Abstract
Diabetes -associated hyperlipidemia is generally attributed to reduced clearance of plasma lipoproteins, especially remnant lipoproteins enriched in cholesterol and triglycerides. Hepatic clearance of remnants occurs via low density lipoprotein receptors and the heparan sulfate proteoglycan, syndecan-1. Previous studies have suggested alterations in heparan sulfate proteoglycan metabolism in rat and mouse diabetic models, consistent with the idea that diabetic dyslipidemia might be caused by alterations in proteoglycan expression in the liver. In this study we analyzed the content and composition of liver heparan sulfate in streptozotocin-induced insulin-deficient diabetic mice that displayed fasting hypertriglyceridemia and delayed clearance of dietary triglyceride-rich lipoproteins. No differences between normal and diabetic littermates in liver heparan sulfate content, sulfation, syndecan-1 protein levels, or affinity for heparin-binding ligands, such as apolipoprotein E or fibroblast growth factor-2, were noted. Decreased incorporation of [35S]sulfate in insulin-deficient mice in vivo was observed, but the decrease was due to increased plasma inorganic sulfate, which reduced the efficiency of labeling of liver heparan sulfate. These results show that hyperlipidemia in insulin-deficient mice is not due to changes in hepatic heparan sulfate composition.
Keywords: Diabetes, Heparan Sulfate, Liver Metabolism, Proteoglycan Synthesis, Triglyceride, Sulfation, Syndecan
Introduction
Hypertriglyceridemia is a significant complication of insulin-dependent diabetes mellitus (IDDM)2 that likely contributes to cardiovascular disease in affected individuals, but its cause remains unknown (1–3). Insulin deficiency suppresses hepatic triglyceride production (4, 5), suggesting that increased plasma triglyceride levels might result from decreased catabolism of triglyceride-rich lipoproteins (TRL). In various diabetic models delayed TRL remnant clearance has been attributed to altered expression of heparan sulfate proteoglycans (HSPGs) in the liver (6–14).
We recently showed that mutant mice lacking the plasma membrane HSPG, syndecan-1, exhibit hypertriglyceridemia due to delayed clearance of TRL remnants from the circulation associated with reduced VLDL binding, uptake, and degradation in isolated hepatocytes (15). Furthermore, mice with undersulfated liver heparan sulfate have the same phenotype (16, 17). One of mutants lacked the enzyme N-acetylglucosamine N-deacetylase/N-sulfotransferase 1 (Ndst1), a biosynthetic enzyme that regulates the overall level of sulfation of heparan sulfate glycosaminoglycans. In vivo studies have suggested that IDDM causes reduced expression of Ndst1 (6, 10, 11, 13, 14, 18), leading to the hypothesis that hypertriglyceridemia was caused by undersulfated heparan sulfate in the liver.
In this work, we show that mice with IDDM exhibit reduced [35S]sulfate incorporation into hepatic heparan sulfate. However, the reduction was caused by changes in plasma sulfate concentration after the onset of diabetes rather than any change in heparan sulfate biosynthesis. The application of mass spectrometry showed that hepatic heparan sulfate did not change in content and composition in diabetic mice, and the binding of apolipoprotein E (apoE) and fibroblast growth factor-2 was unaltered. Similarly, no alteration of syndecan-1 protein level was observed in freshly isolated hepatocytes from diabetic mice. Thus, fasting and postprandial hypertriglyceridemia in IDDM does not correlate with altered hepatic heparan sulfate in mice.
EXPERIMENTAL PROCEDURES
Mice and Induction of Diabetes
Male C57BL/6 mice (4 weeks of age) were purchased from Jackson Laboratory and maintained in a temperature-controlled (25 °C) facility with a 12-h light/dark cycle. The derivation and genotyping of Ndst1f/fAlbCre+ mice have been described previously (16). Mice were fed laboratory rodent chow (Harlan-Teklad) ad libitum except when fasting blood specimens were obtained. Mice were made diabetic by administering 50 mg/kg body weight of streptozotocin (STZ; Sigma) intraperitoneally for 5 consecutive days. Because of variations in plasma triglycerides in females, only male mice were used in this study.
