Abstract
The retinoblastoma tumor suppressor gene (RB-1) is a key regulator of cellular senescence. Expression of the retinoblastoma protein (pRB) in human tumor cells that lack it results in senescence-like changes. The induction of the senescent phenotype by pRB requires the postmitotic kinase CDK5, the best known function of which is in neuronal development and postmitotic neuronal activities. Activation of CDK5 in neurons depends on its activators p35 and p39; however, little is known about how CDK5 is activated in non-neuronal senescent cells. Here we report that p35 is required for the activation of CDK5 in the process of cellular senescence. We demonstrate that: (i) p35 is expressed in osteosarcoma cells, (ii) p35 is required for CDK5 activation induced by pRB during senescence, (iii) p35 is required for the senescent morphological changes in which CDK5 is known to be involved as well as for expression of the senescence secretome, and (iv) p35 is up-regulated in senescing cells. Taken together, these results suggest that p35 is at least one of the activators of CDK5 that is mobilized in the process of cellular senescence, which may provide insight into cancer cell proliferation and future cancer therapeutics.
Keywords: Cell/Cycle, Diseases/Cancer, Enzymes/Kinase, Transcription/Target Genes, Tumor/Suppressor/Rb, Cell/Senescence
Introduction
Cellular senescence was originally described as the process of cell cycle arrest that accompanies the exhaustion of replicative potential in cultured somatic cells (1). Senescent cells display characteristic changes in cell morphology, physiology, gene expression, and typically express a senescent-associated β-galactosidase (SA-β-gal)2 activity (2, 3). Although the term replicative senescence indicates the widely accepted model of a terminal growth arrest because of telomere attrition, an apparently identical process called premature senescence can be acutely produced in response to activated oncogenes, DNA damage, oxidative stress, and suboptimal cell culture conditions (4). These observations imply that senescence is a cellular response to stress that limits the proliferation of damaged cells. Based on such antiproliferative effects, cellular senescence was proposed to be a tumor-suppressive, fail-safe mechanism that shares conceptual and possibly therapeutic similarities with the apoptosis machinery (5–7). There is now substantial evidence that cellular senescence is a bona fide barrier to tumorigenesis and cells must overcome it to progress to full-blown malignancy. For example, recent studies suggest that oncogene-induced senescence occurs and suppresses tumorigenesis in vivo. Together, the findings identify senescent cells in premalignant hyperplastic lesions but not in malignant ones, and show that oncogene-induced senescence potently restricts tumor progression at an early stage. Mutations in certain tumor suppressor genes compromise senescence, thereby contributing to cell immortalization and cancer (8–13). Furthermore, cytotoxic agents used in cancer chemotherapy can induce cellular senescence, and defects in this process contribute to drug resistance in vivo (14–17).
The RB-1 and p53 tumor suppressors are important senescence regulators. p16INK4a/pRB and p14ARF/p53 pathways are typically activated during senescence, and enforced expression of components of either signaling pathway induces senescence in some cell types (18–24). Oncogenic lesions that disable these tumor suppressor systems bypass senescence (25–30). Significantly, the role of p16INK4a/pRB in the senescence of primary cells can be recapitulated in tumor cells. The reintroduction of pRB or p16INK4a into tumor cells that lack either protein induces a premature senescence requiring p21CIP1 or, in the absence of an intact p53 pathway, p27KIP1 (31–33). Intriguingly, cyclin-dependent kinase inhibitors like p14ARF, p21CIP1, and p27KIP1, which are required for senescence, can induce markers of senescence on their own. However, they cannot mediate the senescent shape change, demonstrating that these two processes in senescence are separable (33–35).
Using several model systems of senescence, including long-term passage and acute expression of Ras or pRB, work in our laboratory has shown that cyclin-dependent kinase 5 (CDK5), a serine/threonine kinase that displays kinase activity predominantly in postmitotic neurons, plays a central role in the morphology change of senescent cells (36–38). Expression of pRB in pRB-deficient SAOS-2 cells activates CDK5 during the course of senescence. Induction of CDK5 activity leads to the phosphorylation and activation of the ERM family member, Ezrin, as well as the repression of Rac GTPase activation, which are coincident with acquisition of the pRB-induced senescent phenotypes. However, little is known about how CDK5 is activated in senescent cells induced by pRB.
In this study, we show that p35, one of the known activators of CDK5 in neurons, is required for CDK5 activation and the cell morphology change in pRB-induced SAOS-2 senescence. An increase of p35 at the mRNA level was also detected upon pRB expression in SAOS-2 cells, as well as in senescing IMR90 human diploid fibroblasts after long-term passage. These results further support a role for the CDK5/p35 pathway in regulating cellular senescence, which may provide insight into the regulatory mechanism underlying the induction of the senescent phenotype and its impact on cell proliferation and tumorigenesis.
EXPERIMENTAL PROCEDURES
Cell Culture and Recombinant Vector
The human osteosarcoma cell line SAOS-2 subclone 2.4 (39) was maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 15% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Human U2OS osteosarcoma cells and IMR90 HDFs were maintained in DMEM supplemented with 10% FBS. Cells were cultured in a 5% CO2 incubator at 37 °C. The pSVE and pSVE-Rb expression plasmids have been previously described (39, 40). The lentivirus expression plasmids pZsG and pZsG-Rb were constructed in our laboratory. The pLKO-p35shRNA-17, -18, and -20 constructs were purchased from Open Biosystems (Clone IDs: TRCN0000006217, TRCN0000006218, TRCN0000006220). SAOS-2 cells were transfected at 80% confluency with the indicated plasmids by using Fugene6 (Roche). SAOS-2 transfectants were selected with puromycin (0.5 μg/ml) 24-h post-transfection or infection and maintained under selection for the duration of the experiment.
