Abstract
Chemokines have two essential interactions in vivo, with G protein-coupled receptors, which activate intracellular signaling pathways, and with glycosaminoglycans (GAGs), which are involved in cell surface localization and transport. Although it has been shown that chemokines bind and activate their respective G protein-coupled receptors as monomers, many chemokines oligomerize upon GAG binding, and the ability to oligomerize and bind GAGs is required for in vivo function. In this study, we investigated the structure, dynamics, and oligomerization behavior of cutaneous T-cell-attracting chemokine (CTACK, also known as CCL27) by NMR. 15N relaxation and translational self-diffusion rates indicate that CCL27 oligomerizes, but in contrast to many other chemokines that form relatively discrete oligomers, CCL27 transitions between monomer, dimer, and tetramer species over a relatively narrow concentration range. A three-dimensional structure determination was pursued under conditions where CCL27 is primarily dimeric, revealing the standard motif for a chemokine monomer. Analysis of chemical shift perturbations of 1H-15N HSQC spectra, relaxation-dispersion experiments, and filtered nuclear Overhauser effects suggest that CCL27 does not adopt a discrete CXC or CC dimer motif. Instead, CCL27 has uncommon oligomerization behavior, where several equilibria involving relatively low affinity interactions between different interfaces seem to be simultaneously at work. However, interaction with heparin avidly promotes oligomerization under conditions where CCL27 is monomeric by itself. We hypothesize that the plasticity in the oligomerization state may enable CCL27 to adopt different oligomeric structures, depending on the nature of the GAG binding partner, thereby providing a mechanism for increased diversity and specificity in GAG-binding and GAG-related functions.
Keywords: Chemokines, Chemotaxis, Glycosaminoglycan, NMR, Protein Structure, CCL27, CTACK, Chemokine Oligomerization, G Protein-coupled Receptors
Introduction
The chemokine family consists of small (∼8–12 kDa) proteins that are responsible for controlling the migration of leukocytes in the context of routine immune surveillance and inflammation (1–5). However, inappropriate expression, regulation, or utilization of chemokines and chemokine receptors has been associated with many diseases, including rheumatoid arthritis, multiple sclerosis, asthma, cancer, and AIDS (6, 7). To date, there are ∼50 known human chemokines and 20 receptors (6). Although some chemokines are specific to one receptor, other chemokines bind multiple receptors and many receptors bind multiple chemokines. However, despite this apparent redundancy, there are many potential sources of specificity, including spatial, temporal, and tissue-specific expression of chemokines and receptors (8), different signaling consequences of a given receptor in response to different ligands (6), and of relevance to this work, interactions of chemokines with glycosaminoglycans (GAGs)5 (9).
Chemokines/receptors have been classified into four subfamilies (CC, CXC, CX3C, and XC) based on the relative positions of conserved cysteine residues in the N termini of the ligands. Structures of many chemokines have been solved and reveal a highly conserved tertiary motif consisting of a disordered N-terminal region followed by a 310 helix, three antiparallel β-strands, and a C-terminal α-helix (10–13). Despite their similar monomeric structures, multiple dimeric and tetrameric structures have been reported. Two primary types of chemokine dimers have been observed, and generally CC chemokines adopt one type of dimer while CXC chemokines adopt the second type of dimer (14). In the CC dimers, the disordered N-terminal residues within each monomeric subunit form a two-stranded antiparallel β-sheet in the dimer interface, giving rise to an overall elongated structure. CXC dimers are more compact and interact primarily through the first strand of their β-sheets (supplemental Fig. 1, A and B) (10–13). In addition, several different tetrameric species have been reported. For example, the CCL2 (MCP-1) tetramer has properties of both CC and CXC dimer interfaces (supplemental Fig. 1, C and D) (11, 15), while human CXCL10 (IP-10) adopts multiple tetrameric forms, one similar to CCL2, as well as two entirely different structures (16), and murine CXCL10 (IP-10) assumes yet a fourth tetrameric structure (17).
Although the functional relevance of the oligomeric forms is not entirely understood, there is considerable evidence that chemokine oligomerization is important for in vivo cell migration (11, 18–22). For example, although monomeric variants of CCL2, CCL4, and CCL5 are capable of inducing cell migration in vitro, they failed to cause migration in vivo when injected into the peritoneal cavity of mice (23). The prevailing explanation for these results is that interactions between oligomeric forms of chemokines and GAGs are required to tether chemokines to cell surfaces as a mechanism for their retention near the site of production, so that they do not diffuse away and can thus provide directional cues for migrating cells (9, 20, 23–27). Other studies have also suggested that GAG binding is required for transcytosis of chemokines from the abluminal to luminal surface of endothelial cells in order to encounter receptor-bearing leukocytes on the opposing surface (23). The connection between chemokine oligomerization and GAG binding is supported by biochemical and biophysical studies that show that chemokines oligomerize on GAGs and that oligomeric forms have a higher affinity for GAGs than non-oligomerizing forms (11, 28).
This study focuses on the structure and oligomerization properties of the chemokine CCL27 (CTACK), which is constitutively expressed in the skin and has only one known chemokine receptor, CCR10 (29). CCL27 has been implicated in inflammatory skin diseases such as psoriasis and is believed to play a role in melanoma metastasis (30, 31). The results of the present studies suggest that its oligomerization behavior is unusually dynamic with weak interactions between monomers, dimers, and tetramers but that one or more oligomeric forms are stabilized by GAG binding. This structural plasticity may allow CCL27 to adopt different oligomeric structures depending on the nature of the GAG, which in turn may add to the functional specificity of this chemokine.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Both CCL27 and CCL2 were cloned into the pHUE vector (kindly provided by Rohan Baker) and expressed as insoluble His-ubiquitin fusion proteins (32). For production of unlabeled protein, cells were grown at 37 °C in Luria Broth (LB), and an MOPS-based media was used for expression of labeled protein (see supplemental Experimental Procedures). 15N/13C- and 13C-labeled CCL27 were expressed in media containing 98% 15N ammonium sulfate, and 99% [13C] glucose. 15N-labeled CCL27 for filtered NOE analysis was expressed with 15N-labeled ammonium sulfate and 13C-depeleted glucose (99.98% 12C) (Cambridge Isotope). Proteins were overexpressed by induction with isopropyl β-d-1-thiogalactopyranoside in BL21(DE3)pLysS Escherichia coli cells, after which cells were harvested by centrifugation. They were then purified from the insoluble fraction by nickel-nitrilotriacetic acid affinity chromatography, refolded by dilution in Hampton Fold-it Buffer #13 (FoldIt Screen, Hampton Research), dialyzed, and concentrated (see supplemental materials for more details). The His-ubiquitin tag was cleaved with ubiquitinase, immediately before final purification by reversed-phase high-performance liquid chromatography with a semi-prep C18 column. Chemokine fractions were lyophilized and stored at −80 °C. Purity was confirmed using matrix-assisted laser desorption ionization mass spectrometry and label incorporation was verified by electrospray ionization-mass spectrometry.
NMR Spectroscopy
The sections below outline general aspects of the NMR procedures; more complete details are described under supplemental Experimental Procedures.
PFG Diffusion Measurements
PFG-NMR experiments were run on a Bruker Avance II 600 MHz NMR spectrometer with a 5-mm TCI CryoProbe at 302.3K. Samples were prepared in 50 mm acetate-d6, pH 5.6. The experiments were carried out using the Bruker pulse sequence ledbpgpprwg2s and the Bruker macro diffusion ordered spectroscopy (DOSY). The self-diffusion coefficients (Ds) were calculated using the Bruker DOSY analysis program, with manual integration for peaks at 7.0, 3.0, 2.0, and 0.7 ppm for each proton spectrum. Each sample was run in triplicate, and the Ds values were averaged over the three experiments.
Monomeric Structure Determination
Residue assignments involved a standard battery of three-dimensional NMR experiments (see supplemental materials) (33). The data were processed using the Azara suite of programs (version 2.7, W. Boucher, University of Cambridge). The assignment of 15N/13C-labeled CCL27 was performed by determining sequential connectivities of backbone resonances using a pairwise approach via frequency matching in the 13C dimension. Distance restraints were derived from NOE-based measurements, dihedral angle restraints, and H-bond restraints, and the structure calculations were performed with ARIA (Ambiguous Restraints for Iterative Assignment) (34, 35) interfaced to CNS (Crystallography and NMR System) (36) (37, 38). The mean structure was generated from the ensemble of the 30 lowest energy water-refined structures (from 100). The ensemble and the structure closest to the mean were then analyzed using PROCHECK and PROCHECK-NMR (39, 40).
15N Relaxation Measurements
Relaxation experiments were recorded on 1.0 mm 15N-labeled CCL27 at fields of 500 and 600 MHz. 15N R1 and R2 relaxation rates and 1H-15N heteronuclear NOEs were measured using standard pulse sequences. The transverse cross-correlation rate constant, ηxy, was measured using the methods described in supplemental Experimental Procedures (23, 41). Relaxation data curve-fitting was performed using CcpNmr analysis. Rotational diffusion tensor fitting and analysis of internal mobility were performed using the program TENSOR2 (42).
