Abstract
Protease-activated receptor-2 (PAR-2) mediates pro-inflammatory signals in a number of organs, including enhancing leukocyte recruitment to sites of injury and infection. At the cellular level, PAR-2 promotes activation of the actin filament-severing protein cofilin, which is crucial for the reorganization of the actin cytoskeleton and chemotaxis. These responses require the scaffolding functions of β-arrestins; however, the mechanism by which β-arrestins spatially regulate cofilin activity and the role of this pathway in primary cells has not been investigated. Here, using size-exclusion chromatography and co-immunoprecipitation, we demonstrate that PAR-2 promotes the formation of a complex containing β-arrestins, cofilin, and chronophin (CIN) in primary leukocytes and cultured cells. Both association of cofilin with CIN and cell migration are inhibited in leukocytes from β-arrestin-2−/− mice. We show that, in response to PAR-2 activation, β-arrestins scaffold cofilin with its upstream activator CIN, to facilitate the localized generation of free actin barbed ends, leading to membrane protrusion. These studies suggest that a major role of β-arrestins in chemotaxis is to spatially regulate cofilin activity to facilitate the formation of a leading edge, and that this pathway may be important for PAR-2-stimulated immune cell migration.
Keywords: Cell/Migration, Cytoskeleton/Actin, G Protein-coupled Receptor (GPCR), Actin, Leukocyte, Protease-activated Receptors, Arrestins, Cofilin
Introduction
Protease-activated-receptor-2 (PAR-2)2 is a G-protein-coupled receptor that signals, through β-arrestin-promoted scaffolds, to promote reorganization of the actin cytoskeleton and chemotaxis (1, 2). In vivo, PAR-2 plays an important role in the recruitment of leukocytes to the sites of inflammation, because this is impaired in PAR-2−/− mice and enhanced by administration of PAR-2 agonists (3–8). However, no studies have yet linked β-arrestin-dependent scaffolding of actin assembly proteins to PAR-2-stimulated chemotaxis under physiological conditions.
β-Arrestins are multifunctional proteins that mediate receptor desensitization and internalization and serve as signaling scaffolds. A role for β-arrestin scaffolds in signaling by PAR-2 and other receptors was first identified for the spatial regulation of ERK1/2 activity (9–11). They are now known to scaffold numerous other signaling molecules (12–15), many of which are involved in actin reorganization and chemotaxis (1, 13, 16–19). An attractive hypothesis is that β-arrestins exert spatial control over actin assembly events at the leading edge to promote membrane protrusion and cell migration. A recent advance in this field was the discovery that β-arrestins are required for PAR-2-dependent activation of the actin filament-severing protein, cofilin (14), which binds to the sides of actin filaments, destabilizing them and promoting their severing. Filament severing has two functions: the reorganization of existing filaments and the creation of free actin barbed ends for monomer addition (20). Actin is a polar molecule containing a barbed and pointed end; addition of actin monomers to a growing filament occurs at the barbed end. Although actin monomers spontaneously assemble into filaments very slowly, the generation of multiple small filaments with free barbed ends increases the rate of actin assembly dramatically. Thus, the presence of active cofilin within the leading edge of a migrating cell controls the availability of polymerization competent free actin barbed ends, which in turn is required for membrane protrusion and cell migration (21–24). Spatial control over cofilin activity is essential as either too much or too little actin filament severing activity will inhibit efficient cell migration.
Cofilin activation is controlled by opposing actions of LIMKs (which inactivate it by phosphorylation on Ser3) and cofilin-specific phosphatases (chronophin (CIN) and slingshot) that activate it (25, 26) and by intracellular pH and PIP2 levels (27). We previously showed that PAR-2 promotes rapid cofilin dephosphorylation that is decreased in the absence of β-arrestins or by expression of a dominant negative CIN mutant (14). We hypothesize that PAR-2 activation results in recruitment of CIN and cofilin to β-arrestins into a scaffolding complex to promote localized generation of free actin barbed ends and membrane protrusion. We predict that this complex might be important in physiological scenarios such as leukocyte migration. Here we demonstrate that: 1) β-arrestins cooperate to scaffold CIN with cofilin at the leading edge; 2) this scaffolding complex exists in primary leukocytes and may play a role in PAR-2-stimulated leukocyte chemotaxis; and 3) β-arrestins and CIN are required for PAR-2-stimulated actin barbed end generation and subsequent membrane protrusion.
MATERIALS AND METHODS
Materials and Cell Lines
All chemicals were from Sigma or Fisher Scientific unless stated otherwise. Activating peptide 2fAP (2-furoyl-LIGRL-ornithine-NH2) was synthesized by Genemed Inc. Dominant negative CIN (DN-CIN), His6-myc-tagged CIN and GFP-cofilin plasmids were from Dr. Gary Bokoch (The Scripps Research Institute (TSRI), La Jolla, CA) and have been described previously (14, 26), and FLAG-tagged β-arrestin-1 and -2 plasmids were from Dr. Robert Lefkowitz (Duke University Medical Center, Durham, NC). Mouse embryonic fibroblasts from wild-type mice (MEFwt), β-arrestin-1/2−/− mice (MEFβarrDKO), and MEFβarrDKO cells stably transfected with physiological levels of either β-arrestin-1 or β-arrestin-2 were from Dr. Robert Lefkowitz and have been described previously (14, 28, 29). MDA-MB-468 cells were from ATCC. All cell lines were grown in Dulbecco's modified Eagle's medium and 10% fetal calf serum. Transient transfections were performed on 70–80% confluent cells using Lipofectamine (Invitrogen), and experiments were performed between 24 and 48 h after transfection.
Antibodies
The following antibodies were: rabbit anti-phospho-cofilin (Cell Signaling, 1:1000), mouse anti-total cofilin (BD Biosciences, 1:1000 WB, 5 μg/ml IP), rabbit anti-total cofilin (Chemicon, 1:1000); rabbit anti-mycA14 and anti-myc9E10 to detect transfected CIN (Santa Cruz Biotechnology, 1:1000 WB, 4 μg/ml IP, 1:200 IF), M2anti-FLAG (Sigma, 1:1000 WB, 1:500 IF), rabbit anti-β-arrestin-1 + 2 (A1CT, from Dr. Robert J. Lefkowitz, 1:500, 2 μg/ml IP), goat anti-β-arrestin-2 (Santa Cruz Biotechnology, 1:500), rabbit polyclonal antibody to CIN (from Dr. Gary Bokoch, 1:200). Alexa-dye conjugated secondary antibodies (1:45,000 WB, 1:100 IF), phalloidin (1:100 IF), and streptavidin (1:1000 IF) were from Invitrogen. IR-dye-conjugated secondary antibodies (1:45,000 WB) were from Rockland Biosciences.
Animals
All animal procedures were in accordance with the guidelines on the use and care of laboratory animals set by the National Institutes of Health and approved by the IACUC at University of California Riverside. β-Arrestin1−/− and β-arrestin2−/− in a C57BL/6 background were provided by Dr. Robert Lefkowitz (18). PAR-2−/− mice were from Dr. Robin Plevin (University of Strathclyde, Glasgow, Scotland), and were developed by KOWA Pharmaceuticals (Tokyo, Japan) (8). Wild-type C57BL/6 mice were from Jackson Laboratories. Age- and gender-matched mice (12–16 weeks old) were used for this study.
Bone Marrow Preparation and Quantification
To isolate leukocytes, bone marrow was flushed from femurs of mice (wt, PAR-2−/−, β-arrestin-1−/−, or β-arrestin-2−/−) with 5 ml of RPMI 1640 medium, after which cells were pelleted and resuspended in sterile PBS. Bone marrow cell suspensions were treated with ammonium chloride (0.83% ammonium chloride, 2 min, at 4 °C) to remove contaminating red blood cells. Approximately 105 cells were centrifuged onto a slide using a Shandon Cytospin 3 at 500 rpm for 5 min, and air dried slides were stained using the Hema 3 System; relative quantities of different white blood cell types were determined based on standard morphological criteria.
