Abstract
Structural characterization of glutamate cysteine ligase (GCL), the enzyme that catalyzes the initial, rate-limiting step in glutathione biosynthesis, has revealed many of the molecular details of substrate recognition. To further delineate the mechanistic details of this critical enzyme, we have determined the structures of two inhibited forms of Saccharomyces cerevisiae GCL (ScGCL), which shares significant sequence identity with the human enzyme. In vivo, GCL activity is feedback regulated by glutathione. Examination of the structure of ScGCL-glutathione complex (2.5 Å; R = 19.9%, Rfree = 25.1%) indicates that the inhibitor occupies both the glutamate- and the presumed cysteine-binding site and disrupts the previously observed Mg2+ coordination in the ATP-binding site. l-Buthionine-S-sulfoximine (BSO) is a mechanism-based inhibitor of GCL and has been used extensively to deplete glutathione in cell culture and in vivo model systems. Inspection of the ScGCL-BSO structure (2.2 Å; R = 18.1%, Rfree = 23.9%) confirms that BSO is phosphorylated on the sulfoximine nitrogen to generate the inhibitory species and reveals contacts that likely contribute to transition state stabilization. Overall, these structures advance our understanding of the molecular regulation of this critical enzyme and provide additional details of the catalytic mechanism of the enzyme.
Keywords: Crystal Structure, Enzyme Catalysis, Enzyme Inhibitors, Enzyme Mechanisms, Glutamate, Glutathione, Oxidative Stress, Thiol, X-ray Crystallography
Introduction
Glutamate cysteine ligase (GCL)2 catalyzes the initial and rate-limiting step of glutathione biosynthesis (1, 2). The ATP-dependent mechanism proceeds via a γ-glutamylphosphate intermediate (2–4), with a subsequent nucleophilic attack by the α-amino group of l-cysteine to produce γ-glutamylcysteine (1, 2). There are three distinct families of GCL enzymes: γ-proteobacteria (Group 1), nonplant eukaryotes (Group 2), and α-proteobacteria and plants (Group 3) (5). Despite low sequence conservation between these groups (typically <10% sequence identity), all of the GCL appear to use this general catalytic mechanism. The resulting γ-glutamylcysteine is coupled to l-glycine by glutathione synthetase (1) in an analogous reaction to generate reduced GSH, an abundant cellular reducing agent.
GCL activity is tightly modulated by free l-cysteine availability (6), transcriptional regulation (7), and post-translational modifications (8). In addition, GCL is feedback regulated by the end product, glutathione (9). Glutathione inhibits GCL competitively with respect to l-glutamate, suggesting that the two binding sites are coincident (9). In heterodimeric GCL, such as the Drosophila, rat, and human enzymes, binding of the modifier subunit relieves feedback inhibition both by increasing the Ki for glutathione and decreasing the Km for glutamate (10–13). Further studies with glutathione analogues such as ophthalmic acid, S-methylglutathione, and GSSG have demonstrated that the free thiol group of glutathione is necessary for maximal inhibition (1, 9). However, the precise mode of glutathione binding has not been described.
The central role of GCL in glutathione homeostasis makes it an attractive target for drug design. Increased glutamate cysteine ligase catalytic subunit mRNA levels and GCL activity have been frequently observed in cells derived from human tumors resistant to chemotherapeutic agents (14–16). Increased production of glutathione likely protects against reactive oxygen and nitrogen species (17, 18) and facilitates detoxification of electrophilic xenobiotics by the glutathione S-transferases (19). Drug resistance in tumors can be overcome by the administration of l-buthionine-S,R-sulfoximine (BSO) (20), which inhibits GCL and subsequently depletes GSH, thus sensitizing the cancer cells to radiation treatment and chemotherapy. Administration of BSO has also been shown to prolong the survival of mice infected with the parasite Trypanosoma brucei (21), the causative agent of African sleeping sickness. Similarly, BSO-mediated depletion of glutathione inhibits the development Plasmodium falciparum in red blood cells (22). BSO presumably binds as an l-glutamate analogue with its S-butyl group extending into the l-cysteine-binding site (23). Subsequent ATP-dependent phosphorylation of the sulfoximine nitrogen by GCL leads to the formation of a tightly bound transition state analogue (20, 23).
Recently, we reported the crystal structure of Saccharomyces cerevisiae GCL (ScGCL) in complex with l-glutamate, Mg2+, and ADP (24). As the first structure of a Group 2 glutamate cysteine ligase, examination of the model provided important molecular details of substrate recognition and led to the identification of key catalytic residues. In the current study, we have determined the crystal structures of two inhibited forms of the enzyme. The structure of ScGCL in complex with glutathione reveals the molecular details of feedback inhibition, whereas the ScGCL-BSO complex structure details the mechanism of BSO inhibition. Examination of the available ScGCL structures provides considerable insight in the catalytic mechanism of the enzyme and suggests approaches by which GCL inhibitors with greater selectivity may be attainable.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
ScGCL was expressed in Escherichia coli RosettaTM2(DE3) pLysS cells (Novagen) and purified to homogeneity as described previously (24). Briefly, soluble cell lysates were cleared of debris by centrifugation and ScGCL isolated by affinity chromatography using a HisTrap Chelating HP Column (GE Healthcare). The protein was further purified by size exclusion chromatography using a Sephacryl 200 gel filtration column. Purified ScGCL was dialyzed against 20 mm Tris-HCl, pH 7.4, 2 mm dithiothreitol, concentrated (Amicon stirred cell 8050, 10-kDa cut-off), flash-frozen in liquid nitrogen, and stored at −80 °C. Point mutations were introduced at residue Cys266 (C266S and C266A) by using the QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's protocol. All of the constructs were verified by sequencing at the University of Nebraska Genomics Facility (Lincoln, NE).