Plasma Glucose and Lipid Measurements
Animals were fasted for 4 h in the morning. Blood glucose levels were monitored using a glucose monitor (Abbott) and test strips after a small tail nick. All experiments were carried out in animals with blood glucose levels between 400 and 500 mg/dl. Prior to experiments, plasma was prepared from retroorbital sinus bleeds to analyze glucose, cholesterol, and triglyceride levels enzymatically (Wako kits).
Fat Tolerance Test
Vitamin A fat tolerance was measured essentially as described in Ref. 19. Briefly, 27 μCi of [11,12-3H]retinol (44.4 Ci/mmol; PerkinElmer Life Sciences) in ethanol was mixed with 1 ml of corn oil (Sigma). Each mouse received 200 μl of the emulsion directly into the stomach by gavage. Blood was sampled at the times indicated by retroorbital sinus bleedings, and circulating radioactivity in 10 μl of plasma was measured in triplicate by scintillation counting.
Liver Heparan Sulfate Purification and Analysis
Heparan sulfate was isolated from whole livers, essentially as described (20). After cutting the portal vein, mice were perfused through the left ventricle with 50 ml of phosphate-buffered saline, pH 8, at 7 ml/min with a syringe pump to remove blood from the liver. Livers were excised, homogenized with a razor blade, and digested overnight with Pronase (2 mg/ml; Roche Applied Science) to degrade proteins, followed by purification of glycopeptides by anion exchange chromatography using DEAE-Sephacel (GE Healthcare). The columns (0.5 ml) were washed with low salt buffer (0.3 m NaCl in 20 mm sodium acetate, pH 6, 10 ml) to remove contaminants, and the glycosaminoglycan fraction was eluted with 2.5 ml of 2 m NaCl. Finally, the glycosaminoglycan preparation was treated with chondroitinase ABC to depolymerize chondroitin sulfate chains, and the heparan sulfate fraction was purified by a second round of anion exchange chromatography.
Heparan sulfate fine structure was determined quantitatively by glycan reductive isotope labeling-liquid chromatography/mass spectrometry (GRIL-LC/MS) as described (21). The amount of heparan sulfate was expressed relative to liver wet weight. The molar percentage of each disaccharide was calculated based on the total recovery. In some cases, mice were injected intraperitoneally with 1 mCi of Na35SO44 (25 mCi/ml; PerkinElmer Life Sciences), and liver [35S]heparan sulfate was isolated after 2 h.
Analysis of Hepatocyte Syndecan-1
Primary hepatocytes were isolated from wild-type and diabetic mice as described (15, 22). Proteoglycans were isolated from the cell pellets according to a protocol adapted from Reizes et al. (23) and collected by anion exchange chromatography (DEAE-Sephacel). Samples were digested for 1 h with a mixture of 2 milliunits/ml heparin lyases I and II and 5 milliunits/ml heparin lyase III and chondroitinase ABC. The deglycosylated core proteins were separated by SDS-PAGE (NuPage 4–12% BisTris gel; Invitrogen) and transferred to nitrocellulose. Syndecan-1 was detected with mAb 281-2 (BD Pharmingen) and horseradish peroxidase-conjugated goat anti-rat IgG (Santa Cruz). One band was detected at 140 kDa. This band likely represents a syndecan-1 oligomer (24–26). β-Actin was detected with an anti-β-actin antibody (Cell Signaling Technologies) and a horseradish peroxidase-conjugated anti-rabbit IgG (Cell Signaling Technologies). Quantitative reverse transcription PCR was performed exactly as described previously (15).
Analysis of Plasma Sulfate Concentration
Inorganic sulfate was determined in plasma samples from wild-type and diabetic mice through the University of California San Diego Glycotechnology Core. Samples, blank and standards were hydrolyzed in 0.03 n HCl at 150 °C for 1 h and dried. Each sample was pyrolyzed in an open flame for about 15 s, cooled, and redissolved in water. Sulfate and phosphate anions were separated by a Dionex IonPac AS11HC column, eluted with a gradient mobile phase (2–50 mm KOH at 1 ml/min for 30 min), and quantified by conductivity detection relative to standard solutions.
