Abstract
FK506 (tacrolimus) is a secondary metabolite with a potent immunosuppressive activity, currently registered for use as immunosuppressant after organ transplantation. FK506 and FK520 are biogenetically related natural products that are synthesized by combined polyketide synthase/nonribosomal peptide synthetase systems. The entire gene cluster for biosynthesis of FK520 from Streptomyces hygroscopicus var. ascomyceticus has been cloned and sequenced. On the other hand, the FK506 gene cluster from Streptomyces sp. MA6548 (ATCC55098) was sequenced only partially, and it was reasonable to expect that additional genes would be required for the provision of substrate supply. Here we report the identification of a previously unknown region of the FK506 gene cluster from Streptomyces tsukubaensis NRRL 18488 containing genes encoding the provision of unusual building blocks for FK506 biosynthesis as well as a regulatory gene. Among others, we identified a group of genes encoding biosynthesis of the extender unit that forms the allyl group at carbon 21 of FK506. Interestingly, we have identified a small independent diketide synthase system involved in the biosynthesis of the allyl group. Inactivation of one of these genes, encoding an unusual ketosynthase domain, resulted in an FK506 nonproducing strain, and the production was restored when a synthetic analog of the allylmalonyl-CoA extender unit was added to the cultivation medium. Based on our results, we propose a biosynthetic pathway for the provision of an unusual five-carbon extender unit, which is carried out by a novel diketide synthase complex.
Keywords: Bacterial Genetics, Bacterial Metabolism, Calcineurin, Gene Knockout, Immunosupressor, FK506, Streptomyces tsukubaensis, Metabolic Pathway, Polyketide Synthase, Tacrolimus
Introduction
FK506 (tacrolimus) and its structural analogs FK520 (ascomycin) and rapamycin (sirolimus) are secondary metabolites belonging to a class of macrolactone compounds of great medicinal importance. They are currently registered for use as immunosuppressants after organ transplantation as well as for treatment of inflammatory skin diseases and eczema (1–3). In recent clinical studies, these compounds have also shown great promise for treatment of various forms of cancer, as well as for neurological disorders (4, 5). FK506 and FK520 are biogenetically related natural products that are synthesized by combined polyketide synthase (PKS)2/nonribosomal peptide synthetase systems. The allyl group at carbon 21 of FK506 is replaced by an ethyl group in FK520, this being the only structural difference between the FK506 and FK520 compounds (see Fig. 1). A common feature of these complex polyketides is the involvement of large polyfunctional polyketide synthases comprising multi-fatty acid synthase-like domains arranged in sets of modules (6). Each module is responsible for the recognition and incorporation of a specific carboxylic acid-derived extender unit into the growing polyketide chain as well as for subsequent reductive reactions (7, 8). Compounds such as FK506/520 and rapamycin are formed by incorporation of lysine-derived pipecolic acid by the nonribosomal peptide synthase-like domain through the cyclization step, resulting in the formation of macrolactone, followed by the post-PKS processing reactions catalyzed by enzymes such as methyltransferase and oxido-reductase enzymes (9, 10). Entire gene clusters for rapamycin biosynthesis from Streptomyces hygroscopicus and FK520 from S. hygroscopicus var. ascomyceticus have been cloned and sequenced (see Fig. 2) (11, 12). On the other hand, the FK506 gene cluster from Streptomyces sp. MA6548 (ATCC55098) was sequenced only partially by Merck Research Laboratories (see Fig. 2) (13–15). The published part of the biosynthetic cluster for FK506 biosynthesis only covers the core region containing PKS genes and two additional genes involved in macrolide core structure “editing” (oxidation and methylation), the so-called post-PKS modifications (15). The origin of building blocks necessary for biosynthesis of both FK506 and FK520, such as malonyl-CoA (acetate), methylmalonyl-CoA (propionate), methoxymalonyl-ACP, shikimate-derived starter unit (cyclohexyl ring), and lysine-derived pipecolate was demonstrated mostly through precursor incorporation experiments and molecular biology approaches (16–20). When comparing structurally related compounds FK520 and rapamycin and their corresponding gene clusters to the published parts of the FK506 gene cluster, it was not possible to identify all of the putative genes involved in provision of substrate supply (building blocks for FK506 biosynthesis). Particularly, the origin of the extender unit providing the allyl group at carbon 21 of FK506 has remained entirely unknown. We suspected that genes involved in the formation of the allyl group are very likely located in the vicinity of the published PKS genes, analogously to the FK520 gene cluster (see Fig. 2). Therefore, the aim of this work was to sequence and analyze putative open reading frames (ORFs) at both arms of the FK506 gene cluster and identify gene homologs involved in the provision of the unusual allyl group at position 21 of the FK506 structure. Here we report the identification of the all subcluster, encoding genes involved in provision of the respective five-carbon extender unit located at the left fringe of the FK506 biosynthetic cluster from Streptomyces tsukubaensis. In addition, we propose a novel biosynthetic pathway for provision of this unusual extender unit that is carried out by a novel diketide synthase complex.
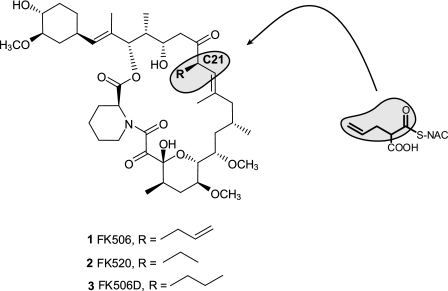
FIGURE 1.
The structures of FK506, FK520, and dihydro-FK506 (FK506D) differ only in the side chain bound to the carbon C21. Incorporation of the allylmalonyl-SNAC extender unit analog (compound 10) into the structure of FK506 is marked (gray areas).
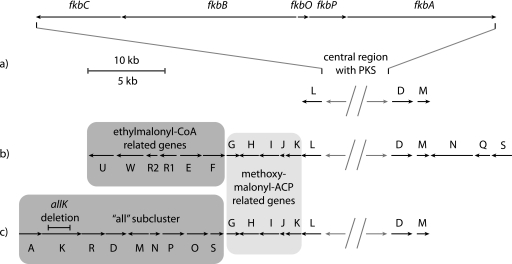
FIGURE 2.
Schematic representation of gene clusters encoding FK506 and FK520 biosynthetic pathways. a, in FK506-producing species Streptomyces sp. MA6548, the PKS core region (above) sequence consists of fkbC, fkbB, fkbO, fkbP, and fkbA PKS genes (13). b, the FK520 biosynthetic cluster PKS core architecture with additional genes encoding biosynthesis of methoxymalonyl-CoA and ethylmalonyl-CoA extender units (12). c, schematic representation of the FK506 biosynthetic cluster from S. tsukubaensis NRRL 18488 with a conserved area of methoxymalonyl-CoA biosynthetic genes and a unique sequence of genes, designated as all subcluster on the left fringe, encoding biosynthesis of five-carbon extender unit leading to the allyl group in position C21. The size and the location of the deleted portion of the allK gene are marked.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Condition
S. tsukubaensis NRRL 18488 wild type strain was used for all genetic manipulations. The ISP4 sporulation agar medium (21) was used for spore stock preparation. S. tsukubaensis strains were cultivated as a confluent lawn on the ISP4 agar medium for 8–14 days at 28 °C. For liquid cultures spores of S. tsukubaensis strains (1% v/v) were inoculated in seed medium VG3 (0.25% (w/v) soy meal, 1% dextrin, 0.1% glucose, 0.5% yeast extract, 0.7% casein hydrolyzate, 0.02% K2HPO4, 0.05% NaCl, 0.0005% MnCl2 × 4H2O, 0.0025% FeSO4 × 7H2O, 0.0001% ZnSO4 × 7H2O, 0.0005% MgSO4 × 7H2O, 0.002% CaCl2, pH 7.0) and incubated at 28 °C and 250 rpm for 24–48 h. 10% (v/v) of the above seed culture was used for the inoculation of a 250-ml Erlenmeyer flask containing 50 ml of production medium PG3 (9% dextrin, 0.5% glucose, 1% soy meal, 1% soy peptone, 1% glycerol, 0.25% l-lysine, 0.1% K2HPO4, 0.15% CaCO3, 0.1% polyethylene glycol 1000, pH 6.5) (22). Cultivation was carried out at 28 °C, 250 rpm for 6–7 days. Thiostrepton (30 μg/ml) and apramycin (50 μg/ml) were added to the solid and liquid media as required.
DNA Sequencing and Annotation of Open Reading Frames
Using 454 sequencing technology (23), we obtained several contigs covering the FK506 biosynthetic cluster of S. tsukubaensis NRRL 18488 strain. BLAST programs (24) and FramePlot software (25) for gene annotation were used to predict ORFs in the newly obtained genomic sequence covering the FK506 biosynthetic cluster. DNA sequence covering the left fringe of the FK506 gene cluster was deposited in GenBankTM with accession number GU067679.
Transformation and DNA Manipulations
Standard methods for isolation and manipulation of DNA were performed as described by Sambrook and Russel (26) and Kieser et al. (27). The Escherichia coli-Streptomyces conjugation procedure (28) was used to introduce plasmid DNA into the S. tsukubaensis strain. Gene disruption/replacement procedures were carried out with plasmids based on the pKC1139 vector, containing thermosensitive replicon as described by Kieser et al. (27).
Disruption of the Target Biosynthetic Gene allK
We designed primers for amplification of the regions flanking the allK gene as follows: allK-F1 primer (AGAATTCGTTACGGGGAGACGGCATCCCGG) containing an EcoRI restriction site and allK-R1 (AGGATCCGGGCGGGCTCGTCGCGGT) were used to amplify the upstream region of the allK gene containing an internal BamHI restriction site. AllK-F2 (TGGATCCGGCGCGTATCGCCAACCGCTAC) containing a BamHI and allK-R2 (AAAGCTTCCCGGTAGTTCGCCATATGTGACCCG) with a HindIII restriction site were used to amplify the downstream region of the allK gene. The allKF1-R1 (upstream fragment) and allKF2-R2 (downstream) DNA fragments of the flanking regions of the allK gene were gel-purified and ligated into the pUC19 vector, and their nucleotide sequence was confirmed by sequencing. The allKF1-R1 fragment was excised with EcoRI and BamHI restriction enzymes, and allKF2-R2 was excised with BamHI and HindIII. Both excised fragments were subcloned simultaneously into pKC1139 previously digested with HindIII and EcoRI to generate pKC1139-allK plasmid construct. This vector was then conjugated into S. tsukubaensis to introduce a genomic deletion by homologous recombination. A 1698-bp in-frame gap was generated between the internal BamHI restriction site and the BamHI site inserted in the allK-F2 primer that caused the deletion of almost entire 2388-bp allK gene, while ensuring a minimum effect on the downstream genes (see Fig. 2c). Manipulation of the Streptomyces strain including conjugation procedure was performed as described previously (27).
Synthesis of Allylmalonyl-SNAC Thioesters
In the first step, allylmalonyl dichloride was prepared according to the procedure published by Mandoli et al. (29). 1.90 g of allylmalonic acid (Fluka, CAS 2583-25-7) was dissolved with a mix of 24 ml of dichloromethane and 150 μl of dimethylformamide in a two-necked flask. The mixture was cooled in an ice bath, and over 1.5 h, 5.3 g of oxalyl chloride was added dropwise while stirring. The mixture was stirred overnight at room temperature. The volatile components were removed by low pressure evaporation. The product, allylmalonyl dichloride, remained in the form of 3.76 g of dark oil. In the second step 3.76 g of the allylmalonyl dichloride was dissolved in 30 ml of tetrahydrofuran and cooled down in an ice bath. During stirring at 0 °C 1.65 g of N-acetyl-cysteamine was slowly added to the mixture. 1.5 ml of triethyl-amine was added dropwise during a 10-min period. The mixture was incubated on ice for additional 2 h. The precipitate was filtered off, and volatile components were removed by evaporation. The remaining oil was mixed with 50 ml of water and extracted three times with 150 ml of dichloromethane. The solvent was evaporated off, and the remaining allylmalonyl-SNAC was recovered in a form of light yellow oil (2.2 g).
Purification of Allylmalonyl-SNAC Thioesters
The theoretical outcome of this first synthesis of allylmalonyl-SNAC thioesters are single (2-((2-acetamidoethylthio)carbonyl)pent-4-enoic acid, compound 10) and double (S,S-bis(2-acetamidoethyl) 2-allylpropanebis (thioate), compound 11) SNAC thioesters of allylmalonic acid. Preparative HPLC was used to purify the two compounds from the crude synthesis product. The samples were dissolved in 1% (v/v) acetic acid, 20% (v/v) acetonitrile, 79% (v/v) H2O to a final concentration of 10 g/liter. Hipersep LAB LC80 (Novasep) combined with Prochrom® dynamic axial compression column (Prochrome LC50 DAC) packed with Kromasil 100-10-C18 was used. Chromatographic conditions were as follows: temperature, 20 °C; detection, UV at 240 nm; flow rate, 118 ml/min; injection volume, 15 ml; pump A, 1% (v/v) acetic acid, 5% (v/v) acetonitrile, and 94% (v/v) H2O; pump B, 1% (v/v) acetic acid, 94% (v/v) acetonitrile, and 5% (v/v) H2O. The isocratic method with 80% pump A, 20% pump B was used. Fractions with retention time 6.2–7.3 min (peak 1) and 9.6–10.8 min (peak 2) from 4 runs were collected (supplemental Fig. S1). The collected fractions (250 and 350 ml, respectively) were subjected to low pressure evaporation to remove acetonitrile followed by extraction (three times with equal volume of dichloromethane). The solvent was removed by evaporation, and 74 mg of peak 1 and 152 mg of peak 2, both in the form of light yellow oil, were obtained.
Analysis of Allylmalonyl-SNAC Thioesters
LC-MS analysis was performed with mass spectrometer Thermo LCQ FLEET (ion trap) equipped with Waters Acquity UPLC system. The chromatographic conditions were: column, Thermo Hypersil Gold aQ 100 × 2.1 mm, 1.9 μm; flow, 0.4 ml/min; temperature, 40 °C; λ = 240 nm; gradient, 0 min with 0.1% acetic acid, 5% acetonitrile, 95% water; 4–5 min with 0.1% acetic acid, 100% acetonitrile, 0% water; 6 min with 0.1% acetic acid, 5% acetonitrile, 95% water; sample injection, 5 μl at 100 mg/liter. The ionization conditions were: ESI+; Spray voltage, 3.5 kV; capillary voltage, −16 V; tube lens voltage, −85.6 V; capillary temperature, 275 °C; sheet gas, 33 AU; auxiliary gas, 5 AU; full scan, normal mode 100–1000 m/z. Analysis of fraction “peak 1” showed a single UV absorption peak at 2.49 min, having [M+H]+ 245.99 as the major m/z detected and additionally [M+Na]+ 268.03, which is consistent with allylmalonyl-SNAC single thioester (compound 10) (supplemental Fig. S2). Isotopic distribution corresponds with the prediction for molecular formula C10H15NO4S. Analysis of fraction “peak 2” showed a single UV absorption peak at 2.90 min, having [M+H]+ 347.04 as the major m/z detected and additionally [M+Na]+ 369.14, which is consistent with allylmalonyl-SNAC double thioester (compound 11) (supplemental Fig. S2). Isotopic distribution corresponds with the prediction for molecular formula C14H22N2O4S2. To further confirm the structure of the synthesized allylmalonyl-SNAC thioesters, 1H NMR analysis was performed. 1H NMR spectral data for peak 1 confirm the structure of allylmalonyl-SNAC single thioester (compound 10). 1H NMR (400 MHz, CDCl3): δ 1.91 (s, 3H), 2.61 (m, 2H), 2.94 (m, 1H), 3.11 (m, 1H), 3.41 (m, 2H), 3.64 (t, J = 7.4 Hz, 1H), 5.04 (m, 2H), 5.69 (m, 1H), 6.12 (br s, 1H), 6.91 (br s, 1H). (supplemental Fig. S3A) 1H analysis of peak 2 corresponds to the structure of allylmalonyl-SNAC double thioester (11). 1H NMR (400 MHz, CDCl3): δ 1.91 (s, 6H), 2.62 (m, 2H), 2.96 (m, 2H), 3.09 (m, 2H), 3.41 (m, 4H), 3.84 (t, J = 7.2 Hz, 1H), 5.04 (m, 2H), 5.62 (m, 1H), 6.17 (br s, 2H) (supplemental Fig. S3B).
Allylmalonyl-SNAC and Allylmalonyl-diSNAC Feeding Experiment
SNAC thioesters of selected carboxylic acids, synthetic analogs of CoA-bound polyketide building blocks, also ensure their efficient transport through bacterial membrane in a manner similar to that described by Stassi et al. (30), where ethylmalonyl-CoA precursor diethylethylmalonate was used to ensure provision of ethylmalonyl extender unit for production of an erythromycin analog (31). As described above, we have synthesized and isolated allylmalonyl-SNAC (compound 10) and allylmalonyl-diSNAC thioesters (compound 11). Although single SNAC thioester structure (compound 10) mimics the natural CoA-bound extender unit, double thioester (compound 11) is thought to undergo partial hydrolysis before loading and processing by a corresponding PKS module (see Fig. 5b). In our experiments, spores of S. tsukubaensis were inoculated in the seed medium VG3 (1% (v/v)) and cultivated for 24–48 h as described earlier. The production medium PG3 was inoculated with the seed medium (10% (v/v)). After 72 h, 10 ml of production phase culture broth was transferred to 250-ml Erlenmeyer flasks containing 20 ml of fresh production medium and 0.15 ml of 10% (w/v) dimethyl sulfoxide solution of either allylmalonyl-SNAC or allylmalonyl-diSNAC was added. The culture was incubated for a further 6 days as described earlier.
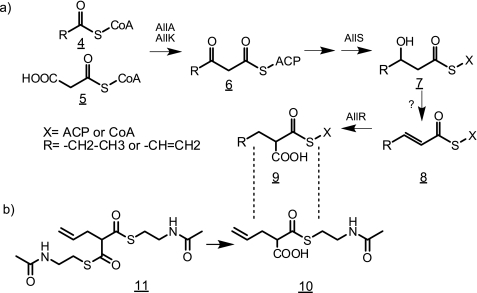
FIGURE 5.
a, proposed biosynthetic pathway for the five-carbon extender unit leading to the allyl group at position C21 of FK506. Firstly, either propionyl-CoA or acryloyl-CoA (compound 4) is condensed with malonyl-CoA (compound 5) by the action of diketide synthase system encoded by allA and allK yielding a five-carbon intermediate most probably bound to ACP. In the next step the 3-keto derivative (compound 6) is reduced to 3-hydroxy compound (compound 7) by the product of allS gene. The dehydration step in the next stage is likely to be performed by a generic dehydratase, not encoded in the cluster. In the last step, the 2-pentenoyl derivative (compound 8) is reduced and carboxylated by the crotonyl-CoA carboxylase/reductase, encoded in allR, to propylmalonyl-CoA or allylmalonyl-CoA (compound 9). The precise moment of transesterification from ACP to CoA and generation of the terminal double bond cannot be established at this time. b, chemical structures of allylmalonyl-SNAC monoester (compound 10) and allylmalonyl-SNAC double ester (compound 11) synthetic analogs of the five-carbon extender unit (compound 9), included into the growing polyketide chain by module 4 of the FK506 PKS. When added to the growth medium, compound 11 probably passes through cell membranes and is then partially hydrolyzed to compound 10, which is incorporated into the growing polyketide chain.
Detection of FK506
After cultivation was completed, the broth was mixed with the equal volume of methanol (1:1), and the soluble phase was applied onto Nucleosil EC100–3 C18, (150 × 4 mm, 3 μm, Machery-Nagel, Germany) reversed phase HPLC column. The mobile phase used for isocratic elution consisted of water, acetonitrile, methyl tert-butyl ether, and phosphoric acid (58.29:34.4:7.29:0.02, v/v/v/v). The column temperature was set to 60 °C, the detection wavelength was set at 210 nm, and the flow rate was set at 1.5 ml/min. The peak corresponding to FK506 was identified using an FK506 external standard (obtained from Lek/Sandoz), and the data were analyzed using ChromQuest software. The identity of FK506 in the allylmalonyl-SNAC and allylmalonyl-diSNAC fed cultivation broths was determined by LC-MS/MS analysis. We used the Agilent 1100 series LC-MS system coupled with Watters Micromass Quattro micro detector using reversed phase column (Gemini C18 column, 5 μm, 150 mm × 2 mm inner diameter) from Phenomenex. The separation was performed at a flow rate of 0.250 ml/min by gradient elution with 0.5% trifluoroacetic acid as solvent A and acetonitrile as solvent B. The gradient program was: 60% A, 0 min; 60–20% A, 0–17 min; 20–60% A, 17–18 min; 60% A, 18–30 min, and the injection volume 10 μl at temperature of the column 45 °C was used. The mass selective detector (Waters, Quattro micro API) was equipped with an electrospray ionization using a cone voltage of 20 V and capillary voltage of 3.5 kV for positive ionization of the analytes. Dry nitrogen was heated to 350 °C, the drying gas flow was 400 liters/h, and collision energy was 20 eV. In the ESI+-positive mode, an ion of m/z = 826.5 that corresponds to a capture of a sodium ion ([M+Na]+) was most intensive, in accordance with the results of other investigators (32). For FK506 identity confirmation, multiple reaction-monitoring mode was used, and the transition FK506 m/z 826.5 [M+Na]+ → m/z 616.4 was recorded.
RESULTS
Newly Identified Region of the FK506 Gene Cluster Located on the Left Side of the PKS Core Region
The newly obtained DNA sequences from S. tsukubaensis NRRL 18488 were initially compared with the previously published sequence of the core region of the FK506 biosynthetic cluster from Streptomyces sp. MA6548. As expected, the central regions of FK506 biosynthetic clusters from both organisms show conserved architecture of ORFs and very high sequence similarity of PKS genes as well as other genes flanking the PKS core region (13). The FK506 biosynthetic cluster from S. tsukubaensis was found to extend 17 kb to the left fringe from the PKS core region, and a total of 15 ORFs were identified in this part of the cluster. The relative position, order, and direction of transcription of the ORFs representing the biosynthetic locus are schematically illustrated in Fig. 2. In addition, the functions of these genes, predicted on the basis of amino acid sequence similarity are listed in Table 1.
TABLE 1.
Organization of the all subcluster involved in allyl side chain-providing extender unit of the FK506 gene cluster
| Gene No. | Gene name | Proposed function | Start | Stop | Length (amino acids) | Closest characterized homolog |
|---|---|---|---|---|---|---|
| 1 | allA | Acyltransferase and acyl-carrier protein | 2775 | 4067 | 431 | Homologs of both genes are found in many species of Burkholderia; none of them is characterized (37) |
| 2 | allK | Ketoacyl synthase | 4078 | 6465 | 796 | |
| 3 | allR | Enoyl-CoA reductase | 6465 | 7799 | 445 | Approximately 60% identity to ccr genes from Streptomyces cinnamonensis and Streptomyces collinus (39) |
| 4 | allD | Acyl-CoA dehydrogenase | 7799 | 8956 | 386 | 32% identical to medium chain acyl-CoA dehydrogenase from Thermus Thermophilus; no close homologs in Streptomyces species (40) |
| 5 | allM | Methionine γ lyase | 10126 | 8975 | 384 | 38% identical to l-methionine γ-lyase from Citrobacter freundii (42) |
| 6 | allN | Transcriptional regulator | 10186 | 10650 | 155 | Homologous to AsnC family of transcriptional regulators present in many bacterial species (33) |
| 7 | allP | P450 monooxygenase | 10757 | 11983 | 409 | 46% identical to Streptomyces coelicolor A3(2) Cyp154c1, a monooxygenase that functionalizes macrolide ring systems (43) |
| 8 | allO | Acyl-CoA oxidoreductase | 12171 | 13358 | 395 | 30% identical to 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione hydroxylase from Rhodococcus jostii; belongs to acyl-CoA dehydrogenase (type 2) superfamily (41) |
| 9 | allS | Acetoacetyl-CoA reductase or acetoacetyl-ACP reductase | 13467 | 14120 | 218 | 45% identical to 3-oxoacyl-[acyl-carrier-protein] reductase from Escherichia fergusonii; also similar to acetoacetyl-CoA reductase from several species (38) |
Architecture of the Newly Identified FK506 Gene Cluster in Part Resembles the FK520 Gene Cluster from S. hygroscopicus var. ascomyceticus ATCC 14891
Not surprisingly, we have noticed a significant similarity of the FK506 cluster ORFs from S. tsukubaensis to the genes located on the left side of the FK520 gene cluster from S. hygroscopicus var. ascomyceticus ATCC 14891, reflecting a very high structural similarity between the two compounds (Ref. 12 and Fig. 1). This similarity is particularly apparent when putative genes for the provision of the common building blocks or enzymes catalyzing post-PKS modifications are compared. As observed in Fig. 2 and analogously to the published sequence of the left side of the FK520 biosynthetic cluster (12), the fkbL gene homolog was found to be located immediately adjacent to the polyketide synthase gene fkbC (Fig. 2). Further to the left, both clusters encode all five contiguous genes providing the 13- and 15-methoxy groups (fkbG, fkbH, fkbI, fkbJ, and fkbK). (Ref. 12 and Fig. 2). These genes have been shown to be involved in methoxymalonyl-ACP extender unit biosynthesis, thus providing the explanation for the origin of the respective carbon atoms derived from metabolites such as erythrose, glycerate, glycolate and glycerol instead of acetate (12). Among others we have also identified a transcription factor, termed allN, which belongs to the AsnC family of regulatory elements, and it is present only in the FK506 gene cluster.
Identification of Putative ORFs Involved in the Synthesis of Carbon 21 Allyl Group-providing Extender Unit
As mentioned earlier, the structures of FK520 and FK506 compounds are almost identical, differing only by the position of carbon C21. Where FK520 bears an ethyl group, the FK506 structure contains an allyl moiety (Fig. 1). Despite this similarity, putative ORFs in the FK506 biosynthetic cluster, located further to the left from methoxymalonyl-ACP biosynthetic gene subcluster (Fig. 2c), are completely unrelated to the published FK520 gene cluster. In the FK520 gene cluster, several putative genes involved in the provision of the ethylmalonyl-CoA extender unit are located in this region (Fig. 2b). Interestingly, no gene homologs for the provision of the ethylmalonyl-CoA extender unit were present at the left extremity of the FK506 gene cluster from S. tsukubaensis, where we have identified a group of ORFs comprised of, among others, two relatively short genes containing domains characteristic of a PKS. Careful annotation of this DNA region clearly revealed that these ORFs encode acyl transferase, ketosynthase, and acyl carrier protein domains (Figs. 2 and 3). The origin of the five-carbon extender unit, which is presumably incorporated in the growing nascent polyketide chain during the biosynthesis of FK506 backbone, has not been elucidated to date. The possible involvement of this small PKS system in the biosynthesis of the five-carbon extender unit was thus considered as a quite unusual but feasible biosynthetic route. We hypothesized that this “minimal” PKS could mediate a Claisen condensation between a three-carbon starter unit and a two-carbon extender unit, a reaction leading to the necessary five-carbon metabolite (3-oxovalerate thioester compound 6) as a possible first step in the biosynthesis of this precursor (see Fig. 5a). Furthermore, additional genes are present in this part of the gene cluster that could enable further processing, involving a series of reduction reactions after the first step to generate the actual extender unit and possibly also to introduce the terminal double bond present in the allyl group in the structure of FK506. We termed the region of the FK506 biosynthetic cluster located to the left from fkbG the allyl or all subcluster. Most of the genes encoded in this subcluster region do not show significant sequence similarity to the genes present in the FK520 biosynthetic cluster and are therefore likely responsible for the structural difference between FK506 and FK520.
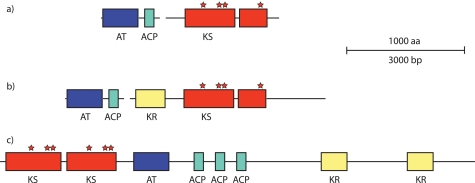
FIGURE 3.
Schematic representation of the genes encoding an unusual PKS-related diketide synthase system at the left side of the all subcluster (a) and comparison with the more complex architectures of homologous systems found in Burkholderia spp. (gi:83650017) (b) and B. marina (gi:87287237) (c). AT, acyltransferase; KR, ketoreductase.
Genes encoded in the all subcluster (left extremity of the FK506 cluster) are listed in Table 1 and schematically presented in Fig. 2c). As mentioned earlier, two genes, allA, encoding an acyltransferase and ACP domains, and allK, encoding two putative ketoacyl synthase (KS) domains, are present at the left extremity of the subcluster in a single transcription unit. The formation of the double bond present in the allyl side chain of FK506 is very likely catalyzed through a series of oxido-reduction reactions carried out by the genes present in the all gene cluster, namely AllR, AllD, AllP, AllO, and AllS (Table 1). Unfortunately, it is not possible to assign exact functions of these genes based on DNA homologies and to deduce putative biosynthetic steps involved in the formation of the double bond. The hypothetic enzymatic steps presumably involved in the formation of the double bond are presented in Fig. 5a and discussed in the following section. Additionally, two more genes, allN and allM, are located in the all subcluster (Table 1). AllN seems to encode an AsnC family transcriptional regulator (33), and allM encodes a methionine γ-lyase homolog. No explanation of their putative function, apart of the regulatory role, can be given at this point based on bioinformatics data.
Inactivation of allK, a Gene Encoding Ketoacyl Synthase Presumably Involved in Provision of the Carbon 21 Allyl Group of the FK506
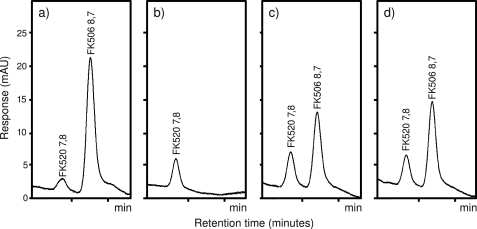
Inactivation of the ketoacyl synthase domain-containing gene allK was carried out to confirm the direct involvement of the newly identified all gene subcluster, particularly the diketide synthase system, in the provision of the extender unit providing the allyl group at position of carbon 21 of the FK506. To avoid any polar effects on the downstream allR and allD genes, which are likely transcribed in the same polycistronic mRNA (Fig. 2c), an in-frame allK gene deletion was carried out using two-step gene deletion as described under “Experimental Procedures.” Indeed, allK in-frame deletion mutants of S. tsukubaensis failed to produce FK506 in detectable amounts (Fig. 4b), thus confirming our hypothesis that the minimal PKS system encoded at the extreme left side of the FK506 biosynthetic cluster is essential for FK506 biosynthesis. Interestingly, although the biosynthesis of the FK506 compound by the mutant strain was completely abolished, the yield of the FK520, which is also produced as a side product by the S. tsukubaensis strain, was still at the wild type level, further supporting the hypothesis that the all subcluster is involved in the formation of the allyl side chain at the carbon 21 position. These data also indicate a parallel biosynthetic route of the ethylmalonyl-CoA extender unit.
FIGURE 4.
HPLC analysis of FK506 and FK520 with the retention time of ∼9 and 8 min, respectively. Wild type strain (a), allK in-frame deletion mutant strain (b), allK in-frame deletion mutant strain fed with allylmalonyl-SNAC thioester (compound 10) (c), and the same strain fed with allylmalonyl-diSNAC double thioester (compound 11) (d). Similar incubation conditions and times (given under “Experimental Procedures”) were used for all experiments.
Feeding of allK Deletion Mutant with Allylmalonyl-SNAC and Allylmalonyl-diSNAC Extender Unit Precursors
The allK in-fame deletion experiment clearly established the involvement of the diketide synthase system located at the left fringe of the FK506 gene cluster (Fig. 2) in FK506 biosynthesis. However, this experiment does not unequivocally confirm the involvement of these PKS-related genes and of the all gene subcluster in the formation of the unusual five-carbon extender unit, which provides the allyl group that is incorporated into the macrolactone polyketide backbone by module 4 of the main PKS-nonribosomal peptide synthase enzyme complex (13). In an attempt to prove this hypothesis, we synthesized allylmalonyl-SNAC and allylmalonyl-diSNAC thioesters (structures 10 and 11), which were fed during the biosynthetic process into the FK506 production medium PG3 to the allK in-frame deletion mutant cultures (Fig. 4b). As previously published, SNAC thioesters of unsubstituted and 2-substituted malonic acid can be added to cultures of polyketide producing microorganisms and are often successfully incorporated into the polyketide chain by the PKS, similarly to native coenzyme A activated extender units (Figs. 1 and 5b) (31, 34, 35). Indeed, when either allylmalonyl-SNAC or allylmalonyl-diSNAC were added to the S. tsukubaensis allK in-frame deletion mutant cultures, the production of FK506 was restored, and significant amounts of FK506 were easily detected by HPLC (Fig. 4). Both compounds showed comparable efficiency in re-establishing FK506 production. The identity of the produced FK506 was confirmed by LC-MS/MS analysis where the transition of FK506 sodium adduct m/z 826.5 [M+Na]+ → m/z 616.4 was monitored (32) (supplemental Fig. S4). This result clearly confirms the essential role of the diketide synthase (allK) in the formation of this unusual extender unit.
DISCUSSION
In this study we have identified a new region of the FK506 gene cluster, assigned as all subcluster, from S. tsukubaensis NRRL18488 involved in the biosynthesis of the carbon C21 allyl group. Previously published data, obtained by 13C-labeled feeding experiments with FK506 producing Streptomyces strains (17), suggested that the extender unit yielding this allyl group is composed of one propionate and one acetate molecule. Therefore, it is reasonable to believe that a five-carbon intermediate should be formed at some stage. The five-carbon unit could potentially emerge as an intermediate during odd chain fatty acid biosynthesis or degradation, bound to ACP or CoA, respectively (36). Until now, it has been difficult to envisage biosynthetic routes that are involved in the provision of this unusual extender unit, based on known primary metabolic pathways present in Actinomyces species. The existence of the all subcluster, encoding a minimal set of PKS-related domains, and data obtained by inactivating the KS domain in this system offered a plausible hypothesis, discussed later. The first two genes of the all subcluster, allA and allK, likely encode a diketide synthase that catalyzes the first step of the biosynthetic pathway, a Claisen condensation of one three-carbon compound (propionyl-CoA or acryloyl-CoA) (Fig. 5a, structure 4) and one two-carbon compound, very likely malonyl-CoA, which undergoes a decarboxylation step (structure 5) to yield 3-oxovaleryl-CoA or 3-oxopent-4-enoyl-CoA (structure 6). It is very difficult to imagine an exact enzymatic mechanism of this diketide synthase biosynthetic machinery without additional experimental data. However, the unusual domain architecture of allA and especially allK genes suggests that this PKS enzyme does not classify into any of the three most common types of PKS (7). Amino acid sequences of both genes have a strikingly high similarity to several short PKS systems of unknown function present in various species of Burkholderia, as revealed by BLASTp searches (Ref. 37 and Fig. 3). A more exhaustive bioinformatic analysis revealed that the S. tsukubaensis system is the most truncated form of what appears to be an unusual PKS gene, found also in a marine organism Blastopirellula marina DSM 3645 with sequences in Burkholderia sp. representing intermediate stages in simplification of its architecture (Fig. 3), thus suggesting that a horizontal transfer may have occurred during evolution. Nevertheless, this seems to be a rather ancient event because the guanine-cytosine content of both genes in S. tsukubaensis (72%) does not differ substantially from the usual GC content characteristic for Streptomyces species. Interestingly, the allK gene encoding the ketoacyl synthase enzymatic activity contains two KS units (domains), analogous to the unusual PKS system from Burkholderia sp. (Fig. 3). When analyzing these KS domains, it could be observed that the N-terminal part of the first KS domain is truncated, whereas in the second KS domain both the central and C-terminal parts are truncated. The first KS domain still contains all the predicted active site residues, whereas only the cysteine residue of the putative active site is present in the second KS domain, suggesting that it might still preserve catalytic function in concert with the KS domain, encoded in the first part of the gene. Whichever the enzymatic mechanism, in-frame deletion of the allK gene resulted in an FK506 nonproducing mutant. The production of FK506 was restored when a SNAC thioester of allylmalonic acid (Fig. 5b) is fed to the production medium, thus clearly confirming the role of the diketide synthase system in the formation of the five-carbon extender unit.
Based on the existing ORFs present in the all subcluster, we propose that in the next biosynthetic step, the thioester derivative of 3-oxovaleric acid (Fig. 5a, structure 6), produced by diketide synthase, is further reduced to a 3-hydroxy compound (structure 7). This reaction could be carried out by the product of the allS gene, located directly upstream from fkbG, which shows relatively high similarity to 3-oxoacyl-ACP reductase and acetoacetyl-CoA reductase homologs (Ref. 38 and Table 1). We have not identified dehydratase homologs in the FK506 gene cluster; therefore, the enzymatic activity responsible for dehydration of the 3-hydroxy compound (structure 7) to pent-2-enoyl thioester (structure 8) could be encoded elsewhere in the genome. We propose that the following biosynthetic steps, by analogy to the conversion of crotonyl-CoA to ethylmalonyl-CoA (39), pent-2-enyl-CoA or pent-2,4-dienoyl-CoA (structure 8) is converted to propylmalonyl-CoA or allylmalonyl-CoA (structure 9) (Fig. 5a) by a crotonyl-CoA reductase/carboxylase-like homolog allR, a gene located immediately downstream from allK, most probably on the same transcription unit. A transesterification step is likely carried out at some stage to convert the ACP-bound five-carbon derivative synthesized by the Claisen condensation to the CoA derivative, which is finally incorporated to the growing nascent polyketide chain by the FK506 PKS module 4. Alternatively, direct protein-protein interaction between small all PKS and large FK506 PKS multimodular complex is also feasible to ensure transfer of the five-carbon extender unit to module 4 of the FK506 PKS system.
The all subcluster contains additional genes whose function can also be partially rationalized in this hypothetical biosynthetic pathway. Two relatively distant members of the acyl-CoA dehydrogenase superfamily, allD and allO (40, 41), are present in the subcluster; however, their function cannot be predicted with confidence at this point using bioinformatics data. These genes could possibly be involved in generation of the terminal double bond of the five-carbon thioester at some stage to yield the allylmalonyl-CoA extender unit (Fig. 5a). The timing and at which stage these biosynthetic steps occur is a question that cannot be answered at this point. The double bond formation could occur either on the propionyl-CoA starter unit before loading onto the diketide synthase or on the five-carbon substrates, possibly still bound to this enzyme. On the other hand, the double bond present in the final FK506 molecule could also arise later on as a post-PKS modification after oxygenation/hydroxylation and subsequent dehydration of the C21 propyl group of the macrolactone (Fig. 5a). Supporting this hypothesis, a P450 monooxygenase homolog allP is located in this part of the all subcluster. Interestingly, the FK506 derivative containing propyl group at carbon 21 of FK506 (FK506D, structure 3) can be detected in the broth as a minor product (13). However, to our knowledge, no other intermediate products containing a three-carbon group at any oxidation stage at carbon 21 has been identified to date. Therefore, neither the timing nor the location of these oxido-reductive reactions on the five-carbon extender unit substrate could be established at this point.
In conclusion, we have identified a previously unknown part of the FK506 gene cluster from S. tsukubaensis strain, which encodes genes involved in the biosynthesis of the five-carbon extender unit, from which the allyl group of the FK506 at carbon 21 is derived. Surprisingly, the gene subcluster involved in the biosynthesis of this unusual extender unit encodes a novel set of PKS-related genes, encoding a diketide synthase enzymatic complex and a series of oxido-reductase genes that represent an entirely novel biosynthetic machinery. We have demonstrated the function of this new diketide synthase system in the formation of the five-carbon extender unit and its involvement in the formation of the allyl group at the position of carbon 21 of FK506. However, it is difficult to rationalize the biosynthetic mechanisms and steps involved the provision of this extender unit at this point, and more work will have to be carried out to understand this small but rather complex biosynthetic machinery, which additionally brings new opportunities in the area of biosynthetic engineering.
Supplementary Material
Acknowledgments
We thank Samo Pirc, Samo Andrenšek, and Borut Gabriel from Lek Pharmaceuticals for advice and help in synthesis, purification, and analysis of SNAC thioesters, used in this work, as well as Tomaž Polak from the Biotechnical Faculty for carrying out LC-MS/MS analyses of FK506. We also thank Alenka Košak, Ivo Zadra, Helmut Haecker, and Sašo Kranjc for review of the manuscript.
This work was supported by Government of Slovenia Ministry of Higher Education, Science and Technology Postdoctoral Fellowship Z4-1054 (to Š. F.) and Grants J4-9331 and L4-7117 (to H. P.) funds from the Ministry of the Economy and the Public Agency of the Republic of Slovenia for Entrepreneurship and Foreign Investments Agency and European Social Fund Contract 102/2008 (to G. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- PKS
- polyketide synthase
- ORF
- open reading frame
- SNAC
- N-acetylcysteamine
- ACP
- acyl carrier protein
- KS
- ketoacyl synthase
- LC
- liquid chromatography
- MS
- mass spectrometry
- contig
- group of overlapping clones
- HPLC
- high pressure liquid chromatography.
REFERENCES
- 1.Husain S., Singh N. (2002) Clin. Infect. Dis. 35, 53–61 [DOI] [PubMed] [Google Scholar]
- 2.Allison A. C. (2000) Immunopharmacology 47, 63–83 [DOI] [PubMed] [Google Scholar]
- 3.Stuetz A., Baumann K., Grassberger M., Wolff K., Meingassner J. G. (2006) Int. Arch. Allergy Immunol. 141, 199–212 [DOI] [PubMed] [Google Scholar]
- 4.Easton J. B., Houghton P. J. (2004) Expert Opin. Ther. Targets 8, 551–564 [DOI] [PubMed] [Google Scholar]
- 5.Graziani E. I. (2009) Nat. Prod. Rep. 26, 602–609 [DOI] [PubMed] [Google Scholar]
- 6.McDaniel R., Welch M., Hutchinson C. R. (2005) Chem. Rev. 105, 543–558 [DOI] [PubMed] [Google Scholar]
- 7.Hopwood D. A. (1997) Chem. Rev. 97, 2465–2498 [DOI] [PubMed] [Google Scholar]
- 8.Staunton J., Weissman K. J. (2001) Nat. Prod. Rep. 18, 380–416 [DOI] [PubMed] [Google Scholar]
- 9.Gregory M. A., Hong H., Lill R. E., Gaisser S., Petkovic H., Low L., Sheehan L. S., Carletti I., Ready S. J., Ward M. J., Kaja A. L., Weston A. J., Challis I. R., Leadlay P. F., Martin C. J., Wilkinson B., Sheridan R. M. (2006) Org. Biomol. Chem. 4, 3565–3568 [DOI] [PubMed] [Google Scholar]
- 10.Motamedi H., Shafiee A., Cai S. J., Streicher S. L., Arison B. H., Miller R. R. (1996) J. Bacteriol. 178, 5243–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwecke T., Aparicio J. F., Molnár I., König A., Khaw L. E., Haydock S. F., Oliynyk M., Caffrey P., Cortés J., Lester J. B., Gunther A. B., Staunton J., Leadlay P. F. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7839–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu K., Chung L., Revill W. P., Katz L., Reeves C. D. (2000) Gene 251, 81–90 [DOI] [PubMed] [Google Scholar]
