Abstract
Cell division protein FtsZ can form single-stranded filaments with a cooperative behavior by self-switching assembly. Subsequent condensation and bending of FtsZ filaments are important for the formation and constriction of the cytokinetic ring. PC190723 is an effective bactericidal cell division inhibitor that targets FtsZ in the pathogen Staphylococcus aureus and Bacillus subtilis and does not affect Escherichia coli cells, which apparently binds to a zone equivalent to the binding site of the antitumor drug taxol in tubulin (Haydon, D. J., Stokes, N. R., Ure, R., Galbraith, G., Bennett, J. M., Brown, D. R., Baker, P. J., Barynin, V. V., Rice, D. W., Sedelnikova, S. E., Heal, J. R., Sheridan, J. M., Aiwale, S. T., Chauhan, P. K., Srivastava, A., Taneja, A., Collins, I., Errington, J., and Czaplewski, L. G. (2008) Science 312, 1673–1675). We have found that the benzamide derivative PC190723 is an FtsZ polymer-stabilizing agent. PC190723 induced nucleated assembly of Bs-FtsZ into single-stranded coiled protofilaments and polymorphic condensates, including bundles, coils, and toroids, whose formation could be modulated with different solution conditions. Under conditions for reversible assembly of Bs-FtsZ, PC190723 binding reduced the GTPase activity and induced the formation of straight bundles and ribbons, which was also observed with Sa-FtsZ but not with nonsusceptible Ec-FtsZ. The fragment 2,6-difluoro-3-methoxybenzamide also induced Bs-FtsZ bundling. We propose that polymer stabilization by PC190723 suppresses in vivo FtsZ polymer dynamics and bacterial division. The biochemical action of PC190723 on FtsZ parallels that of the microtubule-stabilizing agent taxol on the eukaryotic structural homologue tubulin. Both taxol and PC190723 stabilize polymers against disassembly by preferential binding to each assembled protein. It is yet to be investigated whether both ligands target structurally related assembly switches.
Keywords: Antibiotics, Anticancer Drug, Cell Division, Protein Assembly, Tubulin, FtsZ, Taxol
Introduction
Essential cell division protein FtsZ assembles into filaments that form the cytokinetic ring at the division site (1) and recruits the accessory proteins of the bacterial divisome (2–4). FtsZ, with an added membrane anchor, can form constricting rings in liposomes (5). FtsZ is a member of the tubulin family of cytoskeletal GTPases, also including αβ-tubulin, γ-tubulin, bacterial tubulin BtubA/B (3, 6), and divergent TubZs (7). FtsZ and tubulin use the same structural fold and similar head-to-tail protofilaments with GTP bound at the intersubunit association interface (8), yet they have evolved to form different assemblies with specialized functions. Their GTPase sites are completed upon contact of the apical GTP-binding domain of one subunit with the basal GTPase activation domain of the next subunit. GTP hydrolysis renders the polymers metastable and ready for disassembly. There is evidence that unassembled tubulin, unlike other GTPases, is predominantly in an inactive (curved) conformation regardless of which nucleotide is bound and switches into the active (straight) conformation mainly through the assembly contacts and that GTP binding lowers the free energy barrier between both conformations (6, 9–11). Binding of the successful antitumor drug taxol overrides the GTP/GDP regulation, inducing microtubule assembly even with GDP (12, 13). Tubulin promiscuously interacts with small molecules of natural origin that induce or inhibit its assembly. However, there are fewer functional inhibitors for FtsZ.
FtsZ is a relatively simple assembly machine capable of nucleated linear polymerization. Such cooperative behavior of a linear polymer was anticipated by the self-switching assembly concept (14) and suggested to be possible for FtsZ if dimerization caused a conformational change that increases the affinity of the next monomer to bind (3, 15). Cryoelectron microscopy of unsupported samples has shown that FtsZ from Escherichia coli can form semiflexible single-stranded filaments (16). Their cooperative behavior is explained by an unfavorable monomer isomerization (activation switch) between an inactive, assembly incompetent, and active conformation, which is coupled to assembly, creating a nucleation barrier (16–19). On the other hand, FtsZ from Methanococcus jannaschii forms characteristic double-stranded filaments (20, 21). A number of crystal structures of FtsZ did not reveal a nucleotide-induced activation switch (22). The structural flexibility changes coupled to assembly are unknown, requiring determination of an FtsZ filament structure.
A cryoelectron tomography study of the Caulobacter crescentus cytoskeleton revealed a putative FtsZ ring, consisting of a few short (100 nm) apparently single-stranded FtsZ filaments (5-nm wide) below the plasma membrane near the division site, and suggested that these FtsZ polymers generate the force that constricts the membrane for division through iterative cycles of GTP hydrolysis, depolymerization, and repolymerization (23). Lateral FtsZ filament association is also important for Z-ring formation and constriction. Several proteins are known to bundle FtsZ filaments, including ZipA, ZapA, and SepF (4). Helical FtsZ structures remodel by lateral association into the Z-ring in Bacillus subtilis (24), and artificial FtsZ rings coalesce into brighter rings (5). Several theoretical models for the Z-ring have proposed different roles for FtsZ filament condensation and bending (reviewed in Ref. 25).
FtsZ filaments are dynamic, with a subunit half-life of ∼10 s, depending on the GTPase rate (26). Although FtsZ polymers can exchange nucleotides, GDP dissociation may be slow enough for polymer disassembly to take place first, resulting in the subunits of FtsZ polymers recycling with GTP hydrolysis (27, 28). The length dynamics of the small individual FtsZ filaments have not been determined.