Liver Heparan Sulfate-Protein Interaction
Samples of purified [35S]heparan sulfate (104 cpm) were incubated for 20 min with 10 μg of recombinant apoE (Invitrogen) or fibroblast growth factor-2 (Selective Genetics) in physiological saline with and without 100 μg/ml unfractionated heparin (SPL). Bound material was collected on nitrocellulose membranes by fast vacuum filtration as described (27).
Statistics
Statistical analyses were performed using PRISM software (GraphPad Software). All data are expressed as mean values ± S.D. unless otherwise indicated. Significance was determined using an unpaired Student's (two-tailed) t test. Significance was taken as p < 0.05.
RESULTS AND DISCUSSION
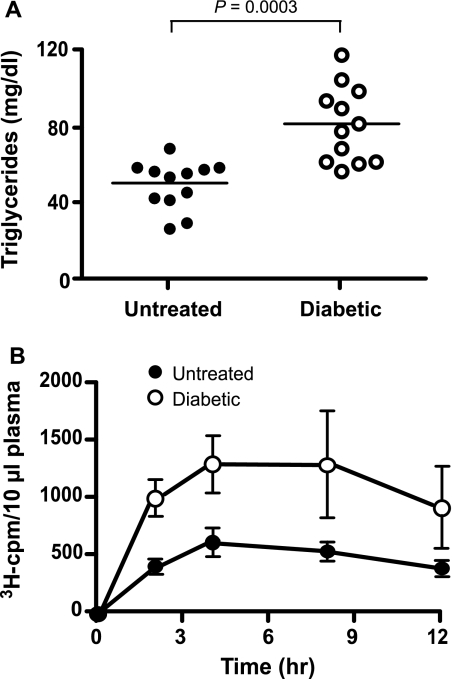
Recent studies linking hypertriglyceridemia to altered hepatocyte heparan sulfate chains and expression of the proteoglycan syndecan-1 led us to investigate hepatocyte heparan sulfate in a mouse model of IDDM (15–17). Prior studies have suggested that insulin deficiency causes alterations in heparan sulfate in STZ-induced diabetes (6–14, 28). To study this problem further, IDDM was induced in C57BL/6 mice by low dose injection of STZ on 5 consecutive days. In a typical study group, STZ-treated mice developed hyperglycemia (>400 mg/dl blood glucose) 6–8 weeks after treatment. Approximately 50% of the diabetic mice exhibited mildly elevated fasting plasma triglycerides (Fig. 1A, 51 ± 13 mg/dl in control mice versus 81 ± 20 mg/dl in STZ-treated mice, n = 12/strain, p < 0.003) with no significant change in plasma cholesterol (data not shown). The majority of the triglycerides were present in lipoproteins of d < 1.019 g/ml, consistent with their identification as very low density lipoproteins and chylomicron remnants.
FIGURE 1.
Hypertriglyceridemia in IDDM mice. A, total fasting plasma triglycerides. STZ-treated diabetic (open circles) or untreated control (filled circles) C57BL/6 mice were fasted for 4 h in the morning, and blood was taken from the retroorbital sinus for triglyceride analysis. Average values ± S.D. were 50 ± 12 mg/dl in control mice versus 81 ± 19 mg/dl in STZ-treated animals, respectively (n = 12). B, chylomicron clearance measurement by retinyl ester excursion. Fasted control mice (closed circles, n = 4) and IDDM mice (open circles, n = 4) were given 200 μl of corn oil containing [3H]retinol by oral gavage. Blood samples were taken at the indicated times, and radioactivity remaining in plasma samples (10 μl) was determined by liquid scintillation counting. The values are expressed as mean ± S.D. (error bars) of three samples and are representative of at least three separate experiments.
