Abstract
A cancer/testis antigen, CAGE, is widely expressed in various cancer tissues and cancer cell lines but not in normal tissues except the testis. In the present study, ectopic expression of CAGE in fibroblast cells resulted in foci formation, suggesting its cell-transforming ability. Using stable HeLa transfectant clones with the tetracycline-inducible CAGE gene, we found that CAGE overexpression stimulated both anchorage-dependent and -independent cell growth in vitro and promoted tumor growth in a xenograft mouse model. Cell cycle analysis showed that CAGE augments the levels of cyclin D1 and E, thereby activating cyclin-associated cyclin-dependent kinases and subsequently accelerating the G1 to S progression. Moreover, increased cyclin D1 and E levels in CAGE-overexpressing cells were observed even in a growth arrested state, indicating a direct effect of CAGE on G1 cyclin expression. CAGE-induced expression of cyclins D1 and E was found to be mediated by AP-1 and E2F-1 transcription factors, and among the AP-1 members, c-Jun and JunD appeared to participate in CAGE-mediated up-regulation of cyclin D1. CAGE overexpression also enhanced retinoblastoma phosphorylation and subsequent E2F-1 nuclear translocation. In contrast, small interfering RNA-mediated knockdown of CAGE suppressed the expression of G1 cyclins, activation of AP-1 and E2F-1, and cell proliferation in both HeLa cervical cancer cells and Malme-3M melanoma cells. These results suggest that the cancer/testis antigen CAGE possesses oncogenic potential and promotes cell cycle progression by inducing AP-1- and E2F-dependent expression of cyclins D1 and E.
Keywords: AP-1 Transcription Factor, Cell Cycle, Cyclins, E2F Transcription Factor, Tumor Marker, Tumor Promoter, CAGE, Cancer/Testis Antigen, Cell Proliferation
Introduction
Cancer/testis (C/T)3 antigens have received particular attention because they are expressed in various malignant tumors but not in normal adult tissues, except for germ cells of the testis (1–4). Because the testis is an immune-privileged site, the cancer-specific expression of C/T antigens makes them ideal candidates for cancer-specific immunotherapy. To date, more than 40 genes encoding C/T antigens, including BAGE, MAGE, NY-SEO-1, and GAGE, have been identified and studied for their expression in numerous cancer types (5–8). Wide expression of C/T antigens in various types of cancer cells suggests that they have a role in cancer development. However, the functional significance of most C/T antigens in tumorigenesis is largely unknown.
We previously identified a novel C/T antigen gene, CAGE, by using SEREX (serological analysis of recombinant cDNA expression library), in which cDNA expression libraries of human testis and gastric cancer cell lines were screened with the sera of patients with gastric cancers (9). Similar to most of the C/T antigens, the CAGE gene displays testis-specific expression among normal tissues, and cancer-specific expression among various human cancer tissues and cell lines, particularly originated from gastric, cervical, lung, liver, kidney, and colon cancers. Also, like many C/T antigens, the CAGE gene is localized to the X chromosome. Additionally, CAGE expression was found to be epigenetically regulated depending on the methylation status of CpG sites of the CAGE promoter (10). Importantly, CAGE expression was found to be induced at the G1 phase preceding the induction of cyclin B, indicating the cell cycle-related expression pattern of CAGE (9). We also recently showed that CAGE transfection increases the growth rate of L929 fibroblast cells as well as cell adhesion and motility (11). Accordingly, the C/T antigen CAGE has been implicated in cancer development and progression, although its biochemical activity/function is largely unknown.
In the present study, we investigated the oncogenic ability of CAGE on the basis of the characteristic loss of growth control displayed by tumor cells. Using HeLa transfectant clones with a tetracycline-inducible expression system of the exogenous CAGE gene, the functional effect of CAGE overexpression on cell proliferation and tumor growth was assessed by an in vitro cell culture system and an in vivo xenograft tumor model, respectively. Here we demonstrate the cell proliferation-stimulating activity of the C/T antigen CAGE and its function in promoting G1 progression in the cell cycle. These results may provide insight into the potential mechanism and role of many other C/T antigens in cancer development and progression.