Migration and Pseudopodia Assays
104 cells were seeded into the upper compartment of Transwell filters with pore sizes of 3 μm (for pseudopodia assays) and 5 μm (for leukocyte migration assays). The 3-μm pore size is large enough for pseudopodia, but too small for the cell bodies to migrate through. Agonist (100 nm 2fAP) was added to the lower chamber for 90 min (protrusion) or 3 h (migration), after which cells were stained with crystal violet. Cell bodies or non-migratory cells removed from the upper side, and cells or protrusions present on the filter underside were imaged and counted under 20× magnification in 8 fields of view with a Nikon phase contrast microscope linked to a 3CCD camera (DAGE-MTI, Michigan City, IN) using PAX-it 5.0 (LECO, St. Joseph, MI). For isolation of pseudopodial proteins, the procedure was the same as for the pseudopodia assay described above except that either cell bodies from the upper side or pseudopodia from the underside were scraped into lysis buffer and analyzed by SDS-PAGE as described in the section on protein analysis.
Video Microscopy
For video microscopy, cells were grown on collagen, and time-lapse imaging was monitored on a Nikon Eclipse TE2000-U microscope (20× objective, respectively) equipped with a 12-bit charge-coupled device camera (model ORCA-AG, Hamamatsu) using Image-Pro software (Media Cybernetics) and 37 °C heated stage. Images were captured at 1-min intervals for 25 min. Cells were monitored for 15 min before adding agonist (1 μm 2fAP in 0.5% agarose with phenol red to monitor diffusion of the agonist) to the upper right corner of the dish, and for an additional 10 min afterward. The experiment was repeated four times for each cell line, and a minimum of 10 cells per experiment was imaged. No migration was observed in response to phenol red alone.
Actin Barbed End Labeling
Cells, seeded onto collagen-coated coverslips, were placed in serum-free medium for 3 h and stimulated with or without 100 nm 2fAP for 1 min. Media was immediately exchanged for actin monomer buffer (20 mm Hepes, pH 7.5, 138 mm KCl, 4 mm MgCl2, 3 mm EGTA, 0.2 mg/ml saponin, 1 mm ATP, 1% bovine serum albumin, 0.3 μm labeled G-actin), and cells were incubated for 1 min to allow incorporation of labeled monomers into cells. Cells were washed in 0.1 m glycine/PBS for 10 min, then fixed, stained, and imaged as described in microscopy section.
Fluorescence Microscopy
Cells were fixed in normal buffered formalin and prepared as described previously (14). Cells were stained with anti-Myc-9E10 followed by Alexa546-conjugated anti-rabbit IgG, Alexa635- or Alexa488-conjugated phalloidin, or Alexa546-conjugated streptavidin (for biotin actin). Cells were imaged using a Zeiss 510 confocal microscope with a 100× oil objective (barbed end labeling) or Leica SP2 confocal microscope with a 63× water objective (cofilin/CIN staining) confocal microscope. High magnification images were taken at 6× zoom (barbed end) and 3× zoom (CIN/cofilin). All image sets were taken at the same laser intensity and gain across cell types and treatments. Settings for Zeiss LSM510 were: 35%/1050 amplifier gain for 488 nm, and 72%/1250 (maximal) amplifier gain for 543 nm. For cells stained with Alexa595 phalloidin, the laser intensity was set at 35% and amplifier gain was 850. Pinhole was always 1 airy unit in each channel, except for imaging of rhodamine actin where the pinhole was 2.5 airy units in the red channel. For images collected in z-sections 2-μm slices were taken. Settings for Leica SP2 were: 30%/540 amplifier gain for 488 nm, 55%/780 amplifier gain for 561 nm, and 80%/900 amplifier gain for 633 nm. Single section images were exported from LSM Image Browser v. 3.5 (Carl Zeiss GmbH Jena) or LCS Lite (Leica Microsystems) as tiff files, and images were assembled in Adobe Photoshop version 5. For quantification, tiff images in each channel were imported into MCID Elite and analyzed using channel linking, so that fluorescence measurements over a defined region of uniform width could be calculated for each label simultaneously (actin and phalloidin in Fig. 5 and CIN, cofilin, and phalloidin in Figs. 6 and 7). Quantification regions were divided into 0.1-μm increments, beginning with a position 0.2 μm outside the cell to 12 μm within the cell, with 0 μm representing the cell edge (defined by phalloidin staining). At least 6 untreated cells and 12 2fAP-treated cells were imaged, and the experiments were repeated 3 times. Average fluorescence intensity as a function of distance was calculated for each experimental group to generate the traces shown in Figs. 5–7.
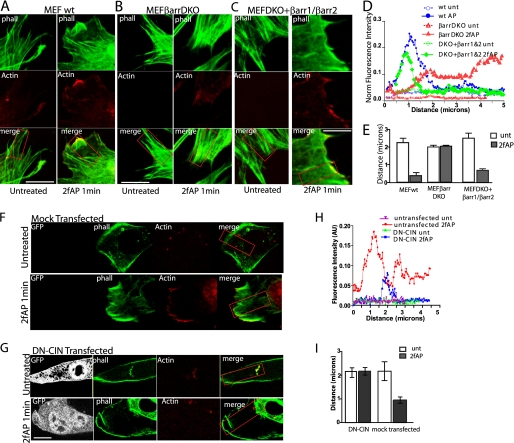
FIGURE 5.
Formation of free actin barbed ends at the cell edge requires β-arrestins. A–C, images (100× magnification) of MEFwt (A), MEFβarrDKO (B), and DKO+βarr-1&2 (C), treated with or without 2fAP for 1 min, in the presence of 300 nm labeled actin monomers (red), fixed and cross-stained with phalloidin (phall, green). Scale bars = 10 μm. D, representative traces of actin monomer incorporation (fluorescence intensity in arbitrary units (AU)), measured along the boxed regions, graphed as a function of distance from the cell edge are shown for MEFwt, MEFβarrDKO, and DKO+βarr1&2. E, mean distance (from the cell edge) at which peak rhodamine actin fluorescence was observed in MEFwt, MEFβarrDKO, and DKO+βarr1&2. F and G, rhodamine actin incorporation in MEFwt, either mock transfected (F) or transfected with DN-CIN (G), was performed as described above. H, representative trace of actin monomer incorporation versus distance from the leading edge in mock transfected and DN-CIN-transfected cells. I, mean distance from cell edge at which peak fluorescence was observed in mock transfected and DN-CIN-transfected cells. (For each group, n = 12 cells from 3 separate experiments.) Error bars indicate mean ± S.E.
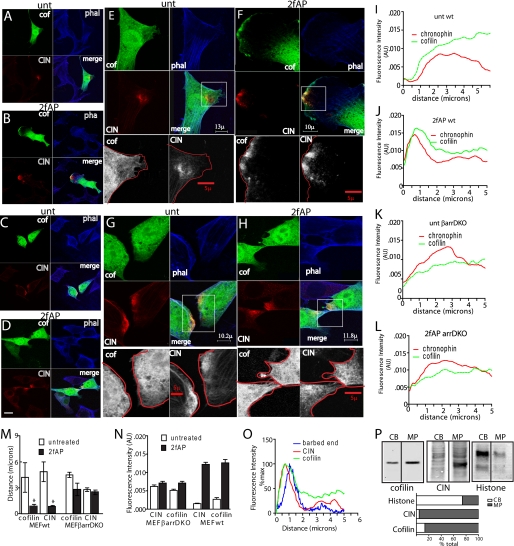
FIGURE 6.
