PDZ domain deletion (24), or empty vector, as indicated, using FuGENE 6 (Roche Applied Science). 48 h after transfection, cells were treated with 100 nm PTH(1–34) for 6 h. Lysates were prepared using Reporter Lysis Buffer (Promega, catalog no. E-3971). Luciferase activity was assayed using the BrightGlo luciferase assay system (Promega, catalog no. E-2620). 10 μl of lysate was added to a single tube. An equal volume of BrightGlo substrate was then added, and luminescence was measured for 10 s in a Turner BioSystems luminometer (model TD-20/20).
Imaging
Myc- and mRFP1-tagged Dvl2 constructs were obtained from T. Kirchhausen (Harvard Medical School). Live cell imaging of the translocation of mRFP1-Dvl2 to the plasma membrane was done by confocal microscopy using a Zeiss LSM5 equipped with a Harvard Biosystems incubation chamber maintained at 37 °C. Images were collected every 20 s.
To measure β-catenin translocation to the nucleus, ROS 17/2.8 cells were treated with 100 nm PTH(1–34) for 8 h, fixed, permeabilized, blocked with 5% goat serum, and incubated with a commercial anti-β-catenin antibody (Millipore). β-Catenin was detected using a TRITC-labeled secondary antibody. The cells were then examined using an Olympus Fluoview 1000 confocal microscope equipped with 405- and 561-nm lasers.
To measure PTH1R internalization, cells were co-transfected with a human EGFP-tagged PTH1R and mRFP-Dvl2. Receptor internalization was measured using total internal reflection fluorescence (TIRF) microscopy as described (25). Briefly, cells expressing both PTH1R-EGFP and mRFP-Dvl2 were identified by epifluorescence. The cells were then challenged with 100 nm PTH(1–34), and the disappearance of the surface-delimited PTH1R-EGFP was determined by TIRF. Data were collected at 20-s intervals for up to 20 min. The rate of internalization was determined from the fit of the collected data to a single exponential (25).
Osteoclastogenesis
Nonadherent bone marrow cells were prepared by removing femurs from 30–90-day-old C57BL/6J mice and flushing the marrow cavity with minimum essential medium (Invitrogen) containing 15% fetal bovine serum (Hyclone). Marrow cells were seeded at a density of 2.5 × 105 cells/cm2 in the same medium and cultured for 48 h. Nonadherent cells were collected and seeded at a density of 2 × 104 cells/cm2 on a cushion of UAMS-32 osteoblastic cells in minimum essential medium containing 10% fetal bovine serum. PTH(1–34) was added at the indicated concentrations, and the co-cultures were maintained at 37 °C in 5% CO2. On day 3, one-half of the medium was replaced with fresh medium. After 6–8 days, cells were fixed and stained for tartrate-resistant acid phosphatase. The plate was scanned, and staining density was determined with Image J (National Institutes of Health, Bethesda, MD).
Statistics
Data are presented as the mean ± S.E., where n indicates the number of independent experiments. Curve fitting and data analysis were performed using GraphPad Prism (GraphPad Software, Inc., San Diego, CA). Multiple comparisons were evaluated by analysis of variance with post-test repeated measures analyzed by the Bonferroni procedure. Differences greater than p ≤ 0.05 were assumed to be significant.
RESULTS
Direct Interactions between PTH1R and Dvl
Recruitment of Dvl to the plasma membrane is mediated by direct interactions between FZD and the PDZ domain of Dvl proteins. These interactions occur primarily through the binding of a critical region (KTXXXW) within the proximal portion of the carboxyl terminus of all 10 FZD receptors (26). Analysis of the PTH1R sequence revealed the presence of a similar motif (KSWSRW; see Fig. 1A). The putative Dvl-binding domain of the PTH1R starts at position 472 in a portion of the intracellular tail topologically comparable with that of FZD. We hypothesized that the PTH1R controls the β-catenin pathway by binding to this domain of Dvl.
FIGURE 1.
Dvl interactions with the PTH1R. A, seven-transmembrane receptor model showing sequence and common location of the Dvl-binding domain in PTH1R and FZD. A PDZ recognition motif is present at the carboxyl terminus of the PTH1R and of FZD1, -2, -4, and -7. B, interactions between PTH1R and Dvl. Coimmunoprecipitation experiments were conducted on CHO-N10 cells stably expressing a human HA-tagged PTH1R construct (23) that were transfected with Myc-Dvl2. Where indicated, cells were treated with 100 nm PTH(1–34) (15 min). The HA-PTH1R construct was immunoprecipitated (IP), and the presence of Myc-Dvl2 in the immunoprecipitate was determined by immunoblotting (IB). C, interactions between PTH1R and Dvl. CHO-N10 cells stably expressing HA-PTH1R were transfected with Myc-Dvl2. Cell lysates were immunoprecipitated with specific anti-Myc antibody, and the presence of the PTH1R in the immunoprecipitate was determined using anti-HA antibodies. D, the interactions of the PTH1R with Dvl2 are mediated by an amino acid sequence contained between residues 470 and 480 of the PTH1R. CHO-N10 cells were transfected with wild-type PTH1R (ETVM), a receptor harboring a carboxyl-terminal mutation that abolishes classical PDZ-PDZ domain interactions (ETVA), or receptor forms truncated at position 480 or 470. Immunoprecipitation and detection were performed as described (23, 51).