Dimer and Hydrodynamic Modeling
Hydrodynamic modeling of the rotational diffusion tensor was performed using the program HYDRONMR (43, 44).
Exchange Broadening
Relaxation-compensated Carr-Purcell-Meiboom-Gill (CPMG) relaxation-dispersion profiles were recorded for 1.0 mm 15N-labeled CCL27 at 500 and 600 MHz (45). In addition to a reference spectrum acquired without a relaxation period, 16 sub-spectra were acquired from νCPMG = 7.7 Hz to νCPMG = 1000 Hz. The data were fit using the program CPMGFit (A. Palmer, Columbia University).
1H-15N HSQC Chemical Shift Perturbation Analysis and Filtered (HC)NH-NOE
Experiments were performed with the same system and sample conditions as the PFG diffusion experiments. All experiments were run at 302.3 K. Spectra were processed and analyzed using the programs NMRPipe (46) and Sparky (SPARKY 3, T. D. Goddard and D. G. Kneller, University of California, San Francisco), respectively. Chemical shift perturbation values were calculated using the equation [(5ΔδHN2) + (ΔδN)2]1/2 (21). To determine NOEs corresponding to the oligomeric interface, two-dimensional 1H-15N versions of a four-dimensional 13C/15N-separated (HC)NH-NOESY experiment were recorded on 3.0 mm and 2.0 mm samples, which were 50% 15N-labeled CCL27 (13C-depleted) and 50% 13C-labeled CCL27 (47).
Heparin Binding Assay
To determine if CCL27 oligomerizes on GAGs, 125I-CCL27 (PerkinElmer Life Sciences) was incubated with heparin immobilized on beads and competed off with unlabeled CCL27 as described by Hoogewerf et al., and Proudfoot et al. (23, 28) (see supplemental materials). Radioactivity was counted with a scintillation plate reader.
Solubility Assays
The protein concentration was determined using a Nanodrop spectrophotometer (Thermo Scientific), via the absorbance at 280 nm. Concentrations were determined in the presence and absence of heparin octasaccharide (Neoparin, Inc., GT8041) after centrifugation to determine the percentage of chemokine remaining in solution.
Chemical Cross-linking
Reactions were performed with the indicated concentrations of CCL27 and heparin decasaccharide (Neoparin, Inc., GT8051) in 50 mm Hepes, pH 7.2, with 5 mm sulfoethyleneglycolbis(sulfosuccinimidylsuccinate) (Sulfo-EGS, Pierce Reagents) for 1 h at 25 °C and quenched with 1 m Tris prior to running on a non-reducing SDS-PAGE gel.
RESULTS
CCL27 Oligomerizes and Forms a Tetramer at Millimolar Concentrations
1H-15N HSQC Spectra
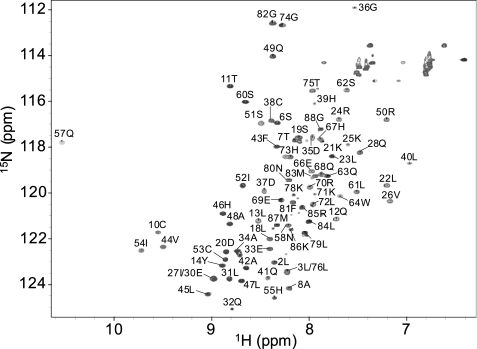
To ascertain optimal solution conditions for characterizing the structure and dynamics of CCL27, a uniformly labeled 15N/13C sample was prepared at 2.1 mm, and 1H-15N HSQC spectra were recorded. The line widths were significantly broader, and the sensitivity much lower than would be expected for a monomeric protein of 10 kDa, or even for a 20-kDa dimer. Because many chemokines form dimers and higher order oligomers, the sample was gradually diluted and resulted in progressive improvement of the signal/noise, until at 1.0 mm the line widths and sensitivity were deemed optimal to proceed with structural studies (Fig. 1). However, due to the functional relevance of chemokine oligomerization, the oligomeric behavior of CCL27 was first further investigated using translational diffusion and 15N relaxation methods.
FIGURE 1.
Assigned 1H-15N HSQC of CCL27 acquired on a Bruker Avance II 600 MHz NMR with a 5-mm TCI CryoProbe. The sample was prepared at a concentration of 0.5 mm, in 50 mm acetate buffer, pH 5.6, at 302.3 K.
Oligomerization Properties of CCL27 by PFG Diffusion NMR
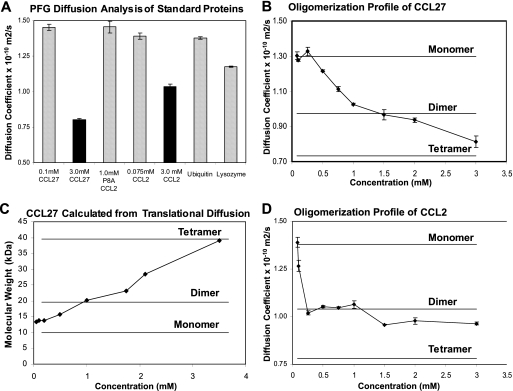
Translational diffusion of CCL27 was measured by PFG 1H NMR spectroscopy to assess the dependence of the oligomerization state on concentration and solution conditions. The self-diffusion coefficient, Ds, is related to the average protein size, and can be extracted from a series of one-dimensional 1H NMR spectra following the attenuation of signal with increasing gradient field strength. Upon oligomerization, the theoretical change in Ds can be calculated from the Stokes-Einstein equation. By approximating the interaction as hard-sphere molecular contacts, the dimer:monomer ratio of the diffusion coefficients was estimated to be 0.75, a value shown to be in good agreement with experimental data for the monomer-dimer equilibrium of CCL2 (48). In addition, the hydrodynamic radius and average molecular weight of the protein in solution may be determined from the measured Ds value (see below). However, because interpretation of diffusion coefficients requires certain assumptions, the Ds values were first compared for several proteins of similar molecular weight to CCL27, and known oligomerization behavior (21, 22, 48) (Fig. 2A). At 0.1 mm, CCL27 (10.1 kDa) has a Ds value similar to that of 0.075 mm CCL2 (8.9 kDa, monomeric at this concentration), P8A CCL2 (an 8.9 kDa monomeric variant of CCL2), and ubiquitin (8.6 kDa) (Ds values are 1.35 × 10−10 m2/s, 1.39 × 10−10 m2/s, 1.45 × 10−10 m2/s, and 1.38 × 10−10 m2/s, respectively). The Ds for lysozyme was somewhat lower (1.2 × 10−10 m2/s), as expected from its molecular mass (15.9 kDa). It can therefore be assumed that at such low concentrations, CCL27 is predominantly monomeric. However, when the CCL27 concentration was increased to 3.0 mm, the Ds value dropped to 0.80 × 10−10 m2/s, indicating an increase in apparent molecular weight relative to the 0.1 mm sample due to oligomerization. This behavior is similar to CCL2, which also oligomerizes at higher concentrations, although the Ds value of CCL27 suggests that it is larger than CCL2 at 3.0 mm (Fig. 2A) (47).
FIGURE 2.
PFG diffusion analysis of CCL27. Assuming the diffusion coefficient at 0.075 mm corresponds to the monomeric form, the theoretical Ds value for the dimer was estimated using the Stokes-Einstein equation as 0.975 × 10−10 m2/s. A, PFG diffusion analysis of standard proteins with known molecular weights and oligomerization states. B, PFG diffusion analysis of CCL27 over a concentration range of 0.050 mm to 3.0 mm in 50 mm acetate, pH 5.6. C, molecular weight of CCL27 at different concentrations, calculated from the Ds values shown in B. D, PFG diffusion profile of WT CCL2 over a range of concentrations from 0.01 mm to 3.0 mm, in 50 mm acetate, pH 5.6.
Because buffer conditions are known to influence chemokine oligomerization (21, 49), the effect of acetate, phosphate, and pH on the apparent molecular weight of CCL27 was evaluated but showed little impact (supplemental Fig. 2). The diffusion profile for CCL27 was then determined over a more extensive concentration range, from 0.075 to 3.0 mm in 50 mm acetate buffer at pH 5.6 (Fig. 2B). The diffusion coefficient for CCL27 is relatively constant from 0.075 to 0.25 mm, with an average value of ∼1.30 × 10−10 m2/s, but decreases gradually above 0.25 mm to 0.813 × 10−10 m2/s at 3.0 mm. This value is less than the expected Ds for a dimer (calculated to be 0.975 × 10−10 m2/s), suggesting the presence of higher order oligomers, most likely tetramers by analogy to other chemokines (the theoretical Ds value for the CCL27 tetramer was calculated to be 0.780 × 10−10 m2/s). Indeed, when the diffusion coefficient is converted into the hydrodynamic radius via the Stokes-Einstein relation, and used to estimate the molecular weight, the data suggest that CCL27 is tetrameric at 3.5 mm (39 kDa estimated versus 40.4 kDa theoretical, Fig. 2C and supplemental Table 1A). Comparing the hydrodynamic radius of CCL27, calculated from the translational diffusion (via HYDRONMR), to hydrodynamic radii for other known chemokine tetramers (calculated using their atomic structures via HYDROPRO), the radius for CCL27 at 3.5 mm is almost identical to that of CCL2, CX3CL1, and CCL14 tetramers (2.68, 2.65, 2.70, and 2.73 nm, respectively, supplemental Table 1B). Together, the data indicates that CCL27 is tetrameric at the higher concentrations of 3.0–3.5 mm.
