Abstract
Iron is an essential cofactor for enzymes involved in numerous cellular processes, yet little is known about the impact of iron deficiency on cellular metabolism or iron proteins. Previous studies have focused on changes in transcript and proteins levels in iron-deficient cells, yet these changes may not reflect changes in transport activity or flux through a metabolic pathway. We analyzed the metabolomes and transcriptomes of yeast grown in iron-rich and iron-poor media to determine which biosynthetic processes are altered when iron availability falls. Iron deficiency led to changes in glucose metabolism, amino acid biosynthesis, and lipid biosynthesis that were due to deficiencies in specific iron-dependent enzymes. Iron-sulfur proteins exhibited loss of iron cofactors, yet amino acid synthesis was maintained. Ergosterol and sphingolipid biosynthetic pathways had blocks at points where heme and diiron enzymes function, whereas Ole1, the essential fatty acid desaturase, was resistant to iron depletion. Iron-deficient cells exhibited depletion of most iron enzyme activities, but loss of activity during iron deficiency did not consistently disrupt metabolism. Amino acid homeostasis was robust, but iron deficiency impaired lipid synthesis, altering the properties and functions of cellular membranes.
Keywords: Amino Acid, Glycolysis, Heme, Iron-Sulfur Protein, Sphingolipid, Sterol, Triacylglycerol, Tricarboxylic Acid Cycle, Diiron
Introduction
With a few notable exceptions, iron is an essential nutrient for virtually every organism. Every cell in the human body has a requirement for iron, yet iron can be toxic when present in excess. Iron deficiency is the most common nutritional disorder in the world (1). Iron deficiency and iron deficiency anemia are especially prevalent among children and women of childbearing age, where they are associated with perinatal mortality. Iron deficiency also impairs neurological development and cognitive function in children, and some of these defects appear to be irreversible (2). Although the pathogenesis of anemia in iron deficiency is well understood, other manifestations of iron deficiency are not understood at the cellular or metabolic level, and it is unlikely that all of the clinical manifestations of iron deficiency can be attributed to a reduction in the oxygen-carrying capacity of the blood. Iron overload is also a common disorder and is associated with hereditary hemochromatosis, iron loading anemias, and chronic inflammatory diseases of the liver (3, 4). Iron overload is typically manifest as an accumulation of iron in organs, especially the liver, heart, and pancreas, leading to organ dysfunction and failure. The mechanisms by which excess iron causes organ failure are unknown.
Iron is an essential nutrient, because iron cofactors activate enzymes involved in most of the major metabolic processes in the cell. This metal is used in the synthesis of organic and inorganic cofactors, such as heme and iron-sulfur clusters, as well as in the metallation of mononuclear and diiron enzymes (5–7). Iron proteins are required for oxygen delivery, for the oxidation of acetyl CoA via the tricarboxylic acid cycle and for the generation of ATP via oxidative phosphorylation. Iron cofactors are required for the biosynthesis of amino acids, proteins, sterols, and fatty acids. They are required for the synthesis and repair of DNA and for the metabolism of xenobiotics. Very little is known about the activity of these enzymes under conditions of iron deficiency. For examples, it is not known whether cells can tolerate large reductions in the activity of any of these enzymes, it is not known whether any particular class of iron enzyme is susceptible to iron deficiency, and it is not known whether individual iron-dependent pathways are disproportionally affected by iron deficiency.
Studies of metal metabolism in budding yeast have yielded important insights into iron metabolism in both humans and pathogenic microorganisms. In determining the cellular response to iron deficiency and iron overload in the yeast Saccharomyces cerevisiae, we and others have focused on changes in the levels of mRNA transcripts and proteins involved in iron homeostasis (8, 9). These studies indicated that S. cerevisiae responds to iron deficiency by (i) activating systems of iron uptake, (ii) mobilizing intracellular stores of iron, and (iii) adjusting metabolism to optimize the use of iron (10).
One metabolic adjustment indicated by the changes in mRNA transcript levels is the redistribution of iron away from some non-essential biosynthetic pathways toward other, essential iron-requiring pathways. Under conditions of iron deficiency, S. cerevisiae relies on two transcription factors, Aft1 and Aft2, to activate the transcription of genes involved in the primary response to iron deficiency (11). Some of this redistribution of iron is apparent in the genes directly activated by Aft1 and -2, and some is suggested by other transcripts that are down-regulated under iron deficiency (12). Many transcripts encoding components of the respiratory cytochromes, which contain several heme cofactors, and some heme biosynthetic genes are down-regulated in iron deficiency (8), while the heme-degrading enzyme encoded by HMX1 (an Aft1 target) is up-regulated (13). This down-regulation of heme synthesis and the respiratory machinery is consistent with the observation that iron-deficient cells cannot grow on non-fermentable carbon sources and thus cannot rely exclusively on respiration for energy production during iron deficiency.
Several transcripts encoding Fe-S cluster proteins involved in glutamate, leucine, and lipoic acid biosynthesis are down-regulated during iron deficiency (8, 9). Multiple transcripts coding for proteins involved in biotin biosynthesis, including the iron-sulfur cluster protein Bio2, are down-regulated in iron deficiency, whereas transporters facilitating the uptake of biotin and biotin precursors are up-regulated by Aft1.
Some of this down-regulation of mRNAs occurs at the transcriptional level while some is controlled post-transcriptionally. CTH1 and CTH2 encode two proteins that are expressed in response to iron deficiency and are under the control of Aft1 and -2 (8, 14). These proteins of the tristetraprolin family of RNA-binding proteins specifically recognize AU-rich sequences in the 3′-untranslated region of many transcripts coding for proteins that contain iron cofactors, such as succinate dehydrogenase, and aconitase. Cth1 and -2 bind to these mRNAs and promote their degradation. Thus, under conditions of iron deficiency, yeast activate the expression of proteins that lead to the degradation of mRNAs encoding proteins that function in iron-dependent pathways, thereby reducing the amount of iron allocated to these pathways. Cth1 and Cth2 also appear to be indirectly involved in the up-regulation of several transcripts that may contribute to an alteration in glucose metabolism and carbohydrate storage that occurs in iron deficiency.
Although these studies have produced some insights into the metabolic response to iron deficiency, they have a major limitation. That is, alterations in protein levels do not always correlate with the flux of metabolites through a pathway, because the flux through a pathway is largely determined by enzyme kinetics and substrate concentrations. Changes in the level of an iron-binding enzyme may alter the amount of iron dedicated to that pathway without producing a significant change in the flux through the pathway. Here we analyzed the metabolomes of yeast grown in iron-rich and iron-poor media to determine the extent to which flux through metabolic pathways is maintained or altered in the face of falling iron availability. We find that in yeast, amino acid homeostatic mechanisms are robust, but that carbon source utilization and lipid biosynthesis are affected by iron deficiency. We found specific alterations in the flux through the ergosterol and sphingolipid biosynthetic pathways at points where heme and diiron enzymes function. We specifically examined the metallation of iron-dependent enzymes and found that, although some enzymes are resistant to iron deficiency, others exhibit loss of their iron cofactors during periods of iron deficiency.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Media
The strain DBY7286 (MATa ura3) was used for all metabolite and microarray analyses. Strains carrying the tandem affinity purification (TAP) tag (MATa leu2Δ0 ura3Δ0 met15Δ0 ORF-TAP::HIS3) were obtained from Open Biosystems, and strains expressing GFP3 fusion proteins were obtained from Invitrogen. OPY113 (fre1Δ::LEU2 fre2Δ::HIS3 ade2–101::pPGK1-FRE1 ADE2) (15) is congenic to YPH499. The ACO1-TAP, LYS4-TAP, and RLI1-TAP strains were converted to leucine prototrophy by transformation with pRS405, linearized with AatII and HpaI, followed by selection on medium lacking leucine. The plasmid pERG6-YFP was constructed by amplifying the coding sequences of ERG6 and YFP and cloning them between the BamHI and XhoI sites of pRS416MET25. All strains were grown in synthetic defined (SD) minimal media containing only the supplements necessary to meet auxotrophic requirements. Defined-iron SD minimal media were prepared with yeast nitrogen base lacking iron and copper, supplemented with 1 μm copper sulfate, 25 mm MES, pH 6.1, 1 mm Ferrozine (Fluka), and the indicated concentrations of ferrous ammonium sulfate. For 55Fe labeling of cells, strains were grown from very low density to late log phase in SD minimal medium containing 2 μm 55Fe2Cl3 at 25–88 mCi/mg, which was reduced with 100 μm ascorbate. Cells were then washed and transferred to SD minimal medium containing 2 μm 55Fe2Cl3, 100 μm ascorbate, and 1 mm Ferrozine (iron-chelated medium) or to medium with 2 μm 55Fe2Cl3, 100 μm ascorbate, and no Ferrozine (iron-replete medium) and grown for 18–24 h to an A600 of 0.5.
Metabolomics and Microarray Analysis
DBY7286 was grown from very low density to mid-log phase (A600 = 0.5, ∼18 h) in defined-iron SD minimal medium containing 10 μm (low iron), 100 μm (optimal iron), and 300 μm (high iron) ferrous ammonium sulfate. For short term growth, cells grown in optimal iron were transferred to low iron or high iron medium for an additional 4 h. All cells were grown at 30 °C with shaking, and six independent cultures were prepared for each growth condition. Metabolomics analysis was carried out in collaboration with Metabolon, Inc. Cells were subjected to glass bead lysis, and metabolites were extracted and analyzed as described (16, 17). The samples were extracted using a series of four solvent extraction steps: 400 μl of tridecanoic acid (2.5 mg/ml) in ethyl acetate:ethyl alcohol (1:1), 200 μl of methanol, 200 μl of methanol:H2O (3:1), and 200 μl of dichloromethane:methanol (1:1). Each solvent-extraction step was performed by shaking for 2 min in the presence of glass beads using a Geno/Grinder 2000 (Glen Mills, Inc., Clifton, NJ). After each extraction the sample was centrifuged, and the supernatant was removed using the MicroLab STAR® robotics system, followed by re-extraction of the pellet. The multiple extract supernatants were pooled and then split into two equal aliquots, one for liquid chromatography/mass spectrometry (LC/MS) and one for gas chromatography/mass spectrometry (GC/MS). Aliquots were placed on a TurboVap® (Zymark) to remove the solvent, frozen, and dried under vacuum overnight. Samples were maintained at 4 °C throughout the extraction process. For LC/MS analysis extract aliquots were reconstituted in 10% methanol and 0.1% formic acid. For GC/MS analysis, aliquots were derivatized using equal parts bistrimethylsilyltrifluoroacetamide and solvent mixture acetonitrile:dichloromethane:cyclohexane (5:4:1) with 5% triethylamine at 60 °C for 1 h.
LC/MS was carried out using a Surveyor HPLC (ThermoElectron Corp., San Jose, CA) with an electrospray ionization (18) source coupled to an LTQ mass spectrometer (ThermoElectron Corp.). The derivatized samples for GC/MS were analyzed on a ThermoFinnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer operated at unit mass resolving power. Compounds were identified by automated comparison to Metabolon's reference library entries. Data normalization and statistical analysis were performed as described previously (16). Data were normalized to correct for variation resulting from instrument inter-day tuning differences. Raw area counts for a compound were divided by the median value, setting the medians equal for each day's run. Missing values were assumed to result from areas being below the limits of detection. Missing values for a given compound were imputed with half the observed minimum after the normalization step. Quantitative values were derived from integrated raw detector counts of the mass spectrometers. Importantly, while peak area comparisons between samples represent relative amounts of each ion detected, different compounds and ions have different ionization potentials. To preserve all of the variation, yet allow compounds of widely different raw peak areas to be compared directly on a similar graphical scale, a simple arithmetic transformation was performed in which each data point was divided by the mean of the entire set for that compound. Statistical analysis was performed on log transformed values with Welch's two-sample t test.
Microarray analysis was performed in quadruplicate (15) on DBY7286 grown in low iron and high iron conditions identical to those used for metabolomics analysis. The full microarray dataset has been deposited at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE19016.
Additional Metabolite Extraction and Analyses
Glucose levels in conditioned media were measured by the glucose oxidase method using a blood glucose monitor (Bayer). Standard curves were generated for low iron and high iron media. Cellular glycogen levels were measured as described before (19).
For amino acid analysis, cells were subjected to glass bead lysis in 10% trichloroacetic acid, and the clarified supernatant was subjected to analysis using an Hitachi Amino Acid Analyzer Model L-8800. Raw data were recorded using EZ Chrom Elite software, and cell extracts were normalized by absorbance at 260 nm.
For sterol analysis, the non-saponifiable sterol fraction was extracted and analyzed on a Hewlett-Packard 5890 series II gas chromatograph (GC) as previously described (20). Sterol intermediates were identified based on retention times and analyses of sterols that accumulate in wild-type and various ergosterol mutant strains. Peaks were identified by comparison to known standards. Neutral lipids were extracted and measured as described (21).
Lipids were extracted in chloroform/methanol (22, 23) then analyzed quantitatively by HPLC. A 3 A600 cell pellet was dried and subjected to acid methanolysis by boiling in 1 n HCl in methanol for 30 min. After cooling, 1 volume of 0.9% NaCl was added, the fatty acid methyl esters were extracted twice with hexane:diethyl ether (1:1), and the extracts were pooled and dried under N2 gas. The pH of the remaining aqueous phase was adjusted with 0.125 volume of 10 n NaOH. The long-chain bases were analyzed as described with the following modifications (24). Long-chain bases were extracted twice with 2 ml of hexane, and the extracts were pooled and dried. The long-chain base extract was redissolved in 80 μl of methanol:190 mm triethylamine (20:3) and 20 μl of AccQ-Fluor Reagent (Waters). Samples were incubated at 23 °C for 60 min, and 60 μl of the sample was injected onto a Genesis C18 4 mm HPLC column (Jones Chromatography) and resolved on an Agilent 1100 series HPLC equipped with a fluorescence detector. The long-chain bases were resolved using an isocratic mobile phase of acetonitrile:methanol:H2O:acetic acid:triethylamine (48:32:16.5:3.0:0.7) at 1.5 ml/min and detected by the AccQ fluorescence (Ex: 244 nm/Em: 398 nm). Phytosphingosine (PHS) concentrations were determined by adding the PHS peak with that of the 1,4-anhydro-PHS peak, which elutes slightly after dihydrosphingosine (DHS). Fatty acid methyl ester extracts were resuspended in 50 μl of hexane, and 1 μl was autoinjected onto a GC/MS (HP 6890/5973) equipped with a 30 m × 0.25 mm × 0.25 μm HP INNOWax column. The fatty acid methyl esters were resolved at 0.5 ml/min constant flow using a temperature gradient as follows: initial oven at 50°, 40°/min to 90° and hold for 2 min, 20°/min to 250°, and hold for 90 min. Fatty acid methyl ester identification was determined by using an MS fragmentation pattern.
Heme was extracted from 55Fe-labeled cells as described (25) and measured by scintillation counting. Ferric reductase assays were performed as described (26).
Western Blotting, Fractionation, and Immunoprecipitations
TAP-tagged strains were lysed as described (15) and subjected to SDS-PAGE, and proteins were detected using peroxidase anti-peroxidase-soluble complex (Sigma) at 1:1000. For subcellular fractionations, cells were lysed as described (15), and lysates were separated on 20–60% continuous sucrose gradients. Fractions were collected and subjected to SDS-PAGE and Western blotting using peroxidase anti-peroxidase, anti-GFP (Roche Applied Science) at 1:1000, and anti-Dpm1 (Molecular Probes) at 1:1000. Immunoprecipitations from 55Fe-labeled cells were performed as described (27) using IgG-Sepharose beads (Sigma). Immune complexes were washed, and 10% of the total washed beads were used for Western blotting, whereas the remainder was subjected to scintillation counting. Immunoprecipitated proteins were detected using peroxidase anti-peroxidase complex and Cy3-labeled goat anti-rabbit at 1:1000. Quantitative fluorescence intensity was determined using a Typhoon imager with ImageQuant software (Amersham Biosciences). As a negative control, protein G-Sepharose was substituted for IgG-Sepharose in the immunoprecipitations of 55Fe-labeled lysates.
RESULTS AND DISCUSSION
Metabolomics and Transcriptomics
Previous studies on iron deficiency in yeast have utilized common laboratory strains of S. cerevisiae that exhibit multiple amino acid auxotrophies. These auxotrophies are rescued with amino acid supplementation of the medium, which typically results in alterations in the expression of amino acid biosynthetic genes. Because the synthesis of amino acids is a major function of intermediary metabolism and because several enzymes involved in amino acid biosynthesis are iron-containing proteins, we used a strain that did not exhibit any amino acid auxotrophies and grew the strain in medium without amino acid supplementation.
We performed metabolite analyses on yeast grown for 18 h (7 to 8 generations) or for short duration (4 h) in low iron and high iron media and compared these metabolite levels to those of yeast grown with optimal iron concentrations (supplemental Table S1). Cells grown in low iron medium exhibited slower growth, with a 20% increase in doubling time compared with optimal-iron medium at the end of the culture period, confirming that iron was limiting for growth in this medium. Cells grown in high iron medium also exhibited slower growth, with a 40% increase in doubling time, confirming that high iron cells were exposed to iron toxicity.
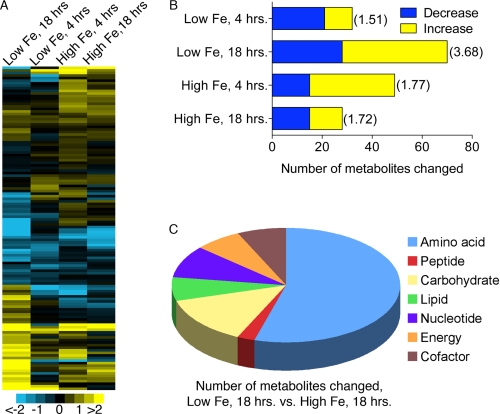
Using both LC/MS and GC/MS, 129 metabolites were detected and identified in the yeast samples, and most of these exhibited some change in either high iron or low iron medium (Fig. 1A). Of the growth conditions tested, the greatest number of metabolite level changes and the highest magnitude changes occurred in the yeast grown for 18 h in low iron medium, with 70 metabolites exhibiting significant changes (Fig. 1B). Yeast grown for 18 h in high iron medium exhibited the smallest changes to the metabolome, and specifically did not show evidence of oxidative stress. For example, levels of oxidized and reduced glutathione did not change. Comparison of the low iron, 18-h yeast to the high iron, 18-h yeast mirrored the changes seen in the comparison of low with optimal iron, but with greater magnitude changes (Fig. 1C), and this comparison is shown in the figures below. Virtually every category of metabolites was affected by iron availability.
FIGURE 1.
Metabolite analysis of iron-deficient and iron-replete yeast. A, heat map of 129 identified metabolites detected in yeast cells grown in high iron or low iron media relative to optimal iron. Metabolite levels were expressed as log2 of the normalized ratio. Shades of yellow and blue indicate an increase or decrease of metabolite, respectively, according to the scale bar. B, largest changes in cells grown 18 h in low iron medium. The number of biochemicals in each growth condition that decreased or increased relative to the levels in cells grown in optimal iron is shown. Significant change was determined using a paired t test of log-transformed, normalized values with p < 0.05. M-scores for each growth condition are shown in parenthesis, M-score = Average |z|. The M-score for a normal distribution is 0.80. C, metabolites affected by iron grouped by category. The 44 metabolites altered in low iron cells are compared with high iron cells, 18 h, grouped by metabolic categories. See also supplemental Table S1.
To correlate changes in metabolite levels with changes in gene transcript levels, we performed microarray analyses on cells grown in low iron and high iron conditions identical to those used for the metabolite analyses. The results of these microarray analyses were very similar those previously published, in that the Aft1 target genes were highly up-regulated in the low iron condition, confirming that our growth conditions produced iron deficiency (data not shown). They differed from previous microarrays in the regulation of genes involved in the uptake and synthesis of amino acids and in the ergosterol biosynthetic pathway.
Glucose Metabolism and Energy Production
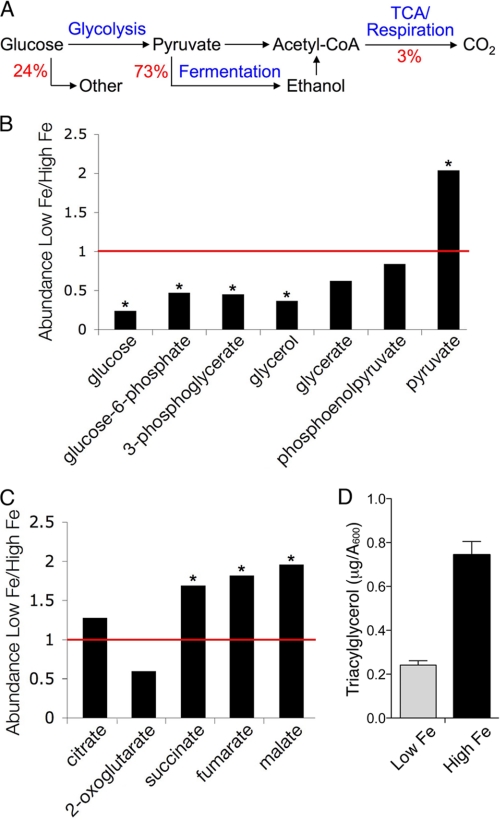
Yeast consume the majority of cellular glucose in the process of energy production. Glucose is metabolized to pyruvate via glycolysis, then pyruvate is either converted to ethanol and CO2 via fermentation, or fully oxidized to CO2 and H2O via the tricarboxylic acid cycle and respiration (Fig. 2A). Even in the presence of oxygen, yeast typically ferment nearly all of the available glucose, with only 3% being metabolized via respiration (28). Although respiration yields far more energy than fermentation, it also requires large quantities of iron, as respiratory complexes II–IV contain numerous heme and Fe-S centers, and two tricarboxylic acid cycle enzymes, aconitase and succinate dehydrogenase, contain Fe-S clusters. Although yeast can grow on carbon sources that can only be metabolized through respiration, they cannot do so under conditions of iron deficiency. Previous studies using microarrays found that iron-deficient yeast down-regulate transcripts involved in the tricarboxylic acid cycle and respiration (8, 9, 29), and we found evidence of similar transcript changes in these studies (supplemental Table S2). Metabolite analysis indicated that levels of intracellular glucose, glycolytic intermediates, and byproducts of glycolysis were significantly depleted in iron-deficient cells while the product of glycolysis, pyruvate, was elevated (Fig. 2B). In contrast, tricarboxylic acid cycle intermediates succinate, fumarate, and malate were significantly elevated (Fig. 2C). Citrate levels rose in cells grown in both low iron medium (2.8-fold) and high iron medium (2.2-fold) when compared with optimal iron cells. Only 2-oxoglutarate levels fell in low iron cells, which may have occurred as large amounts of this intermediate are drawn out of the tricarboxylic acid cycle for the synthesis of amino acids and purines. Intracellular glucose depletion was not associated with depletion of glucose in the growth medium (supplemental Fig. S2A).
FIGURE 2.
Increased glycolysis, diminished respiration, and reduced lipid storage in iron deficiency. A, yeast cells primarily metabolize glucose to ethanol, but can also fully oxidize glucose to CO2 via the tricarboxylic acid cycle and respiration. Levels of glucose and glycolytic intermediates (B) and tricarboxylic acid cycle (C) intermediates were measured in cells grown in low iron and high iron medium and expressed as a ratio. Values < 1 (red bar) indicate depletion in iron-deficient cells, values > 1 indicate accumulation in iron-deficient cells. *, indicates p < 0.05. D, reduced triacylglycerol in lipid particles from iron-deficient cells. Neutral lipids were extracted from cells grown for 24 h in defined-iron medium containing 5 μm (Low Fe) or 300 μm (High Fe) ferrous iron and analyzed by HPLC. See also supplemental Table S2 and supplemental Fig. S2. Error bars indicate ± S.E.
Intermediates from the glycolytic pathway can be diverted for use in carbohydrate storage and lipid biosynthesis. Levels of the storage carbohydrate glycogen were very low, as would be expected in logarithmically growing cells, and did not differ between iron-deficient and iron-replete cells (supplemental Fig. S2B). In contrast, levels of triacylglycerol, a storage form of neutral lipid synthesized from the glycolytic intermediate 3-phosphoglycerate via glycerol, were reduced by 68% in iron-deficient cells (Fig. 2D). The depletion of glucose, glycolytic intermediates, and triacylglycerol coupled with the elevated pyruvate suggested increased flux of glucose through the glycolytic pathway for the purpose of energy production. This would be consistent with a loss of energy production through the tricarboxylic acid cycle and respiration as a consequence of iron deficiency. Thus, even though yeast metabolize only a small fraction of glucose via respiration, this small fraction provides significant energy to the cell. This small amount of respiration was reduced further during iron deficiency, and the metabolic impact on the cell included the diversion of glycolytic intermediates away from lipid biosynthesis.
Amino Acid Biosynthesis
All of the nitrogen-containing compounds in S. cerevisiae can be synthesized from intermediates derived from carbon source metabolism and ammonia (30, 31). The nitrogen of ammonia is exclusively available to the cell through its incorporation into glutamate and glutamine, from which all other amino acids and nucleotides are synthesized. Glutamate is formed from the condensation of ammonium and 2-oxoglutarate. Thus, cells growing in the absence of amino acid supplements devote a substantial amount of their metabolic machinery to the synthesis of amino acids, and must maintain the synthesis of 2-oxoglutarate by maintaining flux through the first three steps of the tricarboxylic acid cycle. Significantly, enzymes catalyzing these first three steps, Cit1, Aco1, and Idh1, were not among those showing >2-fold down-regulation by iron deficiency in our microarrays (supplemental Table S2). Several enzymes involved in amino acid biosynthesis contain Fe-S clusters (5), and, in previous studies, some of these (Glt1 and Leu1) exhibited lowered transcript and protein levels in response to iron deficiency, suggesting that down-regulation of Fe-S-containing proteins is one strategy yeast employ in the adaptation to iron deficiency (8, 9).
In our studies, transcript levels of many genes involved in amino acid synthesis and uptake changed in response to iron deficiency (supplemental Table S3). 23 transcripts involved in the synthesis and uptake of amino acids were up-regulated by iron deficiency. The majority (17 of 23) of these genes are involved in the uptake or synthesis of amino acids that require Fe-S proteins for synthesis. The transcripts of the Fe-S-containing enzymes were consistently down-regulated by iron deficiency, although, with the exception of GLT1, these changes were small, 2-fold or less. Glt1 is also unique among these enzymes in that cells lacking GLT1 can synthesize glutamate via an alternate pathway (30); the other enzymes are required in their respective pathways.
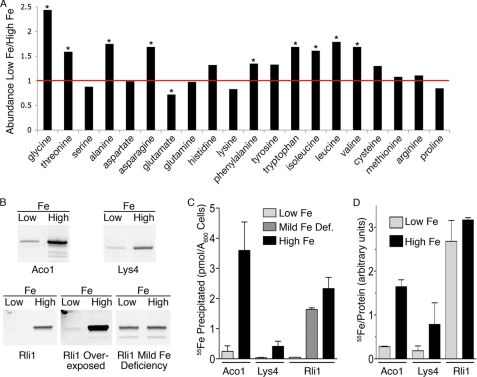
Metabolite analysis revealed a 14-fold accumulation of 2-isopropylmalate in iron-deficient cells (supplemental Table S1). 2-Isopropylmalate is the substrate for the Fe-S enzyme isopropylmalate isomerase, encoded by LEU1 (32), and its accumulation suggested that iron deficiency resulted in either a loss of Leu1 activity or an increased flux of leucine precursors through the biosynthetic pathway. We expected that iron deficiency would result in deficiencies of amino acids dependent on Fe-S enzymes: leucine, isoleucine, valine, lysine, methionine, cysteine, and glutamate. Surprisingly, most amino acids were present at higher levels in iron-deficient cells, and we did not detect severe deficiencies of any amino acids, including leucine, with only glutamate demonstrating a mild, 1.4-fold depletion (Fig. 3A). Only after prolonged iron starvation did a significant amino acid deficiency develop, as lysine levels fell to 55% of those of iron-replete cells (supplemental Fig. S3A). These data indicated that cellular homeostatic mechanisms are robust with respect to amino acids. These data also raise the intriguing possibility that the cellular components that limit growth are amino acids, because depletion of one or a few individual amino acids would have catastrophic effects on new protein synthesis.
FIGURE 3.
Lack of amino acid deficiencies in iron-deficient cells despite loss of iron-sulfur clusters. A, levels of amino acids were measured in cells grown in low iron and high iron media and expressed as a ratio. Values greater than 1 indicate accumulation in iron-deficient cells. *, indicates p < 0.05. B and C, immunoprecipitation of Fe-S enzymes from cells grown in iron-chelated and iron-replete medium. Strains expressing Aco1-TAP, Lys4-TAP, and Rli1-TAP were labeled with 55Fe in iron-replete medium, and then grown in iron-chelated or iron-replete medium containing 55Fe of the same concentration and specific activity. TAP-tagged enzymes with their iron ligands were recovered by immunoprecipitation and analyzed by Western blotting (B). Proteins were quantitated by fluorescent imaging. Iron bound to precipitated protein was measured by scintillation counting (C). The relative amounts of iron bound per unit of precipitated protein were calculated (D). Error bars indicate ±S.E. See also supplemental Table S3 and supplemental Fig. S3.
We questioned how the synthesis of amino acids that require Fe-S enzymes could occur in the setting of iron deficiency. Therefore, we measured the incorporation of Fe-S clusters into Fe-S enzymes under iron-sufficient and iron-deficient growth conditions by affinity purification of enzymes from cells labeled with 55Fe. Strains expressing enzymes with the TAP tag incorporated at the C terminus were used for these assays. We selected two mitochondrial Fe-S proteins, Aco1 and Lys4, as well as the nuclear/cytosolic Fe-S enzyme Rli1 for analysis. The ILV3-TAP and LEU1-TAP strains exhibited iron-dependent isoleucine/valine and leucine auxotrophies and were not analyzed further (data not shown). Cells were labeled with 55Fe in iron-replete or iron-chelated medium. TAP-tagged enzymes with their iron ligands were recovered by affinity purification, and the total protein recovered was measured by Western blotting (Fig. 3B), while the amount of iron retained in the protein was measured by scintillation counting (Fig. 3C).
Growth in iron-chelated medium produced a marked iron deficiency (25- to 41-fold reduction in intracellular iron (supplemental Fig. S3B)), which was associated with a 6-fold reduction of Aco1-TAP and Lys4-TAP, and a 37-fold reduction of Rli1-TAP, with Rli1-TAP becoming almost undetectable (Fig. 3B). Mild iron deficiency (5- to 6-fold reduction in intracellular iron) produced only slight reductions in Rli1-TAP. This loss of Fe-S protein in iron-deficient cells was likely due to instability and degradation of the proteins in the absence of their Fe-S ligands, as transcript levels decreased only slightly under these conditions. We measured the amount of iron associated with the individual enzymes (Fig. 3C and supplemental Fig. S3C) and found that Fe-S proteins from iron-deficient cells exhibited a loss of their iron ligands that was greater than the loss of the polypeptide in the case of Aco1-TAP and Lys4-TAP (14- and 12-fold, respectively), but proportional to the loss of polypeptide in the case of Rli1 (33-fold). Calculation of the amount of iron bound per unit of protein (Fig. 3D) indicated that, when grown in iron-sufficient medium, Rli1-TAP contained more iron than Aco1-TAP, consistent with the incorporation of two 4Fe-4S clusters into Rli1 and one 4Fe-4S cluster into Aco1-TAP. Aco1-TAP and Lys4-TAP exhibited large decreases (4- to 6-fold) in the iron bound per unit of protein after growth in iron-chelated medium, suggesting that, despite the instability of the protein in iron-deficient cells, some protein is expressed without Fe-S clusters. In contrast, although the amount of Rli1-TAP protein decreased, the amount of iron bound per unit of Rli1-TAP did not significantly change in iron-chelated medium, suggesting that little to no Rli1-TAP accumulates without Fe-S clusters.
