Abstract
LDL receptor-related protein (LRP1) is expressed by Schwann cells in vivo mainly after injury to the peripheral nervous system (PNS). Schwann cells in primary culture, which provide a model of Schwann cells in the injured PNS, also express abundant LRP1. Herein, we show that LRP1 gene-silencing or treatment with receptor-associated protein (RAP) promotes Schwann cell adhesion and inhibits cell migration on fibronectin. LRP1 gene-silencing also resulted in the formation of prominent focal adhesions and actin stress fibers. These changes, which were induced by loss of LRP1 expression or activity, were explained mechanistically by an increase in activated RhoA, coupled with a decrease in activated Rac1. Known LRP1 ligands, including matrix metalloprotease-9, tissue-type plasminogen activator, and α2-macroglobulin activated Rac1 in LRP1-expressing Schwann cells. An inhibitor of Rac1 activation promoted Schwann cell adhesion. Conversely, in cells in which LRP1 was silenced, a Rho kinase inhibitor promoted migration and inhibited adhesion. These results demonstrate that direct binding of ligands to LRP1 controls activation of small Rho family GTPases. The effects of LRP1 gene-silencing and RAP implicate autocrine pathways involving endogenously produced LRP1 ligands. Regulation of Schwann cell migration by LRP1 may be important in PNS injury.
Keywords: Cell Adhesion, Cell Migration, Cell Motility, Cell Surface Receptor, Signal Transduction, Rho Family GTPases, Schwann Cell, Low Density Lipoprotein Receptor-related Protein (LRP1)
Introduction
In the mature, uninjured peripheral nervous system (PNS),2 Schwann cells are typically immobile, providing trophic support and in some cases, ensheathing and myelinating axons (1). PNS injury triggers Schwann cell dedifferentiation, allowing these cells to survive, detach from axonal connections, proliferate, and migrate (2). Dedifferentiation is essential for PNS regeneration (3). Many exogenous factors have been reported to stimulate migration of dedifferentiated Schwann cells, including neurotrophin-3 (NT-3), neuregulin-1, and insulin-like growth factor-I (4–6). These factors promote migration, at least in part, by activating the Rho family GTPases, Rac1 and Cdc42. Rac1 promotes dynamic actin remodeling, lamellipodia formation and random cell migration (7, 8). Cdc42 controls cell polarity and allows for directionally persistent cell migration (9). In Schwann cells, the ability of Rac1 to promote cell migration may be counteracted by high levels of activated RhoA and its downstream effector, Rho kinase (10). RhoA activation may be inhibited by Rac1 and Rac1 activation may be inhibited downstream of Rho kinase, allowing for orchestration of these GTPases in the control of cell morphology and migration (11–14). In development, Schwann cell Rac1 facilitates radial axonal sorting and myelination (15).
Gene products that control transformation of the Schwann cell phenotype in PNS injury remain incompletely characterized. We recently identified low density lipoprotein receptor-related protein (LRP1) as a receptor expressed by Schwann cells mainly after PNS injury (16). LRP1 is a 600-kDa type I transmembrane protein in the LDL receptor gene family, originally characterized as an endocytic receptor, but currently recognized also for its role in cell signaling (17, 18). Diverse proteins implicated in PNS injury, including matrix metalloprotease-9 (MMP-9), tissue-type plasminogen activator (tPA), and activated α2-macroglobulin (α2M) bind to LRP1 and activate Akt and ERK/MAP kinase in Schwann cells in vitro and in the injured PNS (19–21). By its effect on cell signaling, LRP1 promotes Schwann cell survival and migration (16, 19).
Although the increase in Schwann cell migration, observed when cells are treated with MMP-9, has been attributed to activation of ERK1/2 and PI3K downstream of LRP1, Schwann cells in culture migrate readily in the absence of added MMP-9 (19). The basal rate of Schwann cell migration, in the absence of added reagents, is inhibited by 90% when LRP1 is silenced (19). This result is intriguing given the importance of Schwann cell migration in PNS injury and the fact that LRP1 is expressed by Schwann cells primarily after nerve injury (16). The goal of the present study was to determine the mechanism by which LRP1 expression controls the basal rate of Schwann cell migration. Our results demonstrate that even in the absence of exogenously added ligands, LRP1 is a major activator of Rac1 and a reciprocal inhibitor of RhoA in Schwann cells. The ability of LRP1 to directly regulate Rho family GTPases explains its activity in regulating the basal rate of Schwann cell migration.
EXPERIMENTAL PROCEDURES
Reagents
The LRP1 antagonist, receptor-associated protein (RAP), was expressed as a GST fusion protein (GST-RAP) as previously described (22). As a control, we expressed GST in bacteria transformed with the empty vector, pGEX-2T. Purified fibronectin (FN), vitronectin (VN), and a monoclonal antibody specific for vinculin (clone hVIN-1) were from Sigma-Aldrich. Rac/Cdc42 assay reagent (PAK-PBD1), which includes residues 67–150 of p21-activated kinase (PAK-1) fused to GST and coupled to glutathione-Sepharose was from Upstate Biotechnology (Lake Placid, NY). Mouse monoclonal antibody that specifically binds Rac1 was from BD Biosciences (San Diego, CA). The Rho assay reagent, a GST-tagged fusion protein corresponding to residues 7–89 of mouse Rhotekin Rho Binding Domain (GST-TRBD) expressed in Escherichia coli and bound to glutathione-Sepharose, was from Millipore (Billerica, MA). This fusion protein specifically binds GTP-Rho. RhoA-specific monoclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). The Rho kinase inhibitor, Y27632, and Rac1 inhibitor, NSC23766, were from EMD Biosciences (San Diego, CA). The hemopexin domain of MMP-9 (PEX) and the α2M receptor binding domain (RBD) were expressed as GST fusion proteins and purified as previously described (19, 20). These GST fusion proteins bind to LRP1 and trigger cell signaling to ERK1/2 and Akt. Catalytically inactive tPA (mtPA) was purchased from Molecular Innovations (Novi, MI). MMP-9 was purchased from R&D Systems (Minneapolis, MN).