To examine the metabolism of dietary triglycerides, fasted mice were given a bolus of corn oil by gavage containing [3H]retinol, which is converted into retinol esters and packaged into chylomicrons (vitamin A fat tolerance test). The amount of [3H]retinol in the plasma was determined at various times (Fig. 1B). In untreated animals, plasma 3H counts rose for 4 h and then decreased over the next 8 h as the retinyl esters containing lipoproteins were cleared in the liver. In STZ-treated animals, plasma counts behaved in a similar manner except the extent of accumulation was greater (Fig. 1B, open circles). The area under the curve was increased 2.3-fold for IDDM mice compared with the controls, indicating that diabetic animals cleared intestinally derived lipoproteins at a slower rate as previously observed (12). Similar results were obtained when plasma triglycerides were assayed (data not shown). Analysis of plasma lipoproteins by ultracentrifugation of samples drawn at 2 h after gavage showed that the majority of the counts and triglycerides were present in large buoyant lipoproteins of d < 1.019 g/ml (data not shown).
Analysis of Hepatocyte Syndecan-1 in IDDM Mice
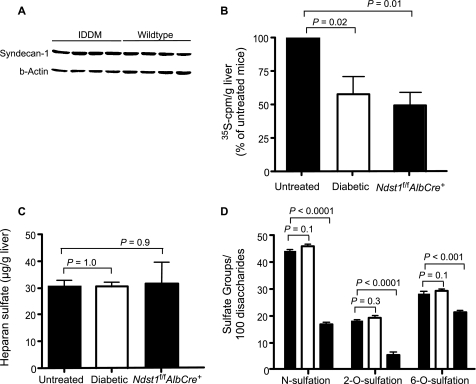
The reduced clearance of plasma triglycerides might reflect decreased expression of syndecan-1 in IDDM mice compared with wild type. Analysis of syndecan-1 mRNA levels in IDDM liver as well as freshly isolated hepatocytes showed no difference compared with wild type (data not shown). Moreover, we did not detect any difference in syndecan core protein levels by Western blotting of extracts prepared from freshly isolated hepatocytes (Fig. 2A). Thus, altered syndecan-1 expression did not account for the delayed clearance of plasma triglycerides in IDDM mice.
FIGURE 2.
Analysis of liver heparan sulfate in IDDM mice. A, Western blot analysis of syndecan-1 expression in IDDM hepatocytes. Syndecan-1 was detected with mAb 281-2 in freshly prepared hepatocytes from untreated and STZ-treated hypertriglyceridemic diabetic mice (n = 4/strain). β-Actin was used as a loading control. B, analysis of endogenous liver [35S]heparan sulfate. Untreated (n = 3) and STZ-treated hypertriglyceridemic diabetic (n = 3) wild-type mice and mutant mice bearing a hepatocyte specific deletion of Ndst1 (n = 3) were given [35S]sulfate intraperitoneally to radiolabel newly made heparan sulfate chains in vivo. Two hours later [35S]heparan sulfate was purified from the liver and quantified by liquid scintillation counting. Results were normalized/g of liver and are presented as a percentage of labeling in untreated control mice (1 ± 0.05 × 105 cpm/g). The values are expressed as mean ± S.D. (error bars) and are representative of three separate studies (n = 9 mice). C, liver heparan sulfate measured chemically. Liver heparan sulfate was purified from untreated (n = 9) and STZ-treated hypertriglyceridemic diabetic (n = 9) wild-type mice and Ndst1 mutant mice (n = 4). The mass of heparan sulfate purified from each animal was determined by GRIL-LC/MS (see “Experimental Procedures”). Results were normalized to the amount of liver that had been analyzed (wet weight). D, sulfation of liver heparan sulfate. The disaccharide composition of liver heparan sulfate isolated in C was determined by GRIL-LC/MS. N-Sulfate and 6-O-sulfate groups in glucosamine moieties and 2-O-sulfate groups in uronic acids were calculated from the recovery of the individual disaccharides (see “Experimental Procedures”).
Analysis of Liver Heparan Sulfate in IDDM Mice
Like diabetic mice, mutant mice lacking the heparan sulfate biosynthetic enzyme Ndst1 in hepatocytes (Ndst1f/fAlbCre+ mice) exhibited delayed clearance of dietary triglycerides and fasting hypertriglyceridemia (16), suggesting that the hypertriglyceridemia observed in IDDM might be due to altered heparan sulfate composition. Previous studies have shown that IDDM leads to ∼50% decreased incorporation of [35S]sulfate into liver heparan sulfate chains in vivo (6, 10–14, 18). Similarly, we observed a reduction (57 ± 27%) in the amount of [35S]heparan sulfate produced in diabetic mouse liver compared with controls (Fig. 2B). A similar level of reduction was also observed in Ndst1f/fAlbCre+ mice, which produce undersulfated heparan sulfate chains (50 ± 13%).
To determine whether the composition of heparan sulfate was altered, we analyzed chemically heparan sulfate in normal and diabetic liver. Surprisingly, the mass of heparan sulfate (micrograms of heparan sulfate/gram of liver) purified from IDDM mouse livers from animals with hypertriglyceridemia was equal to control (n = 9/strain) or Ndst1-deficient liver (n = 3) (Fig. 2C). To determine whether the extent of sulfation changed (glucosamine N-sulfation, uronyl 2-O-sulfation, or glucosamine 6-O-sulfation), we compared the disaccharide composition of the chains obtained from diabetic animals with wild type and Ndst1f/fAlbCre+ mice using GRIL-LC/MS, a quantitative method for analyzing heparan sulfate composition. The total amount of N-sulfated, 2-O-sulfated, and 6-O-sulfated disaccharides derived from IDDM liver heparan sulfate appeared unchanged compared with heparan sulfate obtained from the wild type (Fig. 2D). Furthermore, the composition of individual disaccharides did not vary as well (Table 1). Similar findings were obtained in IDDM glomerular HSPGs as well (29). In contrast, Ndst1-deficient liver contained heparan sulfate chains with reduced N-sulfation and uronic acid 2-O-sulfation, as noted previously (17).
TABLE 1.
Disaccharide analysis of heparan sulfate from wild-type, IDDM, and Ndst1f/fAlbCre+ mouse liver
Heparan sulfate chains were digested with heparin lyases I, II, and III, and the resulting disaccharides were resolved by GRIL-LC/MS (21). Disaccharides are designated by disaccharide structure code, in which D refers to a Δ4,5 unsaturated uronic acid derived from glucuronic acid or iduronic acid and whether sulfate groups are present at C2 of the uronic acid (0 or 2). The next letter designates whether the glucosamine residue is N-sulfated (S) or N-acetylated (A) and the presence or absence of 6-O-sulfate groups (0 or 6) (36). Thus, D0A0 = ΔUA-GlcNAc, D0S0 = ΔUA-GlcNS, D0A6 = ΔUA-GlcNAc6S, D0S6 = ΔUA-GlcNS6S, D2S0 = ΔUA2S-GlcNS, D2A6 = ΔUA2S-GlcNAc6S, and D2S6 = ΔUA2S-GlcNS6S.
| Sample | Disaccharidesa |
||||||
|---|---|---|---|---|---|---|---|
| D0A0 | D0S0 | D0A6 | D0S6 | D2S0 | D2A6 | D2S6 | |
| mol % | |||||||
| Wild type (n = 9) | 40 ± 1 | 20 ± 1 | 11 ± 1 | 7 ± 1 | 7 ± 0 | 0.2 ± 0 | 12 ± 0 |
| IDDM mouse (n = 9) | 41 ± 1 | 20 ± 1 | 12 ± 1 | 7 ± 1 | 8 ± 1 | 0.2 ± 1 | 10 ± 1 |
| Ndst1f/fAlbCre+ (n = 4) | 70 ± 3 | 6 ± 1 | 12 ± 1 | 5 ± 1 | 2 ± 1 | 0 ± 0 | 4 ± 0 |
a Values are expressed as the mole percent relative to the total amount of recovered disaccharides.
These findings contrast studies of experimentally diabetic rats in which insulin resistance was associated with production of HSPGs with reduced negative charge compared with HSPGs from control rats (6, 7). Subsequent studies suggested that reduced sulfation correlated with diminished GlcNAc N-deacetylase activity (7, 8, 10, 11, 13, 14). Although we have not measured Ndst1 expression or enzyme activity, the compositional studies demonstrate clearly that no change in sulfation occurred. The discrepancy between our findings and earlier studies may reflect the animal model under study (rats versus mice). How these results relate to type 1 and type 2 diabetes in humans remains to be determined.
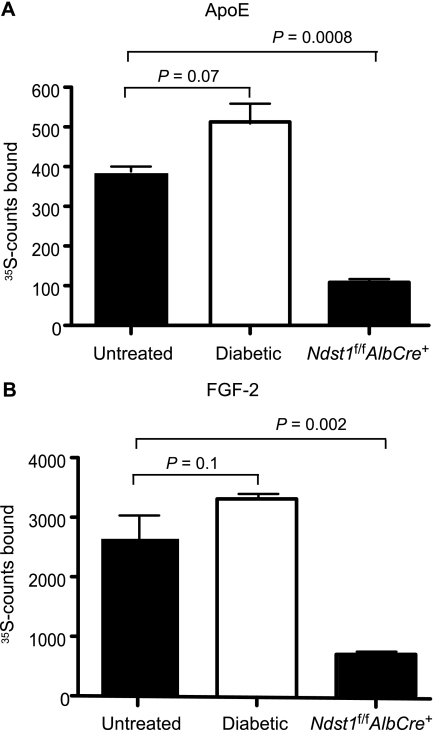
Conceivably, the pattern of sulfated disaccharides might be altered in IDDM liver heparan sulfate in a way that might affect binding to protein ligands. [35S]Heparan sulfate chains were purified from control and IDDM mice and incubated with apoE or fibroblast growth factor-2. The complexes that formed were collected on nitrocellulose membranes. The extent of binding of IDDM liver heparan sulfate to both proteins was equal to control liver heparan sulfate (Fig. 3). In contrast, the extent of binding to heparan sulfate from Ndst1f/fAlbCre+ was dramatically reduced. Binding of these ligands to liver heparan sulfate was completely inhibited by the addition of exogenous heparin, demonstrating the specificity of the interaction (data not shown). Similarly, van den Born and co-workers (29) found that the protein binding properties as well as the disaccharide composition of heparan sulfate derived from control and diabetic glomeruli did not differ.
FIGURE 3.
Liver heparan sulfate binding to protein ligands. Liver [35S]heparan sulfate from diabetic, Ndst1f/fAlbCre+, and control mice (104 cpm) was incubated with 10 μg of recombinant apoE (A) or fibroblast growth factor-2 (FGF-2; B), and [35S]heparan sulfate bound to the proteins was collected by membrane filtration (see “Experimental Procedures”). Unbound [35S]heparan sulfate does not bind to the membrane. Each experiment was done in triplicate, and the average values are shown. The values are expressed as mean ± S.D. (error bars) and are representative of two separate experiments.
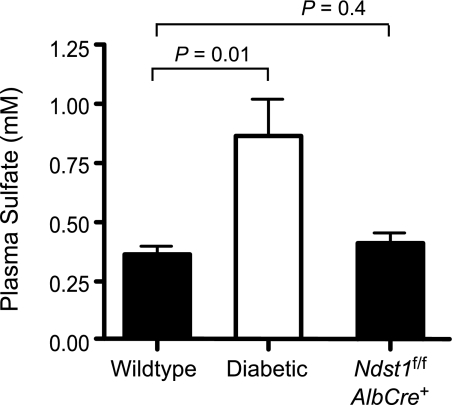
What is the explanation for the decrease in 35SO4 incorporation observed here and in prior studies? Earlier work suggested that diabetes can result in alterations in plasma and tissue sulfate pools (30–32). Thus, the decrease in 35S incorporation might have simply resulted from differences in the radiospecific activity of the 35SO4 in the plasma or in the liver. Indeed, endogenous plasma sulfate concentration in diabetic mice was ∼3-fold greater than in wild-type mice or Ndst1f/fAlbCre+ mice (Fig. 4). The source of elevated plasma sulfate could be caused by altered anion filtration by the kidney due to diabetic nephropathy (33). Alternatively, IDDM is also known to induce gluconeogenesis, which could lead to the increased conversion of cysteine and methionine to pyruvate causing the release of inorganic sulfate from these amino acids. The release of inorganic sulfate from sulfur containing amino acids can be significant, providing all of the sulfate needed for glycosaminoglycan biosynthesis in cultured cells (34).
FIGURE 4.
Plasma sulfate levels in IDDM mice. Sulfate levels were measured in untreated, diabetic, and Ndst1-mutant plasma samples (n = 6, respectively) by high performance liquid chromatography.
In summary, IDDM-induced hypertriglyceridemia in mice is not associated with any obvious changes in liver heparan sulfate content or composition. Although the affinity of the heparan sulfate chains for other factors involved in lipoprotein metabolism might be altered, we believe it more likely that IDDM induces changes in TRL composition that reduce the affinity of the particles for liver heparan sulfate. In support of this hypothesis, it has been shown that TRL in IDDM plasma exhibit reduced apoE and elevated apoC content potentially due to defects in lipoprotein lipase-mediated hydrolysis of triglycerides (35). Thus, we believe that the more likely explanation for elevated plasma triglycerides is a change in the processing of lipoproteins in the extrahepatic circulation rather than clearance mediated by HSPGs. Studies are currently under way to address whether these changes affect the affinity and uptake of lipoproteins by other liver receptors such as low density lipoprotein receptor or low density lipoprotein receptor-related protein.
This was supported by National Institutes of Health Grant GM33063 (to J. D. E.). This work was also supported by an American Heart Association scientist development grant (to J. R. B.).
- IDDM
- insulin-dependent diabetes mellitus
- TRL
- triglyceride-rich lipoprotein
- HSPG
- heparan sulfate proteoglycan
- Ndst1
- N-acetylglucosamine N-deacetylase/N-sulfotransferase 1
- STZ
- streptozotocin
- GRIL-LC/MS
- glycan reductive isotope labeling-liquid chromatography/mass spectrometry
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- apoE
- apolipoprotein E
- FGF-2
- fibroblast growth factor-2.
REFERENCES
- 1.Haffner S. M. (1998) Endocr. Rev. 19, 583–592 [DOI] [PubMed] [Google Scholar]
- 2.Reusch J. E. (2003) J. Clin. Invest. 112, 986–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg R. B. (2003) Cardiol. Clin. 21, 399–413 [DOI] [PubMed] [Google Scholar]
- 4.Bar-On H., Roheim P. S., Eder H. A. (1976) Diabetes 25, 509–515 [DOI] [PubMed] [Google Scholar]
- 5.Ebara T., Hirano T., Mamo J. C., Sakamaki R., Furukawa S., Nagano S., Takahashi T. (1994) Metabolism 43, 299–305 [DOI] [PubMed] [Google Scholar]
- 6.Kjellén L., Bielefeld D., Hook M. (1983) Diabetes 32, 337–342 [DOI] [PubMed] [Google Scholar]
- 7.Unger E., Pettersson I., Eriksson U. J., Lindahl U., Kjellén L. (1991) J. Biol. Chem. 266, 8671–8674 [PubMed] [Google Scholar]
- 8.Oturai P., Deckert M., Rolin B., Jensen T., Kofoed-Enevoldsen A. (1999) Exp. Clin. Endocrinol. Diabetes 107, 453–456 [DOI] [PubMed] [Google Scholar]
- 9.Olsson U., Egnell A. C., Lee M. R., Lundén G. O., Lorentzon M., Salmivirta M., Bondjers G., Camejo G. (2001) Diabetes 50, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 10.Kofoed-Enevoldsen A., Eriksson U. J. (1991) Diabetes 40, 1449–1452 [DOI] [PubMed] [Google Scholar]
- 11.Kofoed-Enevoldsen A. (1992) Kidney Int. 41, 763–767 [DOI] [PubMed] [Google Scholar]