Impairing FtsZ filament dynamics should block bacterial division. FtsZ has been recognized as an attractive target for new antibiotics (29) for emerging resistant pathogens. Expression of ftsZ is more stringently required for bacterial growth than the established antibacterial targets murA and fabl (30). Potentially druggable cavities in FtsZ structures (22) are the apical nucleotide-binding site and a lateral channel between the N- and C-terminal domains. The latter overlaps the binding site of the microtubule-stabilizing antitumor drug taxol in eukaryotic tubulin. Several GTP analogues substituted at C-8 selectively inhibit FtsZ polymerization but support tubulin assembly into microtubules (31), indicating differences in nucleotide binding by each protein that may be exploited to selectively inhibit bacterial FtsZ without poisoning eukaryotic tubulin. FtsZ recently has been validated as the target of an effective antibacterial compound developed as a cell division inhibitor in B. subtilis and the pathogen Staphylococcus aureus, PC190723. Its putative binding site, mapped with resistance mutations, corresponds to the taxol-binding site in tubulin (32).
We aimed to determine the mechanism of functional inhibition of FtsZ by the small antibacterial molecule PC190723, 3-[(6-chloro[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-difluorobenzamide (abbreviated here as PC).3 We have found that PC enhances the cooperative assembly of Bs-FtsZ into filaments, bundles, and other condensates. PC also is a Sa-FtsZ polymer stabilizer. Stabilizing FtsZ polymers should reduce their dynamics, which explains why PC blocks cell division in bacteria analogously to the action of taxol on tubulin.
EXPERIMENTAL PROCEDURES
Methods for Bs-FtsZ, Ec-FtsZ, Sa-FtsZ, and tubulin purification, monitoring assembly with light scattering, electron microscopy, polymer pelleting, binding measurements, GTPase, and a screen of solution conditions for assembly are described under the supplemental “Experimental Procedures.” PC190723 was provided by Prolysis (32). Stock solutions (1 mm) in deuterated dimethyl sulfoxide (Merck Uvasol) were kept frozen and dry. They were diluted directly into the experimental samples. Residual dimethyl sulfoxide was 2% (v/v) unless indicated. DFMBA, 2,6-difluoro-3-hydroxybenzamide, and CTPM were synthesized in this work (supplemental data). Buffers employed were as follows: Hepes50 (50 mm Hepes/KOH, 50 mm KCl, 1 mm EDTA, pH 6.8), Hepes250 (50 mm Hepes/KOH, 250 mm KCl, 1 mm EDTA, pH 6.8) (where indicated, these buffers were made at pH 7.5), and Mes50 (50 mm Mes/KOH, 50 mm KCl, 1 mm EDTA, pH 6.5).
RESULTS
PC190723 induced Bs-FtsZ assembly into bundles and isolated filaments. These processes could be dissected and analyzed, employing different solution conditions, as described below.
PC Induces Bundles of B. subtilis FtsZ Filaments
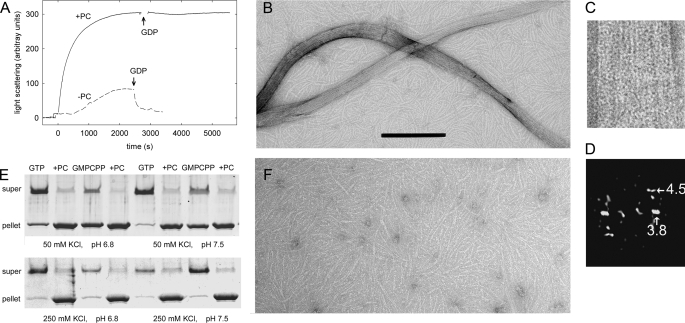
To characterize the effects of PC on FtsZ assembly, full-length, untagged FtsZ from B. subtilis was first expressed and purified, and its reversible assembly was induced by 10 mm MgCl2 and 2 mm GTP in Hepes50, 25 °C (see “Experimental Procedures”) without other additives, which were chosen for convenience as reference conditions. Addition of PC induced a dramatic increase in FtsZ light scattering and stabilized polymers against disassembly induced by GDP (Fig. 1A). Many large PC-induced bundles and ribbons, in addition to FtsZ filaments, were observed by negative stain electron microscopy (Fig. 1B). These bundles and ribbons were several microns long, had a variable ∼30 to ∼60 nm width, and were apparently made of associated FtsZ filaments. Computed diffraction patterns indicated an axial spacing of 4.5 ± 0.5 nm between subunits (Fig. 1, C and D), which is compatible with the 4.3-nm spacing of FtsZ monomers associated head-to-tail in tubulin-like protofilaments (20, 21), and a shorter lateral spacing of 3.8 ± 0.3 nm, suggesting a lateral association of Bs-FtsZ molecules (Protein Data Bank code 2VXY; 32) in contact along their shortest dimension (that is, rotated ∼90° on the longitudinal axis with respect to tubulin protofilaments in a microtubule) (33). PC-induced bundles and ribbons also formed with the slowly hydrolyzable GTP analogue GMPCPP (0.1 mm). Most FtsZ above a small ∼0.3 μm critical concentration (Cr) pelleted upon 20 min of centrifugation at 386,000 × g (Fig. 1E). In the absence of PC, relatively short, ∼4-nm wide filaments were observed (Fig. 1F), which sedimented above a larger Cr ∼5 μm FtsZ. These results indicated that the small molecule PC stabilized Bs-FtsZ filament bundles but did not distinguish whether PC primarily enhances filament formation or bundling.
FIGURE 1.
Assembly of bundles of Bs-FtsZ (10 μm) induced by PC (20 μm). A, light scattering time courses with and without PC in Hepes50 assembly buffer, 10 mm MgCl2, pH 6.8, 25 °C. GTP (2 mm) was added at time 0 and 2 mm GDP as indicated by the arrows. B, electron micrograph of Bs-FtsZ with PC. Bar, 200 nm. C, an enlarged bundle. D, its computed diffractogram. The corresponding spacing of the main equatorial spot and the first layer line are indicated in nm. E, Bs-FtsZ polymer pelleting assays at varying pH and ionic strength. F, Bs-FtsZ without PC, magnified as in B.