EXPERIMENTAL PROCEDURES
Cell Culture, Antibodies, and Reagents
Tet-On sublines of HeLa human cervical cancer cells obtained from Clontech Laboratories (Mountain View, CA) were cultured in DMEM supplemented with 10% Tet system-approved fetal bovine serum (Clontech), 100 units/ml penicillin, 100 μg/ml streptomycin, and 200 μg/ml G418 in 5% CO2 at 37 °C. NIH3T3 mouse fibroblast cells were cultured in DMEM supplemented with 10% calf serum. Preparation of anti-CAGE antibody was described in a previous study (9). Monoclonal antibodies against cyclin D1 (M-20), cyclin E (HE12), cyclin A (BF683), cyclin B (GNS1), CDK4 (C-22), CDK2 (M2), Rb (IF8), p53 (DO-1), E2F-1 (KH95), p15 (K-18), p16 (N-20), p18 (18P118), p19 (DCS-100), p21 (F-5), p27 (F-8), JunB (210), JunD (329), c-Fos (D-1), FosB (C-11), Fra-1 (C-12), Fra-2 (L-15), and p65 (SC-109X) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies specific to phospho-RbSer-795, phospho-c-JunSer-63/Ser-73, and c-Jun were obtained from Cell Signaling Technology (Danvers, MA), and an antibody to histone H3 was obtained from Upstate Chemicon (Temecula, CA). All other reagents were from Sigma unless otherwise indicated.
Generation of Stable Tetracycline-inducible CAGE Transfectant Clones of HeLa Cells
Full-length CAGE cDNA was subcloned into the site downstream of a tetracycline-responsive transactivator-binding promoter of the pTRE2 vector (Clontech) and transfected into HeLa/Tet-On cells using Lipofectamine/PLUS reagent (Invitrogen). CAGE transfectant clones of HeLa/Tet-On cells were isolated by growing the cells in DMEM containing 10% Tet system-approved fetal bovine serum, 400 μg/ml G418, and 200 μg/ml hygromycin. Stable CAGE transfectant clones cultured in the absence or presence of doxycycline (1 μg/ml) were characterized by RT-PCR and immunoblotting analyses for the CAGE expression level. Selected transfectant clones with the doxycycline-inducible CAGE gene were maintained with working concentrations of 200 μg/ml for G418 and 100 μg/ml for hygromycin.
Clonogenic Soft Agar Assay
Tetracycline-inducible CAGE stable transfectant clones of HeLa/Tet-On cells were suspended in DMEM containing 20% fetal bovine serum and 0.3% (w/v) soft agar and overlaid onto the basal layer containing 0.5% agarose. After cells were cultured in the absence or presence of doxycycline (1 μg/ml) for 21 days, the culture plates were stained with 1% crystal violet, and colonies with a diameter greater than 1 mm were counted directly.
Nude Mouse Tumor Xenograft Model
Stable tetracycline-inducible CAGE-transfected HeLa cells were harvested, resuspended in phosphate-buffered saline, and injected subcutaneously into 8-week-old female nude mice (106/mouse). Two independent groups composed of five mice each were established; the groups were divided on the basis of doxycycline treatment. Doxycycline (1 μg/ml) was administered to one group via drinking water, which was changed every other day. At the onset of tumor formation, tumor diameter was measured every other day with calipers. Approximately 35 days after inoculation the mice were euthanized, and tumor weight was assessed.
Cell Cycle Analysis
For double thymidine block, exponentially growing cells were incubated in serum-free medium containing 5 mm thymidine for 14 h. Following rinsing with thymidine-free medium, cells were incubated in complete medium containing 24 μm deoxycytidine for 8 h and then subjected to a second thymidine block. Synchronized cells were released to enter the cell cycle with the addition of complete medium to the cells. For flow cytometric analysis of DNA content, cells were harvested by trypsinization and then fixed with ice-cold 70% ethanol. The cells were resuspended in 1 ml of solution containing 0.4 mm sodium citrate, 25 μg/ml propidium iodide, and 50 μg/ml RNase. The stained cells were analyzed in a FACScan flow cytometer (BD Biosciences) using the ModFit LT program (BD Biosciences). For fluorescence staining of the S phase-entering cells, bromodeoxyuridine (BrdUrd) incorporation assay was performed as described previously (12).
