Abstract
The c-KIT receptor tyrosine kinase is constitutively activated and oncogenic in the majority of gastrointestinal stromal tumors. The identification of selective inhibitors of c-KIT, such as imatinib, has provided a novel therapeutic approach in the treatment of this chemotherapy refractory tumor. However, despite the clinical importance of these findings and the potential it provides as a model system for understanding targeted therapy, this approach has not yielded curative outcomes in most patients, and the biochemical pathways connecting c-KIT inhibition to cell death are not completely understood. Here, we show that inhibition of c-KIT with imatinib in gastrointestinal stromal tumors (GISTs) triggered the up-regulation of the proapoptotic protein BIM via both transcriptional and post-translational mechanisms. The inhibition of c-KIT by imatinib increased levels of the dephosphorylated and deubiquitinated form of BIM as well as triggered the accumulation of the transcription factor FOXO3a on the BIM promoter to activate transcription of BIM mRNA. Furthermore, using RNA interference directed against BIM, we demonstrated that BIM knockdown attenuated the effects of imatinib, suggesting that BIM functionally contributes to imatinib-induced apoptosis in GIST. The identification and characterization of the pathways that mediate imatinib-induced cell death in GIST provide for a better understanding of targeted therapy and may facilitate the development of new therapeutic approaches to further exploit these pathways.
Keywords: Apoptosis, Diseases/Cancer/Therapy, Oncogene, Receptors/Tyrosine Kinase, Signal Transduction, Tumor/Oncogene
Introduction
Therapies that selectively target essential molecular pathways of cancer cells possess the potential to increase the effectiveness of therapy while decreasing the side effects associated with traditional cytotoxic chemotherapy. One example that has evoked considerable interest in both the laboratory and clinic is the identification in GI stromal tumors of activating mutations in the c-KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog) receptor tyrosine kinase gene coupled with the development of targeted c-KIT inhibitors such as imatinib (1). Gastrointestinal stromal tumors (GISTs)2 are the most common mesenchymal tumor of the gastrointestinal tract, and ∼80% of GISTs harbor activating c-KIT mutations (2). While surgical resection of localized disease can be curative, GISTs are poorly responsive to cytotoxic chemotherapy, and for metastatic disease, there were few therapeutic options prior to the advent of targeted c-KIT therapies (2). However, although ∼80% of patients with metastatic GIST respond to imatinib therapy, responses are rarely complete, and patients eventually progress on therapy, often due to the development of secondary, imatinib-resistant c-KIT mutations (3).
Evidence suggests that activated c-KIT drives GIST cell survival through multiple pathways and that the specific c-KIT mutation type may influence the degree of activation of the different downstream signal transduction pathways (4). Accordingly, the key downstream mediators of imatinib-induced cell death in GISTs remain incompletely understood. While imatinib inhibition of c-KIT has been shown to down-regulate survival pathways downstream from c-KIT, such as PI3K-AKT and MAPK (5), other effects of imatinib have also been identified such as soluble histone H2AX up-regulation (6), quiescence activation through modulation of the CDH1-SKP2-p27 signaling axis (7), and possibly autophagy (8). Identifying and characterizing the pathways that mediate imatinib-induced cell death may facilitate the development of synergistic therapies to increase the effectiveness of imatinib as well as salvage therapies to overcome imatinib resistance.
Although there are undoubtedly a number of different biochemical mediators of imatinib-induced cell death, a wide variety of cellular stress or damage signals can converge on the programmed cell death or apoptotic pathway. The intrinsic apoptotic pathway is controlled by the interactions of pro- and antiapoptotic members of the BCL-2 family of proteins and culminates with mitochondrial membrane permeabilization, release of cytochrome c, and activation of caspases (9). We hypothesized that modulating this balance between the pro- and antiapoptotic members of the intrinsic mitochondrial pathway may also be important for the mechanism of action of imatinib in GIST therapy.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
UO126 was purchased from Calbiochem, and imatinib was purchased from LC Laboratories. The pCMV6.BIM-FLAG vector was obtained from Origene. The GIST 882 and 48 cell lines (kindly provided by J. A. Fletcher, Brigham and Women's Hospital, Boston, MA), were cultured as described previously (1, 5).
siRNA Transfection
BIM, Bad, and control (nontargeting) siRNAs were obtained from Dharmacon. Subconfluent GIST 882 cells in 12-well plates were transfected with 5 nm siRNA in RPMI 1640 medium with Hiperfect transfection reagent (Qiagen). After 4 h of incubation, the medium was changed back to the complete culture medium but with only 5% fetal bovine serum. Imatinib or DMSO was added 24 h after transfection, and after the prescribed incubation period, either whole-cell lysates were prepared for Western blot analysis, or cell viability was assessed with the CellTiter-Glo luminescent assay (Promega).