Effects of Solution Conditions, Mg2+ and Nucleotide
To elucidate the mechanism of PC-induced assembly and to characterize the effects of PC under more physiological conditions, pH was increased to 7.5, or ionic strength increased to 250 mm KCl, which reduced the extent of spontaneous Bs-FtsZ assembly with nucleotide and Mg2+. Assembly was efficiently induced by PC in these cases (Fig. 1E), although bundle formation was reduced. Assembly was markedly sensitive to the buffer anion (supplemental “Experimental Procedures”). Disassembly could be monitored by light scattering following assembly of 10 μm Bs-FtsZ with 25 μm GMPCPP (expected to stabilize polymers) and 10 mm MgCl2 in 250 mm KCl (scattering with GTP was very small). This was attributed to GMPCPP hydrolysis and was not observed in the presence of PC. GTP or GMPCPP was required for PC-enhanced assembly. Mg2+ without nucleotide was ineffective. Interestingly, GTP or GMPCPP without Mg2+ induced assembly of Bs-FtsZ filaments, which were not formed with GDP or GMPCP. Mg2+ plus GDP or GMPCP and PC gave a weak polymer formation. These results confirmed that PC stabilizes Bs-FtsZ polymers and showed the requirement of the nucleotide γ-phosphate.
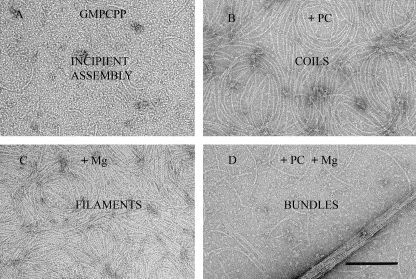
It was possible to separate PC-induced filament assembly from bundle formation by using 250 mm KCl and GMPCPP. Bs-FtsZ assembly was only incipient without Mg2+ (Fig. 2A). Adding only PC induced characteristic filament coils (Fig. 2B), whereas Mg2+ alone induced the formation of filaments (Fig. 2C). Both were inhibited with GMPCP. PC and Mg2+ together induced the formation of filaments and bundles (Fig. 2D). Similar results were obtained with GTP, but with PC and Mg2+, dense coils and no bundles were observed (supplemental Fig. S1).
FIGURE 2.
PC and magnesium-induced Bs-FtsZ assemblies. Electron micrograph of negatively stained polymers formed by Bs-FtsZ (10 μm) in Hepes250 assembly buffer with 0.1 mm GMPCPP (A), 0.1 mm GMPCPP and 20 μm PC (B), 0.1 mm GMPCPP and 10 mm MgCl2 (C), and 0.1 mm GMPCPP and 10 mm MgCl2 and 20 μm PC (D). The bar indicates 200 nm.
PC Induces Bs-FtsZ Single-stranded Filament Assembly
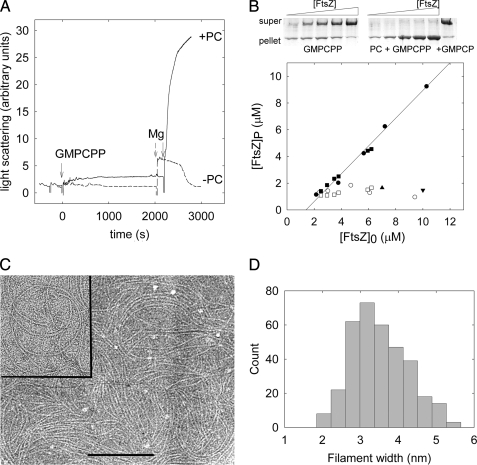
Mg2+-less Hepes250 buffer permitted PC-induced nucleated assembly of single-stranded Bs-FtsZ filaments. Our divalent cation-free solutions typically contain ∼1 μm residual Mg2+ (34) that with 1 mm EDTA in buffer results in ∼4 nm free Mg2+. Incipient assembly of Bs-FtsZ was observed with nucleotide in the absence of PC (no nucleotide-induced light scattering increase or pelleting). However, addition of PC under these conditions resulted in Bs-FtsZ polymerization. There was a small light scattering increase with PC and GMPCPP (Fig. 3A). These PC-induced FtsZ filaments sedimented in 80 min at 386,000 × g, which allowed their quantification. Practically all Bs-FtsZ was pelleted with PC and GTP or GMPCPP above a Cr ∼1 μm (Fig. 3B; GTP, 0.8 ± 0.1 μm; GMPCPP, 1.3 ± 0.3 μm). Insignificant GTP hydrolysis was detected under these conditions. These results indicated a nucleated polymerization mechanism for the PC-induced assembly of Bs-FtsZ filaments, with an apparent free energy change for polymer elongation ΔGapp0 = RTlnCr = −8.2 kcal mol−1 (16). With GTP or GMPCPP and no PC, a background similar to that of PC and GDP or GMPCP was observed (Fig. 3B). Cr decreased to insignificant values in experiments made in parallel with 10 mm MgCl2 (supplemental Fig. S2), indicating a larger effective elongation affinity, although the system was not at chemical equilibrium due to nucleotide hydrolysis. Interestingly, the PC-induced Bs-FtsZ filaments without Mg2+ typically had an ∼100-nm curvature radius and appeared single-stranded. To confirm that these filaments are single-stranded, we performed cryoelectron microscopy under these conditions, using holey carbon film. The sample trapped within the small holes was quickly vitrified and kept under liquid nitrogen temperatures to rule out any effect of the staining agent or the adsorption to a support film (Fig. 3C). Filament width measurements from the cryomicrographs had a distribution with an average 3.5 ± 0.7 nm (Fig. 3D), indicating that these filaments are one Bs-FtsZ molecule wide.
FIGURE 3.
Bs-FtsZ filament assembly induced by PC. A, solid line, light scattering changes induced on Bs-FtsZ (10 μm) with PC (20 μm) by additions of GMPCPP (25 μm) and MgCl2 (10 mm) (arrows) in Hepes250, pH 6.8, 25 °C. Dashed line, same without PC. B, example of sedimentation of Bs-FtsZ polymers formed without Mg2+ (top) and pellet quantification (bottom). Filled circles, PC and GMPCPP (0.1 mm); filled squares, PC and GTP (2 mm); open symbols, without PC; triangle, PC and GDP; inverted triangle, PC and GMPCP. C, cryoelectron micrograph of Bs-FtsZ filaments assembled with PC and GTP under the same conditions without Mg2+. Inset, negatively stained filaments. Bar, 200 nm. D, cryoelectron microscopy filament width distribution, indicating that the filaments are one Bs-FtsZ molecule-wide.
Binding of PC and Its Fragment DFMBA to Bs-FtsZ Polymers
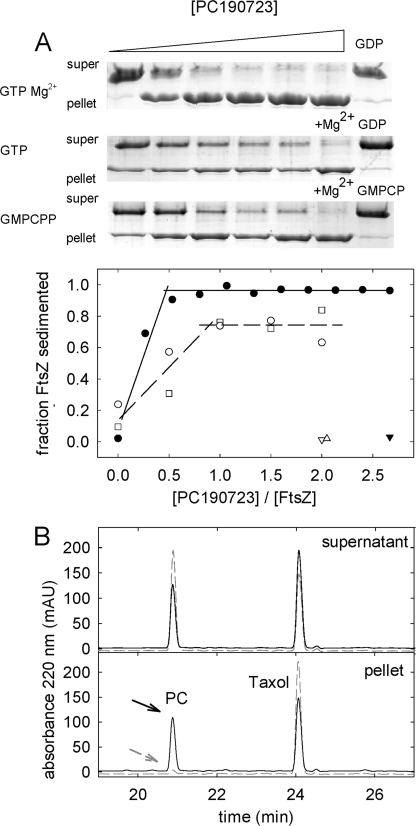
Titrations of Bs-FtsZ polymer formation with PC showed an increasing fraction of sedimented FtsZ, which saturated between 0.5 and 1 PC per FtsZ (Fig. 4A), compatible with PC inducing polymerization by binding to one site in FtsZ. For a ligand to induce protein association, it has to bind more to the protein polymer than the monomer. As a first approach, PC binding to Bs-FtsZ polymers was determined by extraction of the ligand from the pellets and supernatants followed by HPLC. PC cosedimented with the polymers and inhibition of polymerization with GDP also inhibited the PC pulldown. The results were 0.5 PC bound per FtsZ at ∼10 μm free PC, irrespective of ionic strength and magnesium level (0.46 ± 0.14 in Hepes50 with 10 mm MgCl2, 0.40 ± 0.10 in Hepes250 without MgCl2, and 0.54 ± 0.10 in Hepes250 with 10 mm MgCl2). FtsZ was active in polymer formation (Fig. 3B), and saturating the binding to an expected 1 PC per FtsZ was precluded by the free ligand solubility (∼10 μm in our buffers). The simplest interpretation of these results is one PC binding site per polymer FtsZ molecule with an apparent affinity ∼105 m−1.
FIGURE 4.
Binding of PC to Bs-FtsZ polymers. A, Bs-FtsZ (10 μm) sedimentation assays in Hepes250, pH 6.8, 25 °C, and quantification of polymer with increasing concentrations of PC. Open circles, with 2 mm GTP; filled circles, with 2 mm GTP and Mg2+; void squares, with 0.1 mm GMPCPP. Triangles, corresponding data with GDP, GDP, and Mg2+ or GMPCP. B, HPLC analysis of PC (10 μm total) co-sedimentation with Bs-FtsZ polymers. Solid lines and arrow, with GTP and Mg2+; dashed gray lines and arrow, control with GDP and Mg2+. Taxol is an internal standard here. In this experiment, the pelleted PC and FtsZ were 4.8 and 10 μm, respectively. mAU, milliabsorbance units.
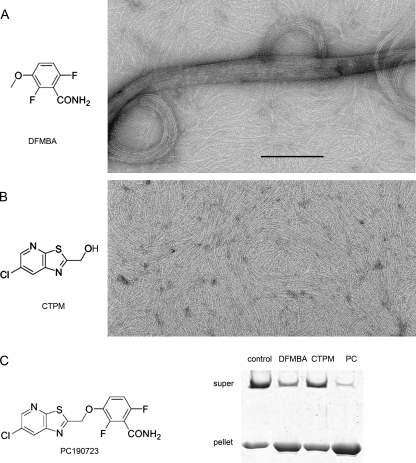
Interestingly, the PC fragment DFMBA (1–4 mm; Fig. 5, A and C), 2,6-difluoro-3-hydroxybenzamide(1–4 mm) and to a lesser extent 3-methoxybenzamide (20 mm), were found to induce large Bs-FtsZ bundles similar to those obtained with PC (20 μm), thus following their relative antibacterial activities (35). The other fragment of the PC molecule, CTPM (1 mm) did not induce Bs-FtsZ bundling (Fig. 5, B and C). These results suggested specific but weak binding of DFMBA to Bs-FtsZ polymers, which could not be accurately measured by pelleting HPLC. DFMBA has an ∼102-fold higher effective concentration than PC, suggesting a two order of magnitude lower affinity. Note that if PC were a nonself-associating ideal bifunctional ligand (36), the rest of the molecule, if isolated, would be expected to bind more weakly than DFMBA and be inactive below ∼0.5 m.
FIGURE 5.
Bs-FtsZ polymers induced by PC fragments in Mes50 assembly buffer with 10 mm MgCl2 and 2 mm GTP. A, 1 mm fragment of DFMBA. B, 1 mm fragment of analogue CTPM (similar to control without fragment, as in Fig. 1F). Bar, 200 nm. C, chemical structure of PC and low speed FtsZ polymers pelleting (15,000 × g, 20 min) under the same conditions (PC, 20 μm). DFMBA also induced a FtsZ light scattering increase that was not observed with CTPM. The Cr for assembly in Hepes50 was reduced from ∼5 μm to ∼1 μm FtsZ with DFMBA (4 mm).