RT-PCR Analysis
Total cellular RNA was purified from the cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. First-strand cDNA synthesis was performed with 1 μg of total RNA using a cDNA synthesis kit (Promega, Madison, WI). For PCR amplification of CAGE cDNA, 5′-ACGCGTATGTCCCACTGGGCCCCAGAGT-3′ was used as the sense primer, and 5′-TTCTAGAACATCATTACCGACTCGATCG-3′ was used as the antisense primer. The reaction mixture was subjected to 25 PCR amplification cycles for 60 s at 94 °C, 60 s at 54 °C, 1 min, and 30 s at 72 °C. PCR primers for amplification of cyclin D1 and cyclin E transcripts were as follows: cyclin D1, 5′-CTTCCTCTCCAAAATGCCAG-3′ and 5′-AGAAAGGGGGAAGGTAGAGA-3′, sense and antisense, respectively; cyclin E, 5′-GGAGCCAGCCTTGGGACAATAATG-3′ and 5′-AAACGTGGTCAAACGCATACACTGT-3′, sense and antisense, respectively. β-Actin amplification was used as an internal PCR control with 5′-GATATCGCCGCGCTCGTCGTCGAC-3′ as the sense primer and 5′-CAGGAAGGAAGGCTGGAAGAGTGC-3′ as the antisense primer.
Transfection of Small Interfering RNA (siRNA)
siRNAs for JunD and E2F-1 were designed and synthesized using the software and SilencerTM siRNA construction kit from Ambion (Austin, TX) according to the manufacturer's instructions. siRNAs for CAGE was synthesized by Bioneer (Daejon, Korea). Specific oligonucleotide sequences for each target gene were as follows: 5′-CUCUGUCAACCUAAGAAGC(dAdT)-3′ (sense) and 5′-GCUUCUUAGGUUGACAGAG(dAdT)-3′ (antisense) targeting CAGE; 5′-CAGCUCGGUGUUCUGGCUUTT-3′ (sense) and 5′-AAGCCAGAACACCGAGCUGTT-3′ (antisense) targeting JunD; and 5′-UCAUAGCGUGACUUCUCCCTT-3′ (sense) and 5′-GGGAGAAGUCACGCUAUGATT-3′ (antisense) targeting E2F-1. The siRNA control was 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense), which bears no homology with the relevant human gene (13). Transfection of siRNA into cells was performed as described previously (14).
Generation of Promoter Deletion Constructs and Promoter Assay
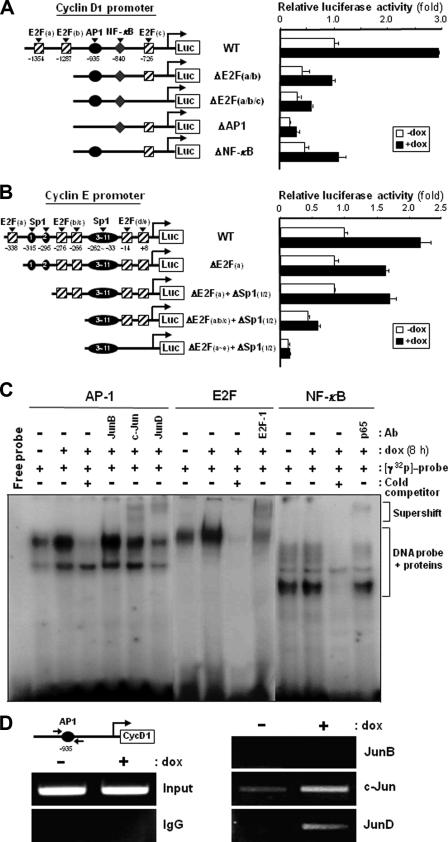
A 1400-bp (−1380 to +20) and 370-bp (−350 to +20) DNA fragment, corresponding to the promoters of the human cyclