Abstract
The mammalian target of rapamycin (mTOR) protein kinase responds to diverse environmental cues to control a plethora of cellular processes. mTOR forms the catalytic core of at least two distinct signaling complexes known as mTOR complexes 1 and 2. Differing sensitivities to the mTOR inhibitor rapamycin, unique partner proteins, distinct substrates, and unique cellular functions distinguish the complexes. Here, we review recent progress in our understanding of the regulation and function of mTOR signaling networks in cellular physiology.
Keywords: Growth Factors, Serine/Threonine Protein Kinase, Signal Transduction, TOR, TOR Complex (TORC)
Introduction
The target of rapamycin (TOR),2 an evolutionarily conserved Ser/Thr protein kinase, forms the catalytic core of at least two functionally distinct complexes, TOR complex 1 (TORC1) and TOR complex 2 (TORC2) (1–6). These complexes contain shared and distinct partner proteins and control a myriad of cellular processes in response to diverse environmental cues. Whereas higher eukaryotes (mammals, flies, worms) contain only one TOR gene (e.g. mTOR, dTOR, ceTOR), yeast (Saccharomyces cerevisiae, Schizosaccharomyces pombe) contain two TOR genes, with Tor1 (or Tor2 in its absence) forming TORC1 and Tor2 forming TORC2 in S. cerevisiae (2, 7). The bacterial macrolide-derived rapamycin (clinically known as sirolimus) interacts with the cellular protein FKBP12, and this complex directly binds to the TOR FKBP12-rapamycin-binding (FRB) domain to allosterically inhibit TOR (8). During acute treatment, rapamycin inhibits assembled mammalian TORC1 (mTORC1) but not assembled mTORC2 (8, 9). Although the mechanism by which rapamycin inhibits mTORC1 remains incompletely defined, rapamycin weakens the interaction between mTOR and raptor (regulatory associated protein of mTOR), an mTORC1 regulatory partner (10), and reduces mTORC1 intrinsic kinase activity (11, 12). Chronic high-dose rapamycin inhibits mTORC2 signaling in certain cell types by impeding mTORC2 assembly, however (8, 9).
The clinical utility of rapamycin as an immunosuppressive agent and cardiology drug that reduces coronary artery stent restenosis underscores the importance of mTOR in organismal physiology. Additionally, many neoplasms, including tuberous sclerosis complex (TSC) tumors, exhibit aberrantly high mTOR signaling (8, 13, 14). Consequently, rapamycin analogs (13, 14) and novel second generation mTOR catalytic inhibitors (8, 15–18) hold therapeutic promise as anticancer agents. Although we have learned a great deal about mTORC1 signaling over the ∼15 years since the identification of TOR, much important biology remains to be deciphered regarding cellular mTORC1 and mTORC2 signaling networks, their functions in physiology and disease, and their potential roles as therapeutic targets. This minireview will focus on mTOR signaling networks (for more information on invertebrate TOR biology, see Ref. 7).
mTORC1 and mTORC2: Composition, Substrates, and Functions
mTORC Composition
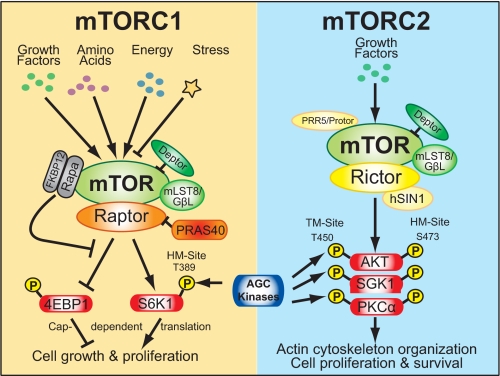
mTORC1 and mTORC2 contain shared and unique partners, the molecular functions of which remain poorly understood. Each complex contains mTOR, mLST8/GβL, and deptor (Fig. 1) (1–6). mLST8/GβL binds the mTOR kinase domain in both complexes but appears more critical for mTORC2 assembly and signaling (19). Deptor functions as an inhibitor of both complexes (4, 20). Other partner proteins distinguish the two complexes. mTORC1 contains exclusively raptor (Kog1 in budding yeast) and PRAS40. Raptor functions as a scaffolding protein that links the mTOR kinase with mTORC1 substrates to promote mTORC1 signaling. PRAS40 functions in an incompletely defined and controversial regulatory capacity as an mTORC1 inhibitor, competitive substrate, or both (3). Thus, disagreement exists in the field as to whether PRAS40 represents a core mTORC1 partner or an interacting substrate. In contrast, mTORC2 contains exclusively rictor (rapamycin-insensitive companion of mTOR) (Avo3 in S. cerevisiae), mSin1 (Avo1 in S. cerevisiae), and PRR5/protor (1–6). Rictor and mSin1 promote mTORC2 assembly and signaling; the function of PRR5/protor remains obscure.
FIGURE 1.
mTORC1 versus mTORC2. Distinct rapamycin sensitivities, partner proteins, substrates, and cellular functions distinguish the two known mTOR signaling complexes, mTORC1 and mTORC2.
mTORC1 Substrates and Functions
mTORC1 senses and integrates diverse extra- and intracellular signals to promote anabolic and inhibit catabolic cellular processes. Growth factors and nutrients (e.g. amino acids, energy) promote mTORC1-dependent protein synthesis, cell growth (increase in cell mass/size), cell proliferation, and cell metabolism (3, 21). Conversely, insufficient levels of these factors, or signals of cell stress, blunt mTORC1 action to maintain cellular biosynthetic rates appropriate for suboptimal cellular conditions (3, 21, 22). Reduced mTORC1 signaling also promotes macroautophagy, a degradative process that enhances cell survival in the face of decreased nutrient availability via the breakdown of cell constituents into amino acids and other small molecules (23). TORC1 in yeast and mammals also promotes “ribosome biogenesis,” a process whereby mTORC1 increases the transcription of ribosomal RNAs and proteins to augment cellular protein biosynthetic capacity (24).
Raptor binds directly to mTOR signaling (TOS) motifs on downstream targets, including S6K1 (ribosomal S6 protein kinase 1) and 4EBP1 (eukaryotic initiation factor (eIF) 4E-binding protein 1) (as well as PRAS40 and Hif1α), thus linking them to the mTOR kinase (2, 3, 25–27). The TOS motif is required for mTOR/raptor-mediated phosphorylation of S6K1 on its hydrophobic motif (HM) site (Thr389) and 4EBP1 on multiple sites (Thr37/46, Thr70, Ser65). Raptor mutation within its raptor N-terminal conserved domain abrogates 4EBP1 binding and mTORC1-mediated 4EBP1 phosphorylation in vitro while retaining mTOR interaction (28), thus underscoring the importance of the raptor-4EBP1 interaction for mTORC1 signaling. As the avidity of the raptor-mTOR interaction increases during nutrient and growth factor insufficiency (when mTORC1 signaling is low) (29–32), raptor may possess opposing cell condition-dependent functions in mTORC1 regulation.
Cell growth, cell cycle progression, and cell proliferation represent evolutionarily conserved TORC1 functions (33). Strikingly, mTORC1-dependent control of cell size extends to control of organ and organismal size (34). mTORC1-mediated signaling to S6K1 and 4EBP1 contributes to mTORC1-driven cell growth and cell cycle progression, as rapamycin-resistant S6K1 rescues these rapamycin-inhibited processes, whereas overexpression of a phosphorylation site-defective mutant of 4EBP1 or a TOS motif mutant of 4EBP1 dominantly inhibits these processes (25, 35, 36). Mechanisms underlying mTORC1-mediated inhibition of cell growth and proliferation remain incompletely defined but likely involve reduced protein synthesis. Indeed, the two best established substrates of mTORC1, S6K1 and 4EBP1, each control unique aspects of translation (21). Inactive S6K1 associates with the eIF3 translation initiation complex at the 5′-methylguanosine cap of mRNAs. Upon activation, mTORC1 is recruited to the eIF3 complex, whereby it directly phosphorylates S6K1 (at Thr389) to enhance translation via poorly defined mechanisms (21, 37). Substrates of S6K1 include other proteins possessing roles in translational control, including the ribosomal protein S6, eIF4B, eEF2K (eukaryotic elongation factor 2 kinase), PDCD4 (programmed cell death 4), CBP80 (cap-binding protein of 80 kDa), and SKAR (S6K1 Aly/REF-like target) (21). Upon activation, mTORC1 remains at the mRNA 5′-cap, where it is well positioned to phosphorylate 4EBP1 (on multiple sites, including Thr37/46, Thr70, and Ser65) (21). When hypophosphorylated, 4EBP1 functions as a translational repressor that binds and inhibits eIF4E, an initiation factor bound to the mRNA 5′-cap. mTORC1-mediated 4EBP1 phosphorylation induces 4EBP1 dissociation from eIF4E, allowing eIF4G and eIF4A to assemble with eIF4E, a complex known as eIF4F, to initiate cap-dependent translation. mTORC1 also promotes lipogenesis by increasing transcriptional peroxisome proliferator-activated receptor γ and sterol regulatory element-binding protein activities (38, 39). Thus, mTORC1 drives cell growth by coordinately promoting both protein and lipid synthesis.
mTORC1-mediated S6K1 phosphorylation and the subsequent S6K1-mediated phosphorylation of the 40 S ribosomal protein S6 were previously assumed to promote translation of 5′-terminal oligopyrimidine mRNAs, which encode ribosomal proteins and translation elongation factors. Thus, their translation prepares cells for high rates of protein synthesis. It is evident, however, that whereas mTOR contributes to 5′-terminal oligopyrimidine translation, the S6 kinases and phosphorylation of the 40 S ribosomal protein S6 do not, although S6 phosphorylation promotes cell, organ, and body size via unknown mechanisms (34).
mTORC2 Substrates and Functions
The serine/threonine protein kinase Akt (also known as protein kinase B) represents the first identified substrate of mTORC2 (40). Akt promotes cell proliferation, cell survival, and cell migration and controls various metabolic processes (41). Full activation of Akt in response to growth factor-mediated phosphatidylinositol 3-kinase (PI3K) signaling requires dual phosphorylation on its activation loop site (Thr308) by PDK1 and HM site (Ser473) by mTORC2 (4, 6, 41). mTORC2 also phosphorylates the HM sites on SGK1 (Ser422) and protein kinase Cα (PKCα; Ser657) (6, 42). As the mTORC1 substrate S6K1 and the mTORC2 substrates Akt, PKCα, and SGK1 represent AGC kinases, an emerging theme in mTOR signaling is that mTORC1 and mTORC2 phosphorylate members of the AGC kinase family (Fig. 1) (2, 6). mTORC2-mediated phosphorylation of the turn motif site (either directly or indirectly) in Akt (Thr450) as well as several PKCs appears to stabilize newly synthesized AGC kinases (6, 43, 44).
Control of actin cytoskeleton organization represents the first noted function of mTORC2 (12, 45). Recently, a role for mTORC2 in control of cell size and cell cycle progression was also reported (46). Recent data suggest that rather than controlling all cellular functions of Akt, mTORC2-mediated HM site phosphorylation may modulate substrate specificity (2, 14): whereas genetic knock-out of rictor, mSin1, or mLST8/GβL in mouse embryonic fibroblasts abrogated Akt Ser473 but not Thr308 phosphorylation, Akt-mediated phosphorylation of FoxO1/3a but not TSC2 or GSK3 was strongly reduced (19, 47). Thus, Akt may require phosphorylation of its HM site (Ser473) to phosphorylate FoxO1/3a but not TSC2 or GSK3. As compensation can occur in the context of chronic knock-out, it will be important to confirm that another AGC kinase does not substitute for Akt to mediate the phosphorylation of TSC2 and GSK3 in this cellular scenario.
Regulation of mTORC Signaling Networks
Diverse environmental cues, including growth factors (e.g. insulin, insulin-like growth factor 1, epidermal growth factor (EGF)) and nutrients (e.g. amino acids, energy), promote mTORC1 signaling (Fig. 2). Although many regulatory molecules have been defined (at least for mTORC1), many questions remain, including the direct molecular mechanisms underlying mTORC regulation.
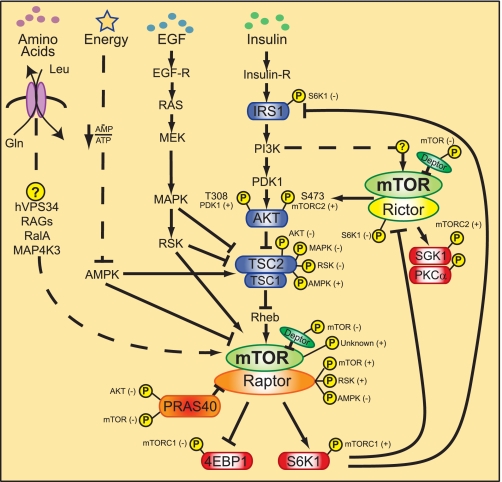
FIGURE 2.
Regulation of mTORC signaling networks. Growth factors/mitogens (insulin, EGF) and nutrients (e.g. amino acids, energy) promote mTORC1 signaling via phosphorylation cascades that converge on TSC and the mTORCs themselves. Insulin signals via its receptor (Insulin-R) to activate the PI3K/Akt/TSC/Rheb pathway; EGF signals via its receptor (EGF-R) to activate the Ras/MEK/MAPK/RSK pathway; amino acid sufficiency signals via hVps34 and the RAG and RalA GTPases; and energy sufficiency suppresses AMPK. Insulin/PI3K signaling likely promotes mTORC2 signaling via an unknown pathway. An mTORC1/S6K1-mediated negative feedback loop signals via two pathways to suppress PI3K/mTORC2/Akt signaling. Arrows versus blocked lines indicate activation or inhibition of protein function, respectively, by an upstream regulator. Phosphorylation events (denoted by circled yellow P) known to modulate protein function are shown. The kinases responsible for phosphorylation events are also indicated, with (+) or (−) denoting activation or inhibition of protein function, respectively.
Growth Factor/Mitogen Sensing by mTORC1
The tumor suppressor TSC, composed of TSC1 (hamartin) and TSC2 (tuberin), integrates diverse mTORC1 regulatory signals to suppress mTORC1 function (48). Genetic inactivation of either TSC1 or TSC2 thus results in constitutively high mTORC1 signaling and causes TSC, a pediatric disease characterized by benign tumors in diverse organ systems (48). TSC2 contains a GTPase-activating protein domain that converts the mTORC1 activator Rheb from a GTP-bound active state to a GDP-bound inactive state. Although the mechanism by which Rheb-GTP activates mTORC1 signaling remains poorly defined, Rheb-GTP provided in vitro augments mTORC1 kinase activity toward substrate (S6K1, 4EBP1) and may promote substrate binding (49, 50).
Growth factor signaling by PI3K and MAPK inhibits TSC function via distinct pathways to activate mTORC1. Insulin/insulin-like growth factor-mediated activation of class I PI3K leads to production of phosphatidylinositol (3,4,5)-trisphosphate on the plasma membrane, followed by recruitment and activation of Akt via cooperative phosphorylation by PDK1 (Thr308) and mTORC2 (Ser473) (2, 14, 41). Although current thinking suggests that activated Akt phosphorylates TSC2 at several sites (Ser939 and Thr1462) to inhibit TSC2 GTPase-activating protein activity, the data supporting such a widely embraced model remain weak (48). Of concern, mutation of the Akt phosphorylation sites on Drosophila TSC2 has no effect on tissue growth or viability (51). EGF-mediated activation of Ras leads to activation of MEK and thus MAPK and its substrate RSK. Both MAPK and RSK phosphorylate TSC2, leading to suppression of TSC function (48). Thus, growth factors activate mTORC1 signaling by suppressing TSC function via parallel PI3K-dependent and PI3K-independent pathways.
Although somewhat controversial, mitogen-stimulated production of the lipid second messenger phosphatidic acid via the enzyme phospholipase D promotes mTORC1 signaling via poorly defined mechanisms, possibly by binding the mTOR FRB domain (52). Moreover, phosphatidic acid may aid mTORC1 and mTORC2 complex assembly and compete with rapamycin for mTOR binding, providing a potential explanation for why cancer cells, which often possess high phospholipase D activity, display variable resistance to rapamycin (53, 54). Wnt ligand, another mTORC1-activating mitogen implicated in cancer, may promote mTORC1 signaling by inhibiting GSK3 because GSK3 phosphorylates TSC2 to augment its inhibitory action (55). Thus, TSC/Rheb integrates signals from diverse mitogens to coordinate mTORC1 action.
Energy, Nutrient, and Stress Sensing by mTORC1
Insufficient cellular energy levels decrease mTORC1 signaling. Elevation of the cellular AMP/ATP ratio, together with activation loop phosphorylation (at Thr172) by LKB1, activates AMP-activated protein kinase (AMPK), a master regulator of cellular energy metabolism (56). AMPK in turn phosphorylates TSC2 (at Ser1345 and potentially other sites) to augment TSC function and suppress mTORC1 signaling (48, 57). Thus, TSC2 phosphorylation either promotes or inhibits TSC function depending upon the sites of phosphorylation. Various forms of cell stress, including hypoxia, genotoxic stress, osmotic stress, and mechanical stress, also reduce mTORC1 signaling by augmenting TSC action (22).
Amino acid sufficiency represents the most ancestral and earliest identified activator of mTORC1 but remains the least understood. Withdrawal of amino acids, particularly the branched chain amino acids leucine and isoleucine, rapidly inhibits mTORC1 signaling, even in the face of abundant growth factors. As mTORC1 remains sensitive to amino acid levels in TSC-deficient cells (48, 58), this amino acid-regulated signal interacts with mTORC1 downstream of TSC. hVPS34, a class III PI3K known to function in vacuolar sorting and autophagy, represents the first identified link between amino acid sufficiency and mTORC1 activation (59, 60). Several GTPases (e.g. RAGs and RalA) have also been shown to promote mTORC1 signaling in response to amino acid sufficiency (61–63). A recent model posits that GTP-loaded RAG heterodimers bind raptor and induce the recruitment of mTORC1 from an ill defined cytoplasmic compartment to a Rab7-positive late endosomal membrane compartment that contains the mTORC1 activator Rheb (62). Such a model explains why growth factor-induced mTORC1 activation absolutely requires amino acids: if mTORC1 is in the wrong place (as during amino acid deprivation), it cannot be activated at the right time (by Rheb). More recently, RalA was reported to promote amino acid-induced mTORC1 activation downstream of Rheb (63), and amino acids were reported to activate the kinase MAP4K3 (64). The molecular mechanisms by which amino acid sufficiency translates to regulation of such signaling intermediates remain unknown, although the bidirectional transport of glutamine (out of the cell) and leucine (into the cell) by the permease SLC7A5-SLC3A2 seems essential (65).
mTORC2 Regulation?
Relative to mTORC1, the regulatory inputs to mTORC2 remain virtually unknown. As insulin signaling via PI3K promotes mTORC2-mediated Akt Ser473 phosphorylation (2, 14, 41) and as pharmacological inhibition of PI3K reduces mTORC2 kinase activity in vitro (66), PI3K presumably lies upstream of mTORC2. Interestingly, the TSC1-TSC2 complex may promote mTORC2 signaling, opposite to its inhibitory mTORC1 action (66).
Feedback Inhibition
Feedback loops complicate the study of mTORC regulation. mTORC1/S6K1 signaling mediates inhibitory Ser/Thr phosphorylation of insulin receptor substrate proteins, which uncouples insulin receptor substrate from PI3K and leads to reduced PI3K signaling (67). Thus, TSC-null cells, which bear constitutive mTORC1 signaling, exhibit attenuated PI3K signaling, which may explain the benign nature of TSC-null tumors (48, 68). As the mTORC1/S6K1 feedback loop also generates a state of cellular insulin resistance and as S6K1 knock-out in the mouse increases whole body insulin sensitivity (69), it is possible that constitutive mTORC1 signaling promoted by excess nutrients (as in states of obesity) may contribute to insulin resistance and type II diabetes (67, 70). Additionally, S6K1 phosphorylates rictor (at Thr1135) to reduce mTORC2 signaling (71–73). Thus, the mTORC1/S6K1 axis operates in at least two negative feedback loops to suppress PI3K and mTORC2.
Novel Insights Gained by Second Generation mTOR Catalytic Inhibitors
Whereas rapamycin completely inhibits mTORC1-mediated S6K1 phosphorylation, rapamycin reduces 4EBP1 phosphorylation, protein synthesis, cell growth, and cell proliferation only incompletely and variably, depending on cell type (8, 74, 75). Moreover, whereas rapamycin strongly promotes autophagy in yeast, it does so only modestly in mammalian cells. In contrast, novel second generation ATP-competitive mTOR inhibitors (e.g. Torin1, PP242, Ku-0063794, WAY-600) (8, 15–18), which inhibit both mTORC1 and mTORC2, profoundly inhibit 4EBP1 phosphorylation, protein synthesis, cell growth, and cell proliferation and strongly promote autophagy. These data suggest that mTOR-dependent but rapamycin-insensitive signaling controls these cellular processes. Although mTORC2 represents an obvious candidate, it does not appear to mediate these effects (15, 16). As raptor knockdown phenocopies the effect of mTOR catalytic inhibitors by ablating 4EBP1 phosphorylation (15), these data unexpectedly reveal that depending on substrate, rapamycin does not inhibit all mTOR/raptor-dependent functions and can signal in a rapamycin-resistant manner. Recently, RhoE overexpression was found to suppress 4EBP1 but not S6K1 phosphorylation (31). Perhaps RhoE functions as an inhibitor of rapamycin-insensitive mTORC1? These findings overturn the long-standing presumption in the field that rapamycin inhibits all mTORC1 function completely (15, 74, 75).
The therapeutic effects of mTORC1-specific rapamycin analogs on human tumors have proven underwhelming and variable thus far (8), possibly due to rapamycin-resistant mTORC1 action or inadvertent promotion of PI3K signaling upon suppression of the mTORC1 negative feedback loop. Hopefully, simultaneous inhibition of both mTORC1 and mTORC2 with mTOR catalytic inhibitors will better battle tumorigenesis.
Direct Molecular Mechanisms of mTORC Regulation
Although many upstream mTORC1 regulators of have been identified, the direct mechanisms that modulate mTORC1 have only begun to emerge recently. To date, numerous and diverse mechanisms of direct mTORC1 regulation have been reported, consistent with the broad and complex network of pathways that converge upon and modulate mTORC1. The avidity of interactions within mTORC1 has emerged as an important point of mTORC1 regulation. mTORC1 activation leads to modest weakening of the mTOR-raptor, mTOR-deptor, and raptor-PRAS40 interactions (20, 29, 30, 32, 76). Although quite controversial, Rheb-GTP reportedly interacts with FKBP38, an endogenous mTORC1 inhibitor that binds to the mTOR FRB domain, thereby blunting the inhibitory FKBP38-mTOR interaction (3, 48, 77). Other groups have failed, however, to experimentally reproduce much of this work (50, 78, 79). Cellular insulin stimulation (80) or Rheb-GTP provided in vitro (50) increases binding of recombinant 4EBP1 substrate to mTORC1.
As reversible protein phosphorylation regulates many components in mTORC signaling networks, recent research has examined a regulatory role for the direct phosphorylation of mTORC components (Fig. 2). Insulin-stimulated activation of PI3K leads to Akt- and mTOR-mediated phosphorylation of PRAS40 (at Thr246 and Ser183/Ser212/Ser221, respectively) (49, 76, 81, 82), which cooperates to reduce PRAS40-mediated suppression of mTORC1. Activated mTOR also phosphorylates deptor, leading to its degradation and thus relieving its mTOR inhibitory action (20). Raptor represents another phosphorylation target. mTORC1 activation by diverse stimuli (e.g. the PI3K/TSC/Rheb pathway, amino acids, EGF, energy sufficiency) promotes the rapamycin-sensitive and thus mTOR-mediated phosphorylation of raptor Ser863, which promotes mTORC1 activity (32, 83). In addition to raptor Ser863, Rheb overexpression promotes raptor phosphorylation at bat least five other sites (e.g. Ser696, Thr706, Ser855, Ser859, and Ser877); strikingly, the phosphorylation of a subset of these sites occurs in a hierarchical manner (32). Activation of the Ras-regulated MAPK pathway leads to RSK-mediated raptor phosphorylation (Ser719, Ser721, and Ser722), which promotes mTORC1 kinase activity (84). In response to energy deprivation, AMPK mediates raptor phosphorylation (Ser722 and Ser792) to inhibit mTORC1 signaling, which activates a metabolic checkpoint to cause cell cycle arrest (56, 85). Most recently, the mitotic kinase Cdc2 was reported to phosphorylate raptor Ser696 and Thr706, with unknown functional significance (86). It is also clear that rictor experiences extensive phosphorylation at multiple sites, although the functional significance of almost all sites remains unknown (71–73). The phosphorylation of raptor and rictor at multiple sites may suggest that partner protein phosphorylation functions as a biochemical rheostat that fine-tunes mTORC1 and mTORC2 signaling in response to diverse environmental cues.
To date, four mTOR phosphorylation sites have been characterized in the literature, Ser2448, Ser2481 (an autophosphorylation site), Thr2446, and Ser1261 (in order of discovery), although only one (Ser1261) has been found to regulate mTOR function (30, 87–89). Although Akt was believed originally to mediate mTOR Ser2448 phosphorylation (87), more recent work identifies S6K1 as the mTOR Ser2448 kinase (90, 91). Prior to the identification of distinct mTOR complexes, mTOR Ser2481 autophosphorylation was believed to be insensitive to rapamycin and amino acid withdrawal, leading to the idea that modulation of mTOR intrinsic kinase activity does not universally underlie mTOR regulation (88). More recent work indicates, however, that rapamycin and amino acid withdrawal reduce mTORC1- but not mTORC2-associated mTOR Ser2481 autophosphorylation, consistent with the known sensitivities or lack thereof of mTORC1 and mTORC2 to these conditions (11, 12). Thus, mTORC1- and mTORC2-associated mTOR Ser2481 autophosphorylation serves as a simple biomarker that monitors intrinsic mTOR catalytic activity. It is important to note that mTOR Ser2481 autophosphorylation was reported by another group to represent an mTORC2-specific event (due to undetectable Ser2481 autophosphorylation in mTORC1) (92). These data may suggest higher stoichiometry of mTOR Ser2481 autophosphorylation in mTORC2 relative to mTORC1. In 3T3-L1 adipocytes, insulin signaling via PI3K increases both mTORC1- and mTORC2-associated mTOR Ser1261 phosphorylation. Importantly, mTOR Ser1261 phosphorylation promotes, at least in part, mTORC1 catalytic activity (as monitored by mTOR Ser2481 phosphorylation) and mTORC1-mediated substrate phosphorylation (e.g. S6K1, 4EBP1) and cell growth (30). Currently, the identity of the mTOR Ser1261 kinase remains unknown. Taken together, the data of many groups indicate that multiple phosphorylation events on mTORC components cooperate to regulate mTORC signaling, both positively and negatively, enabling the mTORCs to function as sensors of diverse physiological cues.
Future Directions
Although the early embryonic lethality of mice lacking mTOR (E5.5), raptor (E5.5), rictor (E10.5), and mLST8/GβL (E10.5) underscores the critical importance of mTORC1 and mTORC2 in embryonic development, these phenotypes provide little information regarding the physiological roles for mTORCs in vivo (19, 70, 93–95). Recent research utilizing mouse tissue-specific knock-out reveals critical roles for mTORC1 in adipose tissue and skeletal muscle metabolism and physiology (70, 96, 97). Future work should reveal the physiological functions for mTORCs in diverse organ systems and should facilitate the development of novel therapeutics to treat mTOR-linked pathologies. It will be important to decipher the molecular pathway by which mTORC1 senses and responds to amino acid availability. A good starting point may be the identification of the guanine nucleotide exchange factors for the RAG GTPases, which remain unknown. As the yeast protein Vam6 functions as the guanine nucleotide exchange factor for the yeast RAG homolog, Gtr1 (98), the mammalian Vam6 ortholog (mVam6) should be investigated as a regulator of amino acid-dependent mTORC1 activation. Finally, future work must continue to elucidate the regulation and function of cellular mTORC1 and mTORC2 signaling, including the identification of direct mTOR substrates (which remain few) and the diverse roles for mTORC component phosphorylation events in mTORC function. As treatment with rapamycin or knock-out of S6K1 in mice extends life span, similar to caloric restriction, mTORC1 inhibition has potential to serve as an elusive “fountain of youth” to delay onset and severity of age-related pathologies (99).
Supplementary Material
Acknowledgments
We thank members of the Fingar lab as well as Kathy A. Martin, Philippe P. Roux, and Martin G. Myers, Jr., for helpful feedback.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-DK 078135 (to D. C. F. and K. G. F.). This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- TOR
- target of rapamycin
- TORC
- TOR complex
- FRB
- FKBP12-rapamycin-binding
- mTORC
- mammalian TORC
- TSC
- tuberous sclerosis complex
- TOS
- mTOR signaling
- eIF
- eukaryotic initiation factor
- HM
- hydrophobic motif
- PI3K
- phosphatidylinositol 3-kinase
- PKCα
- protein kinase Cα
- EGF
- epidermal growth factor
- MAPK
- mitogen-activated protein kinase
- MEK
- MAPK/extracellular signal-regulated kinase kinase
- RSK
- ribosomal S6 kinase
- AMPK
- AMP-activated protein kinase
- E
- embryonic day
- GβL
- G-protein β-subunit-like.
REFERENCES
- 1.Bhaskar P. T., Hay N. (2007) Dev. Cell 12, 487–502 [DOI] [PubMed] [Google Scholar]
- 2.Jacinto E., Lorberg A. (2008) Biochem. J. 410, 19–37 [DOI] [PubMed] [Google Scholar]
- 3.Dunlop E. A., Tee A. R. (2009) Cell. Signal. 21, 827–835 [DOI] [PubMed] [Google Scholar]
- 4.Laplante M., Sabatini D. M. (2009) J. Cell Sci. 122, 3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cybulski N., Hall M. N. (2009) Trends Biochem. Sci. 34, 620–627 [DOI] [PubMed] [Google Scholar]
- 6.Alessi D. R., Pearce L. R., García-Martínez J. M. (2009) Sci. Signal. 2, pe27. [DOI] [PubMed] [Google Scholar]
- 7.Soulard A., Cohen A., Hall M. N. (2009) Curr. Opin. Cell Biol. 21, 825–836 [DOI] [PubMed] [Google Scholar]
- 8.Guertin D. A., Sabatini D. M. (2009) Sci. Signal. 2, pe24. [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 10.Oshiro N., Yoshino K., Hidayat S., Tokunaga C., Hara K., Eguchi S., Avruch J., Yonezawa K. (2004) Genes Cells 9, 359–366 [DOI] [PubMed] [Google Scholar]
- 11.Soliman G. A., Acosta-Jaquez H. A., Dunlop E. A., Ekim B., Maj N. E., Tee A. R., Fingar D. C. (2010) J. Biol. Chem. 285, 7866–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 13.Chiang G. G., Abraham R. T. (2007) Trends Mol. Med. 13, 433–442 [DOI] [PubMed] [Google Scholar]
- 14.Guertin D. A., Sabatini D. M. (2007) Cancer Cell 12, 9–22 [DOI] [PubMed] [Google Scholar]
- 15.Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman M. E., Apsel B., Uotila A., Loewith R., Knight Z. A., Ruggero D., Shokat K. M. (2009) PLoS Biol. 7, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Martínez J. M., Moran J., Clarke R. G., Gray A., Cosulich S. C., Chresta C. M., Alessi D. R. (2009) Biochem. J. 421, 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu K., Toral-Barza L., Shi C., Zhang W. G., Lucas J., Shor B., Kim J., Verheijen J., Curran K., Malwitz D. J., Cole D. C., Ellingboe J., Ayral-Kaloustian S., Mansour T. S., Gibbons J. J., Abraham R. T., Nowak P., Zask A. (2009) Cancer Res. 69, 6232–6240 [DOI] [PubMed] [Google Scholar]
- 19.Guertin D. A., Stevens D. M., Thoreen C. C., Burds A. A., Kalaany N. Y., Moffat J., Brown M., Fitzgerald K. J., Sabatini D. M. (2006) Dev. Cell 11, 859–871 [DOI] [PubMed] [Google Scholar]
- 20.Peterson T. R., Laplante M., Thoreen C. C., Sancak Y., Kang S. A., Kuehl W. M., Gray N. S., Sabatini D. M. (2009) Cell 137, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma X. M., Blenis J. (2009) Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 22.Reiling J. H., Sabatini D. M. (2006) Oncogene 25, 6373–6383 [DOI] [PubMed] [Google Scholar]
- 23.Chang Y. Y., Juhász G., Goraksha-Hicks P., Arsham A. M., Mallin D. R., Muller L. K., Neufeld T. P. (2009) Biochem. Soc. Trans. 37, 232–236 [DOI] [PubMed] [Google Scholar]
- 24.Inoki K., Ouyang H., Li Y., Guan K. L. (2005) Microbiol. Mol. Biol. Rev. 69, 79–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schalm S. S., Fingar D. C., Sabatini D. M., Blenis J. (2003) Curr. Biol. 13, 797–806 [DOI] [PubMed] [Google Scholar]
- 26.Choi K. M., McMahon L. P., Lawrence J. C., Jr. (2003) J. Biol. Chem. 278, 19667–19673 [DOI] [PubMed] [Google Scholar]
- 27.Nojima H., Tokunaga C., Eguchi S., Oshiro N., Hidayat S., Yoshino K., Hara K., Tanaka N., Avruch J., Yonezawa K. (2003) J. Biol. Chem. 278, 15461–15464 [DOI] [PubMed] [Google Scholar]
- 28.Dunlop E. A., Dodd K. M., Seymour L. A., Tee A. R. (2009) Cell. Signal. 21, 1073–1084 [DOI] [PubMed] [Google Scholar]
- 29.Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 30.Acosta-Jaquez H. A., Keller J. A., Foster K. G., Ekim B., Soliman G. A., Feener E. P., Ballif B. A., Fingar D. C. (2009) Mol. Cell. Biol. 29, 4308–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villalonga P., de Mattos S. F., Ridley A. J. (2009) J. Biol. Chem. 284, 35287–35296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foster K. G., Acosta-Jaquez H. A., Romeo Y., Ekim B., Soliman G. A., Carriere A., Roux P. P., Ballif B. A., Fingar D. C. (2010) J. Biol. Chem. 285, 80–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fingar D. C., Blenis J. (2004) Oncogene 23, 3151–3171 [DOI] [PubMed] [Google Scholar]
- 34.Ruvinsky I., Meyuhas O. (2006) Trends Biochem. Sci. 31, 342–348 [DOI] [PubMed] [Google Scholar]
- 35.Fingar D. C., Salama S., Tsou C., Harlow E., Blenis J. (2002) Genes Dev. 16, 1472–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fingar D. C., Richardson C. J., Tee A. R., Cheatham L., Tsou C., Blenis J. (2004) Mol. Cell. Biol. 24, 200–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holz M. K., Ballif B. A., Gygi S. P., Blenis J. (2005) Cell 123, 569–580 [DOI] [PubMed] [Google Scholar]
- 38.Kim J. E., Chen J. (2004) Diabetes 53, 2748–2756 [DOI] [PubMed] [Google Scholar]
- 39.Porstmann T., Santos C. R., Griffiths B., Cully M., Wu M., Leevers S., Griffiths J. R., Chung Y. L., Schulze A. (2008) Cell Metab. 8, 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 41.Manning B. D., Cantley L. C. (2007) Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.García-Martínez J. M., Alessi D. R. (2008) Biochem. J. 416, 375–385 [DOI] [PubMed] [Google Scholar]
- 43.Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A. C., Mao Y., Miao R. Q., Sessa W. C., Qin J., Zhang P., Su B., Jacinto E. (2008) EMBO J. 27, 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. (2008) EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 46.Rosner M., Fuchs C., Siegel N., Valli A., Hengstschläger M. (2009) Hum. Mol. Genet. 18, 3298–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J., Su B. (2006) Cell 127, 125–137 [DOI] [PubMed] [Google Scholar]
- 48.Huang J., Manning B. D. (2008) Biochem. J. 412, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. (2007) Mol. Cell 25, 903–915 [DOI] [PubMed] [Google Scholar]
- 50.Sato T., Nakashima A., Guo L., Tamanoi F. (2009) J. Biol. Chem. 284, 12783–12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schleich S., Teleman A. A. (2009) PLoS One 4, e6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Y., Chen J. (2008) Cell Cycle 7, 3118–3123 [DOI] [PubMed] [Google Scholar]
- 53.Toschi A., Lee E., Xu L., Garcia A., Gadir N., Foster D. A. (2009) Mol. Cell. Biol. 29, 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foster D. A., Toschi A. (2009) Cell Cycle 8, 1026–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K., Wang C. Y., He X., MacDougald O. A., You M., Williams B. O., Guan K. L. (2006) Cell 126, 955–968 [DOI] [PubMed] [Google Scholar]
- 56.Shaw R. J. (2009) Acta Physiol. 196, 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inoki K., Zhu T., Guan K. L. (2003) Cell 115, 577–590 [DOI] [PubMed] [Google Scholar]
- 58.Smith E. M., Finn S. G., Tee A. R., Browne G. J., Proud C. G. (2005) J. Biol. Chem. 280, 18717–18727 [DOI] [PubMed] [Google Scholar]
- 59.Byfield M. P., Murray J. T., Backer J. M. (2005) J. Biol. Chem. 280, 33076–33082 [DOI] [PubMed] [Google Scholar]
- 60.Nobukuni T., Joaquin M., Roccio M., Dann S. G., Kim S. Y., Gulati P., Byfield M. P., Backer J. M., Natt F., Bos J. L., Zwartkruis F. J., Thomas G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14238–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L. (2008) Nat. Cell Biol. 10, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008) Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maehama T., Tanaka M., Nishina H., Murakami M., Kanaho Y., Hanada K. (2008) J. Biol. Chem. 283, 35053–35059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Findlay G. M., Yan L., Procter J., Mieulet V., Lamb R. F. (2007) Biochem. J. 403, 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicklin P., Bergman P., Zhang B., Triantafellow E., Wang H., Nyfeler B., Yang H., Hild M., Kung C., Wilson C., Myer V. E., MacKeigan J. P., Porter J. A., Wang Y. K., Cantley L. C., Finan P. M., Murphy L. O. (2009) Cell 136, 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang J., Dibble C. C., Matsuzaki M., Manning B. D. (2008) Mol. Cell. Biol. 28, 4104–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harrington L. S., Findlay G. M., Lamb R. F. (2005) Trends Biochem. Sci. 30, 35–42 [DOI] [PubMed] [Google Scholar]
- 68.Manning B. D., Logsdon M. N., Lipovsky A. I., Abbott D., Kwiatkowski D. J., Cantley L. C. (2005) Genes Dev. 19, 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. (2004) Nature 431, 200–205 [DOI] [PubMed] [Google Scholar]
- 70.Polak P., Hall M. N. (2009) Curr. Opin. Cell Biol. 21, 209–218 [DOI] [PubMed] [Google Scholar]
- 71.Dibble C. C., Asara J. M., Manning B. D. (2009) Mol. Cell. Biol. 29, 5657–5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Julien L. A., Carriere A., Moreau J., Roux P. P. (2010) Mol. Cell. Biol. 30, 908–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Treins C., Warne P. H., Magnuson M. A., Pende M., Downward J. (2010) Oncogene 29, 1003–1016 [DOI] [PubMed] [Google Scholar]
- 74.Thoreen C. C., Sabatini D. M. (2009) Autophagy 5, 725–726 [DOI] [PubMed] [Google Scholar]
- 75.Choo A. Y., Blenis J. (2009) Cell Cycle 8, 567–572 [DOI] [PubMed] [Google Scholar]
- 76.Vander Haar E., Lee S. I., Bandhakavi S., Griffin T. J., Kim D. H. (2007) Nat. Cell Biol. 9, 316–323 [DOI] [PubMed] [Google Scholar]
- 77.Bai X., Ma D., Liu A., Shen X., Wang Q. J., Liu Y., Jiang Y. (2007) Science 318, 977–980 [DOI] [PubMed] [Google Scholar]
- 78.Wang X., Fonseca B. D., Tang H., Liu R., Elia A., Clemens M. J., Bommer U. A., Proud C. G. (2008) J. Biol. Chem. 283, 30482–30492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uhlenbrock K., Weiwad M., Wetzker R., Fischer G., Wittinghofer A., Rubio I. (2009) FEBS Lett. 583, 965–970 [DOI] [PubMed] [Google Scholar]
- 80.Wang L., Rhodes C. J., Lawrence J. C., Jr. (2006) J. Biol. Chem. 281, 24293–24303 [DOI] [PubMed] [Google Scholar]
- 81.Oshiro N., Takahashi R., Yoshino K., Tanimura K., Nakashima A., Eguchi S., Miyamoto T., Hara K., Takehana K., Avruch J., Kikkawa U., Yonezawa K. (2007) J. Biol. Chem. 282, 20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang L., Harris T. E., Lawrence J. C., Jr. (2008) J. Biol. Chem. 283, 15619–15627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang L., Lawrence J. C., Jr., Sturgill T. W., Harris T. E. (2009) J. Biol. Chem. 284, 14693–14697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carrière A., Cargnello M., Julien L. A., Gao H., Bonneil E., Thibault P., Roux P. P. (2008) Curr. Biol. 18, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 85.Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008) Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gwinn D. M., Asara J. M., Shaw R. J. (2010) PLoS One 5, e9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Navé B. T., Ouwens M., Withers D. J., Alessi D. R., Shepherd P. R. (1999) Biochem. J. 344, 427–431 [PMC free article] [PubMed] [Google Scholar]
- 88.Peterson R. T., Beal P. A., Comb M. J., Schreiber S. L. (2000) J. Biol. Chem. 275, 7416–7423 [DOI] [PubMed] [Google Scholar]
- 89.Cheng S. W., Fryer L. G., Carling D., Shepherd P. R. (2004) J. Biol. Chem. 279, 15719–15722 [DOI] [PubMed] [Google Scholar]
- 90.Holz M. K., Blenis J. (2005) J. Biol. Chem. 280, 26089–26093 [DOI] [PubMed] [Google Scholar]
- 91.Chiang G. G., Abraham R. T. (2005) J. Biol. Chem. 280, 25485–25490 [DOI] [PubMed] [Google Scholar]
- 92.Copp J., Manning G., Hunter T. (2009) Cancer Res. 69, 1821–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gangloff Y. G., Mueller M., Dann S. G., Svoboda P., Sticker M., Spetz J. F., Um S. H., Brown E. J., Cereghini S., Thomas G., Kozma S. C. (2004) Mol. Cell. Biol. 24, 9508–9516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murakami M., Ichisaka T., Maeda M., Oshiro N., Hara K., Edenhofer F., Kiyama H., Yonezawa K., Yamanaka S. (2004) Mol. Cell. Biol. 24, 6710–6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shiota C., Woo J. T., Lindner J., Shelton K. D., Magnuson M. A. (2006) Dev. Cell 11, 583–589 [DOI] [PubMed] [Google Scholar]
- 96.Polak P., Cybulski N., Feige J. N., Auwerx J., Rüegg M. A., Hall M. N. (2008) Cell Metab. 8, 399–410 [DOI] [PubMed] [Google Scholar]
- 97.Bentzinger C. F., Romanino K., Cloëtta D., Lin S., Mascarenhas J. B., Oliveri F., Xia J., Casanova E., Costa C. F., Brink M., Zorzato F., Hall M. N., Rüegg M. A. (2008) Cell Metab. 8, 411–424 [DOI] [PubMed] [Google Scholar]
- 98.Binda M., Péli-Gulli M. P., Bonfils G., Panchaud N., Urban J., Sturgill T. W., Loewith R., De Virgilio C. (2009) Mol. Cell 35, 563–573 [DOI] [PubMed] [Google Scholar]
- 99.Sharp Z. D., Strong R. (2010) J. Gerontol. A Biol. Sci. Med. Sci., in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.