PAR-2-induced co-localization of cofilin and CIN in lamellipodia requires β-arrestins. MEFwt (A and B) and MEFβarrDKO (C and D) were transiently transfected with CIN and cofilin, either untreated (A and C) or treated with 2fAP for 1 min (B and D), fixed, stained, and imaged by confocal microscopy (40× magnification). Panels depict localization of cofilin (green), CIN (red), phalloidin (phal, blue), and a merge of all three (scale bar = 30 μm). E–H, high magnification (63×) of membrane protrusions from cells in A–D. Lower panels in grayscale represent 5× enlarged images of cofilin and CIN in the indicated boxed regions of the protrusions, overlaid with tracings of the cell edge. I–L, representative traces of fluorescence intensity calculated for CIN and cofilin, graphed on the y axis in arbitrary units (AU), as a function of distance in microns from the cell edge. M, redistribution of CIN and cofilin upon PAR-2 activation, as demonstrated by the decrease in mean distance from the cell edge at which peak fluorescence intensity for each was observed. N, mean fluorescence intensity was determined for each protein at the leading edge (defined as 0–2 μm from the cell edge). O, overlay of PAR-2-stimulated CIN, cofilin, and rhodamine actin fluorescence (from Fig. 5A) in PAR-2-activated MEFwt demonstrates overlap between actin barbed end formation, CIN, and cofilin localization. P, membrane protrusions (MP) were physically separated from cell bodies (CB), lysed, and analyzed by Western blotting with cofilin and CIN antibodies. Histone was included to demonstrate valid separation of the two fractions. A bar graph of the relative distribution of each protein is shown under the Western blots. Error bars indicate mean ± S.E. *, a statistically significant decrease in distance between untreated and 2fAP treated cells, p = .01.
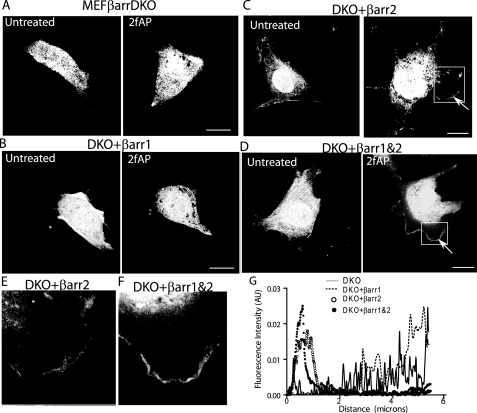
FIGURE 7.
Transfection of β-arrestins rescues cofilin localization to membrane protrusions. MEFβarrDKO (A), DKO+βarr1 (B), DKO+βarr2 (C), or DKO+βarr1&2 (D) were transfected with GFP-cofilin, treated with or without 2fAP for 1 min, fixed, stained, and imaged by confocal microscopy. Arrows indicate cofilin in protrusions. Enlarged images of cofilin in membrane protrusions of DKO+βarr2 (E) and DKO+βarr1&2 (F) are shown. G, representative traces of fluorescence intensity calculated for cofilin in 2fAP-treated cells, graphed on the y axis in arbitrary units (AU), as a function of distance in microns from the cell edge.
Protein Analysis
Cleared lysates were prepared as follows: Cells (either cultured cells or bone marrow leukocytes) were treated with or without 100 nm to 1 μm 2fAP for indicated times and lysed in 0.5 ml of lysis buffer, as described previously (10). 10 μg of protein was analyzed by 10 or 15% SDS-PAGE, transferred to polyvinylidene difluoride and probed with antibodies at concentrations described above, followed by Alexa600- and IR800-conjugated secondary antibodies. For co-immunoprecipitations, cleared lysates were immunoprecipitated with antibody to Myc or to cofilin for 4 h; beads were washed and analyzed by 15% SDS-PAGE followed by Western blotting. Blots were imaged and band intensity determined using a LICOR Odyssey Infrared Imaging System (Li-COR Biosciences).
Gel Filtration
Pooled lysates from 4 confluent 10-cm dishes of cultured cells or from 4 mouse bone marrow preps were loaded onto a column (3 × 150 cm, 400-ml bed volume) of Sephacryl S-300, pre-equilibrated in 1× PBS plus 1% Triton X-100 as previously described (2). Briefly, proteins were eluted in PBS; 7-ml fractions were collected. Protein was precipitated with methanol/chloroform from every other fraction within the included volume and analyzed by SDS-PAGE, followed by Western analysis with antibodies to β-arrestin-1 and -2, total cofilin, and CIN. The band intensity for each protein in a given elution was divided by the sum of the band densities across the included volume and graphed as a function of the Stokes radius. To determine Stokes radii, the column was calibrated with Dextran blue (void volume, Vo), 3 m KCl (total volume, Vt), thyroglobulin (8.5 nm), catalase (5.2 nm), bovine serum albumin (3.5 nm), and myoglobin (1.8 nm). Absorbance of each fraction was measured with Bradford reagent (Bio-Rad). The partition co-efficient (Kd) for each standard was determined as described (Kd = Ve − Vo/Vt − Vo), where Ve is the elution volume of the protein. Kd was then graphed as a function of known Stokes radii for standards, and the Stokes radius of the cofilin-CIN-β-arrestin complex was determined from the standard graph. Predicted Stokes radii for cofilin and β-arrestin were reported in the literature (30, 31).
Data and Statistical Analysis
All graphs and statistical analyses were performed using KaleidaGraph Version 4.0, Microsoft Excel 2003, or GraphPad Prism 5.0. All experiments were performed a minimum of three times. Statistical significance was determined using one-way analysis of variance and Tukey t-tests (to compare between treatment groups).
RESULTS
PAR-2-stimulated Cell Migration Is Decreased in Primary Leukocytes from β-Arrestin Knock-out Mice
We have shown previously that PAR-2-stimulated chemotaxis and cofilin dephosphorylation in multiple cultured cell lines are abolished by small interference RNA depletion of β-arrestins, expression of a dominant negative β-arrestin mutant, or genetic deletion of β-arrestins (1, 2, 14). A major goal of this work is to understand the physiological significance of β-arrestin scaffolding, and, given the wealth of studies demonstrating a role for PAR-2 in mediating leukocyte infiltration during inflammation, we chose primary leukocytes for these initial experiments. We isolated leukocytes from bone marrow of wt, β-arrestin-1−/−, and β-arrestin-2−/− mice and first demonstrated that the PAR-2/β-arrestin/cofilin pathway is relevant in these cells by examining cofilin phosphorylation levels and PAR-2-stimulated cell migration. Bone marrow preparations consisted of 55–60% neutrophils, 24–28% lymphocytes, 14–16% monocytes, and <1% eosinophils; no significant differences were observed in the distribution of these cell types between wt and β-arrestin-1 or -2 knock-out mice (Fig. 1A and supplemental Fig. S1). The amount of active cofilin in leukocytes from wt, PAR-2−/−, β-arrestin-1−/−, and β-arrestin-2−/− mice was determined by Western blotting with antibodies to phosphorylated and total cofilin. Baseline ratios of phosphorylated-cofilin (inactive) to total cofilin were increased in leukocytes from all three knock-out mice, compared with wild-type controls (Table 1 and supplemental Fig. S2). Because baseline phospho-cofilin levels were lower in wild-type than in PAR-2 or β-arrestin knock-out leukocytes, there may be some constitutive activation of PAR-2/β-arrestin/cofilin signaling pathway in vivo.