To determine if the PTH1R interacts with Dvl proteins, we performed co-immunoprecipitation experiments using HA-tagged PTH1R and Myc-tagged Dvl2. As shown in Fig. 1, B and C, the PTH1R co-immunoprecipitates with Dvl2. To identify the structural determinants required for PTH1R interactions with Dvl2, we co-transfected the Myc-Dvl2 construct with various HA-tagged PTH1R mutants and performed additional coimmunoprecipitation studies. The results are shown in Fig. 1D. Wild-type PTH1R (ETVM) co-immunoprecipitated with Dvl2 (lane 1). It should be noted that the PTH1R sequence contains a canonical carboxyl-terminal PDZ ligand (590ETVM). This motif is not responsible for Dvl binding because a PTH1R harboring a mutated carboxyl-terminal PDZ recognition domain (ETVA) bound Dvl2 comparably with the wild type receptor (lane 2). Mutation of ETVM to ETVA is sufficient to abolish PTH1R interaction with the PDZ protein NHERF1 (27). Likewise, a truncated PTH1R lacking the PDZ-binding domain but bearing the putative Dvl recognition domain (480-stop) also co-immunoprecipitated with Dvl2 (lane 3). This finding demonstrates that PTH1R interactions with Dvl are not mediated by the carboxyl-terminal canonical PDZ-binding motif of the PTH1R. Importantly, a PTH1R lacking the putative Dvl-binding domain (470-stop) failed to co-immunoprecipitate with Dvl (lane 4), although it was well expressed (bottom). These findings show that the PTH1R interacts with Dvl through a specific recognition sequence located between residues 470 and 480. This result supports the hypothesis that the 472KSWSRW sequence in PTH1R (Fig. 1A) constitutes the Dvl-binding motif.
PTH Activation and Stabilization of β-Catenin
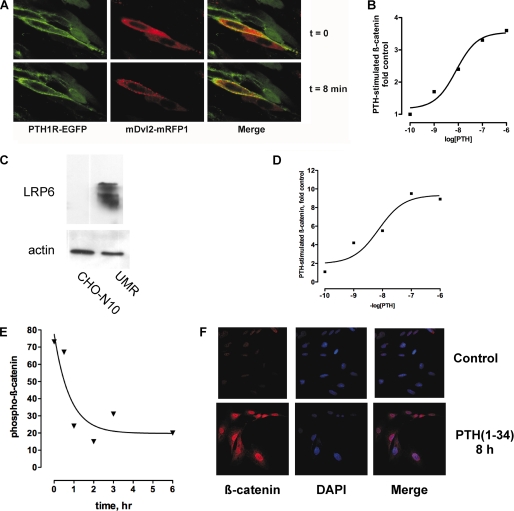
A defining characteristic of Wnt signaling pathways is ligand-induced redistribution and subsequent oligomerization of Dvl proteins (28). Fig. 2A shows that PTH promotes translocation of Dvl to the cell membrane. CHO cells expressing PTH1R-EGFP (green) were transiently transfected with red fluorescent mRFP1-Dvl2. Images were collected at 20-s intervals. Before the addition of PTH(1–34), the PTH1R was membrane-delimited, and Dvl2 was diffusely expressed throughout the cytoplasm (Fig. 2A and supplemental Video 1). Following the addition of PTH, Dvl was recruited to the cell membrane, where it concentrated in discrete puncta a few min after treatment (Fig. 2A, arrows), whereas the PTH1R internalized. Unlike FZD4, which significantly co-localizes with Dvl2 even in the absence of ligand (29), the PTH1R and Dvl2 co-localize only transiently after receptor activation. This result suggests somewhat weaker interactions between PTH1R and Dvl or the presence of additional regulatory proteins. Importantly, Fig. 2A and supplemental Video 1 also show cells expressing only one of the fluorescent constructs. In this situation, PTH treatment failed to induce Dvl2 translocation in cells that did not express the PTH1R. Thus, activation of the PTH1R causes translocation of the Dvl2 to the cell surface.
FIGURE 2.
Recruitment of Dvl and β-catenin activation, stabilization, and nuclear translocation following activation of PTH1R. A, PTH treatment induces translocation of Dvl2 to the plasma membrane. CHO cells expressing EGFP-PTH1R (green) were transiently transfected with mRFP-Dvl2. Confocal images were collected at 20-s intervals after the addition of 100 nm PTH(1–34). The images shown correspond to t = 0 and 8 min. Note the accumulation of red Dvl2 in discrete puncta (arrows, center panels). See supplemental Video 1. B, PTH-dependent β-catenin activation in CHO cells. CHO cells were transfected with HA-tagged human PTH1R and reporter TOP/FOP plasmids as described under “Experimental Procedures.” C, CHO cells do not express LRP6. The expression of LRP6 in UMR-106 cells is shown for comparative purposes. D, concentration-dependent stimulation of β-catenin activation by PTH. MC4 cells were transiently transfected with TOPFlash and 48 h later were studied as described under “Experimental Procedures.” E, stabilized β-catenin (i.e. dephosphorylated β-catenin) in MC4 bone cells after stimulation with PTH(1–34). F, translocation of activated β-catenin to the nucleus. ROS 17/2.8 cells were challenged with 100 nm PTH(1–34) for 8 h and then fixed and stained for dephosphorylated β-catenin and 4′,6-diamidino-2-phenylindole (DAPI), to stain nuclei.
To determine if recruitment of Dvl2 to the cell surface correlates with downstream functions of the PTH1R, we measured β-catenin activation using a TCF/Lef reporter (30). The results shown in Fig. 2B demonstrate that PTH stimulates β-catenin activity in CHO cells transfected with the human PTH1R. Notably, LRP5/6 expression in CHO cells determined by quantitative RT-PCR is negligibly low. mRNA for LRP5 were undetectable, and LRP6 was at background levels (data not shown). Immunoblots with specific antibodies confirm the low expression of LRP6 (Fig. 2C), strengthening the conclusion that β-catenin activation by the PTH1R is independent of LRP5/6 in CHO cells.
The above results demonstrate activation of β-catenin by PTH in heterologous expression systems. These findings were validated in bone cells expressing endogenous levels of the PTH1R. Fig. 2D shows that in MC4 cells, PTH activated β-catenin in a concentration-dependent manner. Activation of β-catenin occurs upon dissociation of the axin-APC-GSK3β complex, which results in reduced phosphorylation and stabilization of non-phosphorylated β-catenin. β-Catenin then accumulates in the cytoplasm before being translocated to the nucleus, where it exerts its transcriptional effects. The results in Fig. 2, E and F, illustrate the time course of β-catenin accumulation and nuclear translocation after challenge with PTH(1–34), respectively. Therefore, stimulation of the PTH1R results in the translocation and oligomerization of Dvl2 at the plasma membrane and the inhibition of β-catenin phosphorylation, which in turn leads to the activation and nuclear translocation of β-catenin. These phenomena are benchmarks of the activation of the canonical Wnt signaling pathway.
Dvl Mediates PTH1R Actions
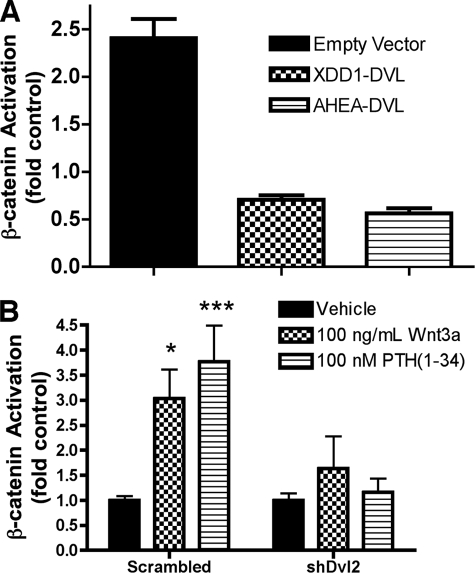
To analyze the role of Dvl proteins in mediating the effects of PTH, we used two different Dvl mutants. Xdd1 is a well characterized dominant negative mutant of Xenopus Dishevelled (Xdsh) lacking amino acids 301–381, which include the core of the Xdsh PDZ domain (31). AHEA-Dvl2 is a double point mutant that blocks FZD4 internalization and planar cell polarity signaling (29). As shown in Fig. 3A, the stimulation of β-catenin signaling by PTH was blocked by expression of either Dvl mutant compared with the empty vector control. These results support the conclusion that the activation of β-catenin by PTH is mediated by the interactions of the PTH1R with Dvl proteins. To confirm the role of Dvl in PTH1R-mediated activation of β-catenin, the endogenous levels of Dvl proteins were manipulated using short hairpin RNA constructs. The expression of the isoforms Dvl1, Dvl2, and Dvl3 in UMR cells was examined using quantitative reverse transcription-PCR, which demonstrated that Dvl2 accounted for >90% of the Dvl message present (supplemental Fig. 1). Therefore, we transfected UMR cells with shRNA constructs that specifically targeted Dvl2. This treatment reduced Dvl2 protein levels by over 50% (supplemental Fig. 2). This number is an underestimate of the actual silencing effects of the shRNA constructs because the data shown were not corrected to account for transfection efficiency. β-Catenin activation was measured in cells co-transfected with Dvl2-targeted shRNA or scrambled control vectors (Fig. 3B). As shown, PTH-dependent β-catenin activity was severely impaired by expression of Dvl2-specific shRNA.
FIGURE 3.
Dvl requirement for PTH1R activation of β-catenin. A, inhibition of β-catenin activation by the expression of dominant negative Dvl constructs, XDD1 and AHEA-Dvl2. UMR-106 cells were co-transfected with TOPFlash reporter (or the control plasmid, FOPFlash) and either empty vector or the specified Dvl mutant (XDD1 or AHEA-Dvl2). β-Catenin activation was determined from chemiluminescence data as described under “Experimental Procedures.” B, Dvl2-specific shRNA inhibits PTH- and Wnt3A-dependent β-catenin activation in UMR-109 cells. In these experiments, Dvl2 expression was inhibited by co-transfecting two plasmids coding for Dvl2-specific shRNA together with the TOPFlash or FOPFlash reporters.
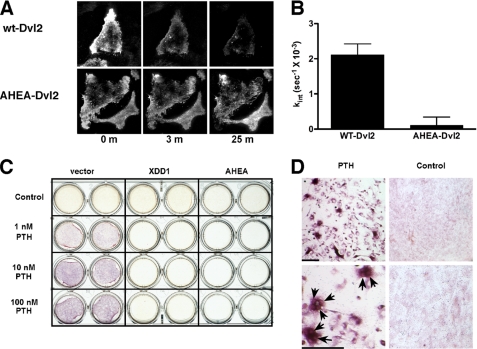
We further examined the effects of Dvl2 mutants on the function of the PTH1R. Neither Xdd1 nor AHEA-Dvl2 interfered with PTH-induced activation of adenylyl cyclase (supplemental Fig. 3). However, overexpression of AHEA-Dvl2 interfered with PTH1R endocytosis (Fig. 4, A and B). Notably, both mutants blocked PTH-induced osteoclastogenesis as determined by tartrate-resistant acid phosphatase (TRAP) staining (Fig. 4, C and D). These results strongly support the hypothesis that some crucial effects of PTH are mediated by direct binding of Dvl proteins by the PTH1R.
FIGURE 4.
Dvl is required for PTH-dependent PTH1R internalization and osteoclastogenesis. A, AHEA-Dvl2 blocks ligand-induced PTH1R internalization. Rat osteosarcoma cells transfected with PTH1R-EGFP and mRFP1-Dvl2 were imaged at 20-s intervals after the addition of 100 nm PTH(1–34) using TIRF. The data obtained with cells that express wild-type Dvl2 are shown in the top panel. The bottom panel shows the results of the expression of AHEA-Dvl2. B, summary of the PTH1R internalization results obtained. The rate constant of internalization of the PTH1R was calculated from TIRF microscopy data. Only cells that expressed both proteins (as determined from green and red fluorescence measurements) were used in these calculations. The data show the average of five different cell plates examined in three independent experiments. C, PTH promotes osteoclastogenesis through a β-catenin-mediated pathway. UAMS-32P cells were co-cultured with non-adherent bone marrow cells (52). PTH(1–34) at increasing concentrations was added in the presence of empty vector, XDD1 dominant negative Dvl, or AHEA dominant negative Dvl. After 6 days, tartrate-resistant acid phosphatase activity was determined using naphthol phosphate as a substrate and fast garnet to label the product as a red-purple precipitate. D, low and high magnification views of the cells shown in C. Tartrate-resistant acid phosphatase staining was primarily localized to multinucleated osteoclasts (arrowheads). Bar, 100 μm.
The present results strongly support the hypothesis that some crucial effects of PTH are mediated by direct binding of Dvl proteins by the PTH1R. We excluded the possibility that the inhibitory actions of the Dvl mutants arose from interfering with PTH1R signaling through adenylyl cyclase by measuring the effects of XDD1 and AHEA-Dvl2 constructs on PTH-stimulated cAMP. Neither Xdd1 nor AHEA-Dvl2 interfered with PTH-induced activation of adenylyl cyclase (supplemental Fig. 3).
DISCUSSION
The findings described here identify Dvl as a molecular router, integrating signals derived from FZD and PTH1R to control osteoclast formation. Regulation of Wnt signaling by GPCRs other than FZD has been described (32–34). However, we demonstrate here for the first time that Dvl proteins play an important role in the activation of β-catenin by classical GPCRs. In all previous studies, the activation of β-catenin has been linked to the activation of classical G-protein mediated pathways. For instance, stimulation of α-adrenergic and endothelin-1 receptors in cardiomyocytes leads to the activation of β-catenin by a mechanism that involves the recruitment of Akt to the β-catenin degradation complex and the subsequent phosphorylation and inactivation of GSK3β (32). Likewise, the lysophosphatidic acid receptors LPA2 and LPA3 activate β-catenin-dependent pathways by a mechanism downstream of the activation of PKC (33). Finally, PGE2 receptors, acting through protein kinase A, stabilize β-catenin in colon cancer cells (34). Importantly, β-arrestins interact with phosphorylated Dvl, and transient co-expression of β-arrestin-1 with either Dvl1 or Dvl2 stimulates a β-catenin reporter even in the absence of Wnt (35). More recently, it has been suggested that β-arrestin is required for the Dvl-mediated disruption of the β-catenin destruction complex (36). The work presented here demonstrates that the PTH1R activates β-catenin signaling by direct interactions with Dvl.
Dvl contains three functional domains: an amino-terminal Dix domain, a centrally located type 1 PDZ domain, and a carboxyl-terminal Dep domain (37). The structural basis for the interaction of FZD with Dvl is somewhat unconventional. Thus, despite the presence of a carboxyl-terminal PDZ binding motif, it has been argued that Dvl interacts with FZD through a non-canonical, internal sequence (38, 39). Interestingly, Dvl binds to the PTH1R through the comparable K(S/T)XXXW sequence despite the presence of a carboxyl-terminal PDZ recognition motif. Thus, Dvl interacted robustly with the PTH1R even following mutation of the PDZ-binding motif or truncation of the PTH1R distal to the putative Dvl-binding sequence.
The mechanism by which the PTH1R promotes the redistribution of Dvl involves direct interactions of the receptor with Dvl. Arrestin-mediated Dvl2 binding can be ruled out because the PTH1R(480-stop) binds Dvl2 with an efficiency comparable with that of the wild type receptor (see Fig. 1D). This truncated receptor is devoid of the core of serines that are phosphorylated by G-protein receptor kinases and mediate the binding of arrestin, located between residues 489 and 501 (40, 41). The direct involvement of Dvl in the signaling pathways leading to β-catenin activation is further supported by the finding that two different Dvl mutants block TCF/β-catenin-dependent transcription and PTH-induced osteoclastogenesis. Were the PTH1R modulating these downstream effects through the activation of protein kinase A or Akt and the phosphorylation of GSK3β, the actions of PTH would be expected to be insensitive to the expression of dominant negative Dvl mutants, and this was not the case.
Recent studies show that PTH can activate Wnt signaling despite overexpression of Dkk1 (42). The results described here are consistent with these findings and may explain the results with Dkk1. Upon stimulation by PTH, the PTH1R recruits Dvl and does so independent of FZD and LRP5/6 (and therefore of Dkk1).
The present work demonstrates for the first time that GPCRs other than the members of the FZD subfamily may interact with and activate Dvl proteins. The possibility that additional receptors may also use this mechanism to activate β-catenin-dependent protein expression remains to be explored.
The dominant negative effects of the AHEA-Dvl2 mutant require additional comment. AHEA-Dvl2 interacts normally with FZD4 but cannot bind AP-2 and, as a consequence, acts as a dominant negative for Wnt5a-induced FZD4 internalization (29). This effect was recapitulated with the PTH1R (Fig. 4, A and B). However, although AHEA-Dvl2 interferes with Wnt5a-dependent JNK phosphorylation, it does not block Wnt5a-induced β-catenin activation in HEK-293 (human embryonic kidney) cells (29). AHEA-Dvl2 clearly disrupted PTH-induced PTH1R osteoclastogenesis and β-catenin-dependent gene transcription in the bone-derived cell models used here. Because AHEA-Dvl2 blocked PTH1R endocytosis, this phenomenon can be explained by a requirement for PTH1R internalization in the activation of the β-catenin pathway. This is consistent with recent reports showing that β-arrestin plays an important role in the coupling of Wnt signaling to β-catenin activation (36).
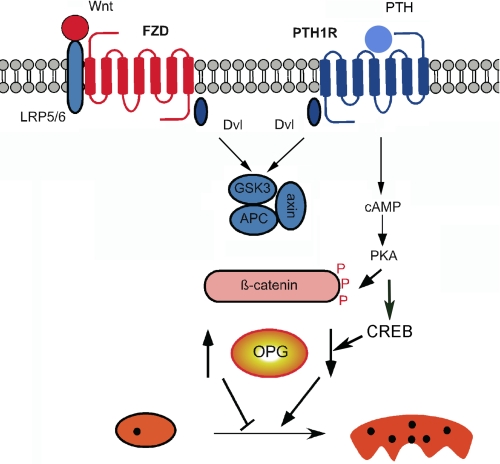
The results described here regarding the effects of PTH versus Wnt activation of β-catenin present an apparent paradox. Although both ligands lead to activation of β-catenin, they have opposite actions on osteoclastogenesis. Although β-catenin mediates convergent actions of PTH1R and FZD upon mineralization, it promotes divergent actions on osteoclastogenesis as we propose here. A model incorporating this scheme is shown in Fig. 5.
FIGURE 5.
Working model for divergent effects of PTH1R and FZD signaling and action on osteoclastogenesis. Activation of FZD and of PTH1R stimulates β-catenin. In the case of FZD, this leads to inhibition of osteoclastogenesis, whereas activation of the PTH1R increases osteoclastogenesis. We propose that the divergent actions of FZD or PTH1R effects are due to protein kinase A-dependent phosphorylation of β-catenin and GSK3β and the activation of CREB. PKA, protein kinase A.
PTH promotes osteoclastogenesis, whereas Wnt stimulation of β-catenin blocks osteoclastogenesis. One crucial difference in the signaling cascades downstream of Wnt ligands and PTH is the activation of cAMP production by the latter. We propose that the modulation of β-catenin signaling by cAMP and protein kinase A during stimulation with PTH contributes significantly to the differences observed between Wnt- and PTH-driven responses. These effects may be due to differential phosphorylation of β-catenin (43, 44) or of GSK3 (45) by protein kinase A. Alternatively, the generation of cAMP activates the cAMP-response element-binding protein (CREB), which in turn interacts with β-catenin and regulates its function (46, 47). Phosphorylation, whether of β-catenin or GSK3, may occur at different sites or may be of different duration (i.e. transient versus sustained). A clear distinction between Wnt- and PTH-dependent β-catenin activation is that the latter proceeds in an environment with elevated cAMP and protein kinase A activity and, perhaps, CREB. We propose that phosphorylation by protein kinase A of β-catenin (43, 44) or of GSK3 (45) modifies the β-catenin response, leading to divergent patterns of gene expression (Fig. 5). Consistent with this theory, independent studies established that even in the same cell type, protein kinase A can promote or prevent β-catenin-dependent effects, depending on the particular agonist (48). Thus, we envision that Wnt or PTH-stimulated β-catenin activation may evoke opposite actions on osteoclastogenesis by eliciting similar β-catenin responses in a different intracellular background.
PTH promotes bone accretion when administered intermittently but promotes bone resorption when continuously applied. Intermittent PTH administration of PTH causes phosphorylation of LRP6 and stabilization of β-catenin in mouse osteoblasts (18). By contrast, continuous PTH treatment failed to stimulate β-catenin. Thus, the anabolic action of PTH may arise in part from the participation of the canonical β-catenin pathway but independent of Wnt.
Other explanations can be advanced for the different effects of β-catenin activation by Wnt and PTH. For instance, divergent effects of PTH and Wnt on β-catenin may arise from the expression patterns of PTH1R and FZD, respectively, during osteoblast maturation (49, 50). Additionally or alternatively, differences in β-catenin actions upon osteoclastogenesis could be due to distinct actions of PTH or Wnt upon apoptosis. Irrespective of the mechanism, it is likely to have important implications for understanding and treating skeletal disorders.
Supplementary Material
Acknowledgment
We appreciate the assistance of Dr. M. Feili-Hariri in isolating marrow stromal cells.
This work was supported, in whole or in part, by National Institutes of Health Grants DK54171 (to P. A. F.) and GM08208 (to D. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Video 1 and Figs. 1–3.
- NFκB
- nuclear factor-κB
- JNK
- Jun kinase
- PTH
- parathyroid hormone
- CHO
- Chinese hamster ovary
- TIRF
- total internal reflection fluorescence
- TRITC
- tetramethylrhodamine isothiocyanate
- EGFP
- enhanced green fluorescent protein
- GPCR
- G protein-coupled receptor
- CREB
- cAMP-response element-binding protein
- shRNA
- short hairpin RNA
- HA
- hemagglutinin.
