Abstract
Spatiotemporal specificity of cAMP action is best explained by targeting protein kinase A (PKA) to its substrates by A-kinase-anchoring proteins (AKAPs). At synapses in the brain, AKAP79/150 incorporates PKA and other regulatory enzymes into signal transduction networks that include β-adrenergic receptors, α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA), and N-methyl-d-aspartic acid receptors. We previously showed that AKAP79/150 clusters PKA with type 5 adenylyl cyclase (AC5) to assemble a negative feedback loop in which the anchored kinase phosphorylates AC5 to dynamically suppress cAMP synthesis. We now show that AKAP79 can associate with multiple AC isoforms. The N-terminal regions of AC5, -6, and -9 mediate this protein-protein interaction. Mapping studies located a reciprocal binding surface between residues 77–108 of AKAP79. Intensity- and lifetime-based fluorescence resonance energy transfer demonstrated that deletion of AKAP7977–108 region abolished AC5-AKAP79 interaction in living cells. The addition of the AKAP7977–153 polypeptide fragment uncouples AC5/6 interactions with the anchoring protein and prevents PKA-mediated inhibition of AC activity in membranes. Use of the AKAP7977–153 polypeptide fragment in brain extracts from wild-type and AKAP150−/− mice reveals that loss of the anchoring protein results in decreased AMPA receptor-associated AC activity. Thus, we propose that AKAP79/150 mediates protein-protein interactions that place AC5 in proximity to synaptic AMPA receptors.
Keywords: Adenylate Cyclase (Adenylyl Cyclase); Cyclic AMP (cAMP); Fluorescence Resonance Energy Transfer (FRET); Glutamate Receptors Ionotropic (AMPA, NMDA); Protein Kinase A (PKA); A-Kinase-anchoring Protein; AC5; AKAP79; FLIM; GluR1
Introduction
Localized activation of PKA2 triggers a plethora of intracellular signaling processes (1). Precise control of these phosphorylation events is often achieved by restricted activation of PKA in discrete microenvironments. AKAPs participate in this process by tethering the kinase close to preferred substrates. AKAPs now represent a family of 43 diverse but functionally related proteins that bind the regulatory subunit dimer of the PKA holoenzyme (2).
AKAPs have been identified in a range of species, tissues, and cellular compartments. The AKAP79/150 group of anchoring proteins is perhaps the best understood member of this class of signal-organizing proteins. AKAP79/150 consists of three orthologs: bovine AKAP75, human AKAP79, and murine AKAP150. Although originally identified in the postsynaptic densities of neurons, this group of anchoring proteins is also expressed in a variety of other tissues. In addition to binding PKA, AKAP79 has the ability to bind protein phosphatase 2B (3) and protein kinase C (PKC) (4, 5). By organizing these signal transduction and signal termination enzymes in the same location, AKAP79 provides a platform to facilitate the bidirectional control of cAMP- and calcium-mediated signaling events.
Although anchoring of PKA with its substrates provides an efficient mechanism for the spatial regulation necessary for selectivity of cAMP signaling, it was not clear how local pools of cAMP are managed. We have shown that AC isoforms can specifically interact with three different AKAP complexes, AKAP79, Yotiao, and mAKAP, to regulate events downstream of cAMP production (6–8). We have also demonstrated that anchoring of AC5 to an AKAP79/150 complex provides negative feedback on AC5 via PKA phosphorylation of AC5 within the complex (6).
Although characterization of the AKAP79-AC5 interaction has shed some light on the advantages gained by localizing different components of cAMP signaling pathways, several key issues remain unresolved. First of all, do other AC isoforms interact with AKAP79 or other anchoring proteins? Secondly, are AC isoforms recruited into larger signaling networks via their protein-protein interactions with AKAP79? AKAP79/150 has been shown to form a multiprotein signaling complex with AMPA and NMDA receptors (9–11), adhesion molecules (12), and cytoskeleton proteins (13), all of which play important roles in synaptic function. Is AC an integral part of this complex, and if so, which AC isoforms dominate?
In this report, we demonstrate that AKAP79 interacts with a distinct subset of AC isoforms. The interaction of AKAP79 with AC5 is direct, involving the N terminus of AC5 and residues 77–108 of AKAP79. Finally, disruption of the AKAP79 and AC5 complex results in a significant decrease in AMPA receptor type 1 subunit (GluR1)-associated brain AC activity.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids AKAP79-GFP-pcDNA3, AKAP79-GFP-pcDNA3-1-ΔB-153, and purified recombinant His6-S-tag-AKAP79 polypeptide fragments (77–153) and (109–290) were previously described (14). Na+,K+-ATPase (NKA) α1-YFP plasmid and protein characterization was previously described in Ref. 15. CFP-tagged AKAP79 was created by subcloning of enhanced cyan fluorescence protein (CFP) into pEGFPN1-AKAP79 vector using the restriction enzymes BamHI and NotI. AKAP79-Myc-pScript polypeptide fragments (1–153, 77–153, and 154–427) were constructed by PCR. AC5-YFP-pcDNA3 was constructed in a two-step procedure by first generating a unique KpnI restriction site by PCR immediately prior to the stop codon of human AC5 in hAC5-pcDNA3, and second, by subcloning in the yellow fluorescence protein (YFP) using the restriction enzymes KpnI and BamHI. Sequences of all modified fragments were confirmed by restriction digests and DNA sequencing.
Antibodies
Antibodies used were mouse α-Myc (Santa Cruz Biotechnology), mouse α-GST (Invitrogen), mouse α-AKAP79 (BD Transduction Laboratories), mouse α-FLAG M2-agarose (Sigma), mouse α-S-tag (Novagen), rabbit α-GluR1 (Millipore), and rabbit α-AKAP150 (VO88).
Transfections
Human embryonic kidney (HEK293) cells were seeded at 5 × 106 cells/10-cm culture dish and grown for 1 day in Dulbecco's modified Eagle's medium with 10% fetal bovine serum prior to transfection with Lipofectamine 2000 (Invitrogen) or jetPEI (Polyplus). Plasmid DNA (10 μg total) used to transfect cells in 10-cm dishes included: AKAP79 (5 μg), AC3 (3 μg), AC5 (2.5 μg), all other ACs (5 μg), and pcDNA3. Cells were used 44–50 h after transfection.