Modulation of the GTPase Activity of Bs-FtsZ by PC
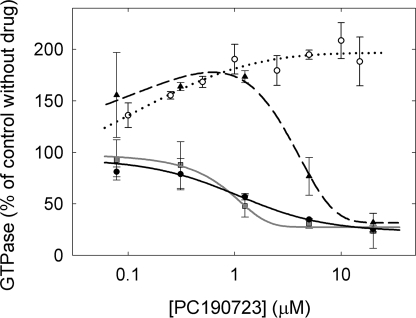
The effects of PC on the GTPase activity of polymerizing Bs-FtsZ solutions was examined with 10 mm MgCl2 in three buffers that give quite different degrees of PC-induced bundling (see above). Control GTPase without PC was dependent on the KCl concentration: 0.61 ± 0.09 min−1 in Hepes250, 0.22 ± 0.02 min−1 in Hepes50, 0.15 ± 0.08 min−1 in Mes50. A partial inhibition of the GTPase rate was observed with PC, to a similar extent (Fig. 6) irrespective of the buffer. The inhibition between ∼1 and ∼10 μm PC roughly paralleled the formation of pelletable polymer (Fig. 4A). Low PC concentrations variably enhanced the relative GTPase activity in Hepes250. This may be a simple consequence of a large increase in FtsZ GTPase upon polymerization relative to the weakly polymerizing control without PC (Fig. 1E) (since the rate measured is per total FtsZ, not per assembled FtsZ). Control measurements showed that 3H[GTP] binding to Bs-FtsZ and to Ec-FtsZ was not modified by PC (supplemental Fig. S3).
FIGURE 6.
Effects of PC on the GTPase activity of FtsZ (10 μm), with 1 mm GTP and 10 mm MgCl2, pH 6.8 at 25 °C. Bs-FtsZ: solid circles, Hepes50; triangles, Hepes250; squares, Mes50. Ec-FtsZ: open circles, Hepes50 (average of three different protein preparations). The lines are drawn solely to show the trend of the data. The increase in GTPase activity of Ec-FtsZ was confirmed by measurements at varying protein concentrations with excess 15 μm PC (not shown).
Formation of Polymorphic Bs-FtsZ Filament Condensates with Different Curvatures
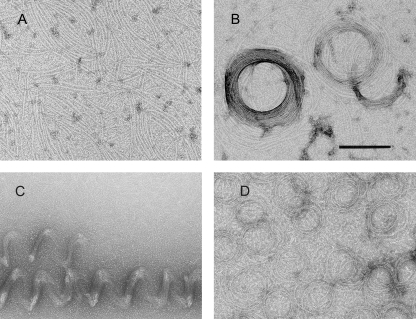
Bs-FtsZ filaments frequently adopted bow shapes and easily coalesced into coils or associated into straight bundles (see above). Other Bs-FtsZ filament condensates also observed include toroids and helical bundles (see supplemental Table S1 for a list of polymorphs and conditions). Briefly, along with filaments formed with GTP and PC (Fig. 7A) and filament coils (similar to Fig. 2B), toroids ∼300 nm in diameter were observed on the same grids (Fig. 7B). Large helical bundles were observed at pH 7.5 with PC, GTP, and Mg2+ (Fig. 7C). Interestingly, Bs-FtsZ condensates could also form in the absence of PC with GMPCPP and Mg2+ as coils, and smaller toroids ∼150 nm in diameter were observed (Fig. 7D).
FIGURE 7.
Polymorphic assemblies of Bs-FtsZ, in addition to bundles and ribbons (see Fig. 1B). A, filaments observed in Hepes50, pH 6.8, with GTP and PC. B, toroids in another field of the same sample. C, helical bundles in Hepes50 at pH 7.5 with GTP, 10 mm MgCl2, and PC. D, coiled filaments and toroids formed without PC or dimethyl sulfoxide in Mes50 with MgCl2 and GMPCPP. Similar polymers were observed in Hepes50, pH 6.8, or with GTP. Bar, 200 nm.
Specificity of FtsZ-PC Interactions
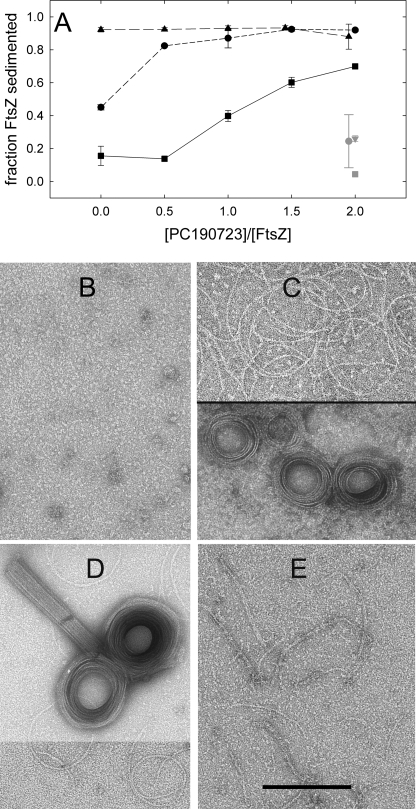
We next compared the effects of PC on the assembly of FtsZ from susceptible (B. subtilis and S. aureus) and nonsusceptible (E. coli) bacteria in Hepes50 (Fig. 8A). The weak formation of pelletable polymer by Sa-FtsZ was markedly enhanced by PC, at concentrations higher than with Bs-FtsZ. A large PC-induced increase in Sa-FtsZ light scattering was observed with nucleotide and Mg2+ (supplemental Fig. S4). Electron micrographs showed a transformation of Sa-FtsZ oligomers (Fig. 8B) into coils, toroids, and straight bundles with PC (Fig. 8, C and D). DFMBA had similar effects at higher concentrations. On the other hand, PC induced bundling was not observed with Ec-FtsZ. Filament morphology was not modified (Fig. 8E), and changes in polymerization were insignificant when employing a GTP-regenerating system (Fig. 8A). Unexpectedly, PC induced a faster disassembly without a GTP regenerating system (supplemental Fig. S4C) and a 2-fold enhancement of Ec-FtsZ GTPase (Fig. 6). HPLC measurements (with 10 μm Ec-FtsZ and 20 μm PC) indicated binding of 0.4 PC per polymerized Ec-FtsZ, although it did not induce bundling. Finally, PC (20 μm) or DFMBA (4 mm) did not affect the assembly of the more distant eukaryotic homologue αβ-tubulin (15 μm) in light scattering tests, or the gross morphology (in electron microscopy) of the microtubular polymers formed (not shown).
FIGURE 8.
Effects of PC on the polymerization of FtsZ (10 μm) from susceptible (B. subtilis and S. aureus) and resistant (E. coli) bacteria. A, FtsZ polymer versus PC concentration. Circles, Bs-FtsZ; squares, Sa-FtsZ; triangles, Ec-FtsZ. The gray symbols are corresponding controls without magnesium. The conditions were Hepes50 buffer with 10 mm MgCl2 and 1 mm GTP, pH 6.8, 25 °C. A GTP regenerating system (1 unit/ml acetate kinase and 15 mm acetyl phosphate) was added for the measurements in A. This was required to reproducibly pellet the Ec-FtsZ polymers in the experiment time and did not modify the pelleting of Bs-FtsZ and Sa-FtsZ. B–D, electron micrographs of Sa-FtsZ with GTP (B), with GTP and 20 μm PC in two fields from the same sample (C) and with 0.1 mm GMPCPP and 20 μm PC (D, upper panel displayed with reduced contrast). E, Ec-FtsZ filaments with GTP and 20 μm PC. Controls without PC gave similar images. Bar, 200 nm.
DISCUSSION
We have shown in this study that binding of the small antibacterial molecule PC190723 enhances the assembly of purified cell division protein Bs-FtsZ into filaments and bundles. Preventing disassembly of FtsZ polymers should inhibit their functional dynamics in vivo, which we propose explains cytokinesis inhibition. This is supported by the observation of FtsZ foci along PC-treated bacilli instead of Z-rings (32).
Our results also give insight into the mechanisms of FtsZ assembly and facilitate the use of PC as a tool to investigate the biological role of FtsZ. PC binding induces Bs-FtsZ filament formation under incipient assembly conditions (250 mm KCl, no Mg2+). These filaments are single stranded, yet they assemble with a cooperative critical concentration behavior, typical of multi-stranded polymers. This may be explained by Bs-FtsZ switching between inactive and active states similarly to Ec-FtsZ (16), indicating that PC induces the polymer Bs-FtsZ conformation, possibly by drugging its assembly switch. However, it cannot be ruled out that the cooperative behavior of Bs-FtsZ with PC might be caused by some filament cyclization in the coils observed or by filament network formation. Direct measurements of PC binding to FtsZ polymers suggest partial saturation of one PC binding site in FtsZ, limited by the solubility of the ligand. PC-induced Bs-FtsZ filament assembly in the absence of magnesium proceeds without GTP hydrolysis, constituting an equilibrium model system of ligand-induced nucleated assembly, analogous to the taxol-induced assembly of GDP-tubulin (12). It would be worth studying in more detail the linkage of PC binding to FtsZ filament assembly.
The FtsZ GTPase site is at the axial interface between consecutive FtsZ molecules in a protofilament (8, 37), so that GTP hydrolysis takes place upon FtsZ assembly. PC modulates the GTPase activity of steady-state polymerizing Bs-FtsZ solutions. The GTPase activation at early stages of assembly remains to be studied. Focusing on the effects of the higher PC concentrations, the partial inhibition of Bs-FtsZ GTPase and the lack of PC effect of GTP binding argue against binding to the same site. This GTPase inhibition is difficult to explain by a bundling effect, due to the coincident extent of inhibition under different bundling conditions (Fig. 6). Mechanistically, PC binding at a site different from the GTP site may either decrease the FtsZ subunit turnover in stabilized filaments and therefore the GTP hydrolysis turnover (27) or allosterically modulate (32) the GTP hydrolysis and exchange in FtsZ filaments.
Depending on the solution conditions, the assembly of Bs-FtsZ varies and so does the effect of PC, with a similar global assembly enhancement. PC binding under Bs-FtsZ filament assembly conditions (50 mm KCl, Mg2+) results in the formation of large condensates of Bs-FtsZ filaments. This may be a consequence of filament accumulation, or PC might induce the condensation of existing filaments, by binding more to condensed than to isolated FtsZ filaments. PC induces, in addition to characteristically curved Bs-FtsZ filaments and straight bundles (Fig. 1), toroids at pH 6.8 and helical bundles at pH 7.5 (Fig. 7), underscoring the flexibility of FtsZ filaments. The observation of Bs-FtsZ coils and toroids formed with nucleotide and Mg2+ in the absence of PC (Fig. 7) indicate that the ability to form curved condensates is an intrinsic property of Bs-FtsZ filaments, which is important for the formation of the Z-ring (24), and is enhanced by PC binding. Curved FtsZ filaments coalescing into the Z-ring have recently been proposed to generate the constriction force (38).
We have employed Bs-FtsZ as a model and confirmed that PC also induces bundling of Sa-FtsZ, but does not induce assembly or condensation of Ec-FtsZ filaments. This is in agreement with the antibacterial spectrum of PC, which has been related to the presence of Val307 in FtsZ from susceptible organisms (32). Mutations conferring resistance to PC cluster at the long channel between the two domains of the FtsZ molecule where PC has been docked (32). This zone is different from the GTP binding site and it overlaps the taxol binding site in β-tubulin. In agreement with the large sequence divergence between both proteins no marked cross reactions have been observed (32; Results). In order to stabilize susceptible FtsZ polymers PC should bind more to them than to monomers. It could be thought that PC does not bind to FtsZ monomers, nor fits well into the binding site of non-susceptible FtsZ (32). However, the GTPase enhancement and HPLC measurements indicate PC binding to Ec-FtsZ. Given that PC did not modify steady-state polymerization, it should bind similarly to the monomers and polymers of this non-susceptible FtsZ. This binding could occur in a different manner (ineffectively or at another site) than in susceptible Bs-FtsZ and Sa-FtsZ polymers. The as yet unresolved atomic structure of an FtsZ-PC complex could give insight into the detailed structural mechanism of PC action, possibly the FtsZ activation switch, and would facilitate the optimization of PC-derived drugs targeting FtsZ in bacterial pathogens. The major determinant of PC binding to FtsZ (Results) and antibacterial activity (35) appears to be its DFMBA moiety.