REFERENCES
- 1.Malbon C. C., Wang H. Y. (2006) Curr. Top. Dev. Biol. 72, 153–166 [DOI] [PubMed] [Google Scholar]
- 2.Gordon M. D., Nusse R. (2006) J. Biol. Chem. 281, 22429–22433 [DOI] [PubMed] [Google Scholar]
- 3.Schulte G., Bryja V. (2007) Trends Pharmacol. Sci. 28, 518–525 [DOI] [PubMed] [Google Scholar]
- 4.Malbon C. C. (2004) Front. Biosci. 9, 1048–1058 [DOI] [PubMed] [Google Scholar]
- 5.Barnes M. R., Duckworth D. M., Beeley L. J. (1998) Trends Pharmacol. Sci. 19, 399–400 [DOI] [PubMed] [Google Scholar]
- 6.Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Crompton A., Chan A. C., Anderson J. M., Cantley L. C. (1997) Science 275, 73–77 [DOI] [PubMed] [Google Scholar]
- 7.Wang S., Raab R. W., Schatz P. J., Guggino W. B., Li M. (1998) FEBS Lett. 427, 103–108 [DOI] [PubMed] [Google Scholar]
- 8.Weinman E. J., Hall R. A., Friedman P. A., Liu-Chen L. Y., Shenolikar S. (2006) Annu. Rev. Physiol. 68, 491–505 [DOI] [PubMed] [Google Scholar]
- 9.Baron R., Rawadi G. (2007) Endocrinology 148, 2635–2643 [DOI] [PubMed] [Google Scholar]
- 10.Krishnan V., Bryant H. U., Macdougald O. A. (2006) J. Clin. Invest. 116, 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmann C. (2006) Trends Cell Biol. 16, 151–158 [DOI] [PubMed] [Google Scholar]
- 12.Bodine P. V., Komm B. S. (2006) Rev. Endocr. Metab. Disord. 7, 33–39 [DOI] [PubMed] [Google Scholar]
- 13.Lanske B., Karaplis A. C., Lee K., Luz A., Vortkamp A., Pirro A., Karperien M., Defize L. H., Ho C., Mulligan R. C., Abou-Samra A. B., Jüppner H., Segre G. V., Kronenberg H. M. (1996) Science 273, 663–666 [DOI] [PubMed] [Google Scholar]
- 14.Kong Y. Y., Yoshida H., Sarosi I., Tan H. L., Timms E., Capparelli C., Morony S., Oliveira-dos-Santos A. J., Van G., Itie A., Khoo W., Wakeham A., Dunstan C. R., Lacey D. L., Mak T. W., Boyle W. J., Penninger J. M. (1999) Nature 397, 315–323 [DOI] [PubMed] [Google Scholar]
- 15.Li J., Sarosi I., Yan X. Q., Morony S., Capparelli C., Tan H. L., McCabe S., Elliott R., Scully S., Van G., Kaufman S., Juan S. C., Sun Y., Tarpley J., Martin L., Christensen K., McCabe J., Kostenuik P., Hsu H., Fletcher F., Dunstan C. R., Lacey D. L., Boyle W. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1566–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkarni N. H., Halladay D. L., Miles R. R., Gilbert L. M., Frolik C. A., Galvin R. J., Martin T. J., Gillespie M. T., Onyia J. E. (2005) J. Cell. Biochem. 95, 1178–1190 [DOI] [PubMed] [Google Scholar]
- 17.Tobimatsu T., Kaji H., Sowa H., Naito J., Canaff L., Hendy G. N., Sugimoto T., Chihara K. (2006) Endocrinology 147, 2583–2590 [DOI] [PubMed] [Google Scholar]
- 18.Wan M., Yang C., Li J., Wu X., Yuan H., Ma H., He X., Nie S., Chang C., Cao X. (2008) Genes Dev. 22, 2968–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan S. D., Karpf D. B., Fowlkes M. E., Hooks M., Bradley M. S., Vuong V., Bambino T., Liu M. Y., Arnaud C. D., Strewler G. J., Nissenson R. A. (1992) J. Biol. Chem. 267, 25202–25207 [PubMed] [Google Scholar]
- 20.Suzuki A., Ozono K., Kubota T., Kondou H., Tachikawa K., Michigami T. (2008) J. Cell. Biochem. 104, 304–317 [DOI] [PubMed] [Google Scholar]
- 21.Bodine P. V., Seestaller-Wehr L., Kharode Y. P., Bex F. J., Komm B. S. (2007) J. Cell. Physiol. 210, 352–357 [DOI] [PubMed] [Google Scholar]
- 22.Yao W., Cheng Z., Shahnazari M., Dai W., Johnson M. L., Lane N. E. (2010) J Bone Miner. Res. 25, 190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B., Bisello A., Yang Y., Romero G. G., Friedman P. A. (2007) J. Biol. Chem. 282, 36214–36222 [DOI] [PubMed] [Google Scholar]
- 24.Hay E., Faucheu C., Suc-Royer I., Touitou R., Stiot V., Vayssière B., Baron R., Roman-Roman S., Rawadi G. (2005) J. Biol. Chem. 280, 13616–13623 [DOI] [PubMed] [Google Scholar]