Protein Purification and Glutathione S-Transferase (GST) Pulldown Assay
Gαs-His6 was expressed in Escherichia coli and activated with [35S]GTPγS (16). GST-tagged proteins were purified on glutathione resin (Amersham Biosciences) in buffers lacking detergents. GST pulldown assays were as described previously (17), utilizing lysates from transfected HEK293 cells (500 μg) or the indicated purified polypeptides (1 μm).
Immunoprecipitation of Adenylyl Cyclase Activity
Immunoprecipitations using anti-FLAG agarose followed by measurement of associated AC activity (IP-AC assays) were performed as described (6). Preparation of brain tissue extracts and subsequent IP-AC assays were performed essentially as described (8). Competing polypeptides were added prior to homogenization. AKAP150 (2 μg) and GluR1 (0.4 μg) antibodies were utilized followed by pulldown of complexes with protein A-Sepharose (30 μl of 50% slurry) and measurement of AC activity. All measurements were performed in duplicate and were normalized to IgG samples with control polypeptide.
Membrane Preparation and Adenylyl Cyclase Activity Measurements
Membranes from transfected HEK293 cells were prepared as described (8). Membranes (30 μg/reaction) were then immediately assayed for adenylyl cyclase activity upon stimulation with the indicated reagents (18).
FRET Experiments
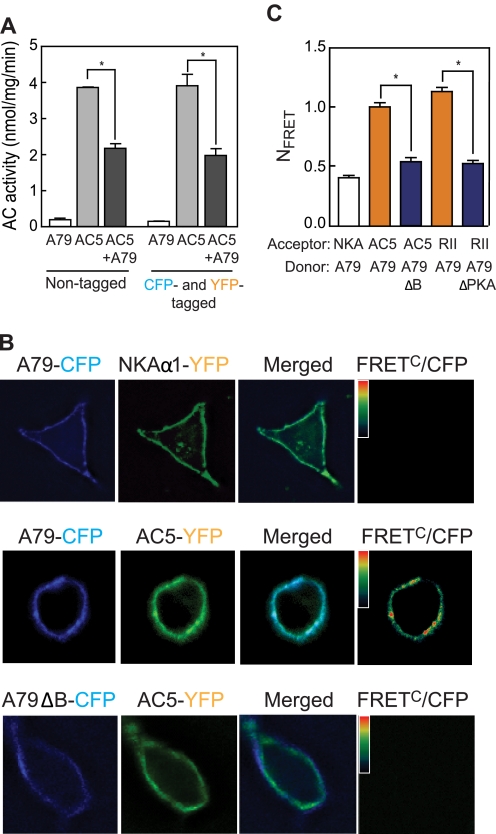
HEK293 cells were plated on coverslips in six-well dishes at 10% confluence. The next day, cells were transfected with 1.8 μg of total DNA, which included 0.4 μg of wild-type or mutant AC5-YFP, 1 μg of AKAP79-CFP, and 0.4 μg of pcDNA3. Before imaging, media were replaced with Tyrode's buffer (10 mm HEPES, pH 7.4, 145 mm NaCl, 4 mm KCl, 1 mm CaCl2, 1 mm MgCl2, and 10 mm glucose). Fluorescence images were acquired after 48 h of transfection using a microscope (TE 2000; Nikon, Tokyo, Japan) with a DG4 xenon light source and two CoolSNAP cameras (Roper Scientific, Trenton, NJ). For FRET determinations, a z-stack of at least seven xy-plains of three images was acquired sequentially (exposure time, 200 ms) using the following filter sets: donor (CFP; excitation, 436/20 nm; emission, 465/30 nm), FRET (CFP/YFP; excitation, 436/20 nm; emission, 535/30 nm), and acceptor (YFP; excitation, 500/20 nm; emission, 535/30 nm). Deconvolution of z-stacks was performed using Slidebook software. Corrected, sensitized FRET (FRETC) was calculated using the equation FRETC = IFRET − (a × ICFP) − (b × IYFP), where IFRET, ICFP, and IYFP correspond to background-subtracted images of cells expressing both CFP and YFP acquired through the FRET, CFP, and YFP channels, respectively. The values a and b are the bleed-through values of CFP and YFP in the FRET channel, respectively. Calibrations of bleed-through were performed in cells expressing either CFP-tagged or YFP-tagged proteins and were calculated as 0.54 (a) and 0.04 (b) for CFP and YFP, respectively. In cells expressing both CFP-tagged and YFP-tagged proteins, FRET values were calculated according to the methods: FRETC/ICFP (19), FRETN = FRETC/(ICFP × IYFP) (20), and NFRET = FRETC/SQRT(ICFP × IYFP) (21). Pseudocolor FRETC/ICFP images were obtained using the Slidebook software and are displayed with deep blue indicating low values and bright red indicating high values of FRET (see Fig. 5). For fluorescence images, the figures show representative images from 16–22 different cells from four different experiments (see Fig. 5).
FIGURE 5.
Cellular interaction of AKAP79-CFP and AC5-YFP in HEK293 cells by intensity-based FRET. A, characterization of fluorescent protein-tagged proteins. Membranes from HEK293 cells expressing AKAP79 and AC5 (with and without fluorescent protein tags) were stimulated with 50 nm Gαs + 100 μm forskolin. Fluorescent protein tags did not affect AC activity and its inhibition by AKAP79. B, FRET analysis of AKAP79 and AC5 in HEK293 cells. Fluorescence microscopy images of HEK293 cells transiently transfected with the indicated proteins were recorded using three different channels: 1) donor, CFPex/CFPem; 2) FRET, CFPex/YFPem; and 3) acceptor, YFPex/YFPem (where “ex” and “em” designate excitation and emission, respectively). A representative cell is shown for each set (top, AKAP79-CFP and NKAα1-YFP; middle, AKAP79-CFP and AC5-YFP; bottom, AKAP79ΔB-CFP and AC5-YFP). Pseudocolor FRETC/CFP images were obtained using Slidebook software. Scale bar, 10 μm. C, quantitative analysis of FRET by NFRET method (n = 4, using images from 16–22 cells, *, p < 0.01). Error bars in A and C indicate mean ± S.E.
Fluorescence Lifetime Imaging Microscopy (FLIM)–FRET Imaging
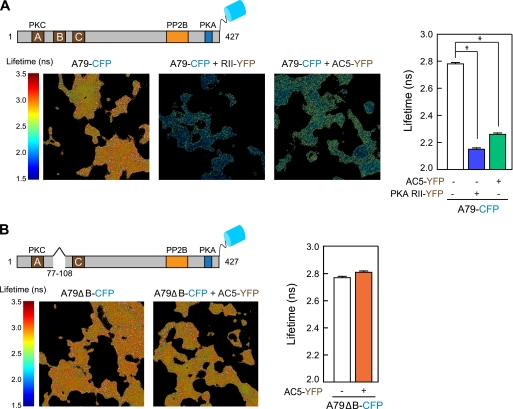
FLIM experiments were performed with a lifetime fluorescence imaging attachment (Lambert Instruments, Leutingewolde, The Netherlands) on an inverted microscope (Nikon). HEK293 cells transiently expressing the donors AKAP79-CFP or AKAP79ΔB-CFP, alone or with AC5-YFP (acceptor), were excited by using a sinusoidally modulated 3-watt 448-nm light-emitting diode at 40 MHz under epi-illumination. Fluorescein at the concentration of 1 μm was used as a lifetime reference standard (4 ns). Cells were imaged with a 60× oil-immersion objective (numerical aperture 1.45) using a CFP filter set. The phase and modulation lifetimes were determined from 12 phase settings by using the manufacturer's software (LIFA). The analysis yields the CFP lifetime of free CFP in donor only (τ1) and the CFP lifetime in donor-acceptor complexes AKAP79-CFP:AC5-YFP (τ2). FRET efficiency was calculated according to formula Feff = 1 − τ2/τ1 (22). FLIM data were averaged on a per cell basis. Three experiments (at least 60 cells in total) were performed for each condition.
Immunohistochemistry
Culture of hippocampal neurons and immunohistochemistry were performed as described previously in Ref. 23. Cells were stained with primary antibodies against AKAP150 (V088, 1:1,000) and PKA RIIβ (BD Biosciences, 1:1,000).
Statistical Analysis
Data were analyzed using a Student t test from an average of at least three experiments, each performed in duplicate or triplicate. Comparison between different experiments groups was determined with the non-paired Student's t test. p < 0.05 is indicated with an asterisk in Figs. 1, 2, and 4–7.
FIGURE 1.
AC types 2, 3, 5, 6, 8, and 9 associate with AKAP79. A, HEK293 cells were transfected with AC isoforms 1 through 9 ± AKAP79-FLAG. Samples were immunoprecipitated with anti-FLAG agarose and assayed for AC activity with 50 nm Gαs and 100 μm forskolin (Fsk). Samples transfected with AKAP79 plus AC2, -3, -5, or -6 (*, p < 0.05) had significant associated Gαs/forskolin-stimulated AC activity as compared with the AC isoform alone. AKAP79 expression was confirmed by Western blotting (supplemental Fig. 1). norm to, normalized to. B, IP-AC assay of AKAP79 and AC9 as performed in A, using 200 nm Gαs to stimulate AC9. Samples transfected with AKAP79 plus AC9 (p < 0.05) had significant associated Gαs-stimulated AC activity as compared with AC9 alone. C, AC8 association with AKAP79 was detected when immunoprecipitates were assayed with 100 μm Ca2+ and 300 nm CaM (*, p < 0.05). Error bars in A–C indicate mean ± S.E.
FIGURE 2.
AKAP79 inhibits AC2, -5, and -6. Membranes from HEK293 cells transfected with vector or AC2, -3, -5, -6, and -9 ± AKAP79 were stimulated with 50 nm Gαs (A) or 200 nm Gαs for AC9 (B). AC2, -5, and -6, but not AC3 or -9, were inhibited in the presence of AKAP79 (*, p < 0.05). Error bars in A and B indicate mean ± S.E.
FIGURE 4.
AKAP7977–153 specifically competes for binding and inhibition of AC5/6-AKAP79 complexes. A, an IP-AC assay was performed as indicated in the legend for Fig. 1A in the presence of 10 μm AKAP7977–153 or control polypeptide AKAP79109–290. The addition of AKAP7977–153 disrupts associations between AKAP79 and AC5 and -6 but not AC2 or -9. Fsk, forskolin. *, p < 0.05. B, membranes from HEK293 cells expressing AC5 ± AKAP79 were incubated with the indicated concentrations of purified AKAP7977–153 prior to stimulation with 50 nm Gαs + 100 μm forskolin. AKAP7977–153 reverses the inhibition of AC5 by AKAP79 in a concentration-dependent manner. C, the dose-response curve from B indicates an IC50 of 1.0 μm for the reversal of AKAP79 inhibition by AKAP7977–153. Error bars in A–C indicate mean ± S.E.
FIGURE 6.
FLIM-FRET of AKAP79-CFP and AC5-YFP. A, HEK293 cells expressing AKAP79-CFP alone or with PKA RII-YFP or AC5-YFP were imaged in the frequency domain in a wide field FLIM microscope. The lifetime of AKAP79-CFP is significantly decreased when co-expressed with PKA RII-YFP or AC5-YFP, as a result of FRET between CFP and YFP. B, the lifetime of AKAP79ΔB-CFP does not change when co-expressed with AC5-YFP. The graphical quantitation of CFP-tagged protein lifetimes is shown for each panel (mean ± S.E.; n = 3; 60–120 cells). *, p < 0.05.
FIGURE 7.