Cell Culture
Schwann cells were isolated from sciatic nerves of 1-day-old Sprague-Dawley rats (Harlan Laboratories) and further separated from other cell types by using anti-Thy1.1 and rabbit complement, as previously described (23). Final preparations consisted of 98% Schwann cells, as determined by immunofluorescence for S100, which is a specific Schwann cell marker. Primary cultures of Schwann cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, 100 μg/ml streptomycin, 21 μg/ml bovine pituitary extract, and 4 μm forskolin (Complete medium) at 37 °C under humidified 5% CO2. Schwann cell cultures were passaged no more than six times before conducting experiments.
LRP1 Gene-silencing
The previously described rat LRP1-specific siRNA (siLRP1, CGAGCGACCUCCUAUCUUUUU) (16) and NTC siRNA were from Dharmacon (Chicago, IL). Primary cultures of Schwann cells (1 × 106) were transfected with LRP1-specific siRNA (25 nm) or with NTC siRNA (25 nm) by electroporation using the Rat Neuron Nucleofector Kit (Amaxa, Gaithersburg, MD). The degree of LRP1 gene-silencing was 92–95%, 24–72-h post-electroporation as determined by quantitative PCR (qPCR). qPCR analysis of gene-silencing was confirmed by immunoblot analysis and RAP ligand blotting, as previously described (16). Cell signaling and cell migration experiments were performed 24–36 h after introducing siRNAs.
Immunoblot Analysis
Schwann cells were transferred to serum-free medium (SFM) and maintained for 1 h. The cells were rinsed twice with ice-cold phosphate-buffered saline (PBS) and extracted in radioimmune precipitation assay buffer (PBS with 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitor mixture, and sodium orthovanadate). The protein concentration in cell extracts was determined by bicinchoninic acid assay (BCA). An equivalent amount of cellular protein (50 μg per lane) was subjected to 10% SDS-PAGE and electrotransferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk in Tris-HCl-buffered saline, pH 7.4 with Tween-20 and incubated with the primary antibodies according to the manufacturer's recommendations. The membranes were washed and treated with horseradish peroxidase-conjugated secondary antibodies for 1 h. Immunoblots were developed using enhanced chemiluminescence (GE Health-Care Biosciences Corp., Piscataway, NJ). Densitometry was performed using NIH Image.
Cell Migration Assays
Migration of Schwann cells was studied using 6.5-mm Transwell chambers with 8-μm pores (Costar, Corning, NY), as previously described (19, 24, 25). The bottom surface of each membrane was coated with 5 μg/ml FN or with 5 μg/ml VN. We selected FN as the primary substratum for our studies because after PNS injury, FN expression is induced to provide a provisional extracellular matrix for nerve regeneration (26). Schwann cells that were transfected with siLRP1 or with NTC siRNA were cultured in Sato medium (27) and treated with Y27632 (25 μm) or vehicle. Wild-type Schwann cells were pretreated with GST-RAP (100 nm) or GST (100 nm) for 15 min at 37 °C prior to adding Y27632. The same reagents were added to both chambers of each Transwell. The bottom chamber contained 10% fetal bovine serum. Cells (105) were added to the top chamber and allowed to migrate at 37 °C. After 4 h, the upper surface of each membrane was cleaned with a cotton swab. The membranes then were stained with Diff-Quik (Dade-Behring, Deerfield, IL). The number of cells on the bottom surface of each membrane was determined. Each condition was studied at least in triplicate. Four fields from each membrane were examined.
Cell Adhesion Assays
96-well plates were coated with various concentrations of FN or VN in PBS overnight at 4 °C, rinsed, and then blocked with 2% (w/v) bovine serum albumin (BSA, Sigma-Aldrich) in PBS for 2 h at 22 °C. Cells transfected with siLRP1 or NTC siRNA and wild-type Schwann cells were suspended at a density of 106 cells/ml in Sato medium, supplemented with 1 mg/ml BSA, and allowed to adhere for 1 h at 37 °C in the presence of Y27632 or the Rac1 inhibitor, NC23766. In some cases, wild-type Schwann cells were pretreated with GST or GST-RAP (100 nm) for 15 min at 37 °C. Non-adherent cells were removed by washing with PBS. Adherent cells were fixed with 4% formaldehyde, rinsed, and stained with 0.2% crystal violet. Cell-associated stain was recovered in 1% SDS. The absorbance at 595 nm was measured. Each value represents the mean of 18 separate replicates, in three separate experiments.
Immunofluorescence Microscopy
siLRP1- or NTC-transfected Schwann cells were dissociated with non-enzymatic dissociation buffer. The cells were plated in Sato medium supplemented with 1 mg/ml of BSA on FN-coated glass coverslips and incubated at 37 °C for 0.5–3 h. The cells were then fixed in 4% formaldehyde. Fixed cells were permeabilized in 0.2% Triton X-100 and incubated with vinculin-specific antibody for 18 h, followed by secondary antibody conjugated with Alexa Fluor 488 and phalloidin conjugated with Alexa Fluor 568. Control cultures were treated equivalently except for the omission of primary antibody. Preparations were mounted on slides using Pro-long Gold with DAPI (Invitrogen, San Diego, CA) and examined using a Leica DMIRE2 fluorescence microscope. Images were obtained with a ×63 oil immersion objective and a Hamamatsu digital camera with SimplePCI software. Deconvolution was performed using Simple PCI software.