BIM Overexpression
GIST 882 and 48 cells were transfected with pCMV6.BIM-FLAG vector using Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions. After 36 h, cell viability was assessed with the CellTiter-Glo luminescent assay (Promega).
Quantitative Reverse Transcription-PCR
Total RNAs were isolated using the RNeasy Plus mini kit (Qiagen). Quantitative real time reverse transcription-PCR was performed with 100 ng total RNA, Quantiscript reverse transcriptase (Qiagen), and iQ SYBR Green Supermix (Bio-Rad) on an ABI 7500 Fast Real Time PCR system. The primers used were 5′-TTGTGGCTCTGTCTGTAGGGAGGTA-3′ (sense) and 5′-GTTCTGAGTGTGACCGAGAAGGTA-3′ (antisense) for BIM and 5′-CTCCATCATGAAGTGTGACGTGGA-3′ (sense) and 5′-CAGGAAAGACACCCACCTTGATCT-3′ (antisense) for β-actin. All reactions were run in triplicate, and the relative expression of BIM was calculated by normalizing BIM mRNA expression to β-actin mRNA expression.
Proliferation and Apoptosis Assays
GIST 882 cells, transfected with BIM or control siRNA as described above, were grown in 96-well plates. After incubation with imatinib or DMSO for the prescribed amount of time, the cells were washed with phosphate-buffered saline, and cell viability was determined using the CellTiter-Glo luminescent assay (Promega) according to the manufacturer's instructions. To assess apoptosis, GIST 882 cells were treated with imatinib or DMSO for the prescribed period of time. Cells were then trypsinized and subsequently stained with annexin V-FITC and propidium iodide and analyzed on a FACSCalibur machine. Viable cells were annexin V- and propidium iodide-negative.
Western Blotting
Whole-cell lysis was performed at 4 °C for 15 min. Proteins were separated by SDS-PAGE and then transferred to nitrocellulose membranes. Antibodies were used to detect the following proteins: BIM (BD Pharmingen), PUMA (Cell Signaling), NOXA (Cell Signaling), BID (Cell Signaling), Bad (Cell Signaling), MCL-1 (Santa Cruz Biotechnology), BCL-2 (Cell Signaling), BCL-xL (Cell Signaling), BAX (Cell Signaling), tubulin (Sigma), cleaved poly(ADP-ribose) polymerase (Cell Signaling), FKHRL1/FOXO3a (Millipore), phospho-FKHRL1/FOXO3a (Ser253; Millipore), phospho-FKHRL1/FOXO3a (Ser32; Millipore), phospho-BIM (Ser69; Cell Signaling), p44/42 MAPK (Erk 1/2; Cell Signaling), phospho-p44/42 MAPK (Erk1/2, Thr202/Tyr204; Cell Signaling), ubiquitin (Cell Signaling), and FLAG (Sigma).
Co-immunoprecipitation of BIM
As described in Ref. 10, whole-cell lysates (∼1000 μg) were incubated for 4 h at 4 °C with 2.5 μg of either anti-BIM antibody (BD Pharmingen) or normal rabbit IgG in 200 μl of immunoprecipitation lysis buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 10 mm sodium phosphate, pH 7.2) containing Protein A/G-agarose (Thermo Fisher Scientific). The immune complexes were collected by centrifugation and washed five times with immunoprecipitation lysis buffer. Western blots were performed as described above, with an antibody directed against ubiquitin (Cell Signaling).
Chromatin Immunoprecipitation Assay
GIST 882 cells were grown in 10-cm plates until they were ∼95% confluent and then either treated with 1 μm imatinib for 48 h or left untreated. Cells were fixed with formalin, and chromatin immunoprecipitation was performed with an antibody directed against FOXO3a as described (11). Subsequent PCR with primers for either BIM promoter or downstream BIM coding sequence, as a control for DNA fragmentation size, was also performed as described (11).