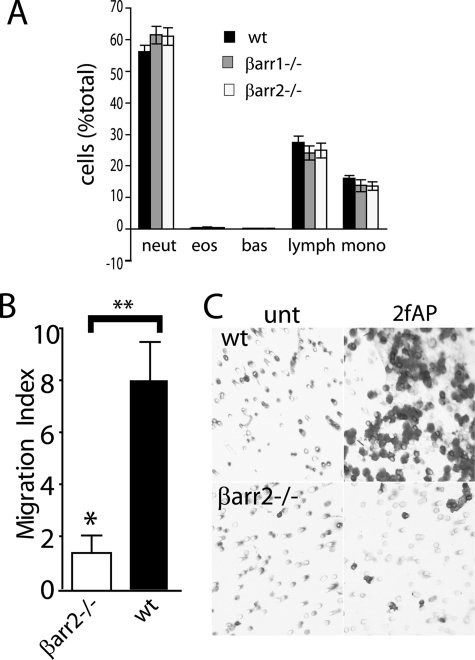
FIGURE 1.
β-Arrestins are required for PAR-2-stimulated primary leukocyte migration. A, percentages of lymphocyte, neutrophil, eosinophil, monocyte precursors, and mature cells were determined from Cytospin preparations of bone marrow leukocytes from wild-type (wt), β-arrestin-1−/−, and β-arrestin-2−/− mice. B, graph showing -fold increase (over PBS-treated controls) in migration of leukocytes from wt and β-arrestin2−/− mice treated with 2fAP (mean ± S.E., n = 3). A statistically significant increase in response to 2fAP versus PBS is shown. C, image of migrated cells on underside of filter.
TABLE 1.
P-cofilin levels in primary leukocytes
Bone marrow leukocytes were lysed, and total lysates were analyzed by SDS-PAGE followed by Western blotting with anti-phospho and anti-total cofilin. Integrated intensities were determined, and the average ratio of phospho-cofilin to total cofilin levels was calculated.
| Mouse strain | Phospho-cofilin/Total cofilin |
|---|---|
| wt mice | 0.369 ± 0.068, n = 10 |
| β-Arrestin1−/− | 0.69 ± 0.07, n = 4a |
| β-Arrestin2−/− | 0.78 ± 0.16, n = 3a |
| PAR-2−/− | 1.3 ± 0.1, n = 6b |
a Tukey t-test results: statistically significant increase compared to wt, p < 0.02.
b Tukey t-test results: statistically significant increase compared to wt, p < 0.0001.
We then addressed whether PAR-2 promotes β-arrestin-dependent cell migration in leukocytes, as it does in other cell lines (1, 2), by seeding them onto Transwell filters and treating with or without the specific PAR-2 peptide agonist, 2fAP (14, 32), for 3 h. Non-migratory cells were removed from the top of the membrane, and cells that had migrated to the filter underside were stained and counted. PAR-2 promoted an 8-fold increase in migration in wild-type leukocytes, and this was reduced to 1.6-fold in the absence of β-arrestin-2 (Fig. 1, B and C). This effect was not restricted to bone marrow leukocytes, because PAR-2 activation also promoted an 8.5-fold increase in cell migration in wild-type neutrophils isolated from the peritoneal cavity 4 h after injection of Thioglycollate, and this was reduced to 2.2- and 2-fold in neutrophils from β-arrestin-1−/− and β-arrestin-2−/− mice, respectively (supplemental Fig. S3).
PAR-2 Promotes the Formation of a β-Arrestin/Cofilin/CIN Scaffold in Primary Leukocytes and Cultured Cells
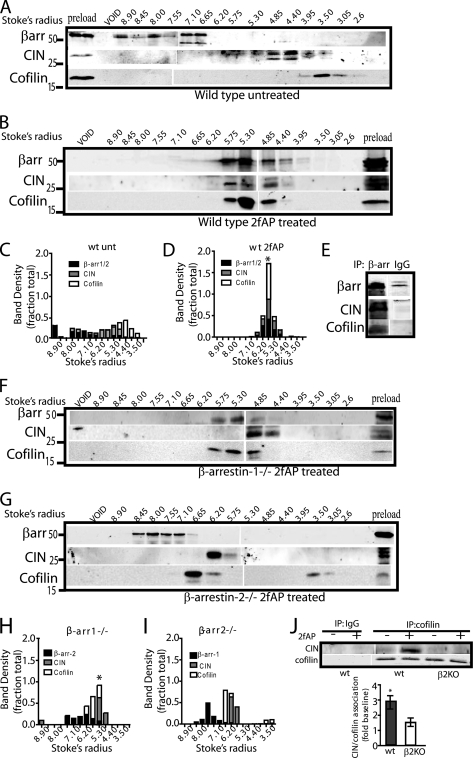
In our previous studies, PAR-2-stimulated cofilin dephosphorylation was abolished by expression of dominant negative CIN, leading us to hypothesize that β-arrestins scaffold CIN with cofilin to promote its dephosphorylation and activation, which is ultimately necessary for migration. To investigate whether β-arrestin forms an oligomeric complex with CIN and cofilin, we performed size-exclusion chromatography (SEC) coupled with co-immunoprecipitations. In previous studies we have used this method to isolate β-arrestin signaling complexes containing components of MAPK and phosphatidylinositol 3-kinase cascades from PAR-2-activated cells (10, 12). Briefly, freshly isolated bone marrow leukocytes were treated with or without 2fAP for 5 min and lysates fractionated by SEC on Sephacryl S300 to identify higher molecular weight fractions containing the putative protein complex; association of the co-eluting proteins was then confirmed by co-immunoprecipitation from these same fractions. In response to PAR-2 activation in leukocytes from wild-type mice, endogenous β-arrestin-1 and -2, cofilin, and CIN co-eluted in fractions corresponding to a Stokes radius of ∼4.85–5.3 nm (Fig. 2, A and B, and supplemental Fig. S4), which is distinctly larger than those reported for either cofilin or β-arrestins alone (supplemental Fig. S4) (30, 31). To better illustrate co-elution, for each protein, the fraction of total protein present in each elution was calculated and graphed as a function of the Stokes radius (Fig. 2, C and D). To prove that co-elution of these proteins reflected their presence in a complex, the fractions corresponding to a 4.5–6 nm Stokes radius were immunoprecipitated with either with anti-β-arrestin-1/2 or IgG, and immune complexes were analyzed by SDS-PAGE followed by Western blotting with anti-CIN, anti-cofilin, and anti-β-arrestin-1/2 (Fig. 2E). The gel-filtration experiment was then repeated in leukocytes from β-arrestin-1−/− and β-arrestin-2−/− mice (Fig. 2, F–I). PAR-2-stimulated co-elution of cofilin with CIN was only severely impaired in β-arrestin-2−/− leukocytes and some co-elution of CIN and cofilin with β-arrestin-2 was still observed in β-arrestin-1−/− leukocytes. These data suggest a major role for β-arrestin-2 in mediating cofilin/CIN association. Consistent with this hypothesis, PAR-2 promoted co-immunoprecipitation of CIN with cofilin in total leukocyte lysates from wild-type mice, and this was reduced by ∼60% in β-arrestin-2−/− leukocytes (Fig. 2J). A similar reduction in PAR-2-induced association of cofilin and CIN was observed in MEFs from β-arrestin knock-out mice compared with wild-type MEFs (not shown). We conclude that β-arrestins are required for PAR-2-stimulated primary leukocyte chemotaxis, and they can recruit cofilin into a scaffolding complex with CIN in these cells.
FIGURE 2.