GluR1 associates with AC5/6 in brain in an AKAP150-dependent manner. A, rat brain extracts were preincubated with either 77–153 or 109–290 AKAP79 polypeptides prior to immunoprecipitation using IgG (control) or antibodies against GluR1 or AKAP150. Immunoprecipitates were stimulated with Gαs/forskolin, and associated AC activity was measured (n = 3). norm. to, normalized to. B, Western blot analysis of brain extracts from wild-type (WT), AKAP150−/−, or WAVE-1−/− mice are shown. C, immunohistochemistry of hippocampal neurons from wild-type or AKAP150−/− (KO) mice indicates the loss of AKAP150. D, IP-AC assays were performed as in A, using brain tissue from wild-type, AKAP150, or WAVE-1 null mice. All samples include the control polypeptide AKAP79109–290, except where AKAP7977–153 is indicated by $ (n = 3). Error bars in A and D indicate mean ± S.E. *, p < 0.05.
RESULTS
AC2, -3, -5, -6, -8, and -9 Associate with AKAP79
Various AC isoforms were screened for association with AKAP79 in an overexpression system. HEK293 cells were transiently transfected with vector, FLAG-tagged AKAP79, and/or ACs 1–9. AKAP79 was immunoprecipitated from cell lysates using anti-FLAG agarose. AC activity was measured in each immune complex (referred to as an IP-AC assay) for detection of direct or indirect binding of AC. For all isoforms except AC9, the immunoprecipitates were stimulated with activated Gαs and forskolin (Fig. 1A). Because AC9 is not sensitive to forskolin, a higher concentration of Gαs was used in these samples (Fig. 1B). AC activity in the cell lysates confirmed that each AC isoform was expressed over background AC activity (supplemental Fig. 1). As shown in Fig. 1, A and B, the AC2, -3, -5, -6, and -9 isoforms associate with AKAP79. AC8 activity was consistently higher in immunoprecipitates of AC8 + AKAP79 versus AC8 alone when assayed with Gαs and forskolin, although the change was not statistically significant. Stimulation with Ca2+/calmodulin (CaM), a potent activator of AC8, resulted in a significant association of AC8 activity with AKAP79 (Fig. 1C).
AKAP79 Inhibits the Activity of AC2, -5, and -6 in Isolated Membranes
To determine the effect of AKAP79 on AC activity, HEK293 cells were transfected with pcDNA3 vector and AC2, -3, -5, -6 or -9 ± AKAP79. Membranes were isolated to measure AC activity. AKAP79 had no effect on the basal activity of any isoform (data not shown). Membranes containing AC2, -5, and -6 were inhibited in the presence of AKAP79 when stimulated with activated Gαs (Fig. 2A). Membranes containing AC9 were not inhibited in the presence of AKAP79 when stimulated with a higher concentration of Gαs (Fig. 2B). AC activity was also inhibited in membranes when stimulated by Gαs + Gβγ (AC2, 46% inhibition, and AC5, 73% inhibition), forskolin alone (AC5, 38% inhibition), or Gαs + forskolin (AC2, 39% inhibition, AC5, 42% inhibition, and AC6, 28% inhibition) (data not shown). Under all stimulation conditions tested, AC3 activity was unaffected by AKAP79 expression.
N Termini of AC5, -6, and -9 Mediate Interactions with AKAP79
We have previously shown that the N terminus of AC2 bound the AKAP Yotiao (8). We therefore screened GST-tagged AC N termini (AC-NT) from all AKAP79-inhibited AC isoforms for interactions with this anchoring protein by GST pulldown assay. AC5, -6, and -9 NT, but not AC2 or -3 NT, bound AKAP79 (Fig. 3A).
FIGURE 3.
Mapping of AC N termini binding domains on AKAP79. A, GST-tagged AC N termini were screened for interactions with AKAP79 by GST pulldown using HEK293 cell lysates expressing AKAP79. AKAP79 bound the N termini of AC5, -6, and -9 but not AC2 or -3. B, schematic diagram of AKAP79 and deletion constructs used in panels C and D. Dotted lines indicate the interaction domain for AC5-NT. C, GST pulldown assay was performed using GST or GST-AC5-NT incubated with HEK293 cell lysates expressing vector or AKAP79 Myc-tagged fragments. D, purified AKAP79 S-tagged fragments 77–153 and 109–290 were pulled down with GST, GST-AC5-NT, or GST-AC5Δ60-NT. S-tag Western blot shows direct binding of AC5-NT with AKAP7977–153.
GST pulldown assays were used to map the AC5 binding site on AKAP79. Cell lysates from HEK293 cells expressing various Myc-tagged truncations of AKAP79 or purified truncations of AKAP79 were incubated with GST-AC5-NT (Fig. 3B). The first 153 residues of AKAP79 and a region that encompasses residues 77–153 bind GST-AC5-NT but not GST alone (Fig. 3C). The AKAP7977–153 polypeptide fragment comprises the second (B) and third (C) polybasic regions of AKAP79; however, neither region alone is sufficient to tether the protein to the plasma membrane (3). To more definitively map the binding site for AC5, we incubated E. coli purified polypeptide fragments encompassing residues 77–153 and 109–290 of the anchoring protein with a GST-tagged N-terminal region of AC5. AKAP7977–153, but not AKAP79109–290, directly binds AC5-NT (Fig. 3D). However, binding is greatly reduced when experiments were repeated with a truncated form of AC5 that lacked the first 60 amino acids (GST-AC5Δ60-NT). Based on the mapping depicted in Fig. 3B, we conclude that the first 60 amino acids of AC5 bind to the second polybasic region of AKAP79.
AKAP7977–153 Is a Selective Disruptor of AC-AKAP79 Interactions
To determine whether AKAP7977–153 can be used as a selective disruptor of AKAP79-AC binding, we performed IP-AC assays in the presence of AKAP7977–153 or a control polypeptide AKAP79109–290. The addition of AKAP7977–153 disrupted association of AC5 and -6 with AKAP79 but not that of AC2 or -9 (Fig. 4A). This result suggests that that AKAP7977–153 selectively disrupts AC5/6 binding to AKAP79 and infers that the AC2 and -9 isoforms interact with different regions of the anchoring protein.
To focus our study, the remainder of the experiments concentrates on the AC5-AKAP79/150 interaction. We have previously shown that anchored PKA phosphorylated AC5/6 to suppress cAMP synthesis (6). In agreement with this finding, we were able to demonstrate that AC5 activity is inhibited upon co-expression with AKAP79 when stimulated with Gαs (Figs. 2A and 4B). To determine whether inhibition of AC5 is reversible, we utilized the AKAP7977–153 polypeptide to disrupt AC5-AKAP79 protein-protein interactions. Membranes prepared from HEK293 cells expressing AC5 ± AKAP79 were assayed for Gαs/forskolin-stimulated AC activity in the presence of purified AC-AKAP disruptor fragment (Fig. 4, B and C). AKAP7977–153 reversed AKAP79-mediated inhibition of AC5 (IC50 = 1.0 μm). Control experiments confirmed that the AC-AKAP disruptor had no effect on AC5 in the absence of AKAP79.
FRET between AC5 and AKAP79
To explore the dynamics of this protein-protein interaction inside cells, we generated fluorescence-tagged versions of each protein. Fluorescent probes containing YFP at the C terminus of AC5 (AC5-YFP) and CFP at the C terminus of AKAP79 (AKAP79-CFP) were produced. In addition, we constructed a mutant form of AKAP79 lacking amino acids 77–108 fused to CFP (AKAP79ΔB-CFP). Initial experiments confirmed that fusion of the fluorescent tags did not alter the AC5 activity with or without AKAP79 (Fig. 5A). Activity measurements confirmed that expression of AC5-YFP stimulated cAMP synthesis and co-expression with AKAP79-CFP attenuated this response (Fig. 5A). In addition, AC5-YFP was localized to the plasma membrane when expressed in HEK293 cells at low levels (Fig. 5B). Higher levels of expression generally produced a particulate cytoplasmic fluorescence that was excluded from the nucleus. Therefore, we have titrated AC expression to levels that produce a largely plasma membrane expression pattern for all fluorescence and activity assays. Previous reports have suggested that AC5 may be present on the nuclear envelope in cardiac myocytes (24). However, we detected no such localization upon expression in HEK293 cells. To analyze AKAP79 interactions with AC5 in living cells, we used an intensity-based FRET approach employing AC5-YFP and AKAP79-CFP. There is a strong inverse distance relationship between FRET and chromophore separation such that FRET between the donor CFP and acceptor YFP molecules only occurs if the two proteins are in proximity (<10 nm). Numerous mathematical methods are used to quantify FRET. We compared the three most commonly used methods: FRETC/ICFP (19), FRETN (20), and NFRET (21). The PKA RII subunit tagged with YFP at C terminus was used as an established FRET partner for AKAP79-CFP, whereas AKAP79 lacking a PKA binding site (AKAP79ΔPKA-CFP) was used as a negative control (25) (Fig. 5C). A second negative control for nonspecific membrane protein interactions was the transmembrane protein NKAα1-YFP. Any FRET signal between AKAP79-CFP and NKAα1-YFP or PKA-RII-YFP and AKAP79ΔPKA-CFP set the baseline for nonspecific membrane interactions. All three computational methods show significant FRET at the plasma membrane for AC5-YFP and AKAP79-CFP relative to all negative controls, and that is lost upon deletion of the second polybasic region of AKAP79 (AKAP79ΔB-CFP) (Fig. 5, B and C, and supplemental Table 1).
FLIM-FRET between AC5 and AKAP79
To complement the intensity-based FRET analysis, we performed FLIM-FRET of HEK293 cells expressing AC5-YFP and AKAP79-CFP. When two chromophores undergo energy transfer, the lifetime of the donor chromophore (CFP) decreases in the presence of an acceptor (YFP) and provides a more quantitative measurement of FRET efficiency (Feff) (22). We observed a significant decrease in CFP lifetime in cells expressing both AC5-YFP and AKAP79-CFP, as compared with those expressing AKAP79-CFP alone (2.26 ± 0.01 versus 2.78 ± 0.01 ns; Feff = 19.2 ± 1.1%, mean ± S.E.) (Fig. 6A). No decrease in CFP lifetime was observed in cells expressing AC5-YFP and AKAP79ΔB-CFP as compared with AKAP79ΔB-CFP alone (2.81 ± 0.01 versus 2.77 ± 0.01) (Fig. 6B). Control experiments monitored FLIM-FRET of AKAP79-CFP with its well characterized binding partner the RII subunit of PKA fused to YFP (Feff = 23.8 ± 1.3%). A negative control for this study was AKAP79ΔPKA-CFP, a form of the anchoring protein that lacks the PKA anchoring domain (Fig. 6A and supplemental Fig. 2). Taken together, the data presented in Fig. 6 provide compelling evidence that AKAP79 functions to dynamically interact with AC5 in living cells.
AMPA Receptor-associated AC Activity Is Mediated in Part by AKAP150 and AC5/6 Interactions
Finally, we investigated the physiological relevance of AKAP79/150-AC interactions with respect to the AMPA receptor. Elegant studies by Hell and colleagues (26, 27) have suggested that ACs are recruited into larger signaling networks that include β-adrenergic receptors, kinases, and ion channels. We have shown that AKAP79 functions to scaffold its cargo of signaling enzymes to synaptic AMPA receptors (9). Furthermore, GluR1 co-immunoprecipitates significant AC activity from rat brain extracts, in accord with previous reports of AC association with a GluR1 complex (27). However, it is unclear which isoform of AC is part of this complex or how it interacts with GluR1. Therefore, it seemed possible that AKAP79 may also function to recruit AC5 into a similar type of multiprotein signaling network. Analyses of the AC5-AKAP79-AMPA receptor interactions were performed in two stages. Initial studies monitored changes in the co-purification of AC5 activity with the GluR1 subunit of the AMPA receptor. Rat brain extracts were incubated with the AKAP79-AC5 disruptor (AKAP7977–153) or a control fragment (AKAP79109–290). The GluR1 subunit of the AMPA channel was immunoprecipitated, and co-purification of AC activity was measured upon stimulation with Gαs and forskolin. AKAP7977–153 decreased GluR1-associated AC activity by an average of 49% (Fig. 7A), suggesting that AKAP150 mediates AC5/6 interactions with GluR1 in rat brain. In addition, disruption of AC5/6 association with the rat ortholog AKAP150 led to the reduction (56%) of AKAP150-associated AC activity (Fig. 7A), indicating that additional AC isoforms may be present in AKAP79/150 complexes.
The next phase of these studies examined this phenomenon in the brains of AKAP null mice. Brain extracts were prepared from two AKAP knock-out mouse strains: AKAP150 and WAVE-1, a neuronal anchoring protein that is concentrated in the dendrites (28, 29). Initial characterization confirmed the ablation of each anchoring protein gene by immunoblotting (Fig. 7B, top and middle panels). Loading controls indicated that equal amounts of protein were loaded in each lane (bottom panel). Immunohistochemical analysis further demonstrated the loss of AKAP150 expression in cultured hippocampal neurons as compared with wild type (Fig. 7C).
To determine the extent to which AKAP79/150 is the sole AKAP that anchors AC to a GluR1 complex, we have repeated these experiments using brain tissue from wild-type and AKAP150 null mice. AKAP150-associated AC activity is completely lost in brain from AKAP150 null mice but not in the knock-out of another AKAP expressed in brain, WAVE-1 (Fig. 7D). As in rat, incubation with AKAP7977–153 led to a 50% loss in GluR1-associated AC activity, once again indicating the presence of AKAP79/150-anchored AC5/6. AC activity associated with GluR1 was also reduced by 70% in AKAP150 null tissue as compared with wild-type control (Fig. 7D). However, a significant amount of GluR1-associated AC activity (30%) remained in AKAP150 null tissue, suggesting some involvement of additional scaffolding proteins in anchoring AC to GluR1. Control experiments using WAVE-1 brains confirmed that this anchoring protein is not involved in anchoring AC to GluR1 as no loss in GluR1-associated AC activity is observed in WAVE-1 knock-out tissue.
DISCUSSION
Organization of the cAMP synthesis machinery with PKA and its preferred substrates is believed to ensure the optimal relay of second messenger signals. AKAPs provide a molecular framework for the relay of this information by bringing together different components of these signaling pathways. Considerable information about how AKAPs function has been gleaned from analysis of the AKAP79/150 group of anchoring proteins (1). In this study, we have employed the methods of protein chemistry and fluorescence microscopy to show that: 1) a subset of AC isoforms can directly interact with AKAP79; 2) this protein-protein interaction proceeds through the N terminus of AC, which docks with the second polybasic region of AKAP79; and 3) AKAP79/150 is responsible for linking AMPA receptors to AC5/6 activity in brain. Collectively, this information provides us with a deeper understanding of how an AKAP signaling complex is assembled.
AKAP79 Selectivity for AC Isoforms
In recent years, AKAP79/150 has been shown to interact with a range of kinases, phosphatases, other scaffolding proteins, and effector proteins. We first became interested in its role as an organizer of the cAMP synthesis machinery when we were able to co-purify AKAP79/150 with AC isoforms 5 and 6 from brain extracts (6). A logical extension of this work was to establish whether other AC isoforms were capable of binding to AKAP79. As shown in Fig. 1, the AC2, -3, -5, -6, -8, and -9 isoforms co-purify with AKAP79. This finding suggests that AC interaction with AKAP79 is a much more general phenomenon than we originally appreciated. However, it is worth noting that many of the associated AC isoforms are also regulated by one or more enzymes that interact with AKAP79. For example, PKA phosphorylates AC5 and -6 to suppress cAMP synthesis, whereas PKC phosphorylates AC2, -5, and -6 to modulate their activities (30–34). Likewise, CaM and protein phosphatase 2B are effectors of AC8 (35) and AC9 (36), respectively. This is reflected in the regulation of cAMP synthesis upon co-expression of AKAP79 of at least a subset of AC isoforms. Thus, the duration and magnitude of the cAMP may be dependent in part on which AC isoform is associated with the AKAP79 complex.
Interaction Domains for AKAP79 and ACs
Our data support a model where multiple AC isoforms have the capacity to interact with AKAP79/150. This would imply that a common motif or structural fold on the AC provides a determinant for AKAP binding. Structurally, C1 and C2 domains of all AC isoforms are highly conserved, whereas the N-terminal intracellular regions are highly variable. Our data suggest that the AKAP79 binding site is located in the variable regions of at least a subset of AC isoforms. Interactions of AC N termini with AKAP79 would possibly explain differences in AKAP79 association/inhibition of ACs with similarities in sequence and regulatory patterns. Of the AC isoforms associated with AKAP79 in Fig. 1A, only AC5, -6, and -9 utilize their N termini as AKAP79 binding sites.
The three polybasic regions in the amino-terminal regions of AKAP79/150 form the binding sites for PKC, CaM, F-actin, phosphatidylinositol 4,5-bisphosphate, and cadherin (4, 11, 12, 14, 37). Any two of these regions are sufficient to confer plasma membrane association and lateral targeting in epithelial membranes with E-cadherin (12, 14). Although the PKC binding site has been further refined to the first polybasic region, additional mapping has not been performed for the other binding partners. Our mapping studies suggest that the second polybasic region of AKAP79 is required for binding to AC5 (Fig. 3). This region was confirmed as key for AC5-AKAP79 interactions because live cell imaging experiments demonstrate that deletion of residues 77–108 from AKAP79 abolishes its ability to interact with AC. This region only appears to encompass a binding site for AC5 and -6 because the disruptor AKAP7977–153 cannot compete for binding with AC2 or -9. It is unclear whether interactions with AC5/6 would alter association of AKAP79 with F-actin or cadherin. In addition, the question remains as to whether AC5/6 binding to AKAP79/150 will be subject to regulation by Ca2+/CaM or PKC phosphorylation. Finally, it is unknown whether AKAP79 retains the ability to interact with AC2 or AC9 when in complex with AC5. These latter AC isoforms clearly utilize different binding sites on AKAP79 that have yet to be defined. These findings suggest that several AC binding sites may reside in AKAP79 and that additional factors such as tissue-specific expression may contribute to the formation of distinct AC-AKAP complexes in vivo. An added complexity may be that other anchoring proteins such as Yotiao and mAKAP may compete for association with the available pool of ACs (7, 8).
AC-AKAP79 Complexes in AMPA Signaling
AKAP79 and AC5 interactions in vivo were confirmed using two FRET-based live cell imaging approaches. Both intensity-based and lifetime-based FRET methodologies demonstrate a strong interaction between AC5 and AKAP79 at the plasma membrane, which was lost upon deletion of AKAP79 residues 77–108. To detect AKAP79-AC complexes in tissues, we utilized AKAP7977–153 as a selective disruptor of AC5/6 and AKAP79. This reagent allowed us to examine the presence of AC5 in AMPA receptor signaling complexes. The GluR1 subunit of the AMPA type glutamate receptor ion channel binds indirectly to AKAP79 via association with the membrane-associated guanylate kinase proteins SAP97 or PSD-95 (9, 38). AKAP79/150-mediated recruitment of PKA facilitates the phosphorylation-dependent regulation of AMPA currents and the surface maintenance of AMPA receptor complexes during glutamate stimulation (23, 39, 40).
In rat brain extracts, both AKAP150 and GluR1 antibodies pull down a significant amount of AC activity over IgG controls, consistent with previous reports of AC association within these complexes (6, 27). Using AKAP7977–153, we show a 56 and 49% decrease in AKAP150- and GluR1-associated AC activity, respectively. Clearly AKAP150 associates with multiple AC isoforms in brain, in addition to AC5/6. Furthermore, a significant percentage of the AC present in the GluR1 complex must be due to AKAP79-AC5/6 associations. However, ∼50% of GluR1-associated AC activity remained after competition with AKAP7977–153. To explore this phenomenon from a different angle, we utilized brain extracts from AKAP150−/− mice. These studies provided us with two valuable pieces of information. First of all, it would appear that the majority of AC5 is recruited to the AMPA channel by AKAP150. Such complexes may be important for mediating dopaminergic control of GluR1 in striatum-dependent learning (41). Secondly, it would appear that the other prominent synaptic anchoring protein WAVE-1 does not participate in these protein-protein interactions as the levels of AC activity isolated with the channel were unaltered when experiments were performed with brain extracts from WAVE-1−/− mice. Finally, ∼30% of AC activity associated with GluR1 remained in the AKAP150 nulls, indicating that a portion of AC activity must also be associated with GluR1 in an AKAP79/150- and WAVE-1-independent manner. These finding also imply that other AC isoforms may exist in complex with GluR1 or its binding partners. In the hippocampus, this may reflect complexes with AC8 or AC9 (36, 42).
Conclusion
We have demonstrated that AKAP79 can interact with a unique set of AC isoforms, playing either a passive scaffolding role or an inhibitory role. We have also identified a unique disrupting agent, specific for AC5/6-AKAP79, that we used to determine the composition of GluR1 complexes in brain. The regulation of GluR1 by AKAP79/150 is clearly important for synaptic transmission and memory retention (23, 43–45). The association to these complexes of calcium-stimulated or calcium-inhibited AC isoforms will set up important feedback or feed-forward pathways to control GluR1 regulation in specific regions of brain.
Supplementary Material
Acknowledgments
We thank Kathryn Hassell for excellent technical assistance, Dr. Mark Dell'Acqua for PKA-RII-YFP and AKAP79ΔPKA-CFP, and Dr. James Broughman for help with microscopy applications.
This work was supported, in whole or in part, by National Institutes of Health Grants GM60419 (to C. W. D.) and GM48231 (to J. D. S.) and a grant from the American Heart Association (to C. W. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
- PKA
- protein kinase A
- PKC
- protein kinase C
- AC
- adenylyl cyclase
- AKAP
- A-kinase-anchoring protein
- AMPA
- α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate
- GluR1
- AMPA receptor type 1 subunit
- CaM
- calmodulin
- CFP
- cyan fluorescence protein
- GFP
- green fluorescent protein
- YFP
- yellow fluorescence protein
- FLIM
- fluorescence lifetime imaging microscopy
- FRET
- fluorescence resonance energy transfer
- GST
- glutathione S-transferase
- HEK
- human embryonic kidney
- NKA
- Na+,K+-ATPase
- NMDA
- N-methyl-d-aspartic acid
- NT
- N terminus
- IP
- immunoprecipitation
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1.Scott J. D., Pawson T. (2009) Science 326, 1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong W., Scott J. D. (2004) Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 3.Dell'Acqua M. L., Dodge K. L., Tavalin S. J., Scott J. D. (2002) J. Biol. Chem. 277, 48796–48802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faux M. C., Rollins E. N., Edwards A. S., Langeberg L. K., Newton A. C., Scott J. D. (1999) Biochem. J. 343, 443–452 [PMC free article] [PubMed] [Google Scholar]
- 5.Klauck T. M., Faux M. C., Labudda K., Langeberg L. K., Jaken S., Scott J. D. (1996) Science 271, 1589–1592 [DOI] [PubMed] [Google Scholar]
- 6.Bauman A. L., Soughayer J., Nguyen B. T., Willoughby D., Carnegie G. K., Wong W., Hoshi N., Langeberg L. K., Cooper D. M., Dessauer C. W., Scott J. D. (2006) Mol. Cell 23, 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapiloff M. S., Piggott L. A., Sadana R., Li J., Heredia L. A., Henson E., Efendiev R., Dessauer C. W. (2009) J. Biol. Chem. 284, 23540–23546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piggott L. A., Bauman A. L., Scott J. D., Dessauer C. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13835–13840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colledge M., Dean R. A., Scott G. K., Langeberg L. K., Huganir R. L., Scott J. D. (2000) Neuron 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 10.Dell'Acqua M. L., Smith K. E., Gorski J. A., Horne E. A., Gibson E. S., Gomez L. L. (2006) Eur. J. Cell Biol. 85, 627–633 [DOI] [PubMed] [Google Scholar]