Rac1 Activation Assays
Affinity precipitation of active Rac1 was performed using PAK1-PBD, which specifically recognizes the GTP-bound form of Rac1 and Cdc42, as previously described (28, 29). Schwann cells were cultured in 10-cm2 plates coated with FN (5 μg/ml) for 18 h and then serum-starved for 4 h in Sato medium supplemented with 1 mg/ml BSA. Cultures were washed with ice-cold PBS and extracted in 1% (v/v) Triton X-100, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 50 mm Tris-HCl, 0.5 m NaCl, 10 mm MgCl2, pH 7.2, supplemented with protease inhibitor mixture and 1 mm sodium orthovanadate. The extracts were incubated with 15 μg of PAK1-PBD coupled to glutathione-Sepharose for 45 min at 4 °C. The glutathione-Sepharose was washed and then treated with SDS-sample buffer to dissociate the PAK1-PBD and associated proteins. Immunoblot analysis was performed to detect active Rac1. Samples of each cell extract were subjected to immunoblot analysis prior to incubation with PAK1-PBD, to determine total Rac1.
RhoA Activity Assays
Affinity precipitation of active RhoA was performed using GST-TRBD, as previously described (30). Schwann cells were cultured in 10-cm2 plates for 18 h, serum-starved for 4 h in Sato medium supplemented with 1 mg/ml BSA, washed with ice-cold PBS, and extracted in 1% (v/v) Triton X-100, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 50 mm Tris-HCl, 0.5 m NaCl, and 10 mm MgCl2 (pH 7.2) supplemented with protease inhibitors and 1 mm sodium orthovanadate. The extracts were incubated with 30 μg of GST-TRBD coupled to glutathione-Sepharose for 45 min at 4 °C. The matrix was washed and then treated with SDS sample buffer to dissociate GST-TRBD and GTP-Rho. Immunoblot analysis was performed to detect active RhoA. Samples of each cell extract were subjected to immunoblot analysis prior to incubation with GST-TRBD to determine total RhoA.
Statistical Analysis
In all studies, replicates refer to separate experiments, typically performed with internal duplicates or triplicates. Results of cell migration, cell adhesion, and cell signaling experiments were subjected to a one way analysis of variance (ANOVA) and Tukey's post hoc analysis to assess differences between greater than three treatment groups. In the case of two treatment means, a Student's t test was performed.
RESULTS
LRP1 Gene-silencing Renders Schwann Cells Immobile
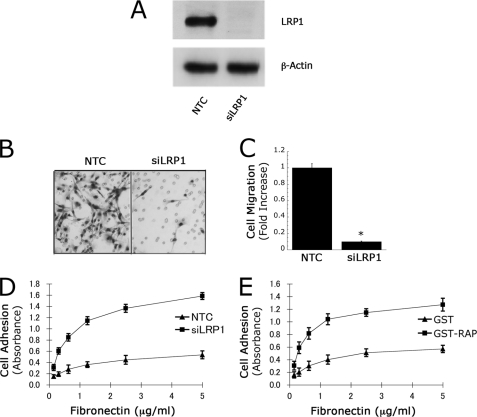
Schwann cells in culture express high levels of LRP1, mimicking the phenotype of Schwann cells in vivo in the injured PNS (16). To study the effects of LRP1 on Schwann cell adhesion and migration, cells were transfected with the previously described LRP1-specific siRNA, siLRP1 (16), or with NTC siRNA. Fig. 1A shows that LRP1 protein was essentially undetectable in gene-silenced cells, as determined by immunoblot analysis. To avoid the previously reported adverse effects of LRP1 gene-silencing on Schwann cell survival, transfected cells were maintained in Sato medium (27). Under these conditions, altered survival was not observed, as determined by MTT assay (results not shown).
FIGURE 1.
LRP1 regulates Schwann cell migration. A, immunoblot analysis of LRP1 in Schwann cells transfected with NTC siRNA or LRP1-specific siRNA (siLRP1). β-Actin was measured as a loading control. B, images of Schwann cells transfected with NTC siRNA or siLRP1 that migrated to the underside surface of Transwell membranes. C, quantification of cell migration results. Cell migration was expressed as the fold-increase compared with NTC-transfected cells (mean ± S.E., n = 6, *, p < 0.01). D and E, cell adhesion to surfaces pretreated with increasing concentrations of FN. In D, Schwann cells were transfected with NTC siRNA or siLRP1. In E, Schwann cells were treated with the LRP-1 antagonist, RAP, or with GST as a control. In all studies, Schwann cells were allowed to adhere for 1 h on various concentrations of FN (mean ± S.E., n = 3).
Cell migration was studied using Transwell units. FN was adsorbed selectively to the lower membrane surface to form a haptotactic gradient. Fetal bovine serum was added to the lower chamber to provide a chemoattractant stimulus. Fig. 1B shows representative images of the lower surfaces of Transwell membranes. When LRP1 was silenced, Schwann cell migration was substantially inhibited. As shown in Fig. 1C, the number of LRP1 gene-silenced cells migrating to the lower membrane surface was decreased by 80–90% compared with the number of control cells (p < 0.01).
In Transwell migration assays, decreased migration may be explained if cells do not adhere to the upper membrane surface. To control for this possibility, we studied Schwann cell adhesion to FN-coated surfaces. Cell culture wells were coated with 0.2–5.0 μg/ml FN. Cells were allowed to adhere for 1 h. Fig. 1D shows that LRP1 gene-silencing did not inhibit cell adhesion but instead, robustly increased cell adhesion. At each FN concentration, the number of adherent cells was increased about 3-fold by LRP1 gene-silencing.
As a second approach to test the effects of LRP1 on Schwann cell adhesion, we treated Schwann cells with RAP, which binds to LRP1 and blocks the binding of other ligands that initiate LRP1-dependent cell signaling (17, 19, 20, 31). Fig. 1E shows that RAP increased Schwann cell adhesion to FN, confirming the results obtained by gene-silencing. Because RAP is expressed as a GST fusion protein, as a control, we examined the effects of purified GST, which did not affect cell adhesion, as anticipated.