An increasing number of proteins are known to modulate FtsZ assembly (4), inhibiting or enhancing FtsZ polymerization and bundling. Small molecules reported to stabilize FtsZ polymers include zantrins 3 and 5 (39), ruthenium red (40) and DAPI (41), and very recently FtsZ bundling by the benzoic acid derivative OTBA (42) has been proposed as a strategy to inhibit bacterial division. It would be of interest to know whether these molecules share the PC site or bind elsewhere in FtsZ. It is known that Ca2+ ions induce FtsZ filament bundling (43, 44) and that FtsZ condensates can form in a nonspecific manner by macromolecular crowding (45) or by interactions with polar molecules at relatively high concentrations (46). Volume exclusion in the bacterial cytosol should favor the formation of FtsZ condensates. An important question is whether there are endogenous regulators sharing the PC binding site of FtsZ.
The biochemical action of PC on FtsZ polymers parallels that of taxol on tubulin (12), since each drug stabilizes its respective protein polymers against disassembly, by binding more to polymers than to monomers. Taxol was the first (47) among antitumor Microtubule Stabilizing Agents, which impair cellular microtubule dynamics and mitosis, triggering cell death (48). Microtubule Stabilizing Agents work by preferential binding to assembled tubulin (49). The structural mechanism coupling taxol binding and microtubule assembly is currently under investigation (50). In the case of the PC-induced FtsZ filaments the problem may be simpler due to the lack of lateral interactions. At a cellular level, multiple microtubule asters form in tumor cells treated with high taxol concentrations, instead of normal spindles (Fig. S5; 51). Multiple mislocalized FtsZ foci, instead of a functional Z-ring, form along PC-treated bacteria, which appear to cause the inhibition of cytokinesis leading to bacterial lysis by PC (32). We speculate that PC is the first of a new class of synthetic antibacterials, the FtsZ Polymer Stabilizing Agents. Our findings raise the question of whether taxol and PC have totally different structural mechanisms of action or, by binding to equivalent sites, to what extent each of them acts on a conserved assembly switch in the tubulin-FtsZ superfamily of proteins.
Supplementary Material
Acknowledgments
We thank N. Stokes and Prolysis for PC190723, M. A. Oliva and J. Löwe for the Bs-FtsZ expression plasmid, J. Jimenez-Barbero for encouragement, P. Chacón and J. F. Díaz for discussions, and D. Juan for technical assistance.
This work was supported by Grants Ministerio de Ciencia (MCINN) BFU2008-00013 (to J. M. A.), Comunidad Autónoma de Madrid S-BIO-0214-2006 (to J. M. A. and O. L.), MCINN SAF2007-67008 (to M. L. L.-R.), S-SAL-249-2006 (to M. L. L.-R.), SAF2008-00451 (to O. L.), Red Temática de Investigación Cooperativa en Cáncer RD06-0020-1001 (to O. L.), Human Frontiers Science Program RGP39-2008 (to O. L.) and contracts Formación de Personal Investigatión (to C. S.-B.), Consejo Superior de Investigaciones Cientificas i3p (to S. H.), and Juan de la Cierva (to A. J. M.-G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Table S1, and Figs. S1–S6.
- PC
- 3-[(6-chloro[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-difluorobenzamide
- Mes
- 4-morpholineethanesulfonic acid
- DFMBA
- 2,6-difluoro-3-methoxybenzamide
- CTPM
- (6-chloro[1,3]thiazolo[5.4-b] prydin-2-yl)methanol
- Bs-FtsZ
- FtsZ from B. subtilis
- Sa-FtsZ
- FtsZ from S. aureus
- Ec-FtsZ
- FtsZ from Escherichia coli
- Cr
- critical protein concentration for polymerization
- HPLC
- high pressure liquid chromatography
- GMPCPP
- guanosine-5′-[(2,β)-methylene]triphosphate.
REFERENCES
- 1.Bi E. F., Lutkenhaus J. (1991) Nature 354, 161–164 [DOI] [PubMed] [Google Scholar]
- 2.Margolin W. (2005) Nat. Rev. Mol. Cell Biol. 6, 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michie K. A., Löwe J. (2006) Annu. Rev. Biochem. 75, 467–492 [DOI] [PubMed] [Google Scholar]
- 4.Adams D. W., Errington J. (2009) Nat. Rev. Microbiol. 7, 642–653 [DOI] [PubMed] [Google Scholar]
- 5.Osawa M., Anderson D. E., Erickson H. P. (2008) Science 320, 792–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buey R. M., Díaz J. F., Andreu J. M. (2006) Biochemistry 45, 5933–5938 [DOI] [PubMed] [Google Scholar]
- 7.Larsen R. A., Cusumano C., Fujioka A., Lim-Fong G., Patterson P., Pogliano J. (2007) Genes Dev. 21, 1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nogales E., Downing K. H., Amos L. A., Löwe J. (1998) Nat. Struct. Biol. 5, 451–458 [DOI] [PubMed] [Google Scholar]
- 9.Rice L. M., Montabana E. A., Agard D. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5378–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebremichael Y., Chu J. W., Voth G. A. (2008) Biophys J. 95, 2487–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett M. J., Chik J. K., Slysz G. W., Luchko T., Tuszynski J., Sackett D. L., Schriemer D. C. (2009) Biochemistry 48, 4858–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Díaz J. F., Menéndez M., Andreu J. M. (1993) Biochemistry 32, 10067–10077 [DOI] [PubMed] [Google Scholar]
- 13.Elie-Caille C., Severin F., Helenius J., Howard J., Muller D. J., Hyman A. A. (2007) Curr. Biol. 17, 1765–1770 [DOI] [PubMed] [Google Scholar]