RESULTS
Up-regulation of BIM in a GIST Cell Line Sensitive to Imatinib
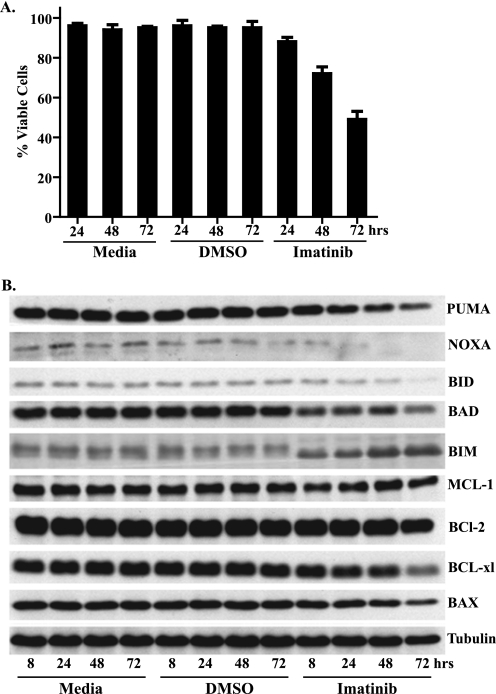
In agreement with previous studies (1, 5), inhibition of c-KIT with imatinib induced apoptosis in a time-dependent fashion in the GIST 882 cell line, which harbors a homozygous, imatinib-sensitive K642E c-KIT mutation (Fig. 1A). To investigate the role of the intrinsic apoptotic pathway in this response to imatinib, we examined the expression of a number of different pro- and antiapoptotic BCL-2 family member proteins following treatment with imatinib or DMSO control (Fig. 1B). Notably, the proapoptotic factor BIM showed a rapid and sustained up-regulation following imatinib treatment. In addition to increased expression, the relative electrophoretic migration of BIM also increased, with a striking change already observed at the earliest time point analyzed (8 h; Fig. 1B). No other apoptotic factor that we probed exhibited as significant a perturbation following imatinib treatment (Fig. 1B and data not shown). Similar up-regulation of BIM was observed following treatment of the GIST 48 cell line with imatinib (supplemental Fig. S1A). The GIST 48 cell line exhibits moderate sensitivity to imatinib as it contains a homozygous, imatinib-sensitive, exon 11 mutation and a heterozygous, imatinib-resistant, exon 17 mutation (5).
FIGURE 1.
Effect of imatinib on GIST 882 cells. A, cell viability following treatment with medium, DMSO, or 1 μm imatinib was assessed by annexin V-FITC and propidium iodide staining followed by flow cytometric analysis. Viable cells lacked annexin V-FITC, propidium iodide, or both annexin V-FITC and propidium iodide staining. Data are mean (±S.D.) of three experiments. B, Western blot analysis of whole-cell lysates prepared from GIST 882 cells following treatment with medium, DMSO, or 1 μm imatinib. Blots were probed with antibodies specific to a number of both pro- and antiapoptotic factors.
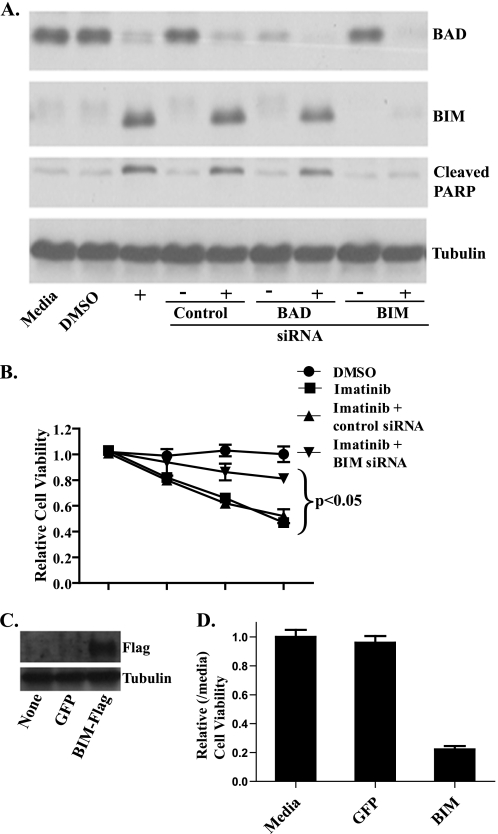
BIM Contributes to Imatinib-induced Apoptosis in GIST 882
We next examined the effects of BIM knockdown on imatinib-induced cell death. Knockdown of another BCL-2 family member, Bad, was also performed as a control as it is another proapoptotic factor but did not show up-regulation by Western blot analysis (Fig. 1B). The siRNAs directed against both BIM and Bad gave efficient knockdown of their respective targets, although despite optimization, slight up-regulation of BIM still occurred following imatinib treatment. After siRNA transfection, the GIST 882 cells were treated with imatinib and assessed for apoptosis by immunoblotting for poly(ADP-ribose) polymerase cleavage (Fig. 2A) and for viability by an ATP-based proliferation assay (Fig. 2B). Imatinib-induced effects on apoptosis and cell proliferation were both attenuated by BIM knockdown (Fig. 2, A and B), suggesting that BIM up-regulation is measurably contributing to imatinib-induced apoptosis in GIST 882. Similar results were obtained with the GIST 48 cell line (supplemental Fig. S1, B and C). The lack of complete abrogation of imatinib-induced effects may, in part, be due to the slight imatinib-induced up-regulation of BIM despite efficient knockdown but also indicates that other mechanisms besides BIM may be important for imatinib-induced cell death.
FIGURE 2.
BIM knockdown by siRNA protects GIST 882 cells from imatinib-induced cell death, and, conversely, BIM overexpression induces cell death. A, GIST 882 cells were treated with control siRNA or siRNA targeting BIM or Bad, followed by treatment with 1 μm imatinib for 48 h prior to whole-cell lysis and Western blot analysis. Blots were probed with antibodies specific to Bad, BIM, tubulin (loading control), and cleaved poly(ADP-ribose) polymerase, a marker for apoptosis. B, GIST 882 cells were treated with siRNA and 1 μm imatinib as in A, and then cell viability was assayed using the CellTiter-Glo ATP luminescence assay after 24, 48, or 72 h. The data were normalized to untreated medium controls and represent mean values (±S.D.) of three experiments. p < 0.05 indicates a significant difference between BIM siRNA and imatinib compared with control siRNA and imatinib or imatinib without siRNA. C, Western blot analysis of whole-cell lysates prepared from GIST 882 cells 24 h following transfection with a BIM-FLAG or green fluorescent protein (GFP) control vector. Blots were probed with antibodies specific to FLAG and tubulin. D, GIST 882 cells were transfected with either BIM-FLAG or green fluorescent protein, and then cell viability was assayed using the CellTiter-Glo ATP luminescence assay after 36 h. The data were normalized to untreated, medium controls.
To further strengthen the role of BIM in imatinib-induced cell death, either BIM or green fluorescent protein, as a control, were overexpressed in GIST 882 and 48 cells and viability assessed after 36 h (Fig. 2C and supplemental Fig. S1, D and E). For both cell lines, BIM overexpression resulted in significant cell death relative to green fluorescent protein control (Fig. 2D and supplemental Fig. S1, D and E) and provides further evidence that BIM can cause apoptosis in GIST cell lines.
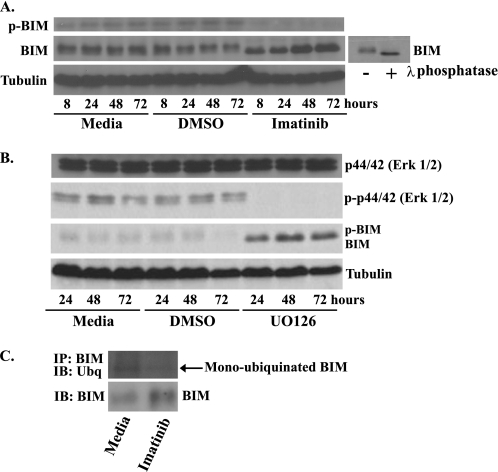
BIM Is Dephosphorylated and Deubiquitinated after Treatment with Imatinib
The MAPK and PI3K-AKT pathways can regulate BIM mRNA expression and protein stability, respectively (11, 12). It seemed likely that either pathway may be participating in imatinib-induced BIM up-regulation in GIST 882 as it has been previously shown that imatinib inhibition of c-KIT causes down-regulation of both the MAPK and PI3K-AKT pathways (5). Erk1/2, of the MAPK pathway, can phosphorylate BIM and target it for rapid proteasomal degradation (12, 13). Accordingly, the amount of phospho-BIM relative to total BIM significantly decreased following imatinib treatment of GIST 882 cells (Fig. 3A). Also consistent with the dephosphorylation of BIM, the relative electrophoretic migration of BIM increased following either imatinib treatment or treatment of whole-cell lysates with λ-phosphatase (Figs. 1B and 3A). Furthermore, treatment of GIST 882 with the MAPK pathway inhibitor UO126 caused a similar shift in the electrophoretic migration of BIM (Fig. 3B). Finally, BIM co-immunoprecipitation and Western blot analysis with ubiquitin antibody demonstrated that the amount of monoubiquitinated BIM decreases after 24 h of imatinib treatment, despite an overall increase in BIM levels (Fig. 3C). Proteosome inhibitors altered the level of c-KIT and, accordingly, were not used in this experiment (data not shown) (14). These data suggest that in GIST 882, the inhibition of c-KIT and subsequent down-regulation of the MAPK pathway increased levels of the dephosphorylated, deubiquitinated, and proteasome-resistant form of BIM.
FIGURE 3.
BIM is dephosphorylated and deubiquitinated after treatment with imatinib in GIST 882 cells. Western blot analysis of whole-cell lysates prepared from GIST 882 cells following treatment with medium, DMSO, 1 μm imatinib (A) or 10 μm MAPK inhibitor UO126 (B). Blots were probed with antibodies specific to BIM, phospho-BIM, phospho-Erk1/2, Erk1/2, and tubulin (loading control). The small blot shows a GIST 882 whole-cell lysate that was treated with λ-phosphatase to generate a standard for the faster migrating, dephosphorylated form of BIM. C, GIST 882 cells were treated for 24 h with 1 μm imatinib, and then BIM was immunoprecipitated (IP) from cell lysates. The degree of BIM ubiquitination was determined by Western blot (IB) followed by probing with an anti-ubiquitin (Ubq) antibody. The amount of BIM in the original cell lysates was also determined by Western blotting.
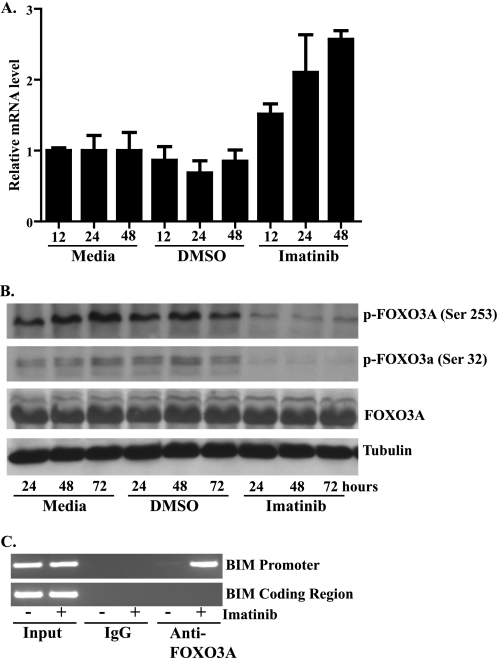
BIM mRNA Expression Increases after Treatment with Imatinib
Quantitative reverse transcription-PCR demonstrated that BIM up-regulation following imatinib treatment also occurred at the mRNA level (Fig. 4A). It has been previously shown that when the transcription factor FOXO3a is phosphorylated, it is exported from the nucleus to the cytoplasm, resulting in the down-regulation of the transcription of target genes including BIM (15). As imatinib inhibition of c-KIT in GIST causes down-regulation of the PI3K-AKT pathway (5), which, in turn, can phosphorylate FOXO3a, we evaluated whether the FOXO3a transcription factor could be involved in the induction of BIM. Treatment of GIST 882 with imatinib significantly decreased levels of the inactive, phosphorylated form of FOXO3a (Fig. 4B). Furthermore, the unchanged levels of total FOXO3a suggest an increase in the dephosphorylated, active form of FOXO3a. To confirm this increase in active FOXO3a, we next performed chromatin immunoprecipitation experiments with anti-FOXO3a antibody both before and after treatment with imatinib. As shown in Fig. 4C, treatment of GIST 882 with imatinib greatly enhanced the efficiency with which the anti-FOXO3a antibody, but not the IgG control, selectively precipitated the region of the BIM promoter containing the FOXO-binding site. These data suggest that imatinib causes the accumulation of FOXO3a on the BIM promoter to activate transcription.
FIGURE 4.
Transcriptional regulation of BIM by FOXO3a contributes to BIM up-regulation following imatinib treatment in GIST 882. A, quantitative reverse transcription-PCR analysis of total RNA isolated from GIST 882 cells following treatment with medium, DMSO, or 1 μm imatinib. BIM mRNA levels were normalized to β-actin. Reactions were performed in triplicate, and data represent mean (±S.D.) of three experiments. B, Western blot analysis of whole-cell lysates prepared from GIST 882 cells following treatment with medium, DMSO, or 1 μm imatinib. Blots were probed with antibodies specific to FOXO3a, phospho-FOXO3a (Ser253), phospho-FOXO 3a (Ser32), or tubulin (loading control). C, chromatin immunoprecipitation assay of GIST 882 cells after 1 μm imatinib treatment for 48 h. Chromatin fragments were immunoprecipitated with antibodies to FOXO3a or IgG (nonspecific control), and DNA was amplified by PCR with primers specific to either the BIM promoter or BIM coding region, to control for chromatin size.
DISCUSSION
c-KIT, a member of the type III receptor tyrosine kinase family, is important for hematopoiesis, melanogenesis, and gametogenesis as well as the development of the interstitial cells of Cajal, which are believed to be the nonmalignant precursor cells of GIST (16). Upon ligand binding, c-KIT dimerizes, undergoes autophosphorylation, and activates a variety of downstream signaling pathways and molecules, including MAPK, PI3K-AKT, Src, and JAK/STAT (17). Dysregulated signaling by c-KIT has been implicated in a number of cancers including GISTs, mastocytosis, germ cell tumors, a subset of melanomas, and acute myelogenous leukemia (17). Targeted inhibition of c-KIT by imatinib has provided a novel therapeutic approach in the treatment of metastatic GIST. However, despite the importance of c-KIT in a variety of cancers and the potential it provides as a model system for understanding targeted therapy, the biochemical pathways connecting c-KIT inhibition to cell death are incompletely understood.
In this study, we have demonstrated that inhibition of constitutively activated c-KIT by imatinib in GIST triggers the up-regulation of the proapoptotic, BCL-2 protein family member BIM. Furthermore, the attenuation of imatinib-induced apoptosis following knockdown of BIM by siRNA supports a functional role for BIM in cell death. The lack of complete rescue following BIM knockdown is consistent with data demonstrating the importance of other pathways, such as soluble H2AX2 up-regulation (6), quiescence (7), and possibly autophagy (8) in the mechanism of action of imatinib in GIST. However, it seems likely that alternative therapies that abrogate c-KIT signaling by mechanisms unique from imatinib, for example, flavopiridol, which inhibits c-KIT transcription (18), and HSP 90 inhibitors, which target c-KIT protein stability (19), may also utilize BIM up-regulation to trigger apoptosis.
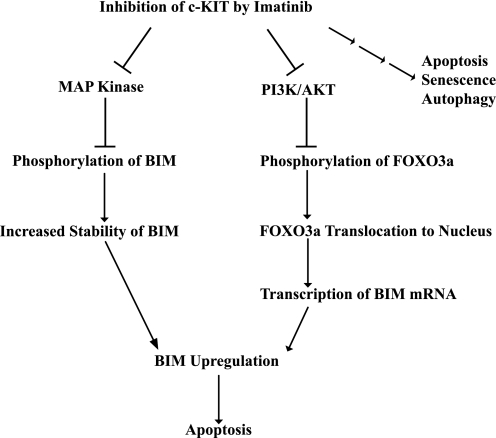
BIM exhibits multifaceted regulation at the levels of transcription, mRNA stability, post-translational modification, proteasomal degradation, and cellular localization (12, 20–23). In this study, we have also demonstrated that after c-KIT inhibition, BIM induction occurs secondary to both transcriptional and post-translational regulation (Fig. 5). Erk1/2 kinases have been shown to phosphorylate BIM and target it for ubiquitination and proteasomal degradation. In agreement, we found that imatinib inhibition increased the dephosphorylated and deubiquitinated form of BIM. In addition to the increase in BIM protein stability, we also found that imatinib induced BIM transcription through the PI3K-AKT-FOXO3a pathway. Upon c-KIT inhibition, the transcription factor FOXO3a is dephosphorylated, which facilitates its entry into the nucleus and localization to the BIM promoter. These modes of BIM regulation are in agreement with data previously showing that c-KIT inhibition caused down-regulation of the MAPK and PI3K-AKT pathways in GIST (5). Furthermore, in the mast cell lineage, both mutant and wild-type c-KIT promoted cell survival by BIM suppression (24, 25) via inhibition of FOXO3a-mediated BIM transcriptional up-regulation and MAPK phosphorylation of BIM (24).
FIGURE 5.
Model for one mechanism by which imatinib inhibition of c-KIT in GIST induces apoptosis. c-KIT inhibition caused the up-regulation of proapoptotic BIM secondary to both transcriptional and post-translational regulation that was mediated by the PI3K-AKT and MAPK pathways, respectively. The three small arrows on the far right of the figure indicate that imatinib effects in GIST are also mediated by other pathways (6–8).
The discovery that dysregulated, constitutively activated tyrosine kinases in cancers can be specifically inhibited has lead to significant advances in the field of cancer therapeutics (26). The results presented here complement and expand the current data on the mechanisms by which oncogenic tyrosine kinase inhibition triggers apoptosis. It has been previously shown that BIM also contributes to apoptosis following EGFR inhibition in nonsmall cell lung cancers (27–30) and BRAF V600E inhibition in melanoma (31–33). Similarly, in BCR/Abl leukemia cells, both BIM and Bad, another proapoptotic BCL-2 family member protein, are required for imatinib-induced cell death (34). As c-KIT in GIST is now another oncogenic tyrosine kinase whose effects of targeted inhibition are mediated significantly by BIM up-regulation, it would suggest that this may be a common mechanism for oncogenic tyrosine kinase inhibitor-induced apoptosis, although lineage-specific distinctions may also exist.
Understanding the common mechanisms of cell death utilized by targeted inhibitors such as imatinib is important for designing new inhibitors for other oncogenic kinases as well as for developing new therapies in the setting of secondary resistance to kinase inhibitors. Despite initial high response rates to imatinib in GIST, complete responses are rare, and disease in the majority of patients will eventually progress. The failure of imatinib can involve a number of different mechanisms, including the development of secondary, imatinib-resistant c-KIT mutations, c-KIT amplification, or the dependence on alternative pathways (35, 36). Furthermore, effective salvage therapies for patients with progressive disease are lacking. Similar acquired resistance has also been encountered in the targeted therapy of BCR/Abl in chronic myelogenous leukemia and EGFR in nonsmall cell lung cancer (26). Based on the data presented here, other strategies to enhance BIM expression in GIST may be effective in the setting of imatinib resistance or to augment the effectiveness of imatinib.
Supplementary Material
Acknowledgments
We thank Dr. J. A. Fletcher (Brigham and Women's Hospital, Boston, MA) for generously providing the GIST 882 and 48 cell lines. We also thank members of the Fisher laboratory for helpful discussions and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant AR043369-14. This work was also supported by the Melanoma Research Alliance (to D. E. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- GIST
- gastrointestinal stromal tumor
- siRNA
- small interfering RNA
- DMSO
- dimethyl sulfoxide
- FITC
- fluorescein isothiocyanate
- MAPK
- mitogen-activated protein kinase
- PI3K
- phosphoinositide 3-kinase.
REFERENCES
- 1.Tuveson D. A., Willis N. A., Jacks T., Griffin J. D., Singer S., Fletcher C. D., Fletcher J. A., Demetri G. D. (2001) Oncogene 20, 5054–5058 [DOI] [PubMed] [Google Scholar]
- 2.Quek R., George S. (2009) Hematol. Oncol. Clin. North Am. 23, 69–78 [DOI] [PubMed] [Google Scholar]
- 3.Verweij J., Casali P. G., Zalcberg J., LeCesne A., Reichardt P., Blay J. Y., Issels R., van Oosterom A., Hogendoorn P. C., Van Glabbeke M., Bertulli R., Judson I. (2004) Lancet 364, 1127–1134 [DOI] [PubMed] [Google Scholar]
- 4.Duensing A., Medeiros F., McConarty B., Joseph N. E., Panigrahy D., Singer S., Fletcher C. D., Demetri G. D., Fletcher J. A. (2004) Oncogene 23, 3999–4006 [DOI] [PubMed] [Google Scholar]
- 5.Bauer S., Duensing A., Demetri G. D., Fletcher J. A. (2007) Oncogene 26, 7560–7568 [DOI] [PubMed] [Google Scholar]
- 6.Liu Y., Tseng M., Perdreau S. A., Rossi F., Antonescu C., Besmer P., Fletcher J. A., Duensing S., Duensing A. (2007) Cancer Res. 67, 2685–2692 [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Perdreau S. A., Chatterjee P., Wang L., Kuan S. F., Duensing A. (2008) Cancer Res. 68, 9015–9023 [DOI] [PubMed] [Google Scholar]
- 8.Miselli F., Negri T., Gronchi A., Losa M., Conca E., Brich S., Fumagalli E., Fiore M., Casali P. G., Pierotti M. A., Tamborini E., Pilotti S. (2008) Transl. Oncol. 1, 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunelle J. K., Letai A. (2009) J. Cell Sci. 122, 437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi X. J., Wildey G. M., Howe P. H. (2006) J. Biol. Chem. 281, 813–823 [DOI] [PubMed] [Google Scholar]
- 11.Essafi A., Fernández de Mattos S., Hassen Y. A., Soeiro I., Mufti G. J., Thomas N. S., Medema R. H., Lam E. W. (2005) Oncogene 24, 2317–2329 [DOI] [PubMed] [Google Scholar]
- 12.Hübner A., Barrett T., Flavell R. A., Davis R. J. (2008) Mol. Cell 30, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luciano F., Jacquel A., Colosetti P., Herrant M., Cagnol S., Pages G., Auberger P. (2003) Oncogene 22, 6785–6793 [DOI] [PubMed] [Google Scholar]
- 14.Bauer S., Parry J. A., Muhlenberg T., Brown M. F., Seneviratne D., Chatterjee P., Chin A., Rubin B. P., Kuan S. F., Fletcher J. A., Duensing S., Duensing A. (2010) Cancer Res. 70, 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Z., Tindall D. J. (2008) Oncogene 27, 2312–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duensing A., Heinrich M. C., Fletcher C. D., Fletcher J. A. (2004) Cancer Invest. 22, 106–116 [DOI] [PubMed] [Google Scholar]
- 17.Lennartsson J., Rönnstrand L. (2006) Curr. Cancer Drug Targets 6, 65–75 [DOI] [PubMed] [Google Scholar]
- 18.Sambol E. B., Ambrosini G., Geha R. C., Kennealey P. T., Decarolis P., O'connor R., Wu Y. V., Motwani M., Chen J. H., Schwartz G. K., Singer S. (2006) Cancer Res. 66, 5858–5866 [DOI] [PubMed] [Google Scholar]
- 19.Bauer S., Yu L. K., Demetri G. D., Fletcher J. A. (2006) Cancer Res. 66, 9153–9161 [DOI] [PubMed] [Google Scholar]
- 20.Abrams M. T., Robertson N. M., Yoon K., Wickstrom E. (2004) J. Biol. Chem. 279, 55809–55817 [DOI] [PubMed] [Google Scholar]
- 21.Dijkers P. F., Medema R. H., Lammers J. W., Koenderman L., Coffer P. J. (2000) Curr. Biol. 10, 1201–1204 [DOI] [PubMed] [Google Scholar]
- 22.Ewings K. E., Wiggins C. M., Cook S. J. (2007) Cell Cycle 6, 2236–2240 [DOI] [PubMed] [Google Scholar]
- 23.Matsui H., Asou H., Inaba T. (2007) Mol. Cell 25, 99–112 [DOI] [PubMed] [Google Scholar]
- 24.Möller C., Alfredsson J., Engström M., Wootz H., Xiang Z., Lennartsson J., Jönsson J. I., Nilsson G. (2005) Blood 106, 1330–1336 [DOI] [PubMed] [Google Scholar]
- 25.Aichberger K. J., Gleixner K. V., Mirkina I., Cerny-Reiterer S., Peter B., Ferenc V., Kneidinger M., Baumgartner C., Mayerhofer M., Gruze A., Pickl W. F., Sillaber C., Valent P. (2009) Blood 114, 5342–5351 [DOI] [PubMed] [Google Scholar]
- 26.Sawyers C. L. (2005) Cold Spring Harb. Symp. Quant. Biol. 70, 479–482 [DOI] [PubMed] [Google Scholar]
- 27.Costa D. B., Halmos B., Kumar A., Schumer S. T., Huberman M. S., Boggon T. J., Tenen D. G., Kobayashi S. (2007) PLoS Med. 4, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cragg M. S., Kuroda J., Puthalakath H., Huang D. C., Strasser A. (2007) PLoS Med. 4, 1681–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng J., Shimamura T., Perera S., Carlson N. E., Cai D., Shapiro G. I., Wong K. K., Letai A. (2007) Cancer Res. 67, 11867–11875 [DOI] [PubMed] [Google Scholar]
- 30.Gong Y., Somwar R., Politi K., Balak M., Chmielecki J., Jiang X., Pao W. (2007) PLoS Med. 4, e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cartlidge R. A., Thomas G. R., Cagnol S., Jong K. A., Molton S. A., Finch A. J., McMahon M. (2008) Pigment Cell Melanoma Res. 21, 534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cragg M. S., Jansen E. S., Cook M., Harris C., Strasser A., Scott C. L. (2008) J. Clin. Invest. 118, 3651–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheridan C., Brumatti G., Martin S. J. (2008) J. Biol. Chem. 283, 22128–22135 [DOI] [PubMed] [Google Scholar]
- 34.Kuroda J., Puthalakath H., Cragg M. S., Kelly P. N., Bouillet P., Huang D. C., Kimura S., Ottmann O. G., Druker B. J., Villunger A., Roberts A. W., Strasser A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14907–14912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corless C. L., Fletcher J. A., Heinrich M. C. (2004) J. Clin. Oncol. 22, 3813–3825 [DOI] [PubMed] [Google Scholar]
- 36.Liegl B., Kepten I., Le C., Zhu M., Demetri G. D., Heinrich M. C., Fletcher C. D., Corless C. L., Fletcher J. A. (2008) J. Pathol. 216, 64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.