Kinetic Assays
Enzymatic activity was measured using an indirect assay that couples ADP production to NADH oxidation, which was monitored at 340 nm (11). The reaction mixture contained 20 mm MgCl2, 5 mm phosphoenolpyruvic acid, 0.2 mm NADH, and 4 units each of pyruvate kinase and lactate dehydrogenase in 1 ml of buffer (100 mm Tris, pH 8.0, 150 mm KCl). The reaction was initiated by the addition of ScGCL. To determine the apparent Km values, two of the three substrates were added to the reaction at a saturating concentration (20 mm l-glutamate, 10 mm l-cysteine, 5 mm ATP), whereas the third was varied. At high concentrations of cysteine or ATP, substrate inhibition was observed.
To examine the mode of inhibition of glutathione, the rates for the enzyme-catalyzed reaction as a function of glutamate concentration were determined in the presence of fixed concentrations of glutathione (0, 2.5, and 5.0 mm). A general mixed model of inhibition was initially selected in Prism (Graph Pad Software) to describe the dependence of rate versus substrate concentration. This global analysis indicated that glutathione was a competitive inhibitor with respect to glutamate. Following this preliminary analysis, the data were reanalyzed designating competitive inhibition (supplemental Fig. S1). For inactivation studies, ScGCL (1.75 μm) was incubated with BSO (Sigma) in 100 mm Tris, pH 8.0, containing 150 mm KCl, 20 mm MgCl2, and 5 mm ATP at 4 °C (20, 25). At the indicated time, an aliquot was removed, and enzymatic activity was measured at saturating substrate concentrations using the coupled assay system. Representative data from three or more determinations are plotted as a function of time with the experimental errors indicated. A single-order decay was used to describe the data using the program Prism (Graph Pad Software).
Crystallization, Data Collection, Structure Determination, and Refinement
Concentrated ScGCL (7 mg/ml) was crystallized in the presence of either 5 mm reduced glutathione and 20 mm MgCl2 or 1 mm BSO, 5 mm ATP, and 20 mm MgCl2. Crystals were grown at 18 °C out of a solution of 12% (w/v) polyethylene glycol 400, 100 mm MES, pH 6.8, with the dimensions 0.15 × 0.15 × 0.15 mm, as described previously (24). Prior to data collection, the crystals were soaked in a stabilizing solution containing 30% polyethylene glycol 400 and the appropriate ligands and then vitrified in liquid nitrogen (26). Diffraction data for the ScGCL-glutathione complex were collected using radiation produced by a Rigaku MicroMax-007 x-ray generator fitted with confocal blue optics and an R-axis IV++ image plate system (λ = 1.54 Å; 100 K). For the ScGCL-BSO complex, diffraction data (λ = 0.9 Å; 100 K) were collected on Beamline 14-BM-C of BioCARS at Argonne National Laboratory Advanced Photon Source. All of the data were processed with the HKL2000 software package (27). The structures of the ScGCL complexes were determined by molecular replacement using the PHENIX software suite (28) with the previously determined ScGCL structure (Protein Data Bank code 3IG5) as the search model. Iterative rounds of model building and refinement were carried out using Coot (29) and Refmac5 (30), respectively. As the protein models neared completion, water molecules obeying proper hydrogen-bonding constraints with electron density greater than 1.0 σ on a 2Fo − Fc map and 4.0 σ on an Fo − Fc map were also included in the final structure. Model geometry was monitored using MOLPROBITY (31), and the figures were produced using Chimera (32).
RESULTS AND DISCUSSION
Kinetic Characterization of ScGCL
Previously reported structural and biochemical data indicate that ScGCL likely functions as a monomer both in vitro and in vivo (24). To investigate its kinetic parameters, ScGCL was purified to homogeneity, and enzymatic activity was assessed using a coupled enzyme system that monitors the production of ADP (11). Apparent kinetic constants for the enzyme-catalyzed formation of γ-glutamylcysteine were determined (Table 1) and are comparable with those reported for other eukaryotic GCL (1, 11–13, 33). Inhibition by glutathione, a feedback inhibitor of ScGCL, was also examined. Glutathione is a competitive inhibitor with respect to the glutamate substrate (supplemental Fig. S1), with an apparent Ki(GSH) of 2.12 ± 0.13 mm, similar to other Group 2 GCL holoenzymes (11–13, 33).
TABLE 1.
Apparent kinetic constants for wild-type ScGCL
| Km | Vmax | Ki | |
|---|---|---|---|
| mm | μmol min−1mg−1 | mm | |
| l-Glutamate | 1.21 ± 0.05 | 10.7 ± 0.17 | |
| l-Cysteine | 0.17 ± 0.01 | 10.9 ± 0.19 | |
| ATP | 0.08 ± 0.01 | 16.1 ± 0.56 | |
| GSH | 12.0 ± 0.18 | 2.12 ± 0.13 |
BSO is one of the most commonly used pharmacological inhibitors of glutathione biosynthesis, and its efficacy with respect to inhibition of ScGCL was examined (Fig. 1). A time-dependent loss of enzymatic activity was observed in the presence of Mg2+ and ATP at each of the BSO concentrations tested (5 μm to 50 μm). A near linear dependence on the inactivation rate as a function of BSO concentration was observed (data not shown). At 50 μm BSO, ScGCL activity was reduced nearly 10-fold in ∼5 min. Unfortunately, reliable rate measurements above this concentration of BSO could not be made because of the limitations of the assay. Nonetheless, BSO is clearly a potent inhibitor of ScGCL. Previous studies of related GCL indicated that l-buthionine-S-sulfoximine is the relevant stereoisomer and that its enzymatic phosphorylation generates a high affinity transition state analogue (23, 34). As discussed below, the ScGCL-BSO structure supports these findings.