The diffusion profile was then determined for CCL2, to compare the CCL27 result to that of a chemokine known to form a stable dimer. Between 0.05 and 0.10 mm, a sharp decrease in the Ds value of CCL2 was observed, corresponding to the formation of a dimer; it then remained relatively constant up to 3.0 mm (Fig. 2D). CCL2 has also been shown to form tetramers at higher concentrations and in the presence of GAGs (15), and the slight decrease in the Ds value at 1.5 mm suggests some population of the tetrameric form. Nevertheless, the overall diffusion profile is what would be expected for a protein that forms a discrete dimer with a Kd distinguishable from the tetrameric state (21, 47). The results for CCL2 contrast with the profile of CCL27, where the Ds value steadily decreased over the entire concentration range investigated, suggesting the simultaneous presence of several oligomeric species (monomer, dimer, and tetramer), with similar dissociation constants (Fig. 2B).
Rotational Diffusion by NMR
As a complement to the PFG experiments, and to assess the dynamics of the protein in solution, 15N R1 and R2 rates, as well as heteronuclear NOE, were measured for the backbone amide groups on a 1.0 mm CCL27 sample at 500 and 600 MHz, and analyzed by the Model-free formalism (supplemental Fig. 3). However, analysis of the data was complicated by significant exchange contributions (>5 Hz) to the transverse relaxation rates for the majority of residues, which were not suppressed under the fast CPMG pulsing conditions applied. Initial attempts to fit the rotational diffusion tensor in TENSOR2 (42) showed that the experimental data could not be reproduced using the calculated diffusion tensor in combination with the experimental uncertainties. Therefore, an alternative approach was adopted, in which the exchange-free transverse relaxation rates, R20, were computed from the measured (CSA)15N/dipole-dipole15N-1H transverse cross-correlated cross-relaxation rate constant, ηxy (50). Comparison of R20 with the R2 rates measured under fast CPMG pulsing conditions confirmed that the majority of residues possess significant residual exchange contributions (supplemental Fig. 3B). Recalculation of the isotropic rotational correlation time using the R20/R1 ratio of residues without significant internal motion yielded values for the rotational correlation time of 9.3 and 9.2 ns for the data collected at 500 and 600 MHz, respectively. These correlation times translate into an estimated molecular mass of ∼21 kDa, almost exactly equal to that of a CCL27 dimer and consistent with the results of the PFG diffusion measurements. Furthermore, diffusion tensor fitting with the new R20/R1 ratios using TENSOR2 indicated that rotational diffusion properties of 1.0 mm CCL27 are best described by an axially symmetric (D‖/D⊥ = 0.69), slightly oblate diffusion tensor (supplemental Table 2).
NMR Assignment and Structure Determination of the Monomeric Form of CCL27
The results of the PFG diffusion measurements suggesting the presence of multiple species in equilibrium, including tetramers, is consistent with the broadening observed in the initial HSQC spectra of 2.1 mm CCL27. Experiments used for resonance assignment and structure determination were therefore recorded on 1.0 mm 15N- and 15N/13C-labeled samples, a concentration that showed translational diffusion behavior consistent with the presence of a predominantly dimeric species. The backbone and side-chain assignment procedure involved a relatively standard approach with triple resonance experiments. Distance restraints were derived from NOE-based measurements, dihedral angle restraints, and H-bond restraints, and the structure calculations were performed using ARIA 2.1 (ambiguous restraints for iterative assignment), interfaced to CNS 1.0 (34, 35) (see supplemental materials).
In the assigned two-dimensional 1H-15N HSQC spectrum, the signals are well dispersed, and all backbone amide resonances are visible with the exception of Arg-15 and Lys-16, indicating that these sites are broadened beyond observation due to chemical exchange on an intermediate timescale (Fig. 1). The line widths for residues C-terminal to Pro-77 suggest that this region is unstructured and exhibits significant flexibility due to fast picosecond internal motion. These residues display a second set of amide peaks in addition to the principal peaks, presumably due to cis-trans isomerization of the peptide bond between Leu-76 and Pro-77. Several residues, such as Asp-35 and Asp-37, exhibit unusually broad 15N-line widths, indicative of large exchange contributions to the 15N transverse relaxation rate. In addition, a number of peaks are significantly overlapped, such as Leu-3/Leu-76, which were ultimately assigned by reference to methyl-selective TOCSY experiments.
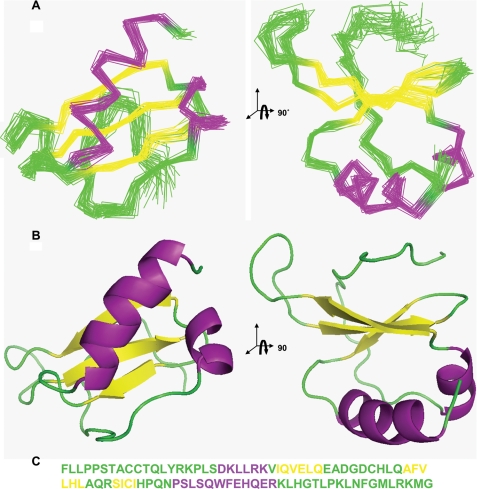
The calculated NMR ensemble is well defined, with a root mean square deviation of ∼0.5 Å over the backbone atoms of the ordered residues, for the 30 lowest energy structures. The calculated structures are in good agreement with the experimental restraints (Table 1). The ensemble of structures of the ordered core of CCL27 is shown in Fig. 3A, along with the corresponding ribbon diagrams (Fig. 3B), and the primary sequence colored to reflect the secondary structure (Fig. 3C). CCL27 adopts a typical chemokine fold, where the flexible N terminus (Phe-1 to Thr-7) is followed by an extended series of bends forming the N-loop, which leads into a 310 helix (Asp-20 to Arg-24), immediately followed by the first of three anti-parallel β-strands. The 30s loop between the first and second β-strands stretches from Glu-33 to Gln-41 and is anchored to the N-loop by the disulfide bridge between Cys-38 and Cys-9. In contrast, the 40s loop, linking the second and third β-strands, is a short type I turn comprising only Gln-48 and Arg-49. The end of the third β-strand is also attached to the N-loop by the second disulfide bond between Cys-53 and Cys-10. A short loop from His-55 to Asn-58 connects the end of the third β-strand to a long C-terminal α-helix, which extends to Arg-70 and lies across one face of the β-strand, whereas the other side of the β-strand faces onto the N terminus and the early part of the N-loop. The C-terminal α-helix is followed by a long, disordered C-terminal region, a feature that distinguishes CCL27 from most other chemokines.
TABLE 1.
Experimental restraints and structural statistics for CCL27
[SA] represents average values for the ensemble, for which the r.m.s.d. values are from the mean structure. [SA]c represents the corresponding values for the structure closest to the mean. EL-J is the Lennard-Jones energy (the Lennard-Jones potential was only used during the water refinement stage).
| No. of experimental restraints | |||||
| Distance restraints from NOEs | intra | seq | med | long | |
| Unambiguous | 1364 | 814 | 339 | 103 | 108 |
| Ambiguous | 1204 | ||||
| Dihedral | 44 | ||||
| H-bond | 22 | ||||
| Coordinate precision (residues 8–71) | [SA] | [SA]c | |||
| r.m.s.d. of backbone atoms, Å | 0.55 ± 0.10 | 0.38 | |||
| r.m.s.d. of heavy atoms, Å | 0.97 ± 0.07 | 0.78 | |||
| r.m.s.d. from experimental restraints | |||||
| NOE distances, Å | 0.0207 ± 0.0025 | 0.0181 | |||
| TALOS dihedral angles, ° | 0.54 ± 0.11 | 0.61 | |||
| r.m.s.d. from idealized geometry | |||||
| Bonds, Å | 0.00393 | 0.00386 | |||
| Angles, Å | 0.548 ± 0.019 | 0.53 | |||
| Improper, Å | 1.58 ± 0.019 | 1.57 | |||
| Final energy | |||||
| EL-J, kJ mol−1 | −3340 ± 50 | −3350 | |||
| Ramachandran analysis | |||||
| Residues in most favored regions | 74.3% | 72.7% | |||
| Residues in additionally allowed regions | 21.3% | 24.7% | |||
| Residues in generously allowed regions | 2.3% | 2.6% | |||
| Residues in disallowed regions | 2.1% | 0.0% | |||
FIGURE 3.
Solution NMR structure of monomeric CCL27, displaying the standard chemokine structural motif: an N-loop (green) followed by a 310 helix (purple), three anti-parallel β-strands (yellow), and a C-terminal α-helix (purple). A, stereo views of the overlaid backbone traces for the 30 water-refined structures (residues Ala-8 to His-71). B, corresponding ribbon representation. C, sequence of CCL27 showing unstructured and loop regions in green, α-helices in purple, and β-strands in yellow.
Characterization of Internal Dynamics
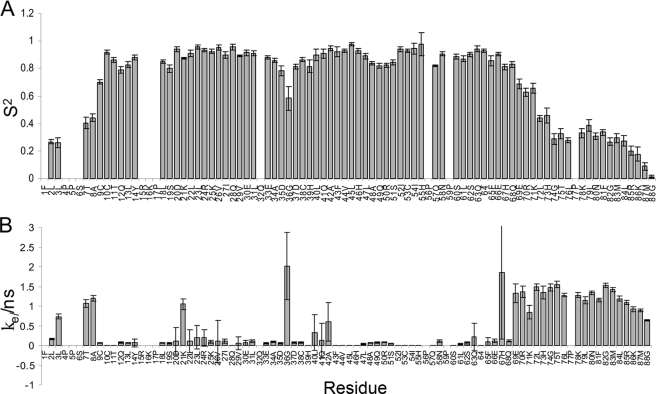
The exchange-free R20 values, described above, enabled the local motion of the majority of amide sites to be accurately described using the classical Model-Free formalism based on R1, R20, and heteronuclear NOE data (51). While the majority of the residues are in well structured regions, the profile of order parameters confirms the flexibility of the N and C termini, with increased internal mobility in the C terminus becoming evident toward the end of the α-helix (Fig. 4A). The affected amide groups exhibit internal correlation times on the order of 1 ns, with S2 values that decrease from 0.6–0.7 at the end of the helix to close to zero for Gly-88. The first eight residues in the N terminus also exhibit significant internal mobility, which is a characteristic of most if not all chemokines where this region plays an important role as a signaling trigger. The extended N-loop, despite its lack of regular secondary structure, is well ordered. For the remainder of the structured core of the protein, the most obvious feature is the relative mobility of the 30s loop, where the order parameters are slightly reduced to ∼0.6 at Gly-36.
FIGURE 4.
Backbone 15N order parameters and internal correlation times from TENSOR2 analysis of relaxation data recorded on 1.0 mm CTACK at 500 MHz. Where the extended model-free formalism was required to fit the relaxation data, the order parameter shown is given by S2 = Ss2Sf2.
Characterization of the CCL27 Dimer Interface Indicates That It Does Not Form a Discrete CC or CXC Dimer
Filtered NOE Analysis of Interfacial Contact Residues
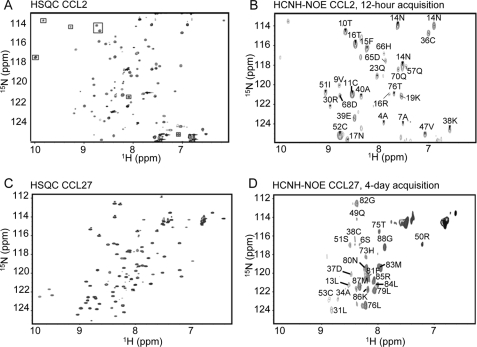
Despite significant effort, it was not possible to unambiguously identify intersubunit NOEs from the 15N-separated or 13C-separated NOESY spectra that were used to calculate the monomer structure of CCL27. To calculate a dimer structure, we therefore utilized a 13C/15N-separated and filtered NOESY experiment on a sample containing 50% 13C-CCL27 and 50% 15N-CCL27 (13C-depleted). In this experiment, NOEs are specifically detected between 1H-13C and 1H-15N protons. The mixed isotope sample maximizes the detection of intersubunit NOEs, as previously demonstrated for CCL2 (47). To initially validate the experiment, and for a direct comparison to a well defined dimer, we first recorded the 1H-15N plane of a two-dimensional version of this (HC)NH-NOE experiment on a 3.0 mm sample of CCL2, suitably labeled as described above, and compared it with the 1H-15N HSQC (Fig. 5, A and B). The resulting spectrum was well dispersed, with intense signals corresponding to structured regions of the protein that were previously identified as comprising the dimer interface (47). A similar experiment was then recorded on the 3.0 mm CCL27 sample and showed positive signals from four different regions of the protein, Ser-6 and Leu-13 in the N terminus, Leu-31, Ala-34, Asp-37, and Cys-38 in the first β-strand/30s loop, Gln-49 to Ser-51, and Cys-53 in the third β-strand/40s loop, and His-73, Thr-75, Leu-76, and Leu-79 to Gly-88 in the C terminus (the positive signals from the unstructured C terminus are likely due to intra-residue NOEs resulting from residual natural abundant 13C in the 15N/13C-depleted sample that appear due to favorable relaxation properties and close distance). Residues were assigned based on comparison to the 1H-15N HSQC (Fig. 5, C and D). When the experiment was repeated on a 2.0 mm CCL27 sample, similar results were observed, with additional signals from the first β-strand and 310 helix that were not present at the higher concentration (supplemental Fig. 4). At 1.0 mm where the dimer dominates, the NOE signals were no longer detectable due to the insensitivity of the experiment (data not shown).
FIGURE 5.
Filtered NOE analysis of a 50:50 15N-labeled plus 13C-labeled sample of CCL2 at 3.0 mm and a 50:50 15N-labeled (13C-depleted) plus 13C-labeled sample of CCL27 at 3.0 mm. A, 1H-15N HSQC of CCL2. B, two-dimensional 1H-15N (HC)NH-NOE, acquired in 12 h and assigned based on the HSQC. This experiment was previously employed to determine the dimeric structure of CCL2 in solution and used here to validate the method and compare with CCL27 (47). C, 1H-15N HSQC of CCL27. D, two-dimensional 1H-15N (HC)NH-NOE CCL27 at 3.0 mm, acquired in 4 days and assigned based on the HSQC.
In contrast to CCL2, the results for CCL27 do not define a discrete CC or CXC dimer interface. A CXC dimer interface would include only the first β-strand, whereas a CC dimer interface would involve predominantly the N-terminal region and the third β-strand, which is linked to the N terminus by a disulfide bond (21, 47), In addition, the experimental sensitivity was considerably weaker for CCL27 than for CCL2 at all concentrations; CCL2 required only a 12-h acquisition time to provide sufficient signal/noise, while CCL27 required an acquisition time of 4 days and ultimately resulted in fewer signals. This is consistent with the formation of a lower affinity oligomer of CCL27 compared with CCL2, or alternatively a larger contribution from higher order oligomers, such as tetramers, both of which are indicated by the PFG diffusion studies. These data are also consistent with the idea that CCL27 does not form a discrete dimeric species, even at the lower concentration where the dimer dominates, but rather has multiple interfaces of similar affinity that ultimately contribute to tetramer structure(s).
Analysis of Chemical Shift Perturbations
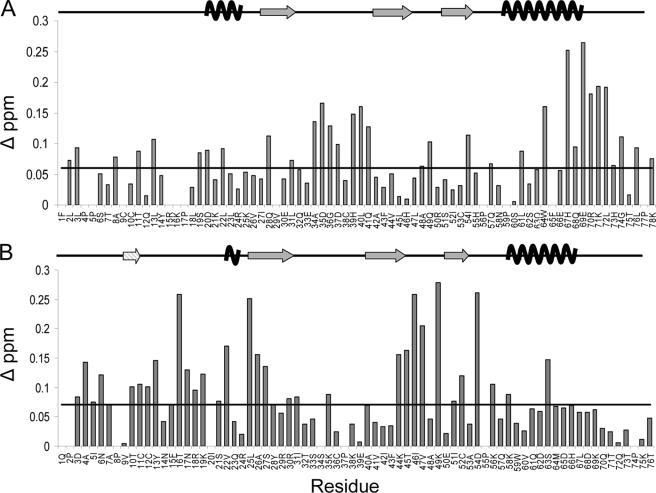
As a second method to identify residues that may be involved in the oligomerization interface(s) and that covers a broader concentration range than was feasible for the (HC)NH-NOE experiment, 1H-15N HSQC chemical shift changes of CCL27 were monitored from 1.0 mm, where the average observed species corresponds to a dimer, to 0.025 mm where CCL27 is predominantly monomeric. The residues that experienced a change in chemical shift corresponded to several regions of the protein, including Leu-2, Leu-3, Ala-8, Thr-11, and Leu-13 in the N terminus, Ser-19, Asp-20, and Leu-22 in the 310 helix, Gln-28 and Leu-31 in the first β-strand, Ala-34 to Asp-37 and His-39 to Gln-41 in the 30s loop, Ala-48 to Gln-49 in the 40s loop, Ile-54 in the third β-strand, Leu-61, Trp-64, and His-67 to Leu-72 from the C-terminal α-helix, and Gly-74, Leu-76, and Lys-78 from the C terminus. The histogram in Fig. 6A shows that the most significant changes clustered to the 30s loop, where Cys-38 forms a disulfide bond with Cys-9 in the N terminus, and the C-terminal end of the α-helix. As was the case for the (HC)NH-NOE experiment, the chemical shift perturbation data suggest that CCL27 does not readily fit the patterns expected for either type of chemokine dimer. Because CCL27 forms a tetramer at higher concentrations, the chemical shift perturbation experiment was repeated from 3.0 mm to 0.025 mm and showed that the regions experiencing shift changes were identical to those at the lower concentration (supplemental Fig. 5).
FIGURE 6.
1H-15N HSQC chemical shift perturbation analysis of CCL27 and CCL2. A, histogram of the change in chemical shift for residues 1–78 of CCL27, comparing data from 1.0 mm with 0.025 mm samples. Residues 79–88 displayed minimal shift changes and were excluded to directly compare CCL27 and CCL2 histograms. The monomeric secondary structural elements are displayed above. B, histogram of the change in chemical shift for each residue of CCL2, comparing data from 1.0 mm and 0.05 mm samples. The secondary structural elements are displayed above, including the N-terminal β-strand formed between the two monomeric subunits in the dimer structure (striped). The lines in both figures represent changes in chemical shift determined to be above baseline.
To compare these results to a known CC dimer, chemical shift changes were also monitored for CCL2 from 1.0 mm, where it forms a stable dimer, to 0.05 mm, where it is monomeric (Fig. 6B). As expected, CCL2 shows some of the most significant changes in the N terminus, in and around the region forming the small β-strand between the monomeric subunits in the dimer structure. The other highly shifted regions include the 310 helix, leading into the first β-strand, the second β-strand leading into the 40s loop and the third β-strand, where a disulfide bond connects Cys-52 to Cys-12 in the N terminus. A comparison of the two histograms in Fig. 6 shows that CCL27 and CCL2 both have similar shift changes in the N terminus. However, unlike CCL27, CCL2 has almost no changes in the 30s loop and C-terminal α-helix. In a similar analysis, Veldkamp et al. used chemical shift perturbations to evaluate the dimeric interface for CXCL12 (21). Their results demonstrate that the regions experiencing the greatest shift changes involved the first β-strand and the C-terminal α-helix (21), which is to be expected for a standard CXC chemokine dimer (16, 52). Comparison to CCL2 and CXCL12 suggests that CCL27 has characteristics of more than one dimer; in general the data are roughly consistent with regions involved in CC and CXC dimers, but could also be novel.
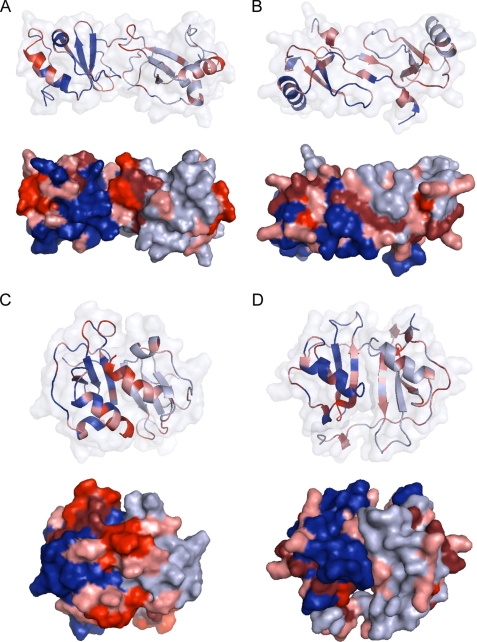
To visualize the chemical shift changes in Fig. 6, Fig. 7 shows model structures of both a CC and CXC chemokine dimer of CCL27, alongside the previously solved dimer structure for CCL2, and the CXC dimer interface of the CCL2 tetramer. In agreement with a CC dimer structure, CCL27 shows significant changes at the dimer interface, including the N terminus and the 30s loop. However, the very large changes to the C-terminal α-helix and C terminus are atypical of such an interface and cannot be rationalized by connections to the N terminus by loops or disulfide bonds (Fig. 7A). For CCL2, a typical CC dimer, the greatest shift changes occur at the N terminus or regions connected to the N terminus by disulfide bonds, such as the 30s loop and the 50s region (Fig. 7B). When these same results are mapped onto the CXC dimer structure, the chemical shift changes clearly do not match this type of interface, which is to be expected, because the CCL2 tetramer forms at higher concentrations (15). However, for CCL27, the C-terminal α-helix, the C terminus, and the 30s loop show substantial chemical shift changes that could be consistent with a CXC dimer interface (Fig. 7, C and D). Thus in contrast to the CC-like dimer observed for CCL2 at 1.0 mm, the data on CCL27 can only be reconciled with more than one dimer interface. These results, together with the gradient diffusion and filtered NOEs, suggest that CCL27 exists as a mixture of oligomeric states throughout the entire concentration range, forming a relatively lose tetramer, with characteristics consistent with both CC and CXC dimerization motifs (although novel interfaces cannot be discounted).
FIGURE 7.
Results from chemical shift perturbation mapped to dimeric forms of CCL27 and CCL2, using a color gradient to depict the intensity of chemical shift change: 0.06–0.09 ppm in pink, 0.10–0.15 in maroon, and >0.16 in red. A, model of CCL27 forming a CC chemokine dimer. B, dimeric solution structure of CCL2. C, model of CCL27 forming a CXC chemokine dimer. D, the CXC dimer-like interface of the tetrameric form of CCL2.
Exchange Broadening by Relaxation Dispersion Experiments
The initial comparison of the transverse cross-correlation rate values with the R2 values recorded under CPMG fast-pulsing conditions revealed many residues containing an exchange contribution (see above). Consequently, we decided to characterize these Rex contributions using CPMG relaxation-dispersion experiments. Information on oligomerization may be extracted from an analysis of exchange contributions to the transverse relaxation. In situations where multiple oligomeric forms are present in solution, exchange of subunits between the different forms will be manifest as increased transverse relaxation rates for those sites whose chemical shifts are dependent on the oligomeric state, such as sites present at the interface between the monomeric subunits in a dimer.
Exchange contributions for 1.0 mm CCL27 were characterized using a TROSY-selected spin-echo experiment at 600 MHz (53). The results, shown in supplemental Fig. 6A, indicate that many residues have significant Rex exchange contributions, with Asp-35, Asp-37, His-39, and Gln-41 in the 30s loop approaching 30 Hz, constituting ∼75% of their overall transverse relaxation rates. In addition, four other exchange “hot spots” include Cys-9, Cys-10, and Leu-13 in the N-loop, Ile-27 to Gln-32 in the first β-strand, Ile-52 to His-55 in the third β-strand, and Gln-57, Ser-62, Phe-65, and Gln-68 to Arg-70 in the C-terminal α-helix, as well as Leu-72 and His-73 in the C terminus. These results were mapped onto the monomeric structure of CCL27 to compare with the chemical shift perturbation data, and to determine whether they correspond to a known chemokine dimer pattern (supplemental Fig. 6B). Similar to chemical shift analysis, these results can only be reconciled with more than one interface in equilibrium, and are qualitatively consistent with the presence of both CC and CXC chemokine dimer types (see supplemental materials).
Comparison of the Rotational Diffusion Tensor to Theoretical CXC and CC Dimer Models
Finally, because the rotational diffusion tensor (calculated above by the 15N relaxation rates) is determined by the size and shape of the molecule, it can in principle, provide information on both the oligomeric state, as well as distinguish between types of oligomers/dimers, if the complexes are significantly different in shape. However, simulated tensor properties of the putative CC and CXC type dimer models discussed previously did not show sufficient agreement with the experimental data, even though the simulated tensors showed clear differences between the CC and CXC models (supplemental Table 3). These results are consistent with the concept that CCL27 does not adopt a discrete chemokine dimer, but rather exists in exchange between different oligomeric forms (see supplemental materials for additional details).
One caveat to most of the above methods is that they reflect overall conformational changes and are, therefore, not restricted to residues at the protein-protein interface. Additionally, the dimer models constructed for comparison of theoretical and experimental diffusion tensors may not be sufficiently accurate. Nevertheless, the results from all of the above methods reach a consistent picture that at 1.0 mm CCL27, where the dimer form predominates, there are interactions between multiple interfaces that ultimately contribute to the tetramer structure, a behavior that contrasts with many chemokines that adopt discrete oligomerization states at a given concentration.
Analysis of Mutants of CCL27 by PFG Diffusion NMR Implicates the N Terminus as Part of the Oligomerization Interface
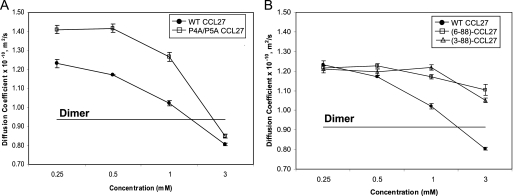
To investigate if CCL27 oligomerization could be altered by the modification of specific interactions, targeted mutagenesis was performed on regions known to be important for the dimerization of other chemokines. A mutant truncated in the C-terminal region was also investigated, as this region is extended in CCL27 relative to most other known chemokines. The Ds values were determined at several concentrations for the mutants using PFG diffusion experiments. The first set of variants involved mutations P4A and P5A near the N terminus of CCL27, because proline to alanine substitutions in the N termini of CCL2 and CCL4 rendered these chemokines incapable of dimerizing even at very high protein concentrations (20). The double mutant, P4A/P5A CCL27, had a larger Ds value at 0.25 mm compared with WT, suggesting an increased population of monomer species, and showed delayed oligomerization, requiring up to a concentration of 1.0 mm protein before a decrease in Ds was observed (Fig. 8A). However, the individual mutations P4A and P5A had no effect (supplemental Fig. 7, A and B).
FIGURE 8.
PFG diffusion analysis of WT and mutant CCL27. A, results for mutations of the double N-terminal proline mutant, P4AP5A. B, PFG diffusion results for N-terminal truncations, resulting in monomeric variants of CCL27 based on theoretical calculations.
As a less residue-biased approach, we also investigated two N-terminal truncation mutants, because the interface of standard CC chemokine dimers involves residues from the N-terminal region (47), and the contribution of Pro-4 and Pro-5 to oligomerization could not be probed by the amide-detected NMR methods employed above. Two mutants were prepared in which the first two residues, (3–88)-CCL27, and the first five residues, (6–88)-CCL27, were deleted. According to the PFG results, neither of these N-terminal truncation mutants was able to efficiently oligomerize, as only a slight decrease in Ds was observed with increasing concentration (Fig. 8B). Finally, because CCL27 has a longer C-terminal region than most chemokines, the C-terminal truncation mutant, (1–73)-CCL27, was evaluated but showed little effect on the overall oligomerization pattern (supplemental Fig. 7, A and C). These results confirm the role of the N terminus in the oligomerization of CCL27, most consistent with its classification as a CC-chemokine. However, the results do not rule out the involvement of other regions such as those important for CXC dimers. Because the associations are weak and likely cooperative, disrupting a single interface may inhibit the overall transition to a tetramer.
The Interaction of CCL27 with Glycosaminoglycans Induces Oligomerization
It has become clear that the ability of certain chemokines to oligomerize is relevant to their function. Although some roles of oligomeric forms in signaling have been reported (22), many chemokines oligomerize upon binding to GAGs, which causes their accumulation and localization on cell surfaces and the formation of haptotactic gradients for cell migration. We therefore investigated whether the oligomerization behavior of CCL27 might be relevant to interaction with GAGs.
Equilibrium Competition Binding on Immobilized Heparin
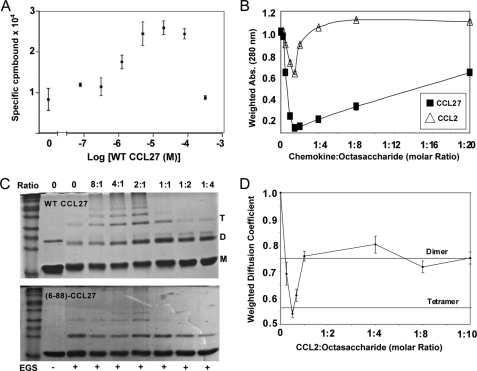
In this assay, heparin-Sepharose beads are incubated with radioactively labeled chemokine, and the labeled chemokine is then competed off with increasing concentrations of unlabeled chemokine (26). In general, one would expect that addition of a competitor would simply compete off the radiolabeled chemokine. However, Fig. 9A shows that, when a fixed amount of 125I-CCL27 is incubated with heparin beads, addition of increasing concentrations of unlabeled WT CCL27 causes the recruitment of additional 125I-CCL27, reflecting oligomerization on the heparin. This result suggests that heparin tends to increase the propensity for CCL27 to oligomerize, as observed for other chemokines like CCL2, CCL5, and CXCL8 (23, 28).
FIGURE 9.
GAG binding analysis of CCL27 and CCL2. A, radioactive heparin-Sepharose binding assay of WT CCL27. The concentration of 125I-CTACK was fixed at 2.0 nm. Specifically bound 125I-CTACK was determined by subtracting Sepharose-bound from heparin-Sepharose-bound counts. B, solubility analysis of 0.1 mm CCL2 and CCL27 in the presence of increasing amounts of heparin octasaccharide. Solubility was determined by measuring the absorbance at 280 nm and normalizing to the value at 0 mm octasaccharide. C, chemical cross-linking of WT and the monomeric mutant (6–88)-CCL27 using Sulfo-EGS in the presence of increasing molar amounts of the GAG heparin decasaccharide. The molar ratios of CCL27:heparin decasaccharide are indicated above the gels and the presence of Sulfo-EGS is indicated below. D, 13C-edited PFG diffusion profile for 0.1 mm CCL2 with increasing concentration of heparin octasaccharide. Diffusion coefficients were normalized to the value at 0 mm octasaccharide, and the resulting ratios were also used to label the lines corresponding to the theoretical dimer and tetramer.
Soluble Heparin Oligosaccharides Induce Oligomerization of CCL27
Ideally, more quantitative information on CCL27-GAG interactions could be obtained from solution methods such as analytical ultracentrifugation, dynamic light scattering, PFG diffusion, and/or fluorescence polarization. Accordingly, we initially attempted PFG diffusion studies with CCL27 and a heparin octasaccharide (Neoporin Inc., GT8041). However, upon GAG addition, a significant attenuation of signal was observed due to precipitation. This same result was seen with heparin hexasaccharide (Neoporin Inc., GT8021). Therefore, as a prelude to any further studies, solubility tests of CCL27 with heparin octasaccharide were conducted to profile its behavior in comparison to CCL2. The results, shown in Fig. 9B, suggest that at 1.5:1 chemokine:octasaccharide (0.1 mm chemokine), the minimum amount of each protein remains in solution. For CCL27, the majority is insoluble, with only 4.5% left in solution. CCL2 only decreases by 40% of its original concentration but is completely resolubilized at a 1:4 molar ratio of CCL2:octasaccharide. CCL27 on the other hand, only resolubilizes up to ∼50% of its original concentration and requires a 20-fold excess of octasaccharide. The results at 25 μm showed an identical pattern (data not shown), suggesting that CCL27 oligomerizes more avidly in the presence of the heparin octasaccharide than CCL2.
At concentrations of 25 and 50 μm, CCL27 was able to remain in solution up to ∼20% in the presence of heparin octasaccharide. Therefore, to determine if the lack of solubility correlated with oligomerization on the GAG, chemical cross-linking of CCL27 was performed using Sulfo-EGS in the presence of increasing amounts of heparin decasaccharide. The non-reducing gel in Fig. 9C suggests that, in the absence of GAGs, CCL27 is predominantly monomeric at 50 μm. However, as the concentration of heparin decasaccharide increases, the intensity of bands corresponding to dimeric, tetrameric, and higher oligomeric forms are apparent. As the heparin concentration exceeds that of CCL27, the higher order oligomers disappear, and monomer and dimer species predominate. This is a consequence of excess GAG competing for binding sites on the chemokine, but is also probably non-physiological. In contrast, cross-linking studies performed with increasing concentration of CCL27 in the absence of GAGs also indicate the presence of dimer and tetramer, but to a significantly lower extent than in the presence of GAGs (supplemental Fig. 8A). Similarly, the monomeric mutant (6–88)-CCL27 showed a decreased propensity to oligomerize on GAGs (Fig. 9C). The mutant also formed predominantly dimers while tetramer formation was significantly diminished, consistent with disruption of a CC-like oligomerization interface.
Finally, the lack of solubility at higher concentrations made it impossible to quantify the extent of CCL27 oligomerization in the presence of GAGs by PFG diffusion NMR. Therefore we examined CCL2, 60% of which remained in solution at a concentration of 0.1 mm in the presence of heparin octasaccharide. To confirm that the insolubility equates to oligomerization, a 13C-edited gradient diffusion experiment was used to determine the effect of the octasaccharide on the diffusion of the CCL2 remaining in solution. The results in Fig. 9D show that at 2:1 CCL2:octasaccharide, the Ds value decreases from monomer in the absence of GAG to the theoretical value of the tetramer in the presence of GAG. This is consistent with previous analytical ultracentrifugation data, which also showed that MCP-1 forms a tetramer in the presence of octasaccharide (54). Furthermore, it suggests that the observed insolubility is associated with increasing size of the complex. Similar to the cross-linking results for CCL27, at a molar ratio of 1:1 and higher, the Ds value of CCL2 corresponds to approximately that of a dimer indicating that the excess GAG dissociates the higher order oligomers, allowing it to be resolubilized. These same results are indicated by the chemical cross-linking studies performed with CCL2 and heparin decasaccharide in the presence of Sulfo-EGS (data not shown). In addition, analytical ultracentrifugation results indicate that, at 30 μm, CCL27 forms a single monomeric species. However, in the presence of a 1:1 molar ratio of heparin tetrasaccharide, there is clear evidence of oligomerization, despite the small size of the GAG (data not shown). The main conclusion to be made is that the insolubility originally observed for CCL27 (and CCL2) correlates with oligomerization in the presence of GAGs. This is clearly demonstrated by the equilibrium competition binding on immobilized heparin, the cross-linking studies, and the analytical ultracentrifugation data.
DISCUSSION
The structure of CCL27 was solved by NMR and revealed the standard tertiary motif for a chemokine. Translational diffusion analysis of the oligomeric properties of CCL27 demonstrates that it oligomerizes on its own in solution, up to a tetramer. At sub-millimolar concentrations, it is monomeric while at low millimolar concentrations, CCL27 oligomerizes, forming dimers and then tetramers over a narrow concentration range of 0.5–3.0 mm. Attempts to characterize the oligomeric interface of CCL27 involved an extensive analysis with 1H-15N HSQC chemical shift perturbation experiments, relaxation dispersion experiments, and filtered (HC)NH-NOEs. The results suggest that, although the oligomeric interface has some characteristics qualitatively consistent with known CC and CXC chemokine dimers, at concentrations where it is primarily dimeric, more than one dimer species co-exist with similar, albeit weak, affinities, and these interfaces ultimately contribute to tetramer formation. These results are most clearly illustrated by mapping concentration-dependent chemical shift perturbations onto CC and CXC dimer models, which show shifted residues at the interface of both structures. However, alternative novel oligomerization motifs can't be discounted.
The oligomeric profile for CCL27 differs with the results seen for other chemokines such as CCL2 and CXCL12 (21), both of which form stable dimers. Thus several experiments were run in parallel with CCL2 to directly compare CCL27 to a chemokine known to form a stable dimer. In contrast to CCL27, CCL2 forms a stable dimer at micromolar concentrations. Furthermore, filtered (HC)NH-NOEs and concentration-dependent chemical shift perturbations are indicative of a discrete CC dimer. Overall, the results indicate that compared with CCL2 and many other chemokines, CCL27 is a more dynamic oligomer.
The full functional relevance of chemokine oligomerization is not entirely understood. However, a direct correlation between the ability of chemokines to oligomerize and to induce in vivo cellular migration has been established. Proudfoot et al. previously demonstrated that monomeric variants of CCL2, CCL4 (MIP-1β), and CCL5 (RANTES (regulated on activation normal T cell expressed and secreted)) were inactive in cellular migration when injected into the peritoneal cavity of mice, even though they were functional in trans-filter assays of cell migration in vitro (23). Similarly, Luster and coworkers demonstrated that a monomeric form of CXCL10 (IP-10) is incapable of inducing cell migration in vivo and transendothelial migration in vitro (19). More recently it was shown that the above monomeric form of CCL2 has anti-inflammatory activity in animal models of experimental arthritis (20) and experimental autoimmune encephalomyelitis (EAE) (55), underscoring the importance of oligomerization in vivo.
It is now believed that, although the monomeric forms of chemokines are involved in binding and activating chemokine receptors, at least to induce cell migration, that oligomeric forms are involved in interactions with GAGs. Indeed, several studies, including this one, have shown that chemokines oligomerize on GAGs (23, 54). Such interactions are believed to be involved in the localization of chemokines to cell surfaces, particularly in the presence of shear forces, as well as in the transcytosis of chemokines across cells (56, 57). Similar to the importance of oligomerization, the critical role of GAG interactions has been demonstrated in studies with GAG binding-deficient chemokines, which maintain the ability to bind and activate receptors in vitro but are inactive in vivo. Such studies have been demonstrated with CCL2, CCL4, and CCL5 (23), as well as CCL7 (56) and CXCL12 (27, 58). Furthermore, these mutants can act as inhibitors of the WT proteins, and in the case of the GAG-deficient mutant [44AANA47]-CCL5, the mechanism was attributed to the formation of non-functional heterodimers with WT CCL5 (9, 27).
Most chemokines studied to date have structures that have been characterized in discrete forms. However, it is becoming clear that, overall, chemokines span a range of oligomeric behaviors from monomers to large oligomers with significant differences in the stabilities of the species. For example, CCL5 forms very large stable oligomers, and although a dimer was solved by NMR, low pH conditions were required to dissociate the oligomer (11, 59). Others like CXCL4 (PF-4) form stable tetramers (60), whereas CXCL1 (GROα) and CCL2 form stable dimers (47, 61) and CCL7 (MCP-3) and CXCL10 (IP-10) are monomers (16, 62). In most cases, these forms are in equilibrium with other states, but specific species dominate at the concentrations used for biophysical studies. Furthermore, it has become apparent that GAGs stabilize the higher order oligomeric forms (54, 63). For example, although CCL2 forms dimers in solution, Lau et al. demonstrated that a heparin octasaccharide induces the formation of a CCL2 tetramer. A detailed analysis of the GAG binding sites then revealed a continuous ring encircling the oligomeric interface, suggesting the octasaccharide binds at the interface, and providing a rationale for how it stabilizes the tetrameric form (54). Similarly human CXCL10 was crystallized in three tetrameric forms, each displaying unique GAG-binding epitopes when mapped onto each structure. These results suggest that different GAGs may stabilize the different oligomeric forms of CXCL10 and conversely that the different oligomeric forms may be involved in recognition of different GAGs (16). A dramatic example of a chemokine that shows this type of structural plasticity is XCL1 (Lymphotactin). XCL1 undergoes a global conformational transition between a canonical chemokine monomeric fold, which binds the receptor XCR1, and a novel dimeric β-sandwich structure, which binds to GAGs (64, 65). The results demonstrated herein suggest that CCL27 falls into the category of chemokines that can readily convert between oligomeric states potentially allowing it to accommodate multiple GAG-binding partners, and thereby providing a mechanism for cell specificity and regulation. Thus future studies of CCL27 will involve a detailed analysis of its GAG-binding epitopes, as well as the ability of different GAGs to stabilize discrete oligomeric forms. However, it is also possible that oligomerization plasticity may have other functions related to kinetics of interactions and even signaling through receptors as recently demonstrated for CXCL12 (22).
Supplementary Material
Acknowledgments
We thank Melinda Hanes, Morgan O'Hayre, and Catherina Salanga for their editing and Nicole Kruse from Bruker-Biospin for assistance with optimization of the PFG diffusion experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-AI37113 and RO1-GM081763 (to T. M. H.) and NIH Molecular Biophysics Training Grant GM08326 (to A. L. J.). The NMR facility of the Biochemistry Department in Cambridge is supported by the Biotechnology and Biological Sciences and Research Council (BBSRC).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, references, Figs. 1–8, and Tables 1–3.
- GAG
- glycosaminoglycan
- MOPS
- 4-morpholinepropanesulfonic acid
- ARIA
- Ambiguous Restraints for Iterative Assignment
- NOE
- nuclear Overhauser effect
- NOESY
- nuclear Overhauser effect spectroscopy
- PFG
- pulsed field gradient
- Sulfo-EGS
- sulfoethyleneglycolbis(sulfosuccinimidylsuccinate)
- HSQC
- heteronuclear single quantum coherence
- WT
- wild type
- CPMG
- Carr-Purcell-Meiboom-Gill.
REFERENCES
- 1.Baggiolini M. (1998) Nature 392, 565–568 [DOI] [PubMed] [Google Scholar]
- 2.Baggiolini M., Dewald B., Moser B. (1997) Annu. Rev. Immunol. 15, 675–705 [DOI] [PubMed] [Google Scholar]
- 3.Charo I. F., Ransohoff R. M. (2006) N. Engl. J. Med. 354, 610–621 [DOI] [PubMed] [Google Scholar]
- 4.Hartley O., Dorgham K., Perez-Bercoff D., Cerini F., Heimann A., Gaertner H., Offord R. E., Pancino G., Debré P., Gorochov G. (2003) J. Virol. 77, 6637–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunkel E. J., Butcher E. C. (2002) Immunity 16, 1–4 [DOI] [PubMed] [Google Scholar]
- 6.Allen S. J., Crown S. E., Handel T. M. (2007) Annu. Rev. Immunol. 25, 787–820 [DOI] [PubMed] [Google Scholar]
- 7.O'Hayre M., Salanga C. L., Handel T. M., Allen S. J. (2008) Biochem. J. 409, 635–649 [DOI] [PubMed] [Google Scholar]
- 8.Bono M. R., Elgueta R., Sauma D., Pino K., Osorio F., Michea P., Fierro A., Rosemblatt M. (2007) Cytokine Growth Factor Rev. 18, 33–43 [DOI] [PubMed] [Google Scholar]
- 9.Johnson Z., Proudfoot A. E., Handel T. M. (2005) Cytokine Growth Factor Rev. 16, 625–636 [DOI] [PubMed] [Google Scholar]
- 10.Blain K. Y., Kwiatkowski W., Zhao Q., La Fleur D., Naik C., Chun T. W., Tsareva T., Kanakaraj P., Laird M. W., Shah R., George L., Sanyal I., Moore P. A., Demeler B., Choe S. (2007) Biochemistry 46, 10008–10015 [DOI] [PubMed] [Google Scholar]