LRP1 Regulates Schwann Cell Spreading and Focal Adhesion Formation
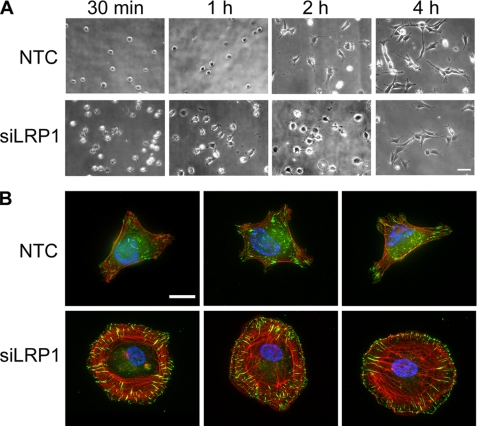
Next, we examined Schwann cell adhesion to FN as a function of time. As is evident by phase contrast microscopy, LRP1 gene-silencing was associated not only with an increase in the rate of adhesion but also with more rapid spreading (Fig. 2A). By 4 h, adhesion of gene-silenced and control cells tended to equalize. Interestingly, although LRP1 gene-silenced cells tended to spread more rapidly, these cells showed fewer and more poorly developed cellular processes at 4 h.
FIGURE 2.
Phase-contrast and immunofluorescence microscopy comparing LRP1 gene-silenced and control Schwann cells. A, phase contrast microscopy of primary Schwann cells transfected with NTC or LRP1-specific siRNA. Cells were plated on FN (5 μg/ml) for 0.5 to 4 h. Magnification is ×200. Images are representative of three independent studies. Scale bar, 50 μm. B, immunofluorescence microscopy imaging vinculin (green) in representative NTC or siLRP1-transfected Schwann cells. The cells were cultured on FN for 2 h. Phalloidin is imaged in red and DAPI in blue. Images are at ×630 magnification (scale bar, 15 μm).
To further examine the effects of LRP1 gene-silencing on Schwann cell spreading, we performed immunofluorescence microscopy studies. Vinculin immunofluorescence was imaged as a marker of focal adhesions (32). Filamentous actin (f-actin) was imaged by phalloidin staining. Fig. 2B shows representative images of control Schwann cells and cells in which LRP1 was silenced. In the gene-silenced cells, prominent focal adhesions were observed, running perpendicular to the plasma membrane near the cell margins. Focal adhesions were fewer and less prominent in control cells. Many LRP1 gene-silenced cells also demonstrated well-defined stress fibers. In other LRP1 gene-silenced cells, the pattern of f-actin was primarily circumferential, near the cell perimeter. This pattern has been observed before and described as a variant of the classic fibroblast stress fiber, formed under the control of Rho and Rho kinase (33, 34).
LRP1 Regulates Activation of Rac1 and RhoA
In Schwann cells, Akt and ERK1/2 are activated when the cells are treated with LRP1 ligands. LRP1 gene-silencing or treatment with RAP decreases Akt activation in the absence of added LRP1 ligands, suggesting that autocrine circuits may be disrupted (16, 19, 20). Because of the effects of LRP1 gene-silencing on Schwann cell adhesion and spreading, we examined activation of Rac1 and RhoA. Rho GTPases cycle between GTP-loaded, activated states, and GDP-loaded, inactive states under the control of guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (35, 36). The assortment of GEFs and GAPs that regulate Rac1 or RhoA is large, allowing for considerable cross-talk in cell signaling.
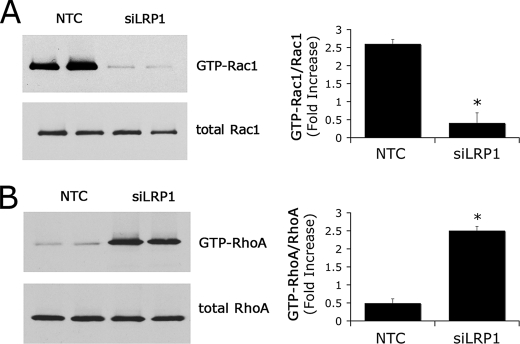
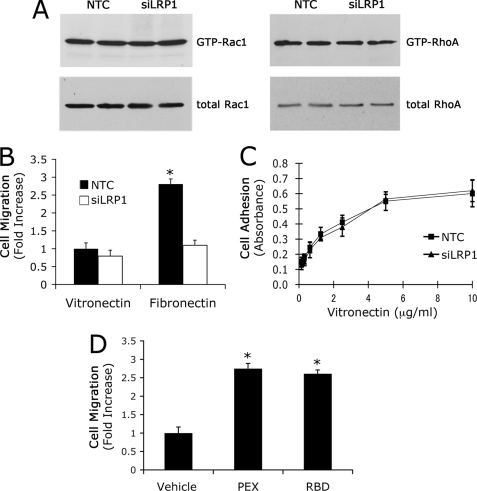
Cell extracts were prepared from confluent cultures of LRP1 gene-silenced and control Schwann cells 22 h after plating on FN-coated plates. For the final 4 h, the cells were maintained in serum-free Sato medium. Fig. 3A shows a representative immunoblot comparing GTP-loaded Rac1 and total Rac1. In LRP1 gene-silenced cells, the fraction of Rac1 that was GTP-loaded was decreased by 85 ± 10% (p < 0.01). At the same time, LRP1 gene-silencing resulted in a substantial increase in GTP-loaded RhoA. As shown in Fig. 3B, the increase was 5.2 ± 0.3-fold (p < 0.01). These changes in small GTPase activity are consistent with the observed changes in Schwann cell phenotype and suggest that LRP1 may control Schwann cell adhesion and the basal rate of cell migration by its effects on Rac1 and RhoA.
FIGURE 3.
LRP1 regulates Schwann cell Rac1 and RhoA. A, levels of activated Rac1 in siLRP1 and NTC-transfected Schwann cells. GTP-bound Rac1 was affinity-precipitated with PAK1-PBD and quantitated by immunoblot analysis. The original cell extracts also were studied by immunoblot analysis using the same antibody to determine total Rac1. Immunoblots were analyzed by densitometry. B, levels of activated RhoA in siLRP1 and NTC-transfected Schwann cells. GTP-bound RhoA was affinity-precipitated with GST-TRBD coupled to glutathione-Sepharose. The original cell extracts were also studied by immunoblot analysis using the same antibody to determine total RhoA. Immunoblots were analyzed by densitometry. Data are expressed as mean ± S.E.; n = 3, *, p < 0.01.