FIGURE 1.
Time-dependent inactivation of ScGCL by the inhibitor BSO. ScGCL was incubated with a given concentration of BSO in the presence of Mg2+ and ATP at pH 8.0 and 4 °C. Relative enzymatic activity was monitored as a function of time. The activity measurements were made in triplicate, and the data for a given BSO concentration fit to a single exponential decay. The curves are shown for the control (filled circles) and six experimental BSO concentrations (5 μm, filled squares; 7.5 μm, filled triangle; 10 μm, filled inverted triangle; 15 μm, filled diamonds; 20 μm, open circles; 50 μm, open squares).
Overall Structures of ScGCL-Glutathione and ScGCL-BSO Complexes
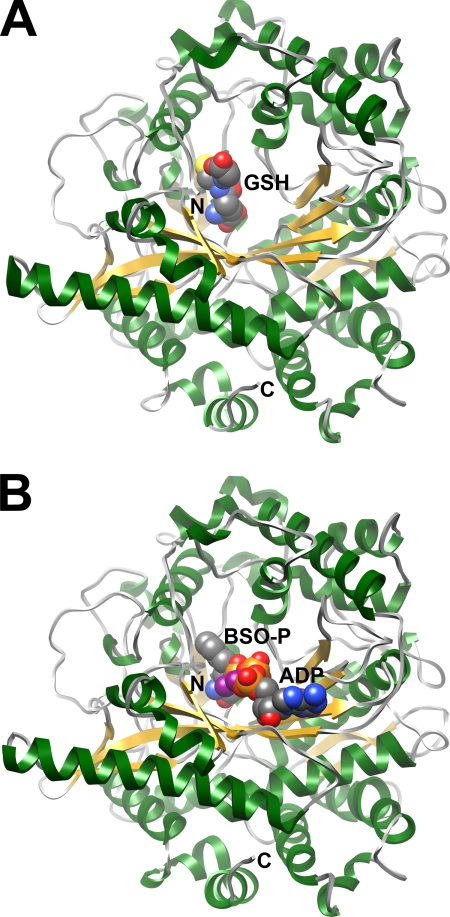
The structures of ScGCL in complex with either glutathione or BSO were determined by molecular replacement using the apo form of ScGCL as a probe (24). In the ScGCL-glutathione complex, reduced glutathione was readily modeled into the strong positive density observed within the enzyme active site (supplemental Fig. S2). The glutamate portion of glutathione is located at the base of the active site funnel (Fig. 2A). The cysteine moiety occupies a relatively hydrophobic binding pocket, whereas the terminal glycine is near the outer edge of the active site and is solvent-exposed. In the ScGCL-BSO complex, the electron density supports the modeling of phosphorylated BSO, ADP, and three Mg2+ ions (supplemental Fig. S3). The adenine ring of ADP is located at the lower lip of the active site cavity and is solvent-exposed (Fig. 2B). The phosphorylated BSO occupies a site comparable but distinct from the glutathione-binding site, as discussed below. The overall ScGCL-glutathione and ScGCL-BSO structures are very similar to that of ScGCL in complex with glutamate and Mg2+ with an root mean square deviation for Cα = ∼0.2 Å (24). Refinement statistics for the final ScGCL-glutathione and ScGCL-BSO models are provided in Table 2. The refined ScGCL-glutathione and ScGCL-BSO structures each have 99.7% of its residues in the allowed regions of the Ramachandran plot.
FIGURE 2.
Ribbon representations of the crystal structures of ScGCL in complex with inhibitors. An ScGCL monomer is contained in the asymmetric unit, and the N- and C-terminal residues are indicated. β-Strands are colored in yellow, and α-helices are depicted in green. A, bound GSH is shown in space filling representation with carbon atoms colored in gray, oxygen atoms are in red, sulfur atoms are in yellow, and nitrogen atoms are in blue. The glutathione-binding site overlaps the glutamate binding site within the active site funnel. B, ADP and the transition state analogue, phosphorylated BSO (BSO-P), are shown in space filling representation. Phosphorus and magnesium atoms are colored in orange and purple, respectively, with the remaining atoms colored as in A.
TABLE 2.