- 25.Wheeler D., Sneddon W. B., Wang B., Friedman P. A., Romero G. (2007) J. Biol. Chem. 282, 25076–25087 [DOI] [PubMed] [Google Scholar]
- 26.Umbhauer M., Djiane A., Goisset C., Penzo-Méndez A., Riou J. F., Boucaut J. C., Shi D. L. (2000) EMBO J. 19, 4944–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sneddon W. B., Syme C. A., Bisello A., Magyar C. E., Rochdi M. D., Parent J. L., Weinman E. J., Abou-Samra A. B., Friedman P. A. (2003) J. Biol. Chem. 278, 43787–43796 [DOI] [PubMed] [Google Scholar]
- 28.Gao C., Chen Y. G. (2010) Cell. Signal. 22, 717–727 [DOI] [PubMed] [Google Scholar]
- 29.Yu A., Rual J. F., Tamai K., Harada Y., Vidal M., He X., Kirchhausen T. (2007) Dev. Cell 12, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 31.Sokol S. Y. (1996) Curr. Biol. 6, 1456–1467 [DOI] [PubMed] [Google Scholar]
- 32.Haq S., Michael A., Andreucci M., Bhattacharya K., Dotto P., Walters B., Woodgett J., Kilter H., Force T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4610–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M., Zhong W. W., Srivastava N., Slavin A., Yang J., Hoey T., An S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castellone M. D., Teramoto H., Williams B. O., Druey K. M., Gutkind J. S. (2005) Science 310, 1504–1510 [DOI] [PubMed] [Google Scholar]
- 35.Chen W., Hu L. A., Semenov M. V., Yanagawa S., Kikuchi A., Lefkowitz R. J., Miller W. E. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14889–14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryja V., Gradl D., Schambony A., Arenas E., Schulte G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6690–6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarz-Romond T., Metcalfe C., Bienz M. (2007) J. Cell Sci. 120, 2402–2412 [DOI] [PubMed] [Google Scholar]
- 38.Wong H. C., Bourdelas A., Krauss A., Lee H. J., Shao Y., Wu D., Mlodzik M., Shi D. L., Zheng J. (2003) Mol. Cell 12, 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.London T. B., Lee H. J., Shao Y., Zheng J. (2004) Biochem. Biophys. Res. Commun. 322, 326–332 [DOI] [PubMed] [Google Scholar]
- 40.Malecz N., Bambino T., Bencsik M., Nissenson R. A. (1998) Mol. Endocrinol. 12, 1846–1856 [DOI] [PubMed] [Google Scholar]
- 41.Tawfeek H. A., Qian F., Abou-Samra A. B. (2002) Mol. Endocrinol. 16, 1–13 [DOI] [PubMed] [Google Scholar]
- 42.Guo J., Liu M., Yang D., Bouxsein M. L., Saito H., Galvin R. J., Kuhstoss S. A., Thomas C. C., Schipani E., Baron R., Bringhurst F. R., Kronenberg H. M. (2010) Cell Metab. 11, 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hino S., Tanji C., Nakayama K. I., Kikuchi A. (2005) Mol. Cell. Biol. 25, 9063–9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taurin S., Sandbo N., Qin Y., Browning D., Dulin N. O. (2006) J. Biol. Chem. 281, 9971–9976 [DOI] [PubMed] [Google Scholar]
- 45.Fang X., Yu S. X., Lu Y., Bast R. C., Jr., Woodgett J. R., Mills G. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11960–11965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takemaru K. I., Moon R. T. (2000) J. Cell Biol. 149, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hecht A., Vleminckx K., Stemmler M. P., van Roy F., Kemler R. (2000) EMBO J. 19, 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogarth D. K., Sandbo N., Taurin S., Kolenko V., Miano J. M., Dulin N. O. (2004) Am. J. Physiol. Cell Physiol 287, C449–C456 [DOI] [PubMed] [Google Scholar]
- 49.Ek E. T., Dass C. R., Choong P. F. (2006) Crit. Rev. Oncol. Hematol. 60, 1–8 [DOI] [PubMed] [Google Scholar]
- 50.Westendorf J. J., Kahler R. A., Schroeder T. M. (2004) Gene 341, 19–39 [DOI] [PubMed] [Google Scholar]
- 51.Wang B., Yang Y., Friedman P. A. (2008) Mol. Biol. Cell 19, 1637–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu Q., Jilka R. L., Manolagas S. C., O'Brien C. A. (2002) J. Biol. Chem. 277, 48868–48875 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.