Diverse LRP1 Ligands Activate Rac1 in Schwann Cells
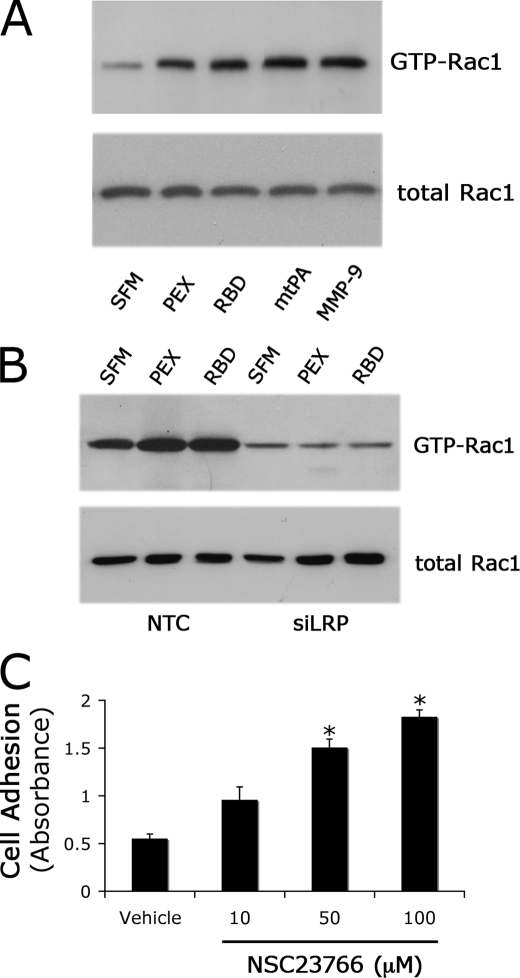
To confirm that LRP1 regulates Rac1 activity, Schwann cells in culture were treated with four structurally diverse LRP1 ligands. MMP-9, the isolated LRP1 binding domain of MMP-9 (PEX), the receptor-binding domain of α2M (RBD), and a mutated form of tPA in which the serine protease active site is deleted (mtPA) activated Rac1 in Schwann cells (Fig. 4A). mtPA-induced cell signaling has not been studied in Schwann cells previously; however, mtPA initiates LRP1-dependent cell signaling in other cell types (21, 31). The extent of Rac1 activation induced by each LRP1 ligand was comparable.
FIGURE 4.
Diverse LRP1 ligands activate Rac1 in Schwann cells. A, levels of activated Rac1 in primary Schwann cells treated with MMP-9-PEX (10 nm), RBD (10 nm), mtPA (10 nm), MMP-9 (10 nm), or vehicle for 10 min. Cell extracts were affinity-precipitated with PAK1-PBD and subjected to by immunoblot analysis to detect GTP-bound Rac1. The original cell extracts were also studied by immunoblot analysis using the same antibody to determine total Rac1. The immunoblot shown here represents two independent experiments. B, levels of activated Rac1 in siLRP1- and NTC-transfected Schwann cells after treatment with RBD (10 nm), PEX (10 nm), or vehicle for 10 min. C, inhibition of Rac1 with NSC23766 increased Schwann cell adhesion in a dose-dependent manner. The results of two independent experiments were averaged to generate the bar graph (mean ± S.E.; *, p < 0.01 compared with vehicle control).
In Schwann cells in which LRP1 was silenced, PEX and RBD failed to activate Rac1 (Fig. 4B). By contrast, in cells that were transfected with NTC siRNA, PEX and RBD activated Rac1, providing further confirmation that the mechanism by which PEX and RBD regulate GTPase activity requires LRP1. We also examined the effects of PEX and RBD on RhoA activation. Because LRP1 expression is associated with a substantial decrease in GTP-RhoA, we hypothesized that PEX and RBD might further decrease GTP-RhoA. In two separate experiments, a further decrease was not observed (results not shown); however, because the level of GTP-loaded RhoA is already very low in LRP1-expressing Schwann cells, our inability to detect a further decrease in GTP-RhoA may have been technical.
To test whether Rac1 inhibits cell adhesion in LRP1-expressing Schwann cells, we treated Schwann cells with increasing concentrations of the Rac1 inhibitor, NSC23766, in the absence of added LRP1 ligands. Fig. 4C shows that the Rac1 inhibitor increased Schwann cell adhesion to FN.
Inhibiting Rho Kinase Rescues the Phenotype of LRP1 Gene-silenced Cells
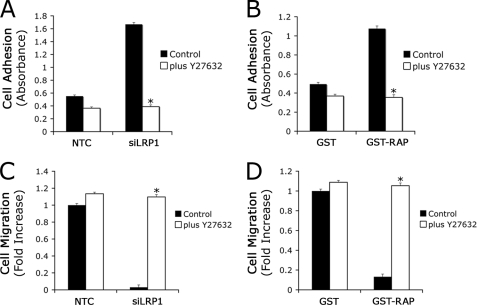
Because RhoA is activated in LRP1 gene-silenced Schwann cells, we tested whether inhibiting Rho kinase reverses the phenotypic changes associated with LRP1 deficiency. Fig. 5A demonstrates that the Rho kinase inhibitor, Y27632 (25 μm), selectively inhibited adhesion of LRP1 gene-silenced cells to FN. In the presence of Y27632, adhesion of LRP1 gene-silenced and control cells was equivalent. We also compared the effects of Y27632 on Schwann cells that were treated with RAP. Again, Y27632 selectively inhibited adhesion of RAP-treated cells so that in the presence of Y27632, adhesion of cells that were treated with RAP and control cells was equivalent (Fig. 5B).
FIGURE 5.