These data indicated that iron deficiency produced large decreases in the metallation and activity of Fe-S enzymes, but that this loss of activity was not associated with the loss of sufficient enzymatic activity to meet biosynthetic needs, as cells were capable of coordinating consumption of amino acids with their synthesis. Because the levels of metallated Aco1, Lys4, and Rli1 were similarly affected by iron deficiency, these data suggest that cells did not have a mechanism to preferentially target Fe-S clusters to essential proteins, such as Rli1. Our observation that amino acid levels are generally not depleted during iron deficiency, despite significant (12- to 33-fold) loss of active, metallated Fe-S proteins, suggests that these Fe-S enzymes are present in excess in iron-replete cells.
Ergosterol Biosynthesis
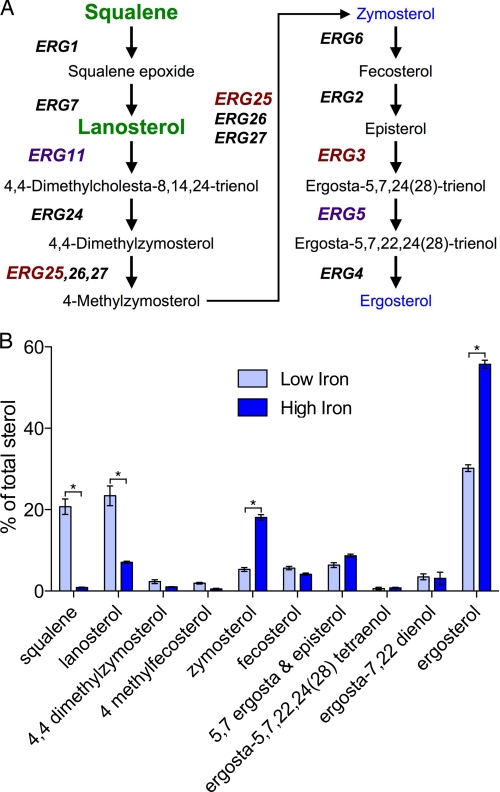
Iron-deficient cells exhibited a 20-fold accumulation of squalene in our initial metabolite analysis (supplemental Table S1), and this led us to examine the sterol biosynthetic pathway. Sterol biosynthesis in yeast is an essential process involving over 20 enzymatic steps (33–35). The second half of the sterol biosynthetic pathway is dedicated to the production of ergosterol, the major oxysterol of yeast, and begins with the condensation of two farnesyl pyrophosphate molecules to form squalene (Fig. 4A). The latter half of the ergosterol pathway contains multiple steps requiring iron-dependent enzymes. Erg11 and Erg5 are heme-dependent enzymes of the cytochrome P450 family and require the hemoprotein Dap1 for full activity (36, 37). Erg25 and Erg3 are oxo-diiron enzymes of the fatty acid hydroxylase/sterol desaturase family. We examined the effects of iron deficiency on ergosterol biosynthesis by measuring the levels of intermediates in cells grown in iron-replete and iron-deficient media (Fig. 4B). Iron-deficient cells exhibited a 24-fold accumulation of squalene and a 3.3-fold accumulation of lanosterol compared with iron-replete cells. They also exhibited depletion of the major oxysterols, zymosterol (3-fold) and ergosterol (2-fold). These data indicated partial biosynthetic blocks at Erg1 and Erg11 and reduced flux through the pathway as a result of iron deficiency. The accumulation of squalene could be explained as a secondary effect of Erg11 deficiency, that is, through the altered equilibrium produced by the accumulation of lanosterol. However, we did not observe accumulation of squalene epoxide, the substrate of Erg7, making this less likely. Alternatively, lanosterol may act as an inhibitor of Erg1.
FIGURE 4.
Disruption of ergosterol biosynthesis in iron-deficient cells. A, ergosterol biosynthetic pathway. Heme-dependent enzymes are in purple; diiron enzymes are in red. Accumulated intermediates are in green, depleted sterols in blue. B, depletion of ergosterol and accumulation of biosynthetic intermediates in iron-deficient cells. Cells were grown in defined-iron medium containing 5–10 μm (Low Iron) or 300 μm (High Iron) ferrous iron, then sterol biosynthetic intermediates were extracted and measured by GC. Intermediates are expressed as a percentage of the total sterol content. Error bars indicate ±S.E. *, indicates p < 0.05.
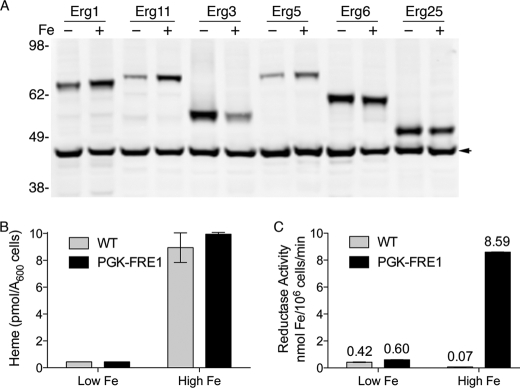
Previous studies have reported that several ergosterol biosynthetic genes are transcriptionally up-regulated during iron starvation (8); however, we observed only small (<2-fold) changes in the transcript levels of ERG genes (data not shown). We examined the effect of iron deficiency on levels of Erg1 and the iron-dependent Erg proteins by growing the TAP-tagged strains in iron-deficient or iron-replete medium and measuring protein levels by Western blotting (Fig. 5A). Erg1 and the heme proteins Erg11 and Erg5 were present at slightly lower levels in iron-deficient cells, whereas the diiron enzymes Erg3 and Erg25 were present at slightly higher levels. These changes were small and may not explain the accumulation of squalene and lanosterol that we observed.
FIGURE 5.
Iron-dependent changes in Erg proteins, intracellular heme, and ferric reductase activity. A, Western blot of Erg proteins from iron-deficient and iron-replete cells. TAP-tagged strains were grown as in Fig. 4B, and equal numbers of cells were subjected to lysis and Western blotting with peroxidase anti-peroxidase and anti-Pgk1 antibodies. Arrowhead indicates Pgk1 loading control. B, loss of intracellular heme in iron-deficient cells. A wild-type strain (YPH499) and the congenic fre1Dfre2D strain expressing FRE1 from the PGK1 promoter were labeled with 55FeCl3, then cells were divided and grown in iron-chelated (Low Fe) or iron-replete medium (High Fe) containing 55FeCl3. 55Fe-labeled heme was quantitatively extracted. C, loss of heme-dependent enzyme activity in iron-deficient cells. Cells were grown as in B using unlabeled iron, and ferric reductase activity was measured. Mean values of activity are indicated above each bar. Error bars represent ±S.E. See also supplemental Fig. S4.
Heme Limitation of Erg11 and Fre1
Erg11 catalyzes the first iron-dependent step in the biosynthetic pathway, as this enzyme requires a heme cofactor for activity (35). We questioned whether the accumulation of lanosterol and loss of Erg11 activity was due to a general loss of cellular heme in the setting of iron deficiency. Previous studies have suggested that heme levels fall in iron-deficient cells (13). Decreased expression of heme-rich respiratory complexes could have accounted for a portion of the decrease in total cellular heme, but whether individual heme proteins lacked cofactors remained unclear. We therefore examined the heme content and activity of a heme protein that was predicted to be active during iron deficiency, Fre1.
The process of iron uptake in yeast is heme-dependent, because the first step in iron uptake is the reduction of Fe(III) to Fe(II) at the cell surface by a family of heme-dependent ferric reductases (10). The genes encoding these enzymes are switched on in iron deficiency, and the preservation of their enzymatic activity in iron deficiency is important for the cell. We constructed a strain in which the major reductases, encoded by FRE1 and FRE2, were deleted, and FRE1 was expressed under the control of the strong PGK1 promoter, which is not regulated by iron, thereby uncoupling the surface reductase activity from transcriptional regulation. We labeled the PGK1-FRE1 strain and the congenic wild-type strain in iron-sufficient and iron-chelated media containing 55Fe, then measured heme content and ferric reductase activity. Both the wild-type and the PGK1-FRE1 strains exhibited a marked (20- to 23-fold) loss of cellular heme after growth in iron-chelated medium (Fig. 5B). This loss of cellular heme was reflected in the 14-fold loss of ferric reductase activity exhibited by the PGK1-FRE1 strain grown in iron-chelated medium (Fig. 5C), indicating that iron deficiency produces a cellular heme deficiency that limits the activity of heme-dependent enzymes. The wild-type strain exhibited a 6-fold increase in reductase activity with iron deficiency, although the level of activity in iron-chelated medium was similar to that of the PGK1-FRE1 strain. This increase in activity was due to the transcriptional activation of the wild-type FRE1 and FRE2 genes in iron-deficient cells (38).
These data indicated that iron deficiency produced a 20-fold drop in cellular heme levels, resulting in heme limitation of Fre1 activity as well as Erg11 activity. Whether iron-dependent enzymes downstream of Erg11 were affected by iron deficiency was unclear in the setting of the upstream block, but inactivation of heme and diiron enzymes in other pathways would suggest that this occurred, contributing to the loss of ergosterol in iron-deficient cells.
Redistribution of Erg1 in Iron Deficiency
Although heme deficiency explained the accumulation of lanosterol and the block in ergosterol synthesis at Erg11, it did not explain the accumulation of squalene and the block at Erg1. The conversion of squalene to squalene epoxide appears to be the rate-limiting step in the latter half of the ergosterol biosynthetic pathway (39, 40), and any loss of activity of Erg1 would be expected to produce an accumulation of squalene. Erg1 exhibits dual localization in the endoplasmic reticulum (ER) and in lipid particles (LPs) (41). Although the majority of ERG proteins are located in the ER, almost all Erg6 and traces of Erg7 exhibit localization to LPs. The Erg1 that is located in LPs is enzymatically inactive, possibly due to the absence of its required cytochrome P450 reductase (42).
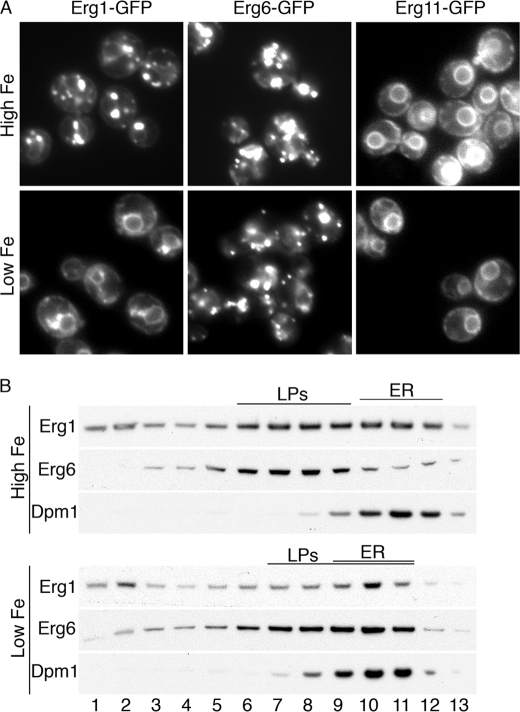
We questioned whether the loss of Erg1 activity in iron-deficient cells could be due to a change in the distribution of Erg1 between the ER and LPs. We grew cells expressing GFP-tagged versions of Erg1, Erg6, and Erg11 in iron-replete or iron-deficient medium and examined the distribution of the proteins within the cells (Fig. 6A). Iron-replete cells exhibited the expected localization of Erg6-GFP to multiple, bright punctate areas of the cytosol that represent LPs. Erg11-GFP was located in peripheral and perinuclear membranes that correspond to the ER. Erg1-GFP exhibited dual localization in both LPs and ER. After growth in iron-deficient medium, Erg6-GFP and Erg11-GFP exhibited no change in localization, but Erg1-GFP exhibited a marked loss of signal in LPs while retaining localization in the ER, indicating a redistribution of Erg1 from LPs to ER. We attempted to confirm this change in localization by separating cellular membranes on sucrose density gradients, and examining the distribution of proteins by Western blotting (Fig. 6B). In iron-replete cells, LPs and ER exhibited distinct patterns of distribution, with Erg1 being distributed in fractions characteristic of both organelles. In iron-deficient cells, however, Erg1 was detected in higher density fractions in a pattern similar to ER membranes. Surprisingly, the LPs also shifted to fractions containing higher density membranes. This shift was likely due to the loss of triacylglycerol from the LPs in iron deficient cells (see Fig. 2D). This redistribution of Erg1 to the ER in iron-deficient cells suggests that, under conditions of heme deficiency and reduced ergosterol synthesis, the cell does not localize Erg1 to a compartment where the enzyme is inactive (the LPs), but retains it in the compartment where it is active (the ER).
FIGURE 6.
Reduced localization of Erg1 to lipid particles and altered lipid particle composition in iron-deficient cells. A, localization of Erg1 to ER in iron-deficient cells. Strains expressing Erg1-GFP, Erg6-GFP, and Erg11-GFP were grown in defined-iron medium containing 5 μm (Low Fe) or 300 μm (High Fe) ferrous iron prior to fluorescence imaging. Erg6 is expressed primarily in lipid particles and Erg11 in the ER. B, altered migration of lipid particles from iron-deficient cells. A strain expressing Erg1-TAP and Erg6-YFP was grown as in A, and lysates were separated on sucrose density gradients. Erg1, lipid particles (Erg6), and endoplasmic reticulum (Dpm1) were detected by Western blotting.
Sphingolipid and Fatty Acid Biosynthesis
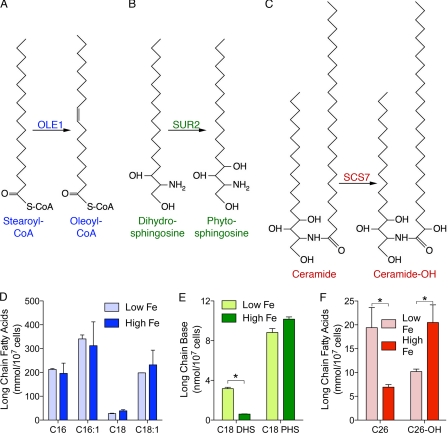
Metabolite analysis also pointed toward a disruption of sphingolipid synthesis in iron-deficient cells, as they exhibited a 9-fold accumulation of sphinganine (also called dihydrosphingosine (DHS)). The C4 hydroxylation of DHS to form PHS is catalyzed by Sur2, a non-essential enzyme with homology to the oxo-diiron enzymes of ergosterol and fatty acid biosynthesis (43). Three enzymes of this family are involved in fatty acid and sphingolipid biosynthesis, Sur2, Ole1, and Scs7 (Fig. 6, A–C). OLE1 encodes the essential δ-9-fatty acid desaturase and contains an N-terminal desaturase domain and a C-terminal cytochrome b5 domain, and, thus, contains both diiron and heme centers (33). The non-essential Scs7 catalyzes the α-hydroxylation of sphingolipid-associated, very long chain fatty acids, and contains both an N-terminal cytochrome b5 domain and a C-terminal desaturase domain (44). Sur2 contains only a diiron-containing desaturase domain, and presumably relies on a separately encoded cytochrome b5 for activity.
We examined the effects of iron deficiency on these enzymes by measuring the levels of substrates and products in iron-deficient and iron-replete cells. Ole1 catalyzes the mono-desaturation of palmitoyl- (C16) and stearoyl- (C18) coenzyme A. Iron-deficient and iron-replete cells exhibited no difference in the amounts of saturated and monounsaturated fatty acids (Fig. 7D), suggesting that iron-deficient cells were not deficient in Ole1 activity. Measurement of C18 DHS and C18 PHS confirmed that iron-deficient cells exhibited a 5-fold accumulation of DHS, indicating a partial loss of Sur2 activity, although there was minimal depletion of PHS in the iron-deficient cells (Fig. 7E). We measured the level of C26 fatty acids (derived from ceramides) and found that iron-deficient cells exhibited both a 3-fold accumulation of the unhydroxylated form and a 2-fold depletion of the hydroxylated form (Fig. 7F), indicating a loss of Scs7 activity.