- 13.Motamedi H., Shafiee A. (1998) Eur. J. Biochem. 256, 528–534 [DOI] [PubMed] [Google Scholar]
- 14.Motamedi H., Cai S. J., Shafiee A., Elliston K. O. (1997) Eur. J. Biochem. 244, 74–80 [DOI] [PubMed] [Google Scholar]
- 15.Shafiee A., Cameron P. M., Boulton D. A., Kaplan L., Motamedi H. (November23, 1993) U. S. Patent 5264355(A)
- 16.Fang A., Demain A. L. (1995) Folia Microbiol. 40, 607–610 [DOI] [PubMed] [Google Scholar]
- 17.Paiva N. L., Demain A. L., Roberts M. F. (1991) J. Nat. Prod. 54, 167–177 [DOI] [PubMed] [Google Scholar]
- 18.Lowden P. A., Wilkinson B., Böhm G. A., Handa S., Floss H. G., Leadlay P. F., Staunton J. (2001) Angew. Chem. Int. Ed. Engl. 40, 777–779 [PubMed] [Google Scholar]
- 19.Gatto G. J., Jr., McLoughlin S. M., Kelleher N. L., Walsh C. T. (2005) Biochemistry 44, 5993–6002 [DOI] [PubMed] [Google Scholar]
- 20.Kato Y., Bai L., Xue Q., Revill W. P., Yu T. W., Floss H. G. (2002) J. Am. Chem. Soc. 124, 5268–5269 [DOI] [PubMed] [Google Scholar]
- 21.Shirling E. B., Gottlieb D. (1966) Int. J. Syst. Bacteriol. 16, 313–340 [Google Scholar]
- 22.Kumar P., Sharma S., Shukla A., Kumar S., Maurya R. K., Katial V., Mitra A., Gigras P. (February2, 2007) U. S. Patent EP1751272(B1)
- 23.Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J., Chen Z., Dewell S. B., Du L., Fierro J. M., Gomes X. V., Godwin B. C., He W., Helgesen S., Ho C. H., Irzyk G. P., Jando S. C., Alenquer M. L., Jarvie T. P., Jirage K. B., Kim J. B., Knight J. R., Lanza J. R., Leamon J. H., Lefkowitz S. M., Lei M., Li J., Lohman K. L., Lu H., Makhijani V. B., McDade K. E., McKenna M. P., Myers E. W., Nickerson E., Nobile J. R., Plant R., Puc B. P., Ronan M. T., Roth G. T., Sarkis G. J., Simons J. F., Simpson J. W., Srinivasan M., Tartaro K. R., Tomasz A., Vogt K. A., Volkmer G. A., Wang S. H., Wang Y., Weiner M. P., Yu P., Begley R. F., Rothberg J. M. (2005) Nature 437, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa J., Hotta K. (1999) FEMS Microbiol. Lett. 174, 251–253 [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 27.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, pp. 161–210, The John Innes Foundation, Norwich, UK [Google Scholar]
- 28.Flett F., Mersinias V., Smith C. P. (1997) FEMS Microbiol. Lett. 155, 223–229 [DOI] [PubMed] [Google Scholar]
- 29.Mandoli A., Orlandi S., Pini D., Salvadori P. (2004) Tetrahedron Asymmetry 15, 3233–3244 [Google Scholar]
- 30.Stassi D. L., Kakavas S. J., Reynolds K. A., Gunawardana G., Swanson S., Zeidner D., Jackson M., Liu H., Buko A., Katz L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7305–7309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuck J. A., McPherson M., Huang H., Jacobsen J. R., Khosla C., Cane D. E. (1997) Chem. Biol. 4, 757–766 [DOI] [PubMed] [Google Scholar]
- 32.Yuan J., Chen J. Q., Xie Z. Y., Zhai J. J., Zhou S. Y. (2008) J. Chromatogr. B Anal. Technol. Biomed Life Sci. 868, 34–41 [DOI] [PubMed] [Google Scholar]
- 33.Kölling R., Lother H. (1985) J. Bacteriol. 164, 310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frykman S., Leaf T., Carreras C., Licari P. (2001) Biotechnol. Bioeng. 76, 303–310 [DOI] [PubMed] [Google Scholar]
- 35.Jacobsen J. R., Hutchinson C. R., Cane D. E., Khosla C. (1997) Science 277, 367–369 [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Florova G., Reynolds K. A. (2005) J. Bacteriol. 187, 3795–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holden M. T., Titball R. W., Peacock S. J., Cerdeño-Tárraga A. M., Atkins T., Crossman L. C., Pitt T., Churcher C., Mungall K., Bentley S. D., Sebaihia M., Thomson N. R., Bason N., Beacham I. R., Brooks K., Brown K. A., Brown N. F., Challis G. L., Cherevach I., Chillingworth T., Cronin A., Crossett B., Davis P., DeShazer D., Feltwell T., Fraser A., Hance Z., Hauser H., Holroyd S., Jagels K., Keith K. E., Maddison M., Moule S., Price C., Quail M. A., Rabbinowitsch E., Rutherford K., Sanders M., Simmonds M., Songsivilai S., Stevens K., Tumapa S., Vesaratchavest M., Whitehead S., Yeats C., Barrell B. G., Oyston P. C., Parkhill J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14240–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iniesta A. A., McGrath P. T., Reisenauer A., McAdams H. H., Shapiro L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10935–10940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W., Reynolds K. A. (2001) J. Bacteriol. 183, 2071–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henne A., Brüggemann H., Raasch C., Wiezer A., Hartsch T., Liesegang H., Johann A., Lienard T., Gohl O., Martinez-Arias R., Jacobi C., Starkuviene V., Schlenczeck S., Dencker S., Huber R., Klenk H. P., Kramer W., Merkl R., Gottschalk G., Fritz H. J. (2004) Nat. Biotechnol. 22, 547–553 [DOI] [PubMed] [Google Scholar]
- 41.McLeod M. P., Warren R. L., Hsiao W. W., Araki N., Myhre M., Fernandes C., Miyazawa D., Wong W., Lillquist A. L., Wang D., Dosanjh M., Hara H., Petrescu A., Morin R. D., Yang G., Stott J. M., Schein J. E., Shin H., Smailus D., Siddiqui A. S., Marra M. A., Jones S. J., Holt R., Brinkman F. S., Miyauchi K., Fukuda M., Davies J. E., Mohn W. W., Eltis L. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15582–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mamaeva D. V., Morozova E. A., Nikulin A. D., Revtovich S. V., Nikonov S. V., Garber M. B., Demidkina T. V. (2005) Acta Crystallogr. Sect. C Cryst. Struct. Commun. 61, 546–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Podust L. M., Bach H., Kim Y., Lamb D. C., Arase M., Sherman D. H., Kelly S. L., Waterman M. R. (2004) Protein Sci. 13, 255–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.