- 11.Czaplewski L. G., McKeating J., Craven C. J., Higgins L. D., Appay V., Brown A., Dudgeon T., Howard L. A., Meyers T., Owen J., Palan S. R., Tan P., Wilson G., Woods N. R., Heyworth C. M., Lord B. I., Brotherton D., Christison R., Craig S., Cribbes S., Edwards R. M., Evans S. J., Gilbert R., Morgan P., Randle E., Schofield N., Varley P. G., Fisher J., Waltho J. P., Hunter M. G. (1999) J. Biol. Chem. 274, 16077–16084 [DOI] [PubMed] [Google Scholar]
- 12.Fernandez E. J., Lolis E. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 469–499 [DOI] [PubMed] [Google Scholar]
- 13.Jin H., Hayes G. L., Darbha N. S., Meyer E., LiWang P. J. (2005) Biochem. Biophys. Res. Commun. 338, 987–999 [DOI] [PubMed] [Google Scholar]
- 14.Jansma A., Handel T. M., Hamel D. J. (2009) Methods Enzymol. 461, 31–50 [DOI] [PubMed] [Google Scholar]
- 15.Lubkowski J., Bujacz G., Boqué L., Domaille P. J., Handel T. M., Wlodawer A. (1997) Nat. Struct. Biol. 4, 64–69 [DOI] [PubMed] [Google Scholar]
- 16.Swaminathan G. J., Holloway D. E., Colvin R. A., Campanella G. K., Papageorgiou A. C., Luster A. D., Acharya K. R. (2003) Structure 11, 521–532 [DOI] [PubMed] [Google Scholar]
- 17.Jabeen T., Leonard P., Jamaluddin H., Acharya K. R. (2008) Acta Crystallogr. D. Biol. Crystallogr. 64, 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baysal C., Atilgan A. R. (2001) Proteins 43, 150–160 [DOI] [PubMed] [Google Scholar]
- 19.Campanella G. S., Grimm J., Manice L. A., Colvin R. A., Medoff B. D., Wojtkiewicz G. R., Weissleder R., Luster A. D. (2006) J. Immunol. 177, 6991–6998 [DOI] [PubMed] [Google Scholar]
- 20.Handel T. M., Johnson Z., Rodrigues D. H., Dos Santos A. C., Cirillo R., Muzio V., Riva S., Mack M., Déruaz M., Borlat F., Vitte P. A., Wells T. N., Teixeira M. M., Proudfoot A. E. (2008) J. Leukocyte Biol. 84, 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veldkamp C. T., Peterson F. C., Pelzek A. J., Volkman B. F. (2005) Protein Sci. 14, 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veldkamp C. T., Seibert C., Peterson F. C., De la Cruz N. B., Haugner J. C., 3rd, Basnet H., Sakmar T. P., Volkman B. F. (2008) Sci. Signal. 1, ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Proudfoot A. E., Handel T. M., Johnson Z., Lau E. K., LiWang P., Clark-Lewis I., Borlat F., Wells T. N., Kosco-Vilbois M. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imberty A., Lortat-Jacob H., Pérez S. (2007) Carbohydr. Res. 342, 430–439 [DOI] [PubMed] [Google Scholar]
- 25.Koopmann W., Krangel M. S. (1997) J. Biol. Chem. 272, 10103–10109 [DOI] [PubMed] [Google Scholar]
- 26.Kuschert G. S., Coulin F., Power C. A., Proudfoot A. E., Hubbard R. E., Hoogewerf A. J., Wells T. N. (1999) Biochemistry 38, 12959–12968 [DOI] [PubMed] [Google Scholar]
- 27.Lau E. K., Allen S., Hsu A. R., Handel T. M. (2004) Adv. Protein Chem. 68, 351–391 [DOI] [PubMed] [Google Scholar]
- 28.Hoogewerf A. J., Kuschert G. S., Proudfoot A. E., Borlat F., Clark-Lewis I., Power C. A., Wells T. N. (1997) Biochemistry 36, 13570–13578 [DOI] [PubMed] [Google Scholar]
- 29.Homey B., Wang W., Soto H., Buchanan M. E., Wiesenborn A., Catron D., Müller A., McClanahan T. K., Dieu-Nosjean M. C., Orozco R., Ruzicka T., Lehmann P., Oldham E., Zlotnik A. (2000) J. Immunol. 164, 3465–3470 [DOI] [PubMed] [Google Scholar]
- 30.Homey B., Alenius H., Müller A., Soto H., Bowman E. P., Yuan W., McEvoy L., Lauerma A. I., Assmann T., Bunemann E., Lehto M., Wolff H., Yen D., Marxhausen H., To W., Sedgwick J., Ruzicka T., Lehmann P., Zlotnik A. (2002) Nat. Med. 8, 157–165 [DOI] [PubMed] [Google Scholar]
- 31.Murakami T., Cardones A. R., Hwang S. T. (2004) J. Dermatol. Sci. 36, 71–78 [DOI] [PubMed] [Google Scholar]
- 32.Catanzariti A. M., Soboleva T. A., Jans D. A., Board P. G., Baker R. T. (2004) Protein Sci. 13, 1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanelis V., Forman-Kay J. D., Kay L. E. (2001) IUBMB Life 52, 291–302 [DOI] [PubMed] [Google Scholar]
- 34.Linge J. P., O'Donoghue S. I., Nilges M. (2001) Methods Enzymol. 339, 71–90 [DOI] [PubMed] [Google Scholar]
- 35.Nilges M., O'Donoghue S. I. (1998) Prog. Nuclear Magn. Reson. Spect. 32, 107–139 [Google Scholar]
- 36.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 37.Colobran R., Pujol-Borrell R., Armengol M. P., Juan M. (2007) Clin. Exp. Immunol. 148, 208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang T. L., Susumu M., Shaka A. J., van Zijl P. C. (1997) J. Am. Chem. Soc. 119, 6203–6204 [Google Scholar]
- 39.Laskowski R. A., MacArthur D. S., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 40.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 41.Ghose R., Prestegard J. H. (1998) J. Magn. Reson. 134, 308–314 [DOI] [PubMed] [Google Scholar]
- 42.Dosset P., Hus J. C., Blackledge M., Marion D. (2000) J. Biomol. NMR 16, 23–28 [DOI] [PubMed] [Google Scholar]
- 43.Carrasco B., García de la Torre J., Pons M. (1999) Biophys. J. 76, 3044–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernadó P., Garcia de la Torre J., Pons M. (2002) J. Biomol. NMR 23, 139–150 [DOI] [PubMed] [Google Scholar]
- 45.Loria J. P., Rance M., Palmer A. G. (1999) J. Am. Chem. Soc. 121, 2331–2332 [Google Scholar]
- 46.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 47.Handel T. M., Domaille P. J. (1996) Biochemistry 35, 6569–6584 [DOI] [PubMed] [Google Scholar]
- 48.Altieri A. S. (1995) J. Am. Chem. Soc. 117, 7566–7567 [Google Scholar]
- 49.Holmes W. D., Consler T. G., Dallas W. S., Rocque W. J., Willard D. H. (2001) Protein Expr. Purif. 21, 367–377 [DOI] [PubMed] [Google Scholar]
- 50.Kroenke C. D. (1998) J. Am. Chem. Soc. 120, 7905–7915 [Google Scholar]
- 51.Clore G. M. (1990) J. Am. Chem. Soc. 112, 4989–4991 [Google Scholar]
- 52.Clore G. M., Appella E., Yamada M., Matsushima K., Gronenborn A. M. (1990) Biochemistry 29, 1689–1696 [DOI] [PubMed] [Google Scholar]
- 53.Wang C., Rance M., Palmer A. G., 3rd (2003) J. Am. Chem. Soc. 125, 8968–8969 [DOI] [PubMed] [Google Scholar]
- 54.Lau E. K., Paavola C. D., Johnson Z., Gaudry J. P., Geretti E., Borlat F., Kungl A. J., Proudfoot A. E., Handel T. M. (2004) J. Biol. Chem. 279, 22294–22305 [DOI] [PubMed] [Google Scholar]
- 55.Brini E., Ruffini F., Bergami A., Brambilla E., Dati G., Greco B., Cirillo R., Proudfoot A. E., Comi G., Furlan R., Zaratin P., Martino G. (2009) J. Neuroimmunol. 209, 33–39 [DOI] [PubMed] [Google Scholar]
- 56.Ali S., Robertson H., Wain J. H., Isaacs J. D., Malik G., Kirby J. A. (2005) J. Immunol. 175, 1257–1266 [DOI] [PubMed] [Google Scholar]
- 57.Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 58.Murphy J. W., Cho Y., Sachpatzidis A., Fan C., Hodsdon M. E., Lolis E. (2007) J. Biol. Chem. 282, 10018–10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung C. W., Cooke R. M., Proudfoot A. E., Wells T. N. (1995) Biochemistry 34, 9307–9314 [DOI] [PubMed] [Google Scholar]
- 60.Mayo K. H., Roongta V., Ilyina E., Milius R., Barker S., Quinlan C., La Rosa G., Daly T. J. (1995) Biochemistry 34, 11399–11409 [DOI] [PubMed] [Google Scholar]
- 61.Fairbrother W. J., Reilly D., Colby T. J., Hesselgesser J., Horuk R. (1994) J. Mol. Biol. 242, 252–270 [DOI] [PubMed] [Google Scholar]
- 62.Kim K. S., Rajarathnam K., Clark-Lewis I., Sykes B. D. (1996) FEBS Lett. 395, 277–282 [DOI] [PubMed] [Google Scholar]
- 63.Yu Y., Sweeney M. D., Saad O. M., Crown S. E., Hsu A. R., Handel T. M., Leary J. A. (2005) J. Biol. Chem. 280, 32200–32208 [DOI] [PubMed] [Google Scholar]
- 64.Kuloğglu E. S., McCaslin D. R., Markley J. L., Volkman B. F. (2002) J. Biol. Chem. 277, 17863–17870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volkman B. F., Liu T. Y., Peterson F. C. (2009) Methods Enzymol. 461, 51–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.