FIGURE 7.
Disruption of sphingolipid synthesis in iron deficiency. Diiron enzymes in fatty acid synthesis. Ole1 (A), Sur2 (B), and Scs7 (C) are heme- and diiron-requiring fatty acid desaturases (Ole1) and hydroxylases (Sur2 and Scs7). D, saturated and unsaturated long chain fatty acids unaffected by iron deficiency. E, accumulation of un-hydroxylated sphingolipid in iron deficiency. F, impaired hydroxylation of ceramides in iron deficiency. Cells were grown in defined-iron medium containing 5 μm (Low Fe) or 300 μm (High Fe) ferrous iron for 24 h, and then fatty acid methyl esters and long chain bases were extracted and analyzed by GC/MS or HPLC, respectively. Error bars indicate ±S.E. *, indicates p < 0.05.
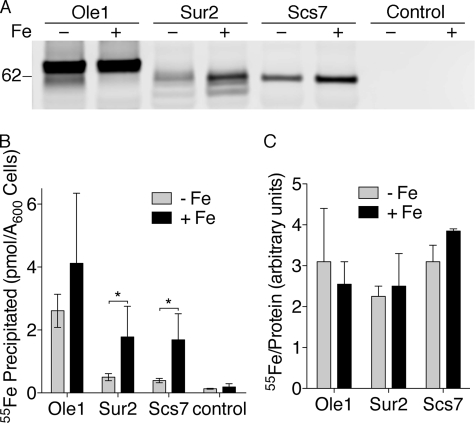
This analysis of cellular lipids suggested that Ole1 was resistant to iron deficiency while Sur2 and Scs7 were sensitive. This surprising result prompted us to more closely examine the expression level and metallation of these enzymes. We examined these enzymes by again labeling the congenic TAP-tagged strains in iron-replete or iron-chelated medium containing 55Fe. TAP-tagged proteins containing iron ligands were recovered by affinity purification, and the amounts of proteins and bound iron were measured. Cells grown in iron-chelated medium exhibited slowed growth and 7- to 14-fold lower levels of cellular iron than cells grown in iron-replete medium, confirming that growth in iron-chelated medium produced iron deficiency. Growth in iron-chelated medium produced little change in the levels of Ole1-TAP, whereas growth in iron-chelated medium produced 60 and 50% reductions in the levels of Sur2-TAP and Scs7-TAP, respectively (Fig. 8A). Similarly, the amount of iron bound to Ole1-TAP did not significantly change when cells were grown in iron-chelated medium, but the iron bound to Sur2-TAP and Scs7-TAP was reduced by 70% (Fig. 8B). Calculation of the amount of iron bound per unit of protein (Fig. 8C) indicated that all three proteins bound similar amounts of iron in both iron-chelated and iron-replete media, suggesting that these proteins were not expressed without their metal cofactors in iron-deficient cells.
FIGURE 8.
Resistance of Ole1 to iron deficiency. A, reduced expression of Sur2 and Scs7, but not Ole1, in iron-deficient cells. Strains expressing TAP-tagged versions of heme/diiron enzymes were labeled with 2 μm 55Fe and then grown in iron-chelated (− Fe) or iron-replete (+ Fe) media as in Fig. 3B. TAP-tagged proteins were quantitatively immunoprecipitated using IgG-Sepharose. Protein levels were measured by Western blotting. Control samples consisted of identically labeled TAP-tagged strains incubated with protein G-Sepharose. B, reduced levels of iron bound to Sur2 and Scs7, but not Ole1, in iron-deficient cells. Iron bound to proteins immunoprecipitated in A was measured by scintillation counting. C, similar metallation of heme/diiron enzymes in iron-deficient and iron-replete cells. Precipitated proteins were quantitated by fluorescent imaging. Relative amounts of iron bound per unit of protein precipitated was calculated. Experiments were replicated three times. Error bars indicate ±S.E. *, indicates p < 0.05.
Implications of Alterations in Iron Deficiency
Ergosterol is enriched in the outer leaflet of the plasma membrane, where it associates with sphingolipids to form detergent-resistant microdomains (45, 46); thus, the loss of ergosterol and hydroxylated sphingolipids associated with iron deficiency would be expected to alter not only the fluidity and permeability of the plasma membrane, but the function of integral membrane proteins that localize to these microdomains. Defects in ergosterol biosynthesis inhibit both receptor-mediated and fluid-phase endocytosis as well as the trafficking of integral membrane proteins, such as transporters, that are sensitive to their lipid environment (35). For example, iron-deficient Candida sp. exhibit an enhanced susceptibility to antifungal agents that has been traced to a reduction in plasma membrane ergosterol and increased membrane fluidity (47). In S. cerevisiae, mutations of ERG genes and loss of ergosterol result in defects in tryptophan uptake due to altered trafficking of the Tat2 transporter (48). Sphingolipids are also required for endocytosis and for the normal trafficking, activity and stability of the plasma membrane H+-ATPase, Pma1, and the general amino acid permease, Gap1 (46). DHS, PHS, and ceramides are also involved in intracellular signaling pathways.
The data presented here suggested that different iron utilization pools (heme, Fe-S cluster, and diiron) are similarly affected by iron deficiency and that yeast do not have a mechanism for preferentially directing iron to a single class of cofactor, to a single subcellular location, or to essential pathways. Transcriptional repression, as observed in the down-regulation of the Fe-S enzyme Glt1 or the heme protein Cyc1, or mRNA degradation via the activities of Cth1 and Cth2, appear to be the only mechanisms through which the cell can specifically divert iron away from a pathway. Conversely, cells may express an essential iron-requiring protein at very high levels, so that if iron deficiency leads to only a small fraction of the protein-acquiring metal cofactors, that small fraction is sufficient to produce the required levels of product. This may explain why cells express very high levels of Leu1, which also loses Fe-S clusters and activity in iron deficiency, yet is sufficient to maintain leucine production. As Leu1 and Aco1 are some of the most abundant enzymes in the cell (roughly 100,000 molecules/cell) (49), these enzymes also represent a large pool of iron that can apparently be depleted without deleterious effects.
Although we found no evidence that cells have the capacity to preferentially direct iron to a specific class of cofactor or enzyme, we did find evidence that Ole1 was resistant to depletion of its iron cofactors during iron deficiency. This resistance to iron deficiency may be an intrinsic property of Ole1 that is shared by some other essential enzymes. Alternatively, other proteins that enhance delivery of iron cofactors or enhance the stability of the metallated enzyme may contribute to this effect. Dap1 is a heme-binding protein that binds to and enhances the activity of the cytochrome P450 family of enzymes involved in ergosterol biosynthesis (36, 37). Although not essential for ergosterol synthesis in iron-replete medium, Dap1 is required for growth in iron-deficient cells (50). A third possibility is that some heme/diiron enzymes are expressed in a subcellular location with enhanced access to iron cofactors, such as a subdomain of the endoplasmic reticulum. However, we found no evidence that the subcellular location of Fe-S proteins affected the delivery of Fe-S clusters in iron deficiency. Iron deficiency could be complicated by the insertion of non-cognate metals into the binding sites of diiron enzymes and may represent a mechanism of metal toxicity in the setting of iron deficiency. In Escherichia coli, copper toxicity was found to cause leucine deficiency due to copper-mediated disruption of iron-sulfur clusters (51). Further studies will be required to determine whether elevated levels of other metals affect the activity of iron-dependent enzymes under conditions of iron deficiency.
Supplementary Material
Acknowledgments
We thank George Poy and WeiPing Chen of the NIDDK Microarray Core Facility.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of NIDDK (to M. S.-E., O. P., W. A. P., and C. C. P.), by NIH Grant NIDDK P30 DK072437 (to J. C.), and by NIH Grant GM62104 (to M. B.). This work was also supported by the Center of Excellence in Molecular Hematology and by United States Public Health Service Grant NS47717 awarded by NINDS, NIH (to T. M. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S2 and S3.