- 11.Gomez L. L., Alam S., Smith K. E., Horne E., Dell'Acqua M. L. (2002) J. Neurosci. 22, 7027–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorski J. A., Gomez L. L., Scott J. D., Dell'Acqua M. L. (2005) Mol. Biol. Cell 16, 3574–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diviani D., Scott J. D. (2001) J. Cell Sci. 114, 1431–1437 [DOI] [PubMed] [Google Scholar]
- 14.Dell'Acqua M. L., Faux M. C., Thorburn J., Thorburn A., Scott J. D. (1998) EMBO J. 17, 2246–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Efendiev R., Cinelli A. R., Leibiger I. B., Bertorello A. M., Pedemonte C. H. (2006) FEBS Lett. 580, 5067–5070 [DOI] [PubMed] [Google Scholar]
- 16.Dessauer C. W., Tesmer J. J., Sprang S. R., Gilman A. G. (1998) J. Biol. Chem. 273, 25831–25839 [DOI] [PubMed] [Google Scholar]
- 17.Sadana R., Dascal N., Dessauer C. W. (2009) Mol. Pharmacol. 76, 1256–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dessauer C. W. (2002) Methods Enzymol. 345, 112–126 [DOI] [PubMed] [Google Scholar]
- 19.Vanderklish P. W., Krushel L. A., Holst B. H., Gally J. A., Crossin K. L., Edelman G. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon G. W., Berry G., Liang X. H., Levine B., Herman B. (1998) Biophys. J. 74, 2702–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia Z., Liu Y. (2001) Biophys. J. 81, 2395–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia Z. G., Refsdal C. D., Merchant K. M., Dorsa D. M., Storm D. R. (1991) Neuron 6, 431–443 [DOI] [PubMed] [Google Scholar]
- 23.Tunquist B. J., Hoshi N., Guire E. S., Zhang F., Mullendorff K., Langeberg L. K., Raber J., Scott J. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12557–12562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boivin B., Lavoie C., Vaniotis G., Baragli A., Villeneuve L. R., Ethier N., Trieu P., Allen B. G., Hébert T. E. (2006) Cardiovasc. Res. 71, 69–78 [DOI] [PubMed] [Google Scholar]
- 25.Oliveria S. F., Gomez L. L., Dell'Acqua M. L. (2003) J. Cell Biol. 160, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davare M. A., Avdonin V., Hall D. D., Peden E. M., Burette A., Weinberg R. J., Horne M. C., Hoshi T., Hell J. W. (2001) Science 293, 98–101 [DOI] [PubMed] [Google Scholar]
- 27.Joiner M. L., Lisé M. F., Yuen E. Y., Kam A. Y., Zhang M., Hall D. D., Malik Z. A., Qian H., Chen Y., Ulrich J. D., Burette A. C., Weinberg R. J., Law P. Y., El-Husseini A., Yan Z., Hell J. W. (2010) EMBO J. 29, 482–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soderling S. H., Guire E. S., Kaech S., White J., Zhang F., Schutz K., Langeberg L. K., Banker G., Raber J., Scott J. D. (2007) J. Neurosci. 27, 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soderling S. H., Langeberg L. K., Soderling J. A., Davee S. M., Simerly R., Raber J., Scott J. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1723–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y., Harry A., Li J., Smit M. J., Bai X., Magnusson R., Pieroni J. P., Weng G., Iyengar R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14100–14104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwami G., Kawabe J., Ebina T., Cannon P. J., Homcy C. J., Ishikawa Y. (1995) J. Biol. Chem. 270, 12481–12484 [DOI] [PubMed] [Google Scholar]
- 32.Jacobowitz O., Iyengar R. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10630–10634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawabe J., Iwami G., Ebina T., Ohno S., Katada T., Ueda Y., Homcy C. J., Ishikawa Y. (1994) J. Biol. Chem. 269, 16554–16558 [PubMed] [Google Scholar]
- 34.Lai H. L., Yang T. H., Messing R. O., Ching Y. H., Lin S. C., Chern Y. (1997) J. Biol. Chem. 272, 4970–4977 [DOI] [PubMed] [Google Scholar]
- 35.Cali J. J., Zwaagstra J. C., Mons N., Cooper D. M., Krupinski J. (1994) J. Biol. Chem. 269, 12190–12195 [PubMed] [Google Scholar]
- 36.Antoni F. A., Palkovits M., Simpson J., Smith S. M., Leitch A. L., Rosie R., Fink G., Paterson J. M. (1998) J. Neurosci. 18, 9650–9661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houbre D., Duportail G., Deloulme J. C., Baudier J. (1991) J. Biol. Chem. 266, 7121–7131 [PubMed] [Google Scholar]
- 38.Leonard A. S., Davare M. A., Horne M. C., Garner C. C., Hell J. W. (1998) J. Biol. Chem. 273, 19518–19524 [DOI] [PubMed] [Google Scholar]
- 39.Hoshi N., Langeberg L. K., Scott J. D. (2005) Nat. Cell Biol. 7, 1066–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y., Allen M., Halt A. R., Weisenhaus M., Dallapiazza R. F., Hall D. D., Usachev Y. M., McKnight G. S., Hell J. W. (2007) EMBO J. 26, 4879–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kheirbek M. A., Britt J. P., Beeler J. A., Ishikawa Y., McGehee D. S., Zhuang X. (2009) J. Neurosci. 29, 12115–12124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong S. T., Athos J., Figueroa X. A., Pineda V. V., Schaefer M. L., Chavkin C. C., Muglia L. J., Storm D. R. (1999) Neuron 23, 787–798 [DOI] [PubMed] [Google Scholar]
- 43.Lu Y., Zhang M., Lim I. A., Hall D. D., Allen M., Medvedeva Y., McKnight G. S., Usachev Y. M., Hell J. W. (2008) J. Physiol. 586, 4155–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snyder E. M., Colledge M., Crozier R. A., Chen W. S., Scott J. D., Bear M. F. (2005) J. Biol. Chem. 280, 16962–16968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tavalin S. J., Colledge M., Hell J. W., Langeberg L. K., Huganir R. L., Scott J. D. (2002) J. Neurosci. 22, 3044–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.