β-arrestins are required to scaffold CIN with cofilin in primary leukocytes. A and B, leukocytes from wild-type mice were treated with or without 2fAP and analyzed by SEC. Protein from every other 7-ml fraction within the included volume was analyzed by SDS-PAGE followed by Western blotting with antibodies to β-arrestins, CIN, and cofilin. For wt cells, both untreated (A) and 2fAP-treated (B) samples are shown. C and D, stacked bar graphs showing distribution of each protein over the column as a function of the Stokes radius (see supplemental Fig. S4) are shown for each treatment group. (The band intensity in each elution was divided by the sum of band intensities in all fractions.) E, fractions from 2fAP-treated wild-type mice (B), in which cofilin/CIN and β-arrestins co-eluted, were pooled and immunoprecipitated with anti-β-arrestin-1/2 or IgG (negative control), followed by Western analysis with anti-β-arrestin1/2, CIN, and cofilin. F and G, SEC was repeated on 2fAP-treated leukocytes from β-arrestin-1−/− (F) and β-arrestin-2−/− (G) knock-out mice. H and I, stacked bar graphs of protein distribution as describe for F and G are shown. J, total lysates from wt or β-arrestin-2−/− bone marrow leukocytes, treated with or without 2fAP, were immunoprecipitated with anti-cofilin or IgG, followed by Western analysis with anti-cofilin and anti-CIN. Integrated intensities of immunoprecipitated CIN and cofilin were calculated. CIN was normalized to the amount of cofilin present in each lane, and the -fold increase in CIN/cofilin co-immunoprecipitated after PAR-2 stimulation, compared with untreated controls, was determined. *, a statistically significant increase in PAR-2-stimulated co-immunoprecipitation, p = 0.025. Co-immunoprecipitation of cofilin with CIN was reduced by 60% in the absence of β-arrestin-2 (p = 0.04), n = 4. Error bars indicate mean ± S.E.
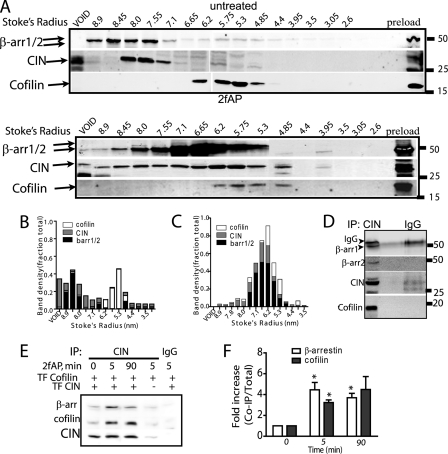
These findings were confirmed in MDA MB-468 cells, the breast cancer cell line in which PAR-2-stimulated, β-arrestin-dependent cofilin activation was originally demonstrated (14). MDA MB-468 cells were treated with or without 2fAP and fractionated by SEC as described for leukocytes. Once again β-arrestins co-eluted with CIN and cofilin in fractions corresponding to a Stokes radius of 5.3–6.2 after 2fAP treatment (Fig. 3A). That these proteins exist in an oligomeric complex was again confirmed by the fact that cofilin and β-arrestins could be co-immunoprecipitated with CIN from the fractions in which they co-eluted (Fig. 3D). The slight variation in size of eluted complexes between cell types may reflect differences in the presence of other proteins, in each complex, differences between human and mouse complexes, or cell-type specific differences in complex composition. Other studies on β-arrestin-src and β-arrestin-ERK complexes have demonstrated a similar disparity in complex size between species and cell type (10, 33). In untreated cells this complex was not detected. In fact, the majority of CIN eluted in early fractions and in the void volume, consistent with its aggregation into very large complexes; a similar phenomenon was observed with src in prior studies (33). Furthermore, β-arrestins and cofilin were co-immunoprecipitated with CIN from total cell lysates, and this co-precipitation was significantly increased upon PAR-2 activation (Fig. 3, E and F). We conclude that PAR-2 promotes the formation of a scaffolding complex containing β-arrestin-1 and -2, CIN, and cofilin in multiple motile cell types.
FIGURE 3.
Formation of a scaffolding complex containing β-arrestins, CIN, and cofilin in breast cancer cells. A, MDA MB-468 cells were transfected with myc-CIN and treated with or without 2fAP for 5 min. Cell lysates were fractionated by SEC followed by Western blotting with antibodies to β-arrestins, cofilin, and myc (to visualize CIN) as described in Fig. 2. B and C, graphs showing distribution of each protein over the column as a function of Stokes radius are shown for untreated (B) and 2fAP-treated (C) samples. D, co-immunoprecipitation of CIN (using anti-myc) with β-arrestins and cofilin was performed on pooled samples from 2fAP-treated cells in which proteins co-eluted. E, co-immunoprecipitation of GFP-cofilin and endogenous β-arrestins with myc-CIN in MDA MB-468 cells, treated with or without 2fAP for 0–90 min. Lanes 4 and 5 are negative controls: anti-myc and anti-IgG IP from cells transfected with GFP-cofilin alone or GFP-cofilin and Myc-CIN. F, graph depicting -fold increase in the amount of β-arrestin and cofilin associated with CIN upon PAR-2 activation. *, a statistically significant increase in PAR-2 stimulated co-immunoprecipitation, p = 0.01, n = 3. Error bars indicate mean ± S.E.
β-Arrestins and CIN Activity Are Required for the Formation of Membrane Protrusions Downstream of PAR-2
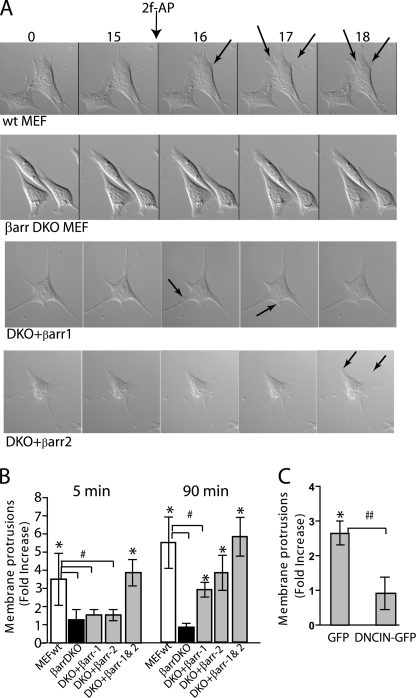
The studies described thus far suggest that β-arrestins are required to bring CIN in contact with its substrate so that it can dephosphorylate and activate cofilin. Because cofilin activity is required for the formation of a leading edge during the initial steps of chemotaxis, we predicted that protrusion formation required both β-arrestins and CIN. We first examined cell morphology in response to directed activation of PAR-2 in embryonic fibroblasts from wild-type (MEFwt) and β-arrestin1/2−/− mice (MEFβarrDKO), or from MEFβarrDKO stably expressing β-arrestin-1 (DKO+βarr1) or β-arrestin-2 (DKO+βarr2). Cells were monitored by time-lapse live video microscopy for 15 min prior to, and for 10 min after, localized PAR-2 activation (Fig. 4A). Within 1–2 min, MEFwt exhibited rapid formation of membrane protrusions toward the agonist, while MEFβarrDKO remained unchanged. DKO+βarr1 and DKO+βarr2 cells constitutively formed small, random protrusions, but no directional protrusions were formed in response to 2fAP (Fig. 4A). To quantify membrane protrusions in MEFwt compared with MEFβarrDKO, we used a previously described pseudopodia assay (1, 34), in which cells were seeded onto membranes with pores too small to allow cell body translocation; 2fAP was added to the bottom chamber, and cell protrusions attached to the underside of the filter were scored. PAR-2 activation resulted in a 3- to 5-fold increase relative to vehicle-treated controls in both rapid membrane protrusions (observed after 5 min of agonist addition) and stable protrusions (observed after 90 min of agonist). In MEFβarrDKO, formation of both rapid and stable protrusions was abolished, and this was rescued by transfection of both β-arrestins 1 and 2 (Fig. 4B). In support of previous observations that both β-arrestins are required for PAR-2-stimulated membrane protrusion and cell migration (1), transfection of either β-arrestin-1 or β-arrestin-2 into MEFβarrDKO partially rescued stable protrusion formation to 65–70% of that observed in MEFwt but did not rescue rapid membrane protrusion formation (Fig. 4B).