- GFP
- green fluorescent protein
- YFP
- yellow fluorescent protein
- TAP
- tandem affinity purification
- ER
- endoplasmic reticulum
- LP
- lipid particle
- DHS
- dihydrosphingosine
- PHS
- phytosphingosine
- SD
- synthetic defined
- GC
- gas chromatography
- LC
- liquid chromatography
- MS
- mass spectrometry
- HPLC
- high-performance liquid chromatography
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Zimmermann M. B., Hurrell R. F. (2007) Lancet 370, 511–520 [DOI] [PubMed] [Google Scholar]
- 2.Beard J. L. (2008) J. Nutr. 138, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alla V., Bonkovsky H. L. (2005) Semin. Liver Dis. 25, 461–472 [DOI] [PubMed] [Google Scholar]
- 4.De Domenico I., McVey Ward D., Kaplan J. (2008) Nat. Rev. Mol. Cell Biol. 9, 72–81 [DOI] [PubMed] [Google Scholar]
- 5.Lill R., Mühlenhoff U. (2008) Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 6.Ozer A., Bruick R. K. (2007) Nat. Chem. Biol. 3, 144–153 [DOI] [PubMed] [Google Scholar]
- 7.Paoli M., Marles-Wright J., Smith A. (2002) DNA Cell Biol. 21, 271–280 [DOI] [PubMed] [Google Scholar]
- 8.Puig S., Askeland E., Thiele D. J. (2005) Cell 120, 99–110 [DOI] [PubMed] [Google Scholar]
- 9.Shakoury-Elizeh M., Tiedeman J., Rashford J., Ferea T., Demeter J., Garcia E., Rolfes R., Brown P. O., Botstein D., Philpott C. C. (2004) Mol. Biol. Cell 15, 1233–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philpott C. C., Protchenko O. (2008) Eukaryot. Cell 7, 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldron K. J., Rutherford J. C., Ford D., Robinson N. J. (2009) Nature 460, 823–830 [DOI] [PubMed] [Google Scholar]
- 12.Li L., Bagley D., Ward D. M., Kaplan J. (2008) Mol. Cell. Biol. 28, 1326–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Protchenko O., Philpott C. C. (2003) J. Biol. Chem. 278, 36582–36587 [DOI] [PubMed] [Google Scholar]
- 14.Puig S., Vergara S. V., Thiele D. J. (2008) Cell Metab. 7, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Protchenko O., Shakoury-Elizeh M., Keane P., Storey J., Androphy R., Philpott C. C. (2008) Eukaryot. Cell 7, 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawton K. A., Berger A., Mitchell M., Milgram K. E., Evans A. M., Guo L., Hanson R. W., Kalhan S. C., Ryals J. A., Milburn M. V. (2008) Pharmacogenomics 9, 383–397 [DOI] [PubMed] [Google Scholar]
- 17.Sreekumar A., Poisson L. M., Rajendiran T. M., Khan A. P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R. J., Li Y., Nyati M. K., Ahsan A., Kalyana-Sundaram S., Han B., Cao X., Byun J., Omenn G. S., Ghosh D., Pennathur S., Alexander D. C., Berger A., Shuster J. R., Wei J. T., Varambally S., Beecher C., Chinnaiyan A. M. (2009) Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 18.Katajamaa M., Oresic M. (2005) BMC Bioinformatics 6, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parrou J. L., François J. (1997) Anal. Biochem. 248, 186–188 [DOI] [PubMed] [Google Scholar]
- 20.Kennedy M. A., Bard M. (2001) Biochim. Biophys. Acta 1517, 177–189 [DOI] [PubMed] [Google Scholar]
- 21.Hanover J. A., Forsythe M. E., Hennessey P. T., Brodigan T. M., Love D. C., Ashwell G., Krause M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11266–11271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertello L. E., Gonçalvez M. F., Colli W., de Lederkremer R. M. (1995) Biochem. J. 310, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung C. Y., Ko Y. G., Thompson G. A., Jr. (1995) Biochem. J. 307, 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lester R. L., Dickson R. C. (2001) Anal. Biochem. 298, 283–292 [DOI] [PubMed] [Google Scholar]
- 25.Stehling O., Smith P. M., Biederbick A., Balk J., Lill R., Mühlenhoff U. (2007) in Mitochondria: Practical Protocols (Leister D., Herrmann J. M. eds) pp. 325–342, Springer, New York [Google Scholar]
- 26.Dancis A., Klausner R. D., Hinnebusch A. G., Barriocanal J. G. (1990) Mol. Cell. Biol. 10, 2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng Y., Guo Y., Watson H., Au W. C., Shakoury-Elizeh M., Basrai M. A., Bonifacino J. S., Philpott C. C. (2009) J. Biol. Chem. 284, 23830–23841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagunas R. (1986) Yeast 2, 221–228 [DOI] [PubMed] [Google Scholar]
- 29.Lesuisse E., Santos R., Matzanke B. F., Knight S. A., Camadro J. M., Dancis A. (2003) Hum. Mol. Genet. 12, 879–889 [DOI] [PubMed] [Google Scholar]
- 30.Magasanik B. (1992) in The Molecular and Cellular Biology of the Yeast Saccharomyces (Jones E. W., Pringle J. R., Broach J. R. eds) pp. 283–317, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31.Magasanik B., Kaiser C. A. (2002) Gene 290, 1–18 [DOI] [PubMed] [Google Scholar]
- 32.Kohlhaw G. B. (2003) Microbiol. Mol. Biol. Rev. 67, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daum G., Lees N. D., Bard M., Dickson R. (1998) Yeast 14, 1471–1510 [DOI] [PubMed] [Google Scholar]
- 34.Espenshade P. J., Hughes A. L. (2007) Annu. Rev. Genet. 41, 401–427 [DOI] [PubMed] [Google Scholar]
- 35.Lees N. D., Bard M. (2003) in Lipid Metabolism and Membrane Biogenesis (Daum G. ed) pp. 213–240, Springer-Verlag, Heidelberg [Google Scholar]
- 36.Hughes A. L., Powell D. W., Bard M., Eckstein J., Barbuch R., Link A. J., Espenshade P. J. (2007) Cell Metab. 5, 143–149 [DOI] [PubMed] [Google Scholar]
- 37.Mallory J. C., Crudden G., Johnson B. L., Mo C., Pierson C. A., Bard M., Craven R. J. (2005) Mol. Cell. Biol. 25, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dancis A., Roman D. G., Anderson G. J., Hinnebusch A. G., Klausner R. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3869–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M'Baya B., Fegueur M., Servouse M., Karst F. (1989) Lipids 24, 1020–1023 [DOI] [PubMed] [Google Scholar]
- 40.Polakowski T., Stahl U., Lang C. (1998) Appl. Microbiol. Biotechnol. 49, 66–71 [DOI] [PubMed] [Google Scholar]
- 41.Müllner H., Zweytick D., Leber R., Turnowsky F., Daum G. (2004) Biochim. Biophys. Acta 1663, 9–13 [DOI] [PubMed] [Google Scholar]
- 42.Leber R., Landl K., Zinser E., Ahorn H., Spök A., Kohlwein S. D., Turnowsky F., Daum G. (1998) Mol. Biol. Cell 9, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haak D., Gable K., Beeler T., Dunn T. (1997) J. Biol. Chem. 272, 29704–29710 [DOI] [PubMed] [Google Scholar]
- 44.Dunn T. M., Haak D., Monaghan E., Beeler T. J. (1998) Yeast 14, 311–321 [DOI] [PubMed] [Google Scholar]
- 45.Bagnat M., Simons K. (2002) Biol. Chem. 383, 1475–1480 [DOI] [PubMed] [Google Scholar]
- 46.Dickson R. C. (2008) J. Lipid Res. 49, 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prasad T., Chandra A., Mukhopadhyay C. K., Prasad R. (2006) Antimicrob. Agents Chemother. 50, 3597–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Umebayashi K., Nakano A. (2003) J. Cell Biol. 161, 1117–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 50.Craven R. J., Mallory J. C., Hand R. A. (2007) J. Biol. Chem. 282, 36543–36551 [DOI] [PubMed] [Google Scholar]
- 51.Macomber L., Imlay J. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.