Data collection and refinement statistics
The values in parentheses are for the highest resolution shell.
| ScGCL-GSH | ScGCL-BSO | |
|---|---|---|
| Data collection statistics | ||
| Protein Data Bank accession code | 3LVW | 3LVV |
| Wavelength | 1.54 Å | 0.90 Å |
| Temperature (K) | 100 | 100 |
| Space group | P43212 | P43212 |
| Cell dimensions (Å) | 118.1, 118.1, 165.8 | 117.9, 117.9, 165.6 |
| Resolution, Å | 20.0-2.50 | 50.0-2.20 |
| Rmerge (%) | 10.1 (53.3) | 5.7 (50.3) |
| Mean I/σI | 9.7 (3.0) | 25.4 (2.8) |
| Completeness (%) | 97.1 (96.1) | 100.0 (100.0) |
| Average redundancy | 8.77 (8.94) | 18.7 (7.0) |
| Refinement statistics | ||
| Resolution, Å | 20.0-2.5 (2.56-2.50) | 50.0-2.20 (2.25-2.20) |
| Number of reflections | 40,046 | 59,891 |
| Rwork/Rfree (%) | 19.9/25.1 (29.0/36.5) | 18.1/23.9 (26.1/31.4) |
| Number of atoms | 5702 | 5811 |
| Protein | 5476 | 5476 |
| Ligand | 50 | 58 |
| Water | 176 | 277 |
| Average B-factors (Å2) | ||
| Protein | 46.2 | 37.7 |
| Ligand | 56.9 | 33.8 |
| Water | 43.3 | 38.8 |
| Root mean square deviations from ideal | ||
| Bond lengths (Å) | 0.02 | 0.02 |
| Bond angles (°) | 1.92 | 1.88 |
| Ramachandran statistics | ||
| Favored (%) | 95.4 | 96.9 |
| Allowed (%) | 99.7 | 99.7 |
The Glutathione-binding Site Is Coincident with the Glutamate- and Cysteine-binding Sites
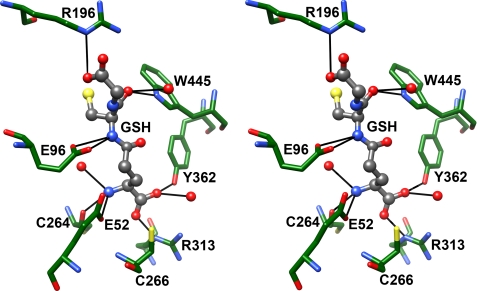
As indicated above, reduced glutathione is a competitive inhibitor of ScGCL with respect to glutamate and a physiologically relevant feedback inhibitor of the enzyme (9). The α-carboxylate of the glutamyl moiety of glutathione is positioned by hydrogen bonds with the side chains of Tyr362 and Arg313 as well as with an ordered water molecule (Fig. 3) that in turn forms a hydrogen bond with the backbone carbonyl of Arg472 (not shown). Cys266 is also positioned above the plane of the carboxylate and may help orient the bound inhibitor. The glutamyl α-amino group is within hydrogen bond distance of the backbone carbonyl of Cys264, the γ-carboxylate of Glu52, and an ordered water molecule. The cysteinyl α-amino and α-carbonyl groups are within hydrogen bond distance of the γ-carboxylate of Glu96 and the indole nitrogen of Trp445, respectively. An ordered water molecule can also form a hydrogen bond with the cysteinyl α-carbonyl group. The side chain of Arg196 is positioned to interact favorably with the terminal carboxylate of GSH, but the glycine portion of glutathione is poorly defined relative to the rest of the inhibitor.
FIGURE 3.
Glutathione occupies the glutamate and presumed cysteine-binding sites of ScGCL. In the stereodiagram, bound glutathione is shown in ball and stick representation, and pertinent active site residues are shown in stick representation. The atoms are colored as in Fig. 2, with the exception of ScGCL carbon atoms, which are colored green. Potential hydrogen bonds were identified in Chimera and are represented as solid black lines.
As compared with the previously described ScGCL/Glu/Mg2+ structure, there are several notable differences in the placement of the γ-glutamyl moiety of glutathione relative to the bound glutamate substrate. In the ScGCL/Glu/Mg2+ structure, the γ-carboxylate of the glutamate substrate occupies one of the coordination sites of the bound M1 Mg2+ (24). However, in the ScGCL-glutathione structure, the γ-carboxylate has been assimilated into the γ-glutamyl peptide bond and can no longer promote Mg2+ binding. Glu52 and Glu96, which also coordinate the Mg2+, maintain comparable positions in both structures, but Glu103 has shifted away from the M1 binding site (not shown). Loss of the M1 binding site causes the γ-glutamyl portion of glutathione to be shifted ∼0.3 Å out from the base of the active site, limiting interactions between its α-carboxylate and the side chain of Arg313. In addition, the M2 and M3 binding sites are not significantly occupied in the absence of ATP or ADP.
An intriguing feature of the γ-glutamyl-binding pocket is the conserved cysteine residue, Cys266, which is in close proximity to the α-carboxylate of glutamate. Previously, mutation of the equivalent cysteine residue in T. brucei GCL to an alanine had little effect on the specific activity or the substrate binding affinity of the enzyme (35). In ScGCL, substitution of this residue with either a serine (C266S) or an alanine residue (C266A) had a modest but reproducible effect on glutamate and glutathione binding (Table 3). For both mutants, the apparent Km(Glu) and the apparent Ki(GSH) increased ∼2-fold relative to the wild-type enzyme. Studies to examine the impact of these mutations on overall glutathione production in S. cerevisiae are ongoing.
TABLE 3.
Apparent kinetic constants for C266S and C266A ScGCL
| Kml-Glu | Vmax | V/K | Ki glutathione | |
|---|---|---|---|---|
| mm | μmol min−1mg−1 | mm | ||
| ScGCL | 1.21 ± 0.05 | 10.7 ± 0.17 | 8.8 | 2.12 ± 0.13 |
| C266S | 2.15 ± 0.07 | 7.58 ± 0.07 | 3.5 | 3.91 ± 0.25 |
| C266A | 1.93 ± 0.07 | 9.22 ± 0.09 | 4.8 | 4.70 ± 0.35 |
Molecular Details of the l-Buthionine-S-sulfoximine-binding Site
In addition to glutathione, all three families of GCL can be inhibited by S-alkyl-l-homocysteine sulfoximines (36). As discussed above, BSO is a potent mechanism-based inhibitor of ScGCL. The enzyme catalyzes the ATP-dependent phosphorylation of BSO to form BSO phosphate and ADP, which mimic the transition state. Phosphorylated BSO binds tightly and dissociates very slowly (20, 37), making this compound pharmacologically important for development of treatments against cancer and certain parasites (21, 22, 38).