FIGURE 4.
β-Arrestins and CIN are required for initial membrane protrusion formation. A, time-lapse images of MEFwt, MEFβarrDKO, DKO+βarr-1, and DKO+βarr2 were observed from 15 min before to 2 min after addition of an agar cube containing the PAR-2 agonist 2fAP, to the upper right corner of the dish. Arrows indicate membrane protrusions. B, protrusion formation in response to PAR-2 activation (mean protrusion number ± S.E.) was quantified using a modified Transwell filter assay in MEFwt and MEFβarrDKO treated with or without 2fAP for 5 and 90 min. Rescue of protrusion formation by β-arrestins was determined in MEFDKO transfected with β-arrestin-1 (DKO+βarr1), β-arrestin-2 (DKO+βarr2), or with both β-arrestins (DKO+βarr1&2). C, protrusion formation was determined in MDA MB-468 cells, transfected with GFP-tagged dominant negative CIN (DN-CIN) or GFP alone (negative control), and treated with or without 2fAP for 90 min. Data are expressed as a -fold change in the number of protrusions in agonist compared with untreated controls. *, statistically significant increase in protrusions (p < 0.01). Statistically significant difference in agonist-induced membrane protrusion between bracketed groups is indicated by # (p < 0.01) or ## (p < 0.04), n = 3.
Our working hypothesis is that β-arrestins sequester cofilin with its upstream activator, CIN, to promote its localized activity, which is known to be essential for both migration and membrane protrusion formation (23, 24). To determine whether CIN was also required for PAR-2-stimulated membrane protrusions, cells were transfected with GFP (negative control) or GFP-tagged dominant negative CIN (DN-CIN). DN-CIN is an inactive point mutant that specifically blocks endogenous CIN activity and was previously shown to inhibit PAR-2-stimulated cofilin dephosphorylation (14, 26). Transfection of dominant negative CIN abolished membrane protrusion formation (Fig. 4C). We conclude that both β-arrestins 1 and 2, and CIN activity, are required for directional membrane protrusion in response to PAR-2 activation.
PAR-2-induced Generation of Free Actin Barbed Ends in Lamellipodia Is Reduced in the Absence of β-Arrestins
Cofilin-induced actin filament severing contributes to chemotaxis by providing free barbed ends at the leading edge for polymerization (24, 35). Actin barbed end formation can be visualized by determining incorporation of fluorescently labeled actin monomers into cells by confocal microscopy. The fluorescence intensity can be quantified over a defined distance to determine the level of actin monomer incorporation at the leading edge (35, 36). Upon treatment of MEFwt with 2fAP for 1 min, actin monomers were incorporated into membrane protrusions in MEFwt, within 0.5–2 μm from the cell edge, but no significant monomer incorporation into MEFβarrDKO was observed (Fig. 5, A, B, D, and E). Lower magnification images showing actin monomer incorporation in the entire cell are shown in supplemental Fig. S5. PAR-2-induced generation of free actin barbed ends at the leading edge could be rescued by transfection of both β-arrestins (Fig. 5, C–E). Transfection of either β-arrestin-1 or -2 alone marginally rescued actin monomer incorporation at the cell edge, although we did not observe as distinct membrane protrusions as with transfection of both β-arrestins (supplemental Fig. S6). Actin monomer incorporation was also decreased in cells expressing DN-CIN, compared with mock transfected controls (Fig. 5, F–I). The minor actin monomer incorporation that is observed near the nucleus of DN-CIN-transfected cells might be due to CIN-independent cofilin activity or Arp2/3-mediated actin nucleation. We conclude that PAR-2-induced generation of new actin barbed ends and subsequent membrane protrusion requires both β-arrestin-1 and 2, and the activity of CIN.
β-Arrestins Facilitate Co-localization of Cofilin with CIN in Membrane Protrusions
PAR-2 promotes redistribution of β-arrestins and cofilin to membrane protrusions (14), and β-arrestins are present in isolated pseudopodia (1); thus, we hypothesized that they might be required for localization of CIN with cofilin at the leading edge. To address this hypothesis, GFP-tagged cofilin and Myc-tagged-CIN were expressed in MEFwt or MEFβarrDKO; cells were treated with 2fAP for 1 min, then fixed and stained with phalloidin and anti-myc. Co-localization of cofilin and CIN was observed by confocal microscopy, and fluorescence intensity of each was again determined as a function of the distance from the cell edge (Fig. 6). In untreated cells, cofilin was diffusely distributed throughout the cytoplasm, whereas CIN exhibited a more punctate cytosolic staining near the membrane (3–5 μm from the cell edge). After treatment of MEFwt with 2fAP, we observed striking redistribution of a pool of cofilin to the membrane protrusions, where it co-localized with CIN (Fig. 6, A, B, E, and F). No cofilin redistribution or co-localization with CIN was observed in MEFβarrDKO (Fig. 6, C, D, G, and H). Quantification of fluorescence intensity as a function of distance from the cell edge revealed that cofilin and CIN co-localization peaked 0.75 μm from the cell edge (Fig. 6, I–M). In contrast the mean fluorescence for cofilin and CIN in both treated and untreated MEFβarrDKO was ∼4 μm from the cell edge, similar to what was observed in untreated MEFwt. Quantification of the mean fluorescence intensity within the zone 0–2 μm from the membrane, reveals that PAR-2 promotes a 12-fold increase in both CIN and cofilin translocation to the leading edge (Fig. 6N). The peak intensity of CIN and cofilin co-localization in MEFwt after PAR-2 activation overlapped with the peak intensity of actin monomer incorporation, consistent with a role for this scaffold in filament severing (Fig. 6O). These microscopic observations were corroborated with a previously described biochemical method for isolating membrane protrusions and cell bodies (1, 2, 34). After inducing extension of protrusions through the 3-μm pores of a Transwell filter as described in Fig. 4, either pseudopodia from the filter underside or cell bodies from the top side were collected, lysed, and analyzed by Western blot. Using this assay, we demonstrated the enrichment of both cofilin and CIN in membrane protrusions (Fig. 6P). Histone is included as a control to demonstrate the restriction of cell body proteins to that fraction.
Similar to what we observed for membrane protrusion and actin barbed end formation, the defect observed in MEFβarrDKO could be rescued by β-arrestin expression. Although cofilin translocation to membrane protrusions was only fully rescued by transfection of both β-arrestins, transfection of β-arrestin-2, but not β-arrestin-1 alone partially rescued the effect (Fig. 7). In contrast, CIN localization to the plasma membrane was only restored upon transfection of both β-arrestins (supplemental Fig. S7). Consistent with these findings, subcellular fractionation demonstrated that PAR-2 activation results in the appearance of cofilin in membrane fractions in wtMEF but not MEFβarrDKO. Transfection of either β-arrestin-2 and to a lesser extent β-arrestin-1 rescues PAR-2-induced redistribution of cofilin to membrane fractions (supplemental Fig. S8). We conclude that PAR-2-stimulated cofilin redistribution and co-localization with CIN at the leading edge requires β-arrestins, but cofilin localization is more dependent on β-arrestin-2 than β-arrestin-1. CIN and cofilin co-localize in the same zone in which free actin barbed ends are generated, which is consistent with the concept that localized cofilin dephosphorylation spatially controls actin filament severing.
DISCUSSION
A requirement for β-arrestins in chemotaxis has been reported for various receptors both in vivo and in vitro (13, 16–18); however, the molecular mechanisms underlying this requirement have remained unclear. Furthermore, a role for β-arrestins in PAR-2-stimulated migration in primary cells has never been demonstrated. This work fills an important gap in the understanding of how β-arrestins regulate actin assembly and cell migration and their role in PAR-2-stimulated chemotaxis, providing a novel mechanism for spatial regulation of cofilin. We demonstrate the following points: 1) PAR-2 promotes the formation of a complex containing β-arrestins, cofilin, and CIN in vivo as well as in cultured cells. PAR-2-stimulated chemotaxis is impaired in primary leukocytes from β-arrestin-2−/− mice, corresponding to a lack of CIN/cofilin association. 2) β-Arrestins and CIN are required for the formation of a leading edge during PAR-2-stimulated chemotaxis. 3) β-Arrestin-dependent scaffolding of cofilin with CIN is required for their localization to leading edge and for the generation of free actin barbed ends. How β-arrestins regulate cell motility has been a topic of debate for some time. Some studies suggest that β-arrestins are essential for signal termination at the trailing edge, allowing for cell polarization in response to different chemotactic signals, while others suggest that they regulate actin-binding proteins and other molecules involved in cell motility (13). These studies are the first to demonstrate a correlation between β-arrestin scaffolding of actin assembly proteins and defective chemotaxis in primary cells, and to directly link CIN and β-arrestins to localized cofilin activity.
Cofilin activity at the leading edge is essential, but when uncontrolled can either inhibit protrusion formation or confer cells with metastatic potential (24, 37, 38). We observed that, in the absence of β-arrestins, cofilin localization to the leading edge and association with CIN is impaired, resulting in decreased generation of free actin barbed ends, defective membrane protrusion, and decreased cell migration. Although other processes besides cofilin activation, such as ARP2/3-mediated nucleation (23, 39), can contribute to the generation of free actin barbed ends, the dependence of PAR-2-stimulated actin monomer incorporation on both β-arrestins and CIN strongly supports our hypothesis that β-arrestin-dependent control of cofilin activity is important for PAR-2-mediated chemotaxis. Expression of β-arrestin-2 in cells lacking both β-arrestins partially restores membrane localization of cofilin, actin barbed end formation at the leading edge, and pseudopodia extension; in contrast, expression of β-arrestin-1 does not. The more dramatic effect of β-arrestin-2 knock-out on PAR-2-stimulated complex formation may reflect an ability to interact with both CIN and cofilin; in fact, we observed direct binding of both proteins to recombinant β-arrestin-2 in vitro, and this is enhanced by addition of recombinant β-arrestin-1.3 Taken together, these data suggest that β-arrestin-2 and to a lesser extent β-arrestin-1 are required to recruit cofilin to the membrane and scaffold it with CIN. Our data also suggest that both β-arrestins are present in a single complex, although in the absence of β-arrestin-1, β-arrestin-2 appears to retain the ability to form this complex.
Although β-arrestin-dependent regulation of CIN is clearly important for PAR-2-stimulated cofilin dephosphorylation and membrane protrusions, there are likely contributions from other pathways as well. For example, we have previously demonstrated that β-arrestin-1 directly binds LIMK and knockdown of β-arrestin-1 increases LIMK activity (14). Thus, β-arrestin-1-dependent inhibition of LIMK activity may contribute to total PAR-2-stimulated cofilin dephosphorylation and chemotaxis, which could explain why cofilin phosphorylation is increased, and protrusion formation and actin barbed end generation are decreased, in the absence of β-arrestin-1 despite its apparent minor contribution to cofilin/CIN association.
A major outcome of this study is the identification of β-arrestin-dependent scaffolding of cofilin and CIN in primary leukocytes, as well as β-arrestin-dependent, PAR-2-induced cell migration in these cells. The fact that phosphorylated (inactive) cofilin levels were elevated in the bone marrow leukocytes from either β-arrestin-1−/−, β-arrestin-2−/−, or PAR-2−/− mice compared with wild-type controls, is suggestive of a general role for the PAR-2/β-arrestin/cofilin signaling axis in vivo (Table 1). PAR-2 has been reported to participate in the recruitment of lymphocytes, neutrophils, and eosinophils to sites of inflammation a variety of disease models, including asthma and inflammatory bowel disease (3, 4, 7). In our present study, leukocytes from β-arrestin knock-out mice exhibited defects in PAR-2-stimulated chemotaxis, pointing to the possible importance of β-arrestins in PAR-2-mediated inflammatory responses.
β-Arrestins may represent a novel means for spatially controlling cofilin activity to generate a localized pool of free actin barbed ends for other receptors besides PAR-2. However, the role of β-arrestins in cell signaling depends on the activating receptor; thus, this mechanism is unlikely to be shared by all receptors. For example, the role of cofilin in chemotaxis of tumor cells, lamellipodia formation, as well as its mechanism of activation, has been well characterized for EGF (23, 35, 40). EGF has been reported to activate and localize cofilin and filament severing, independent of cofilin dephosphorylation (41). Furthermore, EGF stimulates CIN localization to membrane protrusions, but although CIN is required for EGF-stimulated actin barbed end incorporation, it is not required for EGF-stimulated membrane protrusions in MTLn3 cells.4 Studies by other laboratories on AT1-AR demonstrated a requirement for β-arrestin-2 in chemotaxis, but this requirement was not observed for EGF-stimulated chemotaxis (16). Thus, there are likely multiple mechanisms of spatially regulating cofilin activity during chemotaxis and the role of β-arrestins and CIN in this process varies between receptors.
Because β-arrestins are pleiotropic proteins, they are likely to affect cell migration, actin cytoskeletal reorganization, and even cofilin activation on multiple levels. In addition to inhibition of LIMK activity (14), β-arrestins can also regulate Rho-GTPase activity, interact with actin-binding proteins, and regulate MAPK activity at the leading edge (1, 13, 42), all of which are important pathways for cell migration. Still other actin assembly proteins can associate with β-arrestins (13). Future studies investigating the interplay between these other pathways and the cofilin pathway are essential to a complete understanding of the consequences of β-arrestin-dependent scaffolding in PAR-2 signaling, as well as other receptor signaling cascades.
Supplementary Material
Acknowledgments
We thank Dr. Robert Lefkowitz (Duke University Medical Center) for generosity in providing β-arrestin knock-out mice, cells, and antibodies, Dr. Gary Bokoch (TSRI) for generosity in providing CIN reagents, and Dr. Celine DerMardirossian for help with fluorescence measurements.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM066151 and 1R21HL092388.
We dedicate this paper to the memory of Dr. Gary Bokoch, who was a major intellectual force in the field and whose contributions will be sorely missed.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
H. L. Richards and K. A. DeFea, unpublished observations.
C. DerMardirossian and G. Bokoch, personal communication.
- PAR-2
- protease-activated receptor-2
- ERK1/2
- extracellular signal-regulated kinase 1 and 2
- CIN
- chronophin
- 2fAP
- 2-furoyl-LIGRL-ornithine-NH2
- DN
- dominant negative
- wt
- wild type
- PBS
- phosphate-buffered saline
- SEC
- size-exclusion chromatography
- MAPK
- mitogen-activated protein kinase
- MEF
- mouse embryonic fibroblast
- GFP
- green fluorescent protein
- EGF
- epidermal growth factor
- WB
- Western blot
- IF
- immunofluorescence
- IP
- immunoprecipitation.