Inhibition of RhoA activation in LRP1 gene-silenced Schwann cells decreases cell adhesion and promotes cell migration. LRP1 gene-silenced (A) or GST-RAP-treated (B) Schwann cells were allowed to adhere on FN-coated plates for 1 h in the presence of a specific pharmacological inhibitor for RhoA, Y27632 (25 μm). Data are expressed as the mean ± S.E., n = 3, *, p < 0.01 compared with respective controls. LRP1 gene-silenced (C) or GST-RAP-treated (D) Schwann cells were allowed to migrate in Transwells in the presence of Y27632 for 4 h. Cell migration was expressed as the fold-increase relative to NTC-transfected or GST-treated cells in the absence of inhibitor (mean ± S.E., n = 3, *, p < 0.01 compared with respective control).
Next, we compared migration of LRP1 gene-silenced and control cells on FN in the presence and absence of Y27632. The Rho kinase inhibitor selectively and robustly promoted migration of the LRP1 gene-silenced cells, eliminating the difference between gene-silenced and control cells (Fig. 5C). Similarly, Y27632 promoted migration of Schwann cells that were treated with RAP but not cells that were treated with GST (Fig. 5D). As a result, in the presence of Y27632, Schwann cells that were treated with RAP and control cells migrated equivalently. These results demonstrate that inhibiting Rho kinase rescues the phenotype of LRP1-deficient and RAP-treated Schwann cells, restoring migration, and inhibiting adhesion.
LRP1 Regulates Rac1 in an Extracellular Matrix-dependent Manner
LRP1 directly activates cell signaling in response to ligand binding and indirectly regulates cell signaling by controlling the cell surface abundance of other cell signaling receptors (37). In fibroblasts that are plated on VN, LRP1 inhibits Rac1 activation downstream of the urokinase receptor (uPAR) by facilitating uPAR endocytosis (29, 38). Because our studies were performed with Schwann cells cultured on FN and not VN, regulation of uPAR did not contribute to the effects of LRP1 on Rac1 activation.
To explore the balance between direct and indirect pathways by which LRP1 may regulate GTPase activity, Schwann cells were plated on VN instead of FN. Fig. 6A shows that under these conditions, LRP1 gene-silencing did not regulate GTP-loaded Rac1 and RhoA (Fig. 6A). In addition, both cell migration (Fig. 6B) and cell adhesion (Fig. 6C) were unchanged. We interpreted these results to indicate that when Schwann cells are plated on VN, either LRP1 is not active in regulating Schwann cell adhesion and migration or LRP1 gene-silencing induces offsetting activities.
FIGURE 6.
Regulation of Rho GTPases by LRP1 is dependent on the extracellular matrix. A, levels of activated Rac1 and RhoA in siLRP1 and NTC-transfected Schwann cells that were cultured on purified VN. B, migration of siLRP1 and NTC-transfected Schwann cells. Migration was allowed to proceed for 4 h. Data are expressed as mean ± S.E., n = 6. *, p < 0.01 compared with respective controls. C, cell adhesion of siLRP1 and NTC-transfected Schwann cells on various concentrations of VN. Data are expressed as mean ± S.E., n = 2. D, migration of Schwann cells treated with PEX (10 nm), RBD (10 nm) or vehicle and allowed to migrate in Transwell chambers. The membranes were coated with purified VN. Migration is expressed as the fold increase compared with cells that were treated with vehicle (mean ± S.E., n = 3, *, p < 0.01).
To further test the role of LRP1 in regulating Schwann cells on VN, we treated cells with PEX or RBD and studied cell migration in VN-coated Transwells. Fig. 6D shows that both LRP1 ligands promoted Schwann cell migration on VN. These results demonstrate that LRP1 is active in regulating Schwann cell migration even when the cells are plated on VN.
DISCUSSION
In this study, we examined adhesion and migration of Schwann cells on FN because this provisional extracellular matrix protein plays a major role in PNS injury and regeneration (26, 39). Schwann cells express αvβ8, which is responsible for adhesion and migration on FN, unlike many other cells that primarily utilize α5β1 and α4β1 (40–42). Coincident with the accumulation of FN in the injured PNS, Schwann cells dramatically up-regulate expression of LRP1 (16). Thus, the function of LRP1 in Schwann cells cultured on purified FN is a relevant model of PNS injury.
Our results demonstrate that LRP1 is a major regulator of Rho GTPases involved in Schwann cell adhesion and migration. This observation allows us to classify LRP1 together with TrkC and ErbB2, which also control Rac1 and RhoA in Schwann cells (4, 5). The broad ligand binding specificity of LRP1, including many proteases and extracellular mediators present at sites of injury and inflammation (17, 19), makes it highly likely that Schwann cell LRP1 is activated in the injured nerve. This hypothesis is supported by the previous observation that RAP decreases cell survival when injected into the injured sciatic nerve in vivo, presumably by binding to LRP1 and by inhibiting binding of other endogenous ligands that trigger LRP1-dependent cell signaling to PI3K and Akt (16).
The decrease in Schwann cell migration that accompanied LRP1 gene-silencing or treatment with RAP was associated with a substantial decrease in the overall level of activation of Rac1 and an increase in activation of RhoA. We interpret these results as indicating that RAP and LRP1 gene-silencing disrupt autocrine cell signaling pathways involving endogenously produced LRP1 ligands. Rho kinase inhibitor rescued the effects of LRP1 deficiency on Schwann cell adhesion and migration, confirming the importance of this pathway. Although we did not determine a direct linkage between Rac1 and RhoA, inhibition of RhoA downstream of Rac1 has been demonstrated before (11–13).
The transformation in Schwann cell morphology, which resulted from LRP1 gene-silencing, was striking. LRP1 gene-silenced cells spread readily and adopted a flattened appearance with robust stress fiber formation and/or arrangement of f-actin into circumferential wreath-like structures. These changes in morphology may model those that occur in Schwann cells in vivo after nerve injury. Similar changes in Schwann cell morphology have been reported following treatment with lysophosphatidic acid (LPA) (43). RhoA activation plays an important role in the LPA-induced changes. Thus, LRP1 ligands and LPA may regulate overlapping signaling pathways in Schwann cells.