- 12.Ebara T., Conde K., Kako Y., Liu Y., Xu Y., Ramakrishnan R., Goldberg I. J., Shachter N. S. (2000) J. Clin. Invest. 105, 1807–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams K. J., Liu M. L., Zhu Y., Xu X., Davidson W. R., McCue P., Sharma K. (2005) Diabetes 54, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 14.Goldberg I. J., Hu Y., Noh H. L., Wei J., Huggins L. A., Rackmill M. G., Hamai H., Reid B. N., Blaner W. S., Huang L. S. (2008) Diabetes 57, 1674–1682 [DOI] [PubMed] [Google Scholar]
- 15.Stanford K. I., Bishop J. R., Foley E. M., Gonzales J. C., Niesman I. R., Witztum J. L., Esko J. D. (2009) J. Clin. Invest. 119, 3236–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacArthur J. M., Bishop J. R., Stanford K. I., Wang L., Bensadoun A., Witztum J. L., Esko J. D. (2007) J. Clin. Invest. 117, 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanford K. I., Wang L., Castagnola J., Song D., Bishop J. R., Brown J. R., Lawrence R., Bai X., Habuchi H., Tanaka M., Cardoso W. V., Kimata K., Esko J. D. (2010) J. Biol. Chem. 285, 286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams K. J. (2008) J. Clin. Invest. 118, 3247–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishibashi S., Perrey S., Chen Z., Osuga J., Shimada M., Ohashi K., Harada K., Yazaki Y., Yamada N. (1996) J. Biol. Chem. 271, 22422–22427 [DOI] [PubMed] [Google Scholar]
- 20.Grobe K., Inatani M., Pallerla S. R., Castagnola J., Yamaguchi Y., Esko J. D. (2005) Development 132, 3777–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence R., Olson S. K., Steele R. E., Wang L., Warrior R., Cummings R. D., Esko J. D. (2008) J. Biol. Chem. 283, 33674–33684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seglen P. O. (1976) Methods Cell Biol. 13, 29–83 [DOI] [PubMed] [Google Scholar]
- 23.Reizes O., Lincecum J., Wang Z., Goldberger O., Huang L., Kaksonen M., Ahima R., Hinkes M. T., Barsh G. S., Rauvala H., Bernfield M. (2001) Cell 106, 105–116 [DOI] [PubMed] [Google Scholar]
- 24.Miettinen H. M., Jalkanen M. (1994) J. Cell Sci. 107, 1571–1581 [DOI] [PubMed] [Google Scholar]
- 25.Asundi V. K., Carey D. J. (1995) J. Biol. Chem. 270, 26404–26410 [DOI] [PubMed] [Google Scholar]
- 26.Choi S., Lee E., Kwon S., Park H., Yi J. Y., Kim S., Han I. O., Yun Y., Oh E. S. (2005) J. Biol. Chem. 280, 42573–42579 [DOI] [PubMed] [Google Scholar]
- 27.Kreuger J., Lindahl U., Jemth P. (2003) Methods Enzymol. 363, 327–339 [DOI] [PubMed] [Google Scholar]
- 28.Cohen P. M., Surma M. L. (1981) J. Lab. Clin. Med. 98, 715–722 [PubMed] [Google Scholar]
- 29.van den Born J., Pisa B., Bakker M. A., Celie J. W., Straatman C., Thomas S., Viberti G. C., Kjellen L., Berden J. H. (2006) J. Biol. Chem. 281, 29606–29613 [DOI] [PubMed] [Google Scholar]
- 30.Cohen M. P., Surma M. L. (1984) Diabetes 33, 8–12 [DOI] [PubMed] [Google Scholar]
- 31.Spiro M. J. (1987) Diabetologia 30, 259–267 [DOI] [PubMed] [Google Scholar]
- 32.Fan M. Y., Templeton D. M. (1992) Diabetes Metab. 18, 98–103 [PubMed] [Google Scholar]
- 33.Blazquez-Medela A. M., López-Novoa J. M., Martínez-Salgado C. (2010) Curr. Diabetes Rev. 6, 68–87 [DOI] [PubMed] [Google Scholar]
- 34.Esko J. D., Elgavish A., Prasthofer T., Taylor W. H., Weinke J. L. (1986) J. Biol. Chem. 261, 15725–15733 [PubMed] [Google Scholar]
- 35.Levy E., Shafrir E., Ziv E., Bar-On H. (1985) Biochim. Biophys. Acta 834, 376–385 [DOI] [PubMed] [Google Scholar]
- 36.Lawrence R., Lu H., Rosenberg R. D., Esko J. D., Zhang L. (2008) Nat. Methods 5, 291–292 [DOI] [PubMed] [Google Scholar]