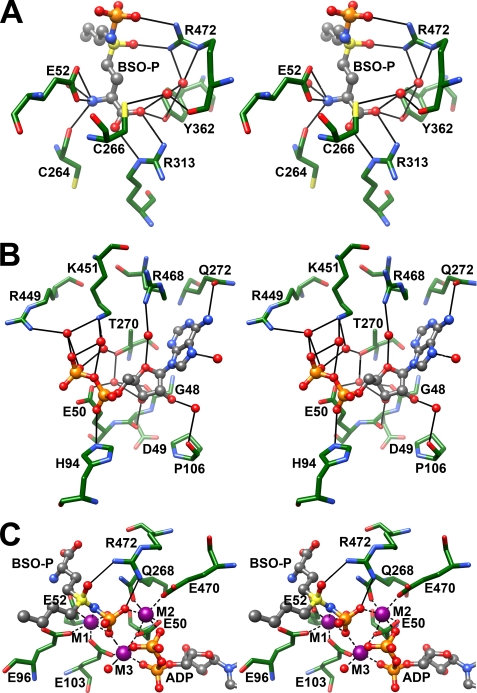
Phosphorylated BSO occupies the l-glutamate and the presumed l-cysteine-binding sites of ScGCL (Fig. 4A). The α-carboxylate and α-amino groups of BSO are virtually superimposable on the comparable functional groups of the glutamate substrate (not shown). BSO is phosphorylated on the sulfoximine nitrogen, and the S-butyl group of BSO mimics l-cysteine, occupying a relatively hydrophobic pocket within the enzyme active site. Arg472 is within hydrogen bond distance of the sulfoximine oxygen and an oxygen of the newly added phosphate group and likely stabilizes the transition state. In support of a direct role in catalysis, mutation of the equivalent arginine, Arg491 in T. brucei GCL, decreased enzymatic activity by 70-fold (39). Phosphorylation by ATP and subsequent tight inhibitor-enzyme interaction is dependent on the metal ion binding (1). The precise locations of the three metal-binding sites are discussed below.
FIGURE 4.
Analysis of the x-ray structure of the ScGCL-BSO complex reveals details of catalysis. In the stereodiagrams, bound ligands are shown in ball and stick representation, and pertinent active site residues are shown in stick representation. Atoms are colored as in Fig. 2, with potential hydrogen bonds represented as solid black lines. A, phosphorylated BSO mimics the transition state. The sulfoximine nitrogen is phosphorylated and is within hydrogen bond distance of Arg472, which may facilitate catalysis by stabilizing the transition state. B, additional details of the ADP binding site. Examination of a previously reported ScGCL structure led to the identification of several protein/ligand interactions (24). The 2.2 Å resolution structure of the BSO-inhibited enzyme reveals additional contributions to ADP binding. Most notably, the imidazole ring of His94 is in close proximity to the α-phosphate, and the side chain of Arg468 is within hydrogen bond distance of an ordered water molecule that helps position the ribose ring of ADP. C, stereodiagram of Mg2+-binding sites in the refined model of ScGCL in complex with phosphorylated BSO (BSO-P) and ADP. In the stereodiagram, bound ADP and phosphorylated BSO are shown, with potential hydrogen bonds between a catalytic arginine residue, Arg472, and phosphorylated BSO represented as solid black lines. Three Mg2+ ions, designated as M1, M2, and M3, are shown as purple spheres, and dashed black lines illustrate their likely coordination (interatomic distances of <2.2 Å).
The crystal structures of E. coli (40) and Brassica juncea (41) GCL in complex with alkyl sulfoximine inhibitors have also been reported. Comparison with the ScGCL-BSO complex reveals a dramatic conservation of active site functionality across bacteria, plants, and nonplant eukaryotes. In these three structures, the γ-glutamyl-binding sites are superimposable, with the α-carboxylate adjacent to a conserved arginine residue (Arg313 in ScGCL) and the α-amino group within hydrogen bond distance of a bound water, the backbone carbonyl of residue 264, and the carboxylate of Glu52 (Fig. 4A). The proposed catalytic arginine residue, Arg472 in ScGCL, is also conserved and suggests that all three enzymes function using a similar mechanism.
Description of the ADP-binding Site of ScGCL
Previously, we described the structure of ScGCL in complex with glutamate, ADP, and Mg2+ to 2.7 Å resolution (24). The current ScGCL-BSO complex structure has been refined to significantly higher resolution (2.2 Å) and provides additional details regarding ADP binding (Fig. 4B). As described previously, the 2′ and 3′ hydroxyls of the ribose are involved in an extended hydrogen bond network. The oxygen of the furanose ring forms a hydrogen bond with an ordered water molecule that is positioned by the side chain of Arg468. Substitution of the equivalent residue in T. brucei GCL, Arg487, with an alanine increases the Km(ATP) > 15-fold (39). The C6 amino group and N7 nitrogen of the adenine ring are within hydrogen bond distance of the side chain of Gln272 and an ordered water molecule, respectively. Through bridging water molecules, Thr270, Arg449, and Lys451 interact with the pyrophosphate group of ADP, and these residues are likely important binding determinants. In T. brucei GCL, mutation of Thr323 (Thr370 in ScGCL) to an alanine dramatically increased the apparent Km for ATP (39). Interestingly, the imidazole ring of His94 moves ∼1.4 Å toward the ADP molecule and forms hydrogen bonds with an α-phosphate oxygen and the γ-carboxylate of Glu103 (not shown). Three bound Mg2+ molecules provide additional stabilizing interactions as described below.