Immunoblotting
Cells were lysed in 100–200 μl of lysis buffer (50 mm HEPES pH 8.0, 150 mm NaCl, 1 mm EDTA, 0.1% Nonidet P-40) plus protease, and phosphatase inhibitors (1 mg of aprotinin/ml, 1 μg of leupeptin/ml, 100 μg of phenylmethylsulfonyl fluoride/ml, 4 mm sodium orthovanadate, 2 mm sodium PPi) per 10-cm plate. Protein concentrations of the cell lysates were determined by the Bradford assay (Bio-Rad). For immunoblotting, 30 μg of protein was separated by SDS-PAGE and transferred to nitrocellulose membrane in a trans-blotting buffer (25 mm Tris, 192 mm glycine, 20% (v/v) methanol). Immunoblot analysis was performed as described previously (36, 39). Antibodies used for immunoblotting include: anti-Cdk5 monoclonal J-3, polyclonal C-8, and anti-p35 polyclonal C-19 antibodies (Santa Cruz Biotechnology), anti-pRB monoclonal 245 (Pharmingen), anti-Ezrin monoclonal 3C12 (NeoMarkers), anti-GAPDH monoclonal MAB374 (Chemicon), anti-actin monoclonal C-2 (Santa Cruz Biotechnology), and anti-α-tubulin monoclonal DM1A (Calbiochem). Horseradish peroxidase-conjugated donkey anti-mouse or anti-rabbit secondary antibodies (Jackson Immunosciences) were used, and signal was detected by ECL (PerkinElmer).
Immunoprecipitation and in Vitro Kinase Assays
An in vitro CDK5-associated histone H1 kinase activity (CDK5 kinase activity) assay was carried out as described by Zheng et al., with slight modifications (37, 41). Immunoprecipitation for CDK5 was performed by incubating 100 μg of cell lysate with 1 μg of anti-Cdk5 C-8 antibody (Santa Cruz Biotechnology) and 30 μl of protein A-Sepharose CL-4B beads (Amersham Biosciences) overnight at 4 °C. The beads were washed with lysis buffer, and the immunocomplexes were used to determine the kinase activity at 37 °C for 30 min, using 1 μg of histone H1 as a substrate.
Immunofluorescence
SAOS-2 cells seeded on coverslips were transfected with the indicated plasmids using Fugene6 (Roche). Cells fixed with 4% formaldehyde were incubated with phosphate-buffered saline containing 0.1% Triton X-100 and 2% bovine serum albumin for 1 h at room temperature, followed by primary antibody or rhodamine phalloidin (Cytoskeleton) incubation overnight at 4 °C. Cells were then washed with phosphate-buffered saline plus 0.1% Triton X-100 and incubated with fluorophore-conjugated secondary antibody. After staining, coverslips were mounted using Fluoromount G and visualized using a Nikon Eclipse 80i fluorescence microscope.
Quantitative Real-time PCR
Total RNA was extracted from cells transfected with pRB using TRIzol (Invitrogen). 100 ng of DNase I-treated RNA was used for first-strand cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. Quantitative PCR was carried out by employing the QuantiTect SYBR green PCR kit (Qiagen) and using 1 μl of the cDNA per reaction. The primer sequences are as follows: p35: 5′-AAGAACGCCAAGGACAAGAA-3′ and 5′-TCATTGTTGAGGTGCGTGAT-3′; GAPDH: 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GTGCGGCTGCTTCCATAA-3′; IL-6: 5′-AACCTGAACCTTCCAAAGATGG-3′ and 5′-TCTGGCTTGTTCCTCACTACT-3′; IL-8: 5′-ACTGAGAGTGATTGAGAGTGGAC-3′ and 5′-AACCCTCTGCACCCAGTTTTC-3′; MMP2: 5′-CCGTCGCCCATCATCAAGTT-3′ and 5′-CTGTCTGGGGCAGTCCAAAG-3′; MMP3: 5′-ATGGACAAAGGATACAACAGGGA-3′ and 5′-TGTGAGTGAGTGATAGAGTGGG-3′; PAI-1: 5′-GCTTGTCCAAGAGTGCATGGT-3′ and 5′-AGGGCTGGTTCTCGATGGT-3′. Relative quantification of gene expression was carried out by the comparative C(T) method (42).
Senescence-associated β-Galactosidase Staining Assay
SAOS-2 cells were cotransfected with the indicated plasmids. Transfectants were selected with puromycin (0.5 μg/ml) 24-h post-transfection for the duration of the experiment. The staining for perinuclear SA-β-gal activity was performed as described (2). Briefly, cells were washed in phosphate-buffered saline and fixed in 2% formaldehyde/0.2% glutaraldehyde. Cells were then washed and incubated at 37 °C overnight with fresh senescence-associated-β-galactosidase staining solution (1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside per ml, 40 mm citric acid/sodium phosphate (pH 6), 150 mm NaCl, 2 mm MgCl2, 5 mm potassium ferrocyanide, 5 mm potassium ferricyanide).
RESULTS
Expression of p35 in SAOS-2 and U2OS Cells
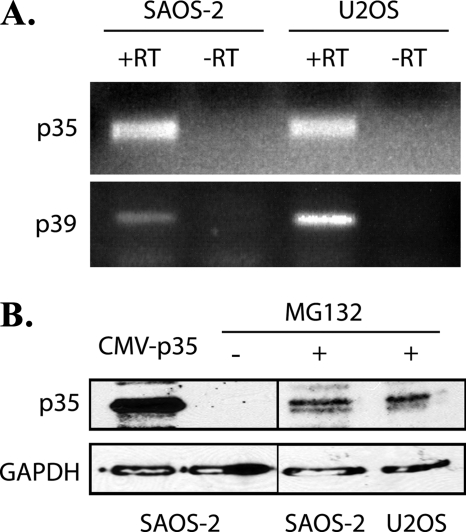
By using several model systems of senescence, we previously showed that cyclin-dependent kinase 5 (CDK5) activation is up-regulated in senescing cells (36). CDK5 kinase activity is stimulated by either one of two non-cyclin regulatory proteins, p35 and p39. Although CDK5 expression is ubiquitous in mammalian tissues, the expression of its activators is largely restricted to post-mitotic neurons (43). No other activators are known. Thus, we asked if p35 and p39 are also expressed in the osteosarcoma cell lines used as senescence models in our study. The expression of mRNA encoding p35 and p39 in SAOS-2 (pRB-deficient) and U2OS (wtpRB) cells was detected by RT-PCR (Fig. 1A). Immunoblotting for p35 was then performed with α-p35 (C19) polyclonal antibody. Because of a short half-life of less than 30 min (44), p35 only became detectable when U2OS and SAOS-2 cells were treated with the proteasome inhibitor MG132 (20 μm) for 16 h (Fig. 1B). These experiments indicate that both p35 and p39 are candidates for CDK5 activation in senescing cells. Because of a lack of appropriate reagents for analyzing p39, the following experiments focused on p35.
FIGURE 1.
Expression of p35 and p39 in SAOS-2 and U2OS cells. A, RT-PCR for p35 and p39 from the total RNA of SAOS-2 and U2OS cells. Lane -RT, negative control using RNA without reverse transcription. B, SAOS-2 and U2OS cells were treated with MG132 (20 μm) for 16 h before harvest, whole cell extracts (30 μg of protein per lane) were analyzed by immunoblotting with anti-p35 antibody, the ectopically expressed p35 was used as positive control.
Requirement of p35 for pRB-induced CDK5 Activation in SAOS-2
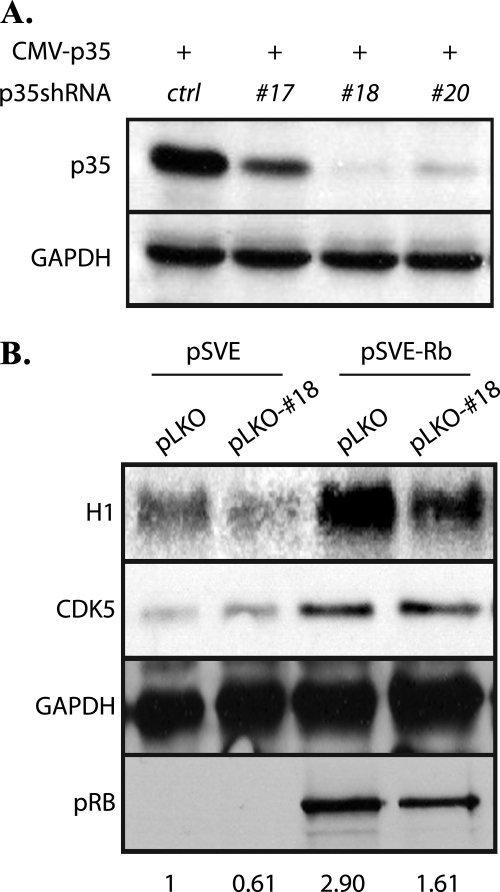
Both the protein levels and activity of CDK5 were shown to be up-regulated in pRB-transfected SAOS-2 cells (36). We therefore investigated the involvement of p35 in CDK5 activation. Using both retrovirus (data not shown) and lentivirus-based p35 shRNA constructs (Fig. 2A), the ectopically expressed p35 was shown to be efficiently knocked-down in SAOS-2 cells. SAOS-2 cells were then cotransfected with a pRB expression vector and a p35 shRNA construct or control empty vectors. Cells were harvested for cell lysates 5 days after pRB reintroduction. Performance of a CDK5 immunoprecipitation and a subsequent in vitro kinase assay using histone H1 as a substrate showed that the activity of CDK5 decreased upon knocking down p35, despite the continued presence of pRB (Fig. 2B). The data suggest that p35 is an activator of CDK5 in pRB-induced senescence in SAOS-2 cells.
FIGURE 2.
Knockdown of p35 decreases CDK5 activity in SAOS-2 Cells. A, efficacy of lentivirus-based shRNAs for p35 was tested by cotransfecting pCMV-p35 and pLKO-p35shRNA-17, -18, -20, or control empty vector into SAOS-2 cells. Cell lysates were collected 2 days after transfection and were immunoblotted for p35. B, pSVE or pSVE-Rb was cotransfected with pLKO-shp35–18 or pLKO vector into SAOS-2 cells. Cells were selected with puromycin (0.5 μg/ml) 2 days after transfection, cell lysates were collected on day 5 and immunoprecipitated with anti-Cdk5 antibody to perform an in vitro kinase assay using histone H1 as the substrate. Cell lysates were also analyzed for CDK5 and pRB expression by immunoblotting. Numbers at the bottom of panels show ratio of kinase activity in each lysate to control lysate.
Impact of p35 on Senescent Morphology Change in SAOS-2 and U2OS Cells
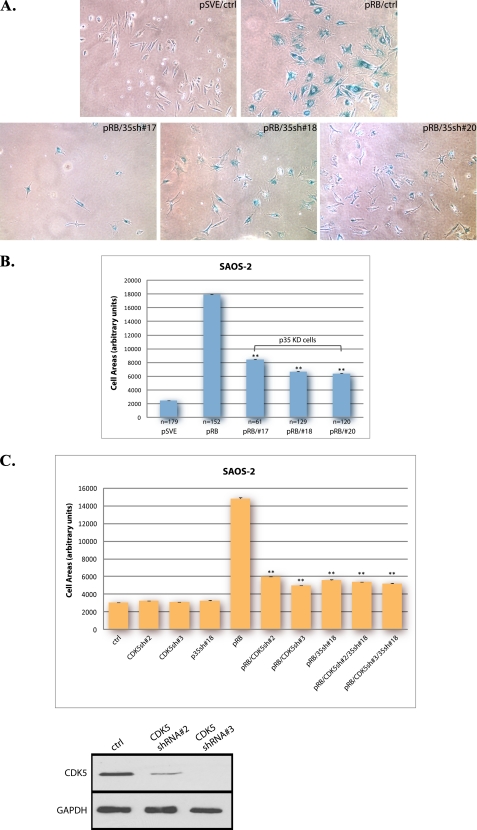
Ectopic expression of pRB in the SAOS-2 osteosarcoma cell line produces a response that displays many characteristics of senescence in primary diploid cells, including the distinct morphological change termed flat cell formation, typified by an increased cell area and a flattened appearance. This phenotype appears identical to that observed during classical senescence and is taken as an indicator of the senescent state (31, 33, 39). Because we previously found that CDK5 was required for this aspect of senescent cell morphology (36), we next investigated the role of p35 in the senescence phenotype. SAOS-2 cells cotransfected with a pRB expression vector and p35 shRNA constructs were examined for senescence by SA-β-gal assay 10 days post-transfection. Cells transfected with empty vectors were used as a control (Fig. 3A). Knockdown of p35 antagonized the formation of pRB-induced senescent flat cells, and cells in the knockdown groups were generally smaller. To better analyze the shape change, cell area was measured in each group and subjected to statistical analysis (Fig. 3B). The result showed that by knocking down p35, the average area of pRB-induced senescent cells was reduced, and this difference was statistically significant, supporting a role for p35 as a regulator of the senescent phenotype. To further confirm the specificity of the p35 knockdowns, shape changes were analyzed in pRB-transfected cells in which CDK5, p35, or both were subjected to knockdown (Fig. 3C). By measuring cell area, cells in each knockdown group were shown to have significantly reduced flat cell formation compared with pRB control cells, but there were no significant differences among all knockdown groups. In the absence of CDK5, changes in p35 levels did not confer further reduction in cell area, suggesting that the effects of p35 on senescence are mediated through CDK5.
FIGURE 3.
Knockdown of p35 reduces pRB-induced flat cell formation in SAOS-2 cells. A, SAOS-2 cells were cotransfected with pSVE-Rb and pLKO-p35shRNA-17, -18, -20, or control empty vector, cells transfected with empty vectors were use as control. 16 h after transfection, cells were selected with puromycin at a final concentration of 0.5 μg/ml. 10 days post-transfection, cells were assayed for SA-β-gal expression, and the morphology change in these cells is shown. All images were obtained by using phase contrast microscopy at a magnification of ×10. B, cell area in pRB-induced senescent SAOS-2 cells was measured using ImageJ. Means and S.E. of data collected in each group are presented in the column chart. To compare the difference in size between pRB-transfectant cells and p35-knockdown cells, p values were determined using both unpaired two-tailed Student's t test and one-way ANOVA. A p value < 0.001 is indicated by **. The experiment was repeated three times. C, pSVE or pSVE-Rb was transfected into SAOS-2 cells with MKO-CDK5 shRNA2, -3, and/or pLKO-p35shRNA-18. Cell area in each group was measured using ImageJ. Means and S.E. of three experiments are presented in the column chart. A p value < 0.001 versus pRB control is indicated by **. Bottom panels, the efficacy of shRNAs for CDK5 was tested by transfecting MKO-CDK5shRNA-2, -3, or control empty vector into SAOS-2 cells. Cell lysates were collected 2 days after transfection and were immunoblotted for CDK5.
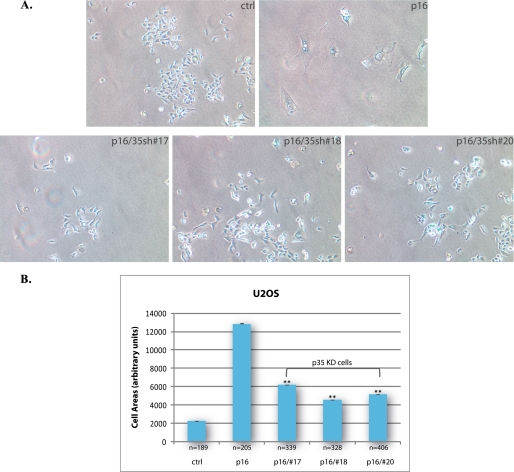
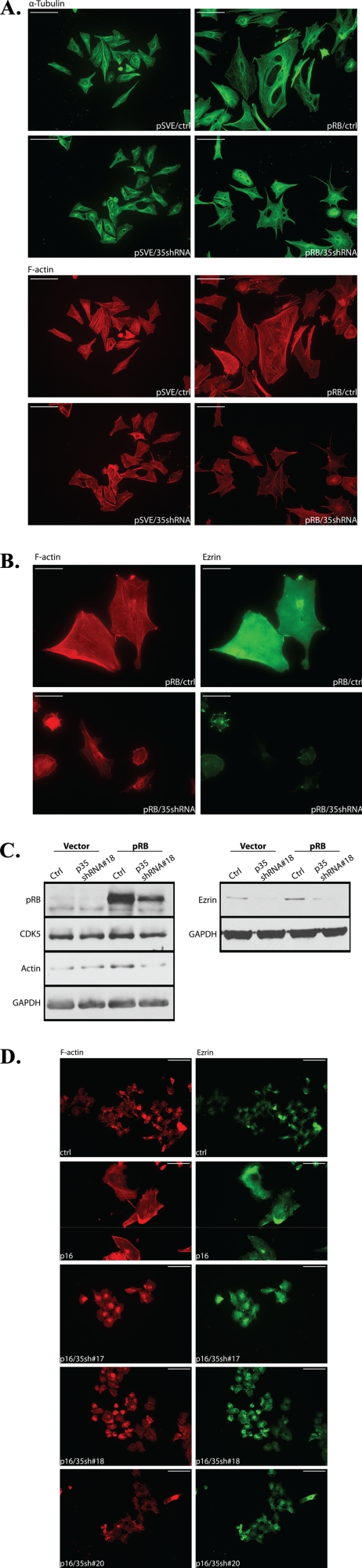
To demonstrate that similar results can be observed in other cells that lack a functional pRB pathway, we used the human osteosarcoma cell line U2OS, which is p16-deficient. By reintroducing p16 into these cells, a premature senescence response can also be induced as in SAOS-2 (32). Firstly, U2OS cells cotransfected with a p16 expression vector and p35 shRNA constructs were examined for senescence by SA-β-gal assay 10 days post-transfection, using cells transfected with empty vectors as a control (Fig. 4A). Secondly, shape changes were quantified by measuring cell area in each group (Fig. 4B). The results showed that knockdown of p35 in U2OS significantly suppressed the senescent morphology change induced by p16 overexpression.
FIGURE 4.
Knockdown of p35 reduces p16-induced flat cell formation in U2OS cells. A, U2OS cells were cotransfected with CMV-p16 and pLKO-p35shRNA-17, -18, -20, or control empty vector, cells transfected with empty vectors were use as control. 16 h after transfection, cells were selected with puromycin at a final concentration of 2 μg/ml. 10 days post-transfection, cells were assayed for SA-β-gal expression and the morphology change in these cells is shown. All images were obtained by using phase contrast microscopy at a magnification of ×10. B, cell area in each group of cells was measured using ImageJ. Means and S.E. of three experiments are presented in the column chart. A p value < 0.001 versus p16 control is indicated by **.
p35 Regulates Cytoskeletal Organization in Senescent SAOS-2 and U2OS Cells
CDK5-dependent flattening of senescent cells suggests that extensive changes must occur to the cellular cytoskeletons as cells undergo senescence. CDK5 has known roles in regulating cell shape through actin reorganization. With this in mind, we investigated the effect of p35 on the changes in cytoskeleton structure as cells underwent senescence. SAOS-2 cells cotransfected with a pRB expression vector and a p35 shRNA construct or control vectors were fixed with 4% formaldehyde 10 days post-transfection and immunofluorescence staining for cytoskeleton organization in the samples was performed by using rhodamine phalloidin or α-tubulin antibody. No significant change in microtubule arrangement was observed (Fig. 5A, upper panels), whereas knockdown of p35 interfered with actin polymerization. In the knockdown group, cells exhibited less intense F-actin fiber staining, along with a reduction in size (Fig. 5A, lower panels). Further, immunoblotting showed that the level of actin was reduced by p35 knockdown (Fig. 5C).
FIGURE 5.
Knockdown of p35 affects cytoskeletal organization in senescent SAOS-2 and U2OS cells. A, SAOS-2 cells were seeded on coverslips and cotransfected with pSVE or pSVE-Rb and pLKO-p35shRNA-18 or control pLKO vector. 16 h after transfection, cells were selected with puromycin at a final concentration of 0.5 μg/ml. Samples were collected 10 days post-transfection and fixed with 4% formaldehyde. Immunofluorescence staining was performed using anti-α-tubulin antibody (DM1A) (upper panels) and rhodamine-phalloidin (lower panels), to show the structure of microtubules and F-actin filaments, respectively. Scale bars: 100 μm. B, immunofluorescence staining for F-actin (red) and Ezrin (green) in SAOS-2 cells cotransfected with pSVE-Rb and pLKO-p35shRNA-18 or control vectors following 8 days of selection. Scale bars: 100 μm. C, immunoblotting for actin and Ezrin in SAOS-2 cells transfected with pSVE-Rb and pLKO-p35shRNA-18 or control vectors following 5 days of selection. D, immunofluorescence staining for F-actin (red) and Ezrin (green) in U2OS cells cotransfected with CMV-p16 and pLKO-p35shRNA-17, -18, -20, or control vectors following 8 days of puromycin selection (2 μg/ml). Scale bars: 100 μm.
Previous studies in our laboratory have shown that the expression of the ERM family member, Ezrin, an actin-binding protein involved in membrane-cytoskeletal signaling, is increased upon pRB-induced senescence. The activation of Ezrin appears to be the consequence of direct phosphorylation of Thr-235 of the protein by CDK5, which is activated in response to pRB expression (37). We thus also checked the status of Ezrin in SAOS-2 cells transfected with pRB and p35 shRNA constructs or control vectors. p35 knockdown cells showed reduced immunofluorescence staining for both F-actin and Ezrin (Fig. 5B), compared with control senescent cells induced by pRB, that was accompanied by an overall decrease in Ezrin levels (Fig. 5C). Furthermore, similar results were observed in U2OS cells (Fig. 5D). p35 knockdown cells showed less intense staining for both F-actin and Ezrin when compared with p16-induced senescent cells. The localization of Ezrin in p35 knockdown cells also remained mostly cytosolic as in control cells, whereas in senescent cells Ezrin was relocated to the membrane periphery. Taken together, these data suggest that p35 is required for the acquisition of the senescent morphology in pRB-induced senescence, which is consistent with our previous finding that CDK5 activity is required in such changes.
Expression of p35 Is Increased during Cellular Senescence
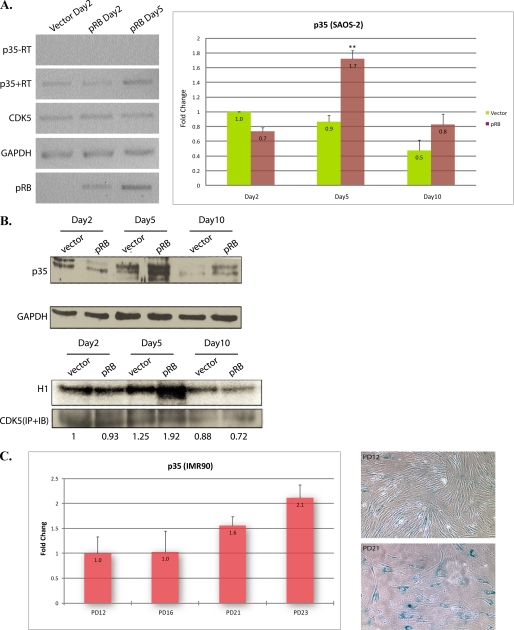
To understand if the regulation of p35 mRNA or protein levels is a significant determinant of CDK5 activity during pRB-induced senescence, SAOS-2 cells infected with pRB lentivirus or control GFP virus were collected at 2, 5, and 10 days post-infection. Both semi-quantitative RT-PCR and quantitative real-time PCR were performed to examine p35 mRNA levels in these samples. Compared with control, pRB infectants showed an approximate 2-fold increase in p35 levels at day 5 and a slight increase at day 10 (Fig. 6A). Concomitantly, both endogenous p35 protein levels and CDK5 activity increased 5 days after pRB infection (Fig. 6B).
FIGURE 6.
p35 is increased in senescing SAOS-2 and IMR90 cells. A, SAOS-2 cells were infected with pRB or control GFP lentiviruses. 16 h after infection, cells were selected with puromycin at a final concentration of 0.5 μg/ml. p35 mRNA levels at 2, 5, and 10 days post-transfection were examined by semi-quantitative RT-PCR (left), -RT, negative control using RNA samples not subjected to reverse transcription, or quantitative real-time PCR in triplicate (right), error bars show S.D. **, p < 0.001 versus day 2 empty vector control. B, cell lysates were prepared from cells infected with pRB or control GFP lentiviruses, followed by MG132 (20 μm) treatment for 16 h before harvest. Immunoblotting was performed using anti-p35 antibody (upper panels), and immunoprecipitation with anti-Cdk5 antibody was performed followed by an in vitro kinase assay using histone H1 as the substrate (lower panels). Numbers at the bottom of panels show ratio of kinase activity in each lysate to control lysate of day2. C, real-time PCR (in triplicate) for p35 (left) was performed with total mRNA prepared from early and middle passage IMR90 cells, error bars show S.D. SA-β-gal assay (right) was performed on IMR90 cells at early and middle passage.
We have also shown that CDK5 is activated in different model systems of senescence, including long-term passage and acute expression of Ras or pRB (36). To see if the increase of p35 expression is a general phenomenon in cells undergoing senescence, we induced senescence in IMR90 HDFs by long-term passage. The p35 mRNA levels were examined by real-time PCR in early and mid passages of IMR90 human diploid fibroblasts (Fig. 6C, left panel). This showed that expression of p35 mRNA was also up-regulated in senescing IMR90 HDFs. Concomitantly, SA-β-gal staining showed more senescent cells at later passage (Fig. 6C, right panel). These findings strongly suggest that p35 regulates CDK5 activity during the process of senescence.
p35 Regulates Senescence Secretome in SAOS-2 Cells
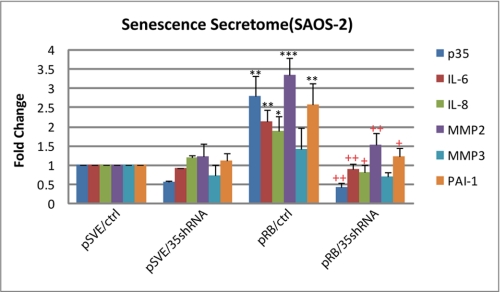
Our previous work and that shown above clearly implicate p35/cdk5 in the senescent cell shape change. However, the physiological significance of this shape change and its impact on the microenvironment of the senescent cell is unclear. To determine if p35/cdk5 could mediate senescent cell processes that may directly affect tissue homeostasis and/or tumor formation, we chose to analyze the impact of p35 knockdown on the expression of genes encoding secreted factors. This analysis was based on increasing evidence that senescent cells show altered expression of many secreted gene products that regulate proliferation (i.e. increased production of PAI-1), impact the immune response (i.e. increased amounts of IL-6, IL-8), and influence remodeling of the extracellular stroma or matrix (ECM), (i.e. increased production of matrix metalloproteinase 1(MMP1) and MMP3) (45). To understand if the regulation of p35 mRNA affects the senescence secretome in pRB-induced senescent SAOS-2 cells, cells cotransfected with a pRB expression vector and a p35 shRNA construct or control vectors were subjected to quantitative real-time PCR for examining changes in a variety of secreted factors in these samples (Fig. 7). Compared with control, pRB transfectants showed a significant increase in all the factors analyzed except MMP3, whereas upon p35 knockdown, the production of these factors were suppressed. This suggests that p35 not only participates in the regulation of cytoskeletal structures but affects other aspects of senescence as well.
FIGURE 7.
Knockdown of p35 affects senescence secretome in SAOS-2 cells. SAOS-2 cells were cotransfected with pSVE or pSVE-Rb and pLKO-p35shRNA-18 or control pLKO vector. 16 h after transfection, cells were selected with puromycin at a final concentration of 0.5 μg/ml. Samples were collected 5 days post-transfection and mRNA levels for p35, IL-6, IL-8, MMP-2, MMP3, and PAI-1 were examined by quantitative real-time PCR (in triplicate). Error bars show S.D. *, p < 0.05 versus control; **, p < 0.01 versus control; ***, p < 0.001 versus control; +, p < 0.05 versus pRB; ++, p < 0.01 versus pRB.
DISCUSSION
We have previously reported the activation of CDK5 in human primary and tumor cells induced to senesce by a variety of stimuli, and the activity of CDK5 is necessary for proper acquisition of the cytoskeletal changes accompanying senescence. Discovery of this role for CDK5 was unexpected, because its activity has primarily been associated with post-mitotic neurons. However, given that CDK5 is ubiquitously expressed in mammalian tissues, an increasing body of evidence has established CDK5 kinase activity and functions in non-neuronal cells. In this report we show the presence of the CDK5 activators, p35 and p39, in our model systems of senescence. Up-regulation of the expression of p35 is concomitant with CDK5 activation in senescing SAOS2 and IMR90 cells. Knockdown of p35 by shRNA markedly suppresses the morphologic changes of senescent SAOS-2 cells induced by pRB, which coincides with a decrease in CDK5 activity. The polymerization of actin filaments is inhibited in the p35 knockdown cells, and the levels of actin and the F-actin-associated Ezrin are reduced as well. These findings underscore a role for CDK5/p35 activity in mediating the cytoskeletal reorganization in the non-neuronal senescent cells. Furthermore, the use of model systems in which senescence is induced by pRB reintroduction in p16Ink4a/pRB-deficient tumor cells strongly implicates the pRB pathway in CDK5/p35 up-regulation at least in part through transcriptional up-regulation of p35. Although the pRB pathway is essential for the transcriptional repression of loci in senescent cells (46), its role in the induction of gene expression in senescence is poorly understood. Because pRB/E2F complexes are usually repressive, they most likely do not directly regulate the genes that are highly expressed by senescent cells, although they could indirectly control such genes, for example, by silencing a repressor. At present, we have been unable to test for a direct role for pRB in regulating p35 mRNA production, as reporter constructs containing the p35 promoter do not respond to pRB (data not shown). It is possible that pRB effects on the p35 promoter, whether direct or indirect, require an appropriate chromatin context as observed for differentiation-specific promoters (47).