Exogenously added MMP-9, which promotes Schwann cell migration, also increased Rac1 activation; however, the increase in Rac1 activation and the promigratory activity of MMP-9 may not be linked. Instead, exogenously added MMP-9 promotes Schwann cell migration by activating ERK1/2 (19). This result may be explained by the fact that cell migration does not increase linearly as a function of the extent of total cellular Rac1 activation. In fact, in certain circumstances, decreasing overall Rac1 activation may promote cell migration by increasing directional persistence and by decreasing formation of multiple unaligned lamellipodia (7, 8). We propose a model in which LRP1-initiated cell-signaling regulates Schwann cell migration by different mechanisms, depending on the degree of LRP1 ligation. At low ligation levels, probably attained as the result of autocrine circuits established in Schwann cell cultures, regulation of GTPases may be instrumental in converting the Schwann cell phenotype from immobile to mobile. At higher degrees of LRP1 ligation, the effects of LRP1 on Schwann cell migration are explained by activation of ERK1/2.
The opposing effects of Rac1 and RhoA in controlling cell migration have been recognized previously (44). In response to NT-3, Rac1 promotes and RhoA opposes Schwann cell migration (10). In neurons, Rac1 promotes and RhoA inhibits neurite extension (36). However, studies that have examined activation of GTPases on a spatial and temporal level have demonstrated that pulsatile activation of RhoA in lamellipodia may in fact support cell migration (45). The experiments performed here, which detected changes in total cellular Rho GTPase activity, were not designed to detect subcellular changes in activation of Rac1 or RhoA, which also may be important. In Schwann cells, the ability of Rac1 to regulate lamellipodia formation and process extension is linked to its effects on myelination and axonal sorting (15, 46, 47).
In addition to the direct pathway, under study here, in which LRP1 ligands activate cell signaling, LRP1 also indirectly regulates cell signaling by controlling the cell surface abundance of other receptors, such as uPAR (29, 48) and TNF receptor-1 (49). Direct and indirect pathways may provide opposing signals. For example, binding of tPA to LRP1 in astrocytes activates NF-κB (50). By contrast, in macrophages, LRP1 indirectly inhibits cell signaling to NF-κB by down-regulating cell surface TNF receptor-1 (51).
A similar paradigm exists for Rac1. Binding of ligands to LRP1, in Schwann cells, activates Rac1; however, in fibroblasts, LRP1 inhibits Rac1 activation by decreasing the cell surface abundance of uPAR (28, 29, 38). The indirect pathway, in which LRP1 regulates Rac1 downstream of uPAR is active only when cells are cultured on VN. Thus, the absence of an effect of LRP1 gene-silencing on Schwann cell GTPase activity, migration, and adhesion on VN may be explained by offsetting effects of direct LRP1-initiated cell signaling and regulation of uPAR. Confirmation of this model will require analysis of the uPAR system in Schwann cells. Because PEX and the RBD promoted Schwann cell migration on VN, the direct pathway in which LRP1 ligands trigger cell signaling and regulate Schwann cell physiology is operational on this substratum. Whether direct or indirect pathways for regulation of cell signaling by LRP1 predominate in a given cell type probably depends on whether the cell type of interest expresses an LRP1-regulated receptor, such as uPAR, and on the availability of ligands for both LRP1 and the regulated receptor.
Taken together with our earlier studies (16, 19, 20), the work presented here demonstrates that regulating LRP1 expression in Schwann cells controls important cellular properties such as cell survival and cell migration, which are key in the Schwann cell response to PNS injury. Our results support a model in which LRP1 is a key gene product that drives phenotypic modulation in Schwann cells in the response to PNS injury.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 NS57456 and R01 NS54671.
- PNS
- peripheral nervous system
- MMP
- matrix metalloprotease
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- GST
- glutathione S-transferase
- LRP
- low density lipoprotein receptor-related protein
- RAP
- receptor-associated protein
- f-actin
- filamentous actin
- FN
- fibronectin
- VN
- vitronectin
- RBD
- receptor-binding domain of α2M
- PAK
- p21-activated kinase
- NTC
- non-targeting control.
REFERENCES
- 1.Jessen K. R., Mirsky R. (1999) Adv. Exp. Med. Biol. 468, 3–12 [DOI] [PubMed] [Google Scholar]
- 2.Ide C. (1996) Neurosci. Res. 25, 101–121 [DOI] [PubMed] [Google Scholar]
- 3.Chen Z. L., Yu W. M., Strickland S. (2007) Annu. Rev. Neurosci. 30, 209–233 [DOI] [PubMed] [Google Scholar]
- 4.Yamauchi J., Chan J. R., Miyamoto Y., Tsujimoto G., Shooter E. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5198–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamauchi J., Miyamoto Y., Chan J. R., Tanoue A. (2008) J. Cell Biol. 181, 351–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng H. L., Steinway M., Delaney C. L., Franke T. F., Feldman E. L. (2000) Mol. Cell. Endocrinol. 170, 211–215 [DOI] [PubMed] [Google Scholar]
- 7.Pankov R., Endo Y., Even-Ram S., Araki M., Clark K., Cukierman E., Matsumoto K., Yamada K. M. (2005) J. Cell Biol. 170, 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrie R. J., Doyle A. D., Yamada K. M. (2009) Nat. Rev. Mol. Cell Biol. 10, 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etienne-Manneville S., Hall A. (2001) Cell 106, 489–498 [DOI] [PubMed] [Google Scholar]