REFERENCES
- 1.Ge L., Ly Y., Hollenberg M., DeFea K. (2003) J. Biol. Chem. 278, 34418–34426 [DOI] [PubMed] [Google Scholar]
- 2.Ge L., Shenoy S. K., Lefkowitz R. J., DeFea K. A. (2004) J. Biol. Chem. 279, 55419–55424 [DOI] [PubMed] [Google Scholar]
- 3.Ebeling C., Forsythe P., Ng J., Gordon J. R., Hollenberg M., Vliagoftis H. (2005) J. Allergy Clin. Immunol. 115, 623–630 [DOI] [PubMed] [Google Scholar]
- 4.Cenac N., Coelho A. M., Nguyen C., Compton S., Andrade-Gordon P., MacNaughton W. K., Wallace J. L., Hollenberg M. D., Bunnett N. W., Garcia-Villar R., Bueno L., Vergnolle N. (2002) Am. J. Pathol. 161, 1903–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin A. C., Lee W. Y., Nusrat A., Vergnolle N., Parkos C. A. (2008) J. Immunol. 181, 5702–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyun E., Andrade-Gordon P., Steinhoff M., Vergnolle N. (2008) Gut 57, 1222–1229 [DOI] [PubMed] [Google Scholar]
- 7.Schmidlin F., Amadesi S., Dabbagh K., Lewis D. E., Knott P., Bunnett N. W., Gater P. R., Geppetti P., Bertrand C., Stevens M. E. (2002) J. Immunol. 169, 5315–5321 [DOI] [PubMed] [Google Scholar]
- 8.Ferrell W. R., Lockhart J. C., Kelso E. B., Dunning L., Plevin R., Meek S. E., Smith A. J., Hunter G. D., McLean J. S., McGarry F., Ramage R., Jiang L., Kanke T., Kawagoe J. (2003) J. Clin. Invest. 111, 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luttrell L. M., Ferguson S. S., Daaka Y., Miller W. E., Maudsley S., Della Rocca G. J., Lin F., Kawakatsu H., Owada K., Luttrell D. K., Caron M. G., Lefkowitz R. J. (1999) Science 283, 655–661 [DOI] [PubMed] [Google Scholar]
- 10.DeFea K. A., Zalevsky J., Thoma M. S., Déry O., Mullins R. D., Bunnett N. W. (2000) J. Cell Biol. 148, 1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tohgo A., Pierce K. L., Choy E. W., Lefkowitz R. J., Luttrell L. M. (2002) J. Biol. Chem. 277, 9429–9436 [DOI] [PubMed] [Google Scholar]
- 12.Wang P., DeFea K. A. (2006) Biochemistry 45, 9374–9385 [DOI] [PubMed] [Google Scholar]
- 13.DeFea K. A. (2007) Annu. Rev. Physiol. 69, 535–560 [DOI] [PubMed] [Google Scholar]
- 14.Zoudilova M., Kumar P., Ge L., Wang P., Bokoch G. M., DeFea K. A. (2007) J. Biol. Chem. 282, 20634–20646 [DOI] [PubMed] [Google Scholar]
- 15.DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K. (2007) Annu. Rev. Physiol. 69, 483–510 [DOI] [PubMed] [Google Scholar]
- 16.Hunton D. L., Barnes W. G., Kim J., Ren X. R., Violin J. D., Reiter E., Milligan G., Patel D. D., Lefkowitz R. J. (2005) Mol. Pharmacol. 67, 1229–1236 [DOI] [PubMed] [Google Scholar]
- 17.Fong A. M., Premont R. T., Richardson R. M., Yu Y. R., Lefkowitz R. J., Patel D. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7478–7483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker J. K., Fong A. M., Lawson B. L., Savov J. D., Patel D. D., Schwartz D. A., Lefkowitz R. J. (2003) J. Clin. Invest. 112, 566–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagane B., Chow K. Y., Balabanian K., Levoye A., Harriague J., Planchenault T., Baleux F., Gunera-Saad N., Arenzana-Seisdedos F., Bachelerie F. (2008) Blood 112, 34–44 [DOI] [PubMed] [Google Scholar]
- 20.Cooper J. A., Schafer D. A. (2000) Curr. Opin. Cell Biol. 12, 97–103 [DOI] [PubMed] [Google Scholar]
- 21.Delorme V., Machacek M., DerMardirossian C., Anderson K. L., Wittmann T., Hanein D., Waterman-Storer C., Danuser G., Bokoch G. M. (2007) Dev. Cell 13, 646–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponti A., Machacek M., Gupton S. L., Waterman-Storer C. M., Danuser G. (2004) Science 305, 1782–1786 [DOI] [PubMed] [Google Scholar]
- 23.Mouneimne G., DesMarais V., Sidani M., Scemes E., Wang W., Song X., Eddy R., Condeelis J. (2006) Curr. Biol. 16, 2193–2205 [DOI] [PubMed] [Google Scholar]
- 24.Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D. S., Condeelis J. S. (2004) Science 304, 743–746 [DOI] [PubMed] [Google Scholar]
- 25.Nagata-Ohashi K., Ohta Y., Goto K., Chiba S., Mori R., Nishita M., Ohashi K., Kousaka K., Iwamatsu A., Niwa R., Uemura T., Mizuno K. (2004) J. Cell Biol. 165, 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gohla A., Birkenfeld J., Bokoch G. M. (2005) Nat. Cell Biol. 7, 21–29 [DOI] [PubMed] [Google Scholar]
- 27.Bamburg J. R. (1999) Annu. Rev. Cell Dev. Biol. 15, 185–230 [DOI] [PubMed] [Google Scholar]
- 28.Kumar P., Lau C., Wang P., Mathur M., DeFea K. A. (2007) Am. J. Physiol. Cell Physiol. [DOI] [PubMed] [Google Scholar]
- 29.Kohout T. A., Lin F. S., Perry S. J., Conner D. A., Lefkowitz R. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1601–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milano S. K., Kim Y. M., Stefano F. P., Benovic J. L., Brenner C. (2006) J. Biol. Chem. 281, 9812–9823 [DOI] [PubMed] [Google Scholar]
- 31.Matsuzaki F., Matsumoto S., Yahara I., Yonezawa N., Nishida E., Sakai H. (1988) J. Biol. Chem. 263, 11564–11568 [PubMed] [Google Scholar]
- 32.McGuire J. J., Saifeddine M., Triggle C. R., Sun K., Hollenberg M. D. (2004) J. Pharmacol. Exp. Ther. 309, 1124–1131 [DOI] [PubMed] [Google Scholar]
- 33.DeFea K. A., Vaughn Z. D., O'Bryan E. M., Nishijima D., Déry O., Bunnett N. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho S. Y., Klemke R. L. (2002) J. Cell Biol. 156, 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouneimne G., Soon L., DesMarais V., Sidani M., Song X., Yip S. C., Ghosh M., Eddy R., Backer J. M., Condeelis J. (2004) J. Cell Biol. 166, 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan A. Y., Raft S., Bailly M., Wyckoff J. B., Segall J. E., Condeelis J. S. (1998) J. Cell Sci. 111, 199–211 [DOI] [PubMed] [Google Scholar]
- 37.Sidani M., Wessels D., Mouneimne G., Ghosh M., Goswami S., Sarmiento C., Wang W., Kuhl S., El-Sibai M., Backer J. M., Eddy R., Soll D., Condeelis J. (2007) J. Cell Biol. 179, 777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W., Mouneimne G., Sidani M., Wyckoff J., Chen X., Makris A., Goswami S., Bresnick A. R., Condeelis J. S. (2006) J. Cell Biol. 173, 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 40.Chan A. Y., Bailly M., Zebda N., Segall J. E., Condeelis J. S. (2000) J. Cell Biol. 148, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song X., Chen X., Yamaguchi H., Mouneimne G., Condeelis J. S., Eddy R. J. (2006) J. Cell Sci. 119, 2871–2881 [DOI] [PubMed] [Google Scholar]
- 42.Barnes W. G., Reiter E., Violin J. D., Ren X. R., Milligan G., Lefkowitz R. J. (2005) J. Biol. Chem. 280, 8041–8050 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.