Senescent cells show striking changes in gene expression. Interestingly, many changes in gene expression appear not to be directly related to growth arrest. Despite the universality of morphological changes observed in a wide variety of senescent cells, little is known about the potential contribution of this phenotype to the establishment or maintenance of the irreversible growth arrest that accompanies senescence. Significant additional studies of the role of cytoskeletal rearrangement in the biochemical and proliferative aspects typical of senescence are needed to fully appreciate the role of CDK5/p35 in this process. The role of CDK5/p35 activity in regulating the differentiation of monocytes might provide some clues. In promyelocytic HL60 cells, treatment with 1,25-dihydroxyvitamin D3 (1,25D3) results in the up-regulation of the Egr1 gene, which in turn activates p35 transcription and expression, and subsequently enhances CDK5 activity. p35-associated CDK5 phosphorylates MEK1 on Thr-286, preventing MAPK/ERK phosphorylation and cell proliferation. However, MEK1 phosphorylation at Thr-286 requires prior Ser-218 and Ser-222 phosphorylation and activation by Raf1 induced by growth factors or cytokines. These events are suggested to result in the up-regulation of p27 and eventually, monocytic differentiation (48, 49). Given the established role of pRB in osteoblast differentiation (47), it is possible that complex mechanisms such as these are stimulated by pRB in senescing mesenchymal cells that are unable to properly differentiate.
There is now substantial evidence that induction of senescence constitutes an important block to tumor progression. It has also become clear that senescent cells have characteristic alterations in secreted growth factors, inflammatory cytokines, extracellular-matrix components, and matrix-degrading enzymes, which could influence the growth of adjacent neighboring tumor cells by altering the tissue microenvironment (4, 45). We find that p35/cdk5 can influence this senescence secretome, implicating cdk5 in the physiological response to this aspect of senescence. Further, recent studies suggest that, as with CDK5 in neurons, the CDK5 activity in non-neuronal cells may influence phenotypic changes mostly through its direct or indirect effect on the organization of cytoskeletal structures. CDK5 regulates cellular processes such as cell-cell and cell-matrix adhesion, cell migration and wound healing in a variety of tissues (49). Thus the induction of senescent shape change may also impact the neighboring tumor cells and their microenvironment.
Tumor cells are exposed to many sources of stress, especially those derived from the aberrant proliferative signals of oncogenes. As one of the cellular responses to these stresses, senescence has emerged as a compelling target for future cancer therapeutics and may determine the response of tumor cells to chemotherapeutic drugs. Because senescence effector mechanisms are still largely unknown, understanding the molecular mechanisms through which CDK5 affects cellular senescence may give clues to potential therapeutic approaches for cancer and age-related diseases. Finally, the intriguing relationship of this aspect of senescence to neuronal and non-neuronal differentiation (48, 50, 51) may help to unravel the role of senescence effectors in the physiology of tissues that accumulate a senescent cell burden following acute or chronic stress.
Acknowledgment
We thank Li-Huei Tsai for critical CDK5-related reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants CA104322 and AG020208 (to P. W. H.).
- SA-β-gal
- senescence-associated β-galactosidase
- RB-1
- retinoblastoma gene
- pRB
- retinoblastoma protein
- CDK5
- cyclin-dependent kinase 5
- HDF
- human diploid fibroblasts
- MMP
- matrix metalloproteinase.
REFERENCES
- 1.Hayflick L. (1965) Exp. Cell Res. 37, 614–636 [DOI] [PubMed] [Google Scholar]
- 2.Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O., et al. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shelton D. N., Chang E., Whittier P. S., Choi D., Funk W. D. (1999) Curr. Biol. 9, 939–945 [DOI] [PubMed] [Google Scholar]
- 4.Campisi J., d'Adda di Fagagna F. (2007) Nat. Rev. Mol. Cell Biol. 8, 729–740 [DOI] [PubMed] [Google Scholar]
- 5.Campisi J. (2001) Trends Cell Biol. 11, S27–31 [DOI] [PubMed] [Google Scholar]
- 6.Mathon N. F., Lloyd A. C. (2001) Nat. Rev. Cancer 1, 203–213 [DOI] [PubMed] [Google Scholar]
- 7.Braig M., Schmitt C. A. (2006) Cancer Res. 66, 2881–2884 [DOI] [PubMed] [Google Scholar]
- 8.Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A. H., Schlegelberger B., Stein H., Dörken B., Jenuwein T., Schmitt C. A. (2005) Nature 436, 660–665 [DOI] [PubMed] [Google Scholar]
- 9.Chen Z., Trotman L. C., Shaffer D., Lin H. K., Dotan Z. A., Niki M., Koutcher J. A., Scher H. I., Ludwig T., Gerald W., Cordon-Cardo C., Pandolfi P. P. (2005) Nature 436, 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A. J., Barradas M., Benguriá A., Zaballos A., Flores J. M., Barbacid M., Beach D., Serrano M. (2005) Nature 436, 642. [DOI] [PubMed] [Google Scholar]
- 11.Michaloglou C., Vredeveld L. C., Soengas M. S., Denoyelle C., Kuilman T., van der Horst C. M., Majoor D. M., Shay J. W., Mooi W. J., Peeper D. S. (2005) Nature 436, 720–724 [DOI] [PubMed] [Google Scholar]
- 12.Lazzerini Denchi E., Attwooll C., Pasini D., Helin K. (2005) Mol. Cell Biol. 25, 2660–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L. V., Kolettas E., Niforou K., Zoumpourlis V. C., Takaoka M., Nakagawa H., Tort F., Fugger K., Johansson F., Sehested M., Andersen C. L., Dyrskjot L., Ørntoft T., Lukas J., Kittas C., Helleday T., Halazonetis T. D., Bartek J., Gorgoulis V. G. (2006) Nature 444, 633–637 [DOI] [PubMed] [Google Scholar]
- 14.Chang B. D., Broude E. V., Dokmanovic M., Zhu H., Ruth A., Xuan Y., Kandel E. S., Lausch E., Christov K., Roninson I. B. (1999) Cancer Res. 59, 3761–3767 [PubMed] [Google Scholar]
- 15.Schmitt C. A., Fridman J. S., Yang M., Lee S., Baranov E., Hoffman R. M., Lowe S. W. (2002) Cell 109, 335–346 [DOI] [PubMed] [Google Scholar]
- 16.te Poele R. H., Okorokov A. L., Jardine L., Cummings J., Joel S. P. (2002) Cancer Res. 62, 1876–1883 [PubMed] [Google Scholar]
- 17.Roberson R. S., Kussick S. J., Vallieres E., Chen S. Y., Wu D. Y. (2005) Cancer Res. 65, 2795–2803 [DOI] [PubMed] [Google Scholar]
- 18.Stein G. H., Beeson M., Gordon L. (1990) Science 249, 666–669 [DOI] [PubMed] [Google Scholar]
- 19.Lin A. W., Barradas M., Stone J. C., van Aelst L., Serrano M., Lowe S. W. (1998) Genes Dev. 12, 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein G. H., Drullinger L. F., Soulard A., Dulić V. (1999) Mol. Cell Biol. 19, 2109–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin A. W., Lowe S. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5025–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly-Spratt K. S., Gurley K. E., Yasui Y., Kemp C. J. (2004) PLoS Biol. 2, E242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimri G. P., Itahana K., Acosta M., Campisi J. (2000) Mol. Cell Biol. 20, 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferbeyre G., de Stanchina E., Lin A. W., Querido E., McCurrach M. E., Hannon G. J., Lowe S. W. (2002) Mol. Cell Biol. 22, 3497–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shay J. W., Pereira-Smith O. M., Wright W. E. (1991) Exp. Cell Res. 196, 33–39 [DOI] [PubMed] [Google Scholar]
- 26.Brown J. P., Wei W., Sedivy J. M. (1997) Science 277, 831–834 [DOI] [PubMed] [Google Scholar]
- 27.Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997) Cell 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 28.Brookes S., Rowe J., Ruas M., Llanos S., Clark P. A., Lomax M., James M. C., Vatcheva R., Bates S., Vousden K. H., Parry D., Gruis N., Smit N., Bergman W., Peters G. (2002) EMBO J. 21, 2936–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rheinwald J. G., Hahn W. C., Ramsey M. R., Wu J. Y., Guo Z., Tsao H., De Luca M., Catricalà C., O'Toole K. M. (2002) Mol. Cell Biol. 22, 5157–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beauséjour C. M., Krtolica A., Galimi F., Narita M., Lowe S. W., Yaswen P., Campisi J. (2003) EMBO J. 22, 4212–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu H. J., Zhou Y., Ji W., Perng G. S., Kruzelock R., Kong C. T., Bast R. C., Mills G. B., Li J., Hu S. X. (1997) Oncogene 15, 2589–2596 [DOI] [PubMed] [Google Scholar]
- 32.Dai C. Y., Enders G. H. (2000) Oncogene 19, 1613–1622 [DOI] [PubMed] [Google Scholar]
- 33.Alexander K., Hinds P. W. (2001) Mol. Cell Biol. 21, 3616–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dulić V., Beney G. E., Frebourg G., Drullinger L. F., Stein G. H. (2000) Mol. Cell Biol. 20, 6741–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collado M., Medema R. H., Garcia-Cao I., Dubuisson M. L., Barradas M., Glassford J., Rivas C., Burgering B. M., Serrano M., Lam E. W. (2000) J. Biol. Chem. 275, 21960–21968 [DOI] [PubMed] [Google Scholar]
- 36.Alexander K., Yang H. S., Hinds P. W. (2004) Mol. Cell Biol. 24, 2808–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H. S., Hinds P. W. (2003) Mol. Cell 11, 1163–1176 [DOI] [PubMed] [Google Scholar]
- 38.Yang H. S., Hinds P. W. (2006) Cancer Res. 66, 2708–2715 [DOI] [PubMed] [Google Scholar]
- 39.Hinds P. W., Mittnacht S., Dulic V., Arnold A., Reed S. I., Weinberg R. A. (1992) Cell 70, 993–1006 [DOI] [PubMed] [Google Scholar]
- 40.Mittnacht S., Weinberg R. A. (1991) Cell 65, 381–393 [DOI] [PubMed] [Google Scholar]
- 41.Zheng M., Leung C. L., Liem R. K. (1998) J. Neurobiol. 35, 141–159 [DOI] [PubMed] [Google Scholar]
- 42.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 43.Dhavan R., Tsai L. H. (2001) Nat. Rev. Mol. Cell Biol. 2, 749–759 [DOI] [PubMed] [Google Scholar]
- 44.Patrick G. N., Zhou P., Kwon Y. T., Howley P. M., Tsai L. H. (1998) J. Biol. Chem. 273, 24057–24064 [DOI] [PubMed] [Google Scholar]
- 45.Adams P. D. (2009) Mol. Cell 36, 2–14 [DOI] [PubMed] [Google Scholar]
- 46.Narita M., Nũnez S., Heard E., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. (2003) Cell 113, 703–716 [DOI] [PubMed] [Google Scholar]
- 47.Thomas D. M., Carty S. A., Piscopo D. M., Lee J. S., Wang W. F., Forrester W. C., Hinds P. W. (2001) Mol. Cell 8, 303–316 [DOI] [PubMed] [Google Scholar]
- 48.Chen F., Wang Q., Wang X., Studzinski G. P. (2004) Cancer Res. 64, 5425–5433 [DOI] [PubMed] [Google Scholar]
- 49.Rosales J. L., Lee K. Y. (2006) Bioessays 28, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 50.Lazaro J. B., Kitzmann M., Poul M. A., Vandromme M., Lamb N. J., Fernandez A. (1997) J. Cell Sci. 110, 1251–1260 [DOI] [PubMed] [Google Scholar]
- 51.Sarker K. P., Lee K. Y. (2004) Oncogene 23, 6064–6070 [DOI] [PubMed] [Google Scholar]