Three Bound Mg2+ Ions Contribute to the Formation and Binding of the Transition State Analogue
In the ScGCL-BSO structure, three octahedrally coordinated Mg2+ ions are observed (Fig. 4C). The first metal-binding site, M1, is formed by the side chain carboxylates of Glu52, Glu96, and Glu103; the sulfoximine nitrogen; an oxygen of the covalently attached phosphate group; and an ordered water molecule. The M2 site is fashioned from the side chains of Gln268, Glu50, and Glu470, as well as from oxygen atoms from the β phosphate of ADP and the phosphoryl group of the transition state analogue. The M3 site is in contact with oxygen atoms from each of the three phosphate groups, the carboxylates of Glu50 and Glu103, and a bound water molecule. This constellation of Mg2+-binding sites facilitates the binding of ATP and positions the γ-phosphate of ATP for in-line nucleophilic attack by the γ-carboxylate of the glutamate substrate. As mentioned above, Arg472 is likely a key residue in this initial step of catalysis.
The coordination of these critical Mg2+ ions appears to be highly conserved. A similar arrangement of active site Mg2+ ions is observed in the equivalent E. coli GCL structure (40), despite less than 10% sequence identity between the Group 1 and 2 enzymes. Mutation of glutamate residues 55 and 100 in T. brucei GCL (equivalent to Glu52 and Glu103 in ScGCL) to alanine led to a striking loss of enzyme activity, suggesting that Glu52 and Glu103 are indispensable for catalysis (42). Substitutions at either residue likely result in the loss of Mg2+ binding at the M1 site. Interestingly, mutation of Glu93 in T. brucei GCL to alanine (equivalent to Glu96 in ScGCL) resulted in an enzyme capable of ATP hydrolysis. However, the E93A mutant could not catalyze the peptide bond formation between l-glutamate and l-aminobutyrate (a surrogate for l-cysteine), suggesting that this glutamate residue may instead facilitate the nucleophilic attack of l-cysteine on the γ-glutamylphosphate intermediate (42).
Identification of the Cysteine-binding Site of ScGCL
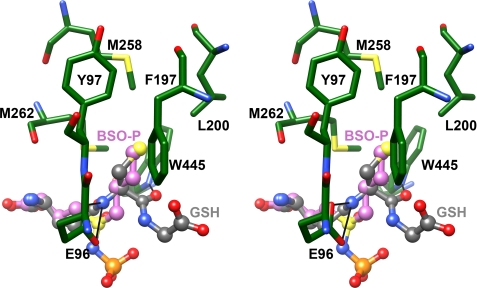
Attempts to crystallize a pseudo-Michaelis complex have been unsuccessful. In each case, the electron density for the cysteine substrate has been quite poor, precluding the direct identification of the cysteine-binding pocket. To overcome this limitation, the ScGCL-glutathione and ScGCL-BSO structures were superimposed, and the environment surrounding the cysteine or cysteine mimic was examined (Fig. 5). The thiol group of glutathione and the S-butyl group of BSO overlay reasonably well and are located in a hydrophobic pocket lined by Tyr97, Phe197, Leu200, Met258, Met262, and Trp445. The cysteinyl amino and carbonyl groups of glutathione are within hydrogen bond distance of the γ-carboxylate of Glu96 and the indole nitrogen of Trp445. These two residues likely orient the incoming cysteine substrate, and Glu96 may facilitate the nucleophilic attack of the α-amino group of cysteine on the γ-glutamylphosphate, leading to the displacement of the phosphate group and the formation of the γ-glutamyl peptide bond (1–4). In addition, the guanidinium group of Arg196 may coordinate the α-carboxylate of cysteine, similar to the arrangement seen for binding of the glycine portion of glutathione (Fig. 3). In support of this assertion, mutagenesis studies of T. brucei GCL indicated that Arg179 (Arg196 in ScGCL) is required for efficient binding of the cysteine analogue, l-aminobutyrate (39).
FIGURE 5.
Superpositioning of the glutathione and BSO binding sites indicate the location of the cysteine-binding site. Bound ADP and BSO are shown in ball and stick representation, and pertinent active site residues are shown in stick representation in the stereodiagram. Atoms are colored as in Fig. 2, with the exception of carbon atoms in BSO (colored in magenta). The S-butyl group of BSO and the thiol group of glutathione occupy a comparable hydrophobic pocket in the ScGCL active site.
Comparison of the ScGCL-BSO structure with that of E. coli GCL in complex with the related mechanism-based inhibitor, (2S)-2-amino-4-[(2S)-2-carboxybutyl-(R)-sulfonimidoyl]butanoic acid (40), suggests potential differences in cysteine binding. In E. coli GCL, the cysteine pocket is formed concurrently with a conformation change in a switch loop (residues 240–249). As a result, the carboxyl group of the inhibitor cysteine moiety is positioned to form a hydrogen bond with Tyr300, as well as Tyr131. In ScGCL there are no significant conformational changes in the backbone of the enzyme upon inhibitor binding, and Tyr300 and Tyr131 of the E. coli enzyme appear to be functionally replaced by Trp445 and Arg196. Interestingly, the cysteine-binding pocket of B. juncea GCL (41) more closely resembles that of ScGCL. However, both enzyme structures were determined in complex with BSO, which lacks the functional equivalent of the α-carboxylate of the cysteine substrate. Perhaps additional conformational changes would occur if this moiety were present.