- 14.Caspar D.L. (1980) Biophys. J. 32, 103–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dajkovic A., Lutkenhaus J. (2006) J. Mol. Microbiol. Biotechnol. 11, 140–151 [DOI] [PubMed] [Google Scholar]
- 16.Huecas S., Llorca O., Boskovic J., Martín-Benito J., Valpuesta J. M., Andreu J. M. (2008) Biophys. J. 94, 1796–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dajkovic A., Mukherjee A., Lutkenhaus J. (2008) J. Bacteriol. 190, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miraldi E. R., Thomas P. J., Romberg L. (2008) Biophys. J. 95, 2470–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan G., Dajkovic A., Wirtz D., Sun S. X. (2008) Biophys. J. 95, 4045–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löwe J., Amos L. A. (1999) EMBO J. 18, 2364–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliva M. A., Huecas S., Palacios J. M., Martín-Benito J., Valpuesta J. M., Andreu J. M. (2003) J. Biol. Chem. 278, 33562–33570 [DOI] [PubMed] [Google Scholar]
- 22.Oliva M. A., Trambaiolo D., Löwe J. (2007) J. Mol. Biol. 373, 1229–1242 [DOI] [PubMed] [Google Scholar]
- 23.Li Z., Trimble M. J., Brun Y. V., Jensen G. J. (2007) EMBO J. 26, 4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monahan L. G., Robinson A., Harry E. J. (2009) Mol. Microbiol. 74, 1004–1017 [DOI] [PubMed] [Google Scholar]
- 25.Erickson H. P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9238–9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stricker J., Maddox P., Salmon E. D., Erickson H. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huecas S., Schaffner-Barbero C., García W., Yébenes H., Palacios J. M., Díaz J. F., Menéndez M., Andreu J. M. (2007) J. Biol. Chem. 282, 37515–37528 [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., Erickson H. P. (2009) Biochemistry 48, 6664–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollmer W. (2006) Appl. Microbiol. Biotechnol. 73, 37–47 [DOI] [PubMed] [Google Scholar]
- 30.Goh S., Boberek J. M., Nakashima N., Stach J., Good L. (2009) PLoS One 4, e6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Läppchen T., Pinas V. A., Hartog A. F., Koomen G. J., Schaffner-Barbero C., Andreu J. M., Trambaiolo D., Löwe J., Juhem A., Popov A. V., den Blaauwen T. (2008) Chem. Biol. 15, 189–199 [DOI] [PubMed] [Google Scholar]
- 32.Haydon D. J., Stokes N. R., Ure R., Galbraith G., Bennett J. M., Brown D. R., Baker P. J., Barynin V. V., Rice D. W., Sedelnikova S. E., Heal J. R., Sheridan J. M., Aiwale S. T., Chauhan P. K., Srivastava A., Taneja A., Collins I., Errington J., Czaplewski L. G. (2008) Science 321, 1673–165 [DOI] [PubMed] [Google Scholar]
- 33.Nogales E., Whittaker M., Milligan R. A., Downing K. H. (1999) Cell 96, 79–88 [DOI] [PubMed] [Google Scholar]
- 34.Rivas G., López A., Mingorance J., Ferrándiz M. J., Zorrilla S., Minton A. P., Vicente M., Andreu J. M. (2000) J. Biol. Chem. 275, 11740–11749 [DOI] [PubMed] [Google Scholar]
- 35.Czaplewski L. G., Collins I., Boyd E. A., Brown D., East S. P., Gardiner M., Fletcher R., Haydon D. J., Henstock V., Ingram P., Jones C., Noula C., Kennison L., Rockley C., Rose V., Thomaides-Brears H. B., Ure R., Whittaker M., Stokes N. R. (2009) Bioorg. Med. Chem. Lett. 19, 524–527 [DOI] [PubMed] [Google Scholar]
- 36.Andreu J. M., Timasheff S. N. (1982) Biochemistry 21, 534–543 [DOI] [PubMed] [Google Scholar]
- 37.Oliva M. A., Cordell S. C., Löwe J. (2004) Nat. Struct. Mol. Biol. 11, 1243–1250 [DOI] [PubMed] [Google Scholar]
- 38.Osawa M., Anderson D. E., Erickson H. P. (2009) EMBO J. 28, 3476–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margalit D. N., Romberg L., Mets R. B., Hebert A. M., Mitchison T. J., Kirschner M. W., RayChaudhuri D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11821–11826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santra M. K., Beuria T. K., Banerjee A., Panda D. (2004) J. Biol. Chem. 279, 25959–25965 [DOI] [PubMed] [Google Scholar]
- 41.Nova E., Montecinos F., Brunet J. E., Lagos R., Monasterio O. (2007) Arch. Biochem. Biophys. 465, 315–319 [DOI] [PubMed] [Google Scholar]
- 42.Beuria T. K., Singh P., Surolia A., Panda D. (2009) Biochem. J. 423, 61–69 [DOI] [PubMed] [Google Scholar]
- 43.Yu X. C., Margolin W. (1997) EMBO J. 16, 5455–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marrington R., Small E., Rodger A., Dafforn T. R., Addinall S. G. (2004) J. Biol. Chem. 279, 48821–48829 [DOI] [PubMed] [Google Scholar]
- 45.González J. M., Jiménez M., Vélez M., Mingorance J., Andreu J. M., Vicente M., Rivas G. (2003) J. Biol. Chem. 278, 37664–37671 [DOI] [PubMed] [Google Scholar]
- 46.Popp D., Iwasa M., Narita A., Erickson H. P., Maéda Y. (2009) Biopolymers 91, 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiff P. B., Fant J., Horwitz S. B. (1979) Nature 277, 665–667 [DOI] [PubMed] [Google Scholar]
- 48.Jordan M. A., Wilson L. (2004) Nat. Rev. Cancer 4, 253–265 [DOI] [PubMed] [Google Scholar]
- 49.Diaz J. F., Andreu J. M., Jimenez-Barbero J. (2009) Topics Curr. Chem. 286, 121–149 [DOI] [PubMed] [Google Scholar]
- 50.Xiao H., Verdier-Pinard P., Fernandez-Fuentes N., Burd B., Angeletti R., Fiser A., Horwitz S. B., Orr G. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10166–10173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abal M., Souto A. A., Amat-Guerri F., Acuña A. U., Andreu J. M., Barasoain I. (2001) Cell Motil. Cytoskeleton 49, 1–15 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.