- 10.Yamauchi J., Chan J. R., Shooter E. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8774–8779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nimnual A. S., Taylor L. J., Bar-Sagi D. (2003) Nat. Cell Biol. 5, 236–241 [DOI] [PubMed] [Google Scholar]
- 12.Sander E. E., ten Klooster J. P., van Delft S., van der Kammen R. A., Collard J. G. (1999) J. Cell Biol. 147, 1009–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenfeldt H., Castellone M. D., Randazzo P. A., Gutkind J. S. (2006) J. Mol. Signal 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayama M., Goto T. M., Sugimoto M., Nishimura T., Shinagawa T., Ohno S., Amano M., Kaibuchi K. (2008) Dev. Cell 14, 205–215 [DOI] [PubMed] [Google Scholar]
- 15.Feltri M. L., Suter U., Relvas J. B. (2008) Glia 56, 1508–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campana W. M., Li X., Dragojlovic N., Janes J., Gaultier A., Gonias S. L. (2006) J. Neurosci. 26, 11197–11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strickland D. K., Gonias S. L., Argraves W. S. (2002) Trends Endocrinol. Metab. 13, 66–74 [DOI] [PubMed] [Google Scholar]
- 18.Rebeck G. W. (2009) Sci. Signal 2, pe28. [DOI] [PubMed] [Google Scholar]
- 19.Mantuano E., Inoue G., Li X., Takahashi K., Gaultier A., Gonias S. L., Campana W. M. (2008) J. Neurosci. 28, 11571–11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mantuano E., Mukandala G., Li X., Campana W. M., Gonias S. L. (2008) J. Biol. Chem. 283, 19904–19911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y., Mantuano E., Inoue G., Campana W. M., Gonias S. L. (2009) Sci. Signal 2, ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herz J., Goldstein J. L., Strickland D. K., Ho Y. K., Brown M. S. (1991) J. Biol. Chem. 266, 21232–21238 [PubMed] [Google Scholar]
- 23.Campana W. M., Hiraiwa M., O'Brien J. S. (1998) Faseb J. 12, 307–314 [DOI] [PubMed] [Google Scholar]
- 24.Nguyen D. H., Catling A. D., Webb D. J., Sankovic M., Walker L. A., Somlyo A. V., Weber M. J., Gonias S. L. (1999) J. Cell Biol. 146, 149–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen D. H., Hussaini I. M., Gonias S. L. (1998) J. Biol. Chem. 273, 8502–8507 [DOI] [PubMed] [Google Scholar]
- 26.Inoue G., Gaultier A., Li X., Mantuano E., Richardson G., Takahashi K., Campana W. M. (2010) Glia 58, 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bottenstein J. E., Sato G. H. (1980) Exp. Cell Res. 129, 361–366 [DOI] [PubMed] [Google Scholar]
- 28.Kjøller L., Hall A. (2001) J. Cell Biol. 152, 1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Z., Thomas K. S., Webb D. J., Moravec R., Salicioni A. M., Mars W. M., Gonias S. L. (2002) J. Cell Biol. 159, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren X. D., Kiosses W. B., Schwartz M. A. (1999) EMBO J. 18, 578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu K., Yang J., Tanaka S., Gonias S. L., Mars W. M., Liu Y. (2006) J. Biol. Chem. 281, 2120–2127 [DOI] [PubMed] [Google Scholar]
- 32.Demali K. A. (2004) Trends Biochem. Sci. 29, 565–567 [DOI] [PubMed] [Google Scholar]
- 33.Kawkitinarong K., Linz-McGillem L., Birukov K. G., Garcia J. G. (2004) Am. J. Respir. Cell Mol. Biol. 31, 517–527 [DOI] [PubMed] [Google Scholar]
- 34.Wiggan O., Shaw A. E., Bamburg J. R. (2006) Cell Signal. 18, 1501–1514 [DOI] [PubMed] [Google Scholar]
- 35.Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 36.Luo L. (2000) Nat. Rev. Neurosci. 1, 173–180 [DOI] [PubMed] [Google Scholar]
- 37.Gonias S. L., Wu L., Salicioni A. M. (2004) Thromb. Haemost. 91, 1056–1064 [DOI] [PubMed] [Google Scholar]
- 38.Jo M., Thomas K. S., O'Donnell D. M., Gonias S. L. (2003) J. Biol. Chem. 278, 1642–1646 [DOI] [PubMed] [Google Scholar]
- 39.Akassoglou K., Yu W. M., Akpinar P., Strickland S. (2002) Neuron 33, 861–875 [DOI] [PubMed] [Google Scholar]
- 40.Carstanjen D., Gross A., Kosova N., Fichtner I., Salama A. (2005) Transfusion 45, 1192–1200 [DOI] [PubMed] [Google Scholar]
- 41.Leiss M., Beckmann K., Girós A., Costell M., Fässler R. (2008) Curr. Opin. Cell Biol. 20, 502–507 [DOI] [PubMed] [Google Scholar]
- 42.Milner R., Wilby M., Nishimura S., Boylen K., Edwards G., Fawcett J., Streuli C., Pytela R., ffrench-Constant C. (1997) Dev. Biol. 185, 215–228 [DOI] [PubMed] [Google Scholar]
- 43.Weiner J. A., Fukushima N., Contos J. J., Scherer S. S., Chun J. (2001) J Neurosci 21, 7069–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pertz O., Hodgson L., Klemke R. L., Hahn K. M. (2006) Nature 440, 1069–1072 [DOI] [PubMed] [Google Scholar]
- 45.Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G. L., Hahn K. M., Danuser G. (2009) Nature 461, 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan J. R. (2007) J. Cell Biol. 177, 953–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krause S., Stendel C., Senderek J., Relvas J. B., Suter U. (2008) J. Peripher. Nerv. Syst. 13, 188–199 [DOI] [PubMed] [Google Scholar]
- 48.Webb D. J., Nguyen D. H., Gonias S. L. (2000) J. Cell Sci. 113, 123–134 [DOI] [PubMed] [Google Scholar]
- 49.Gaultier A., Arandjelovic S., Niessen S., Overton C. D., Linton M. F., Fazio S., Campana W. M., Cravatt B. F., 3rd, Gonias S. L. (2008) Blood 111, 5316–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X., Polavarapu R., She H., Mao Z., Yepes M. (2007) Am. J. Pathol. 171, 1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaultier A., Wu X., Le Moan N., Takimoto S., Mukandala G., Akassoglou K., Campana W. M., Gonias S. L. (2009) J. Cell Sci. 122, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]