Implications for Catalysis and Inhibitor Design
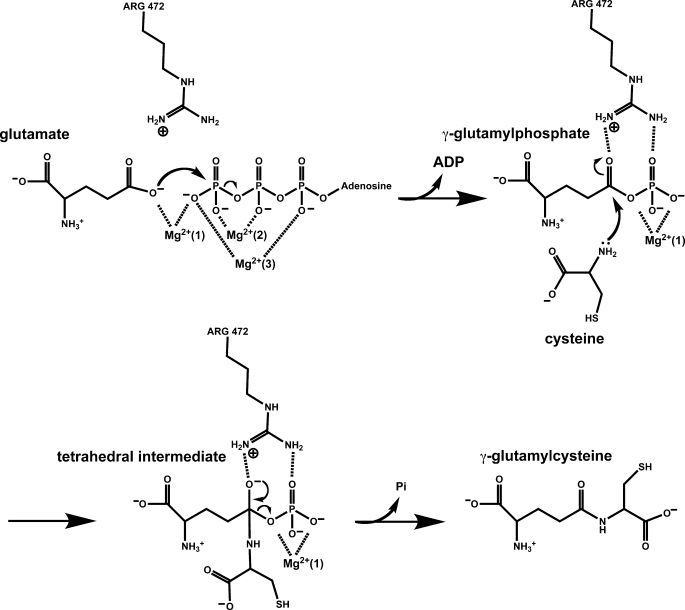
The available biochemical and structural data provide many of the details of the catalytic mechanism of the enzyme (Fig. 6). Glutamate binds at the base of the enzyme active site with its side chain carboxylate occupying one of the coordination sites of the M1 Mg2+. The nucleophilicity of the γ-carboxylate is likely increased by the adjacent Mg2+ as well as the side chain of Arg472. The addition of Mg2+/ATP leads to the formation of two additional magnesium-binding sites, M2 and M3, which orient the phosphate groups of ATP, placing the γ-phosphate in position for in-line attack by the activated glutamate substrate. This leads to the formation of a γ-glutamyl phosphate intermediate, which is tightly anchored in the enzyme active site, and the eventual displacement of ADP. The incoming cysteine nucleophile is potentially activated by the side chain carboxylate of Glu96, and the developing negative charge on the γ-carboxylate oxygen of the glutamate substrate is stabilized by the side chain of Arg472. Collapse of the tetrahedral intermediate leads to the expulsion of the phosphate group and the formation of the γ-glutamyl peptide bond. Additional biochemical and mutational studies to examine this proposed mechanism are ongoing. However, the essential features of catalysis appear to be conserved in related enzymes such as glutamine synthetase (43, 44), glutathione synthetase (45–47), and homoglutathione synthetase (48).
FIGURE 6.
Proposed Catalytic mechanism of ScGCL. The proposed catalytic mechanism depicted is based on available biochemical and structural data for ScGCL as discussed in the text. Arg472 of ScGCL, the residue proposed to stabilize the anionic transition state, is also shown. Additional biochemical and kinetic studies will be required to validate the mechanism, particularly with regard to activation of the nucleophilic cysteine.
Elucidation of the detailed catalytic mechanism of GCL in conjunction with the structural studies of the inhibited ScGCL may lead to improved glutathione biosynthesis inhibitors. The alkyl sulfoximine-based inhibitors are excellent transition state mimics that dramatically reduce enzymatic activity. Examination of the ScGCL-BSO complex suggests that additional functionalities may be engineered to increase selectivity. ScGCL and human GCL share >40% sequence identity, with nearly complete conservation of active site architecture (24), suggesting that the insights garnered from the study of ScGCL will facilitate the development of improved therapeutics that modulate glutathione production in mammalian systems.
Supplementary Material
Acknowledgments
We thank the BioCARS staff for assistance in x-ray data collection, Dr. Mark Wilson (University of Nebraska) for helpful discussions, and Dr. Melanie Simpson (University of Nebraska) for thoughtful insights and review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R01 GM077289 (to J. J. B.) and RR07707. This work was also supported by United States Department of Energy, Basic Energy Sciences, Office of Science Contract W-31-109-Eng-38.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 3LVV and 3LVW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GCL
- glutamate cysteine ligase
- ScGCL
- S. cerevisiae GCL
- BSO
- l-buthionine-S,R-sulfoximine
- MES
- 2-(N-morpholino)ethanesulfonic acid.
REFERENCES
- 1.Griffith O. W., Mulcahy R. T. (1999) Adv. Enzymol. Relat. Areas Mol. Biol. 73, 209–267, xii [DOI] [PubMed] [Google Scholar]
- 2.Orlowski M., Meister A. (1971) J. Biol. Chem. 246, 7095–7105 [PubMed] [Google Scholar]
- 3.Strumeyer D. H., Bloch K. (1960) J. Biol. Chem. 235, PC27. [PubMed] [Google Scholar]
- 4.Yip B., Rudolph F. B. (1976) J. Biol. Chem. 251, 3563–3568 [PubMed] [Google Scholar]
- 5.Copley S. D., Dhillon J. K. (2002) Genome Biol. 3, research0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tateishi N., Higashi T., Shinya S., Naruse A., Sakamoto Y. (1974) J. Biochem. 75, 93–103 [DOI] [PubMed] [Google Scholar]
- 7.Wild A. C., Mulcahy R. T. (1999) Biochem. J. 338, 659–665 [PMC free article] [PubMed] [Google Scholar]
- 8.Lu S. C. (2009) Mol. Aspects Med. 30, 42–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richman P. G., Meister A. (1975) J. Biol. Chem. 250, 1422–1426 [PubMed] [Google Scholar]
- 10.Huang C. S., Anderson M. E., Meister A. (1993) J. Biol. Chem. 268, 20578–20583 [PubMed] [Google Scholar]
- 11.Huang C. S., Chang L. S., Anderson M. E., Meister A. (1993) J. Biol. Chem. 268, 19675–19680 [PubMed] [Google Scholar]
- 12.Fraser J. A., Saunders R. D., McLellan L. I. (2002) J. Biol. Chem. 277, 1158–1165 [DOI] [PubMed] [Google Scholar]
- 13.Misra I., Griffith O. W. (1998) Protein Expr. Purif. 13, 268–276 [DOI] [PubMed] [Google Scholar]
- 14.Godwin A. K., Meister A., O'Dwyer P. J., Huang C. S., Hamilton T. C., Anderson M. E. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3070–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulcahy R. T., Bailey H. H., Gipp J. J. (1994) Cancer Chemother. Pharmacol. 34, 67–71 [DOI] [PubMed] [Google Scholar]
- 16.Mulcahy R. T., Bailey H. H., Gipp J. J. (1995) Cancer Res. 55, 4771–4775 [PubMed] [Google Scholar]
- 17.Anderson M. E. (1998) Chem. Biol. Interact 111–112, 1–14 [DOI] [PubMed] [Google Scholar]
- 18.Meister A., Anderson M. E. (1983) Annu. Rev. Biochem. 52, 711–760 [DOI] [PubMed] [Google Scholar]
- 19.Townsend D. M., Tew K. D. (2003) Oncogene 22, 7369–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffith O. W. (1982) J. Biol. Chem. 257, 13704–13712 [PubMed] [Google Scholar]
- 21.Arrick B. A., Griffith O. W., Cerami A. (1981) J. Exp. Med. 153, 720–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lüersen K., Walter R. D., Müller S. (2000) Biochem. J. 346, 545–552 [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell E. B., Hayward M. L., Griffith O. W. (1991) Anal. Biochem. 194, 268–277 [DOI] [PubMed] [Google Scholar]
- 24.Biterova E. I., Barycki J. J. (2009) J. Biol. Chem. 284, 32700–32708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jez J. M., Cahoon R. E., Chen S. (2004) J. Biol. Chem. 279, 33463–33470 [DOI] [PubMed] [Google Scholar]
- 26.Rodgers D. W. (1994) Structure 2, 1135–1140 [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta. Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K. (2004) Acta. Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta. Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 31.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 33.Huang C. S., Moore W. R., Meister A. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 2464–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mårtensson J., Jain A., Stole E., Frayer W., Auld P. A., Meister A. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9360–9364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brekken D. L., Phillips M. A. (1998) J. Biol. Chem. 273, 26317–26322 [DOI] [PubMed] [Google Scholar]
- 36.Griffith O. W., Meister A. (1979) J. Biol. Chem. 254, 7558–7560 [PubMed] [Google Scholar]
- 37.Griffith O. W. (1999) Free Radic. Biol. Med. 27, 922–935 [DOI] [PubMed] [Google Scholar]
- 38.Rappa G., Gamcsik M. P., Mitina R. L., Baum C., Fodstad O., Lorico A. (2003) Eur. J. Cancer 39, 120–128 [DOI] [PubMed] [Google Scholar]
- 39.Abbott J. J., Ford J. L., Phillips M. A. (2002) Biochemistry 41, 2741–2750 [DOI] [PubMed] [Google Scholar]
- 40.Hibi T., Nii H., Nakatsu T., Kimura A., Kato H., Hiratake J., Oda J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15052–15057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hothorn M., Wachter A., Gromes R., Stuwe T., Rausch T., Scheffzek K. (2006) J. Biol. Chem. 281, 27557–27565 [DOI] [PubMed] [Google Scholar]
- 42.Abbott J. J., Pei J., Ford J. L., Qi Y., Grishin V. N., Pitcher L. A., Phillips M. A., Grishin N. V. (2001) J. Biol. Chem. 276, 42099–42107 [DOI] [PubMed] [Google Scholar]
- 43.Liaw S. H., Eisenberg D. (1994) Biochemistry 33, 675–681 [DOI] [PubMed] [Google Scholar]
- 44.Krajewski W. W., Jones T. A., Mowbray S. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10499–10504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrera K., Cahoon R. E., Kumaran S., Jez J. (2007) J. Biol. Chem. 282, 17157–17165 [DOI] [PubMed] [Google Scholar]
- 46.Gogos A., Shapiro L. (2002) Structure 10, 1669–1676 [DOI] [PubMed] [Google Scholar]
- 47.Polekhina G., Board P. G., Gali R. R., Rossjohn J., Parker M. W. (1999) EMBO J. 18, 3204–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galant A., Arkus K. A., Zubieta C., Cahoon R. E., Jez J. M. (2009) Plant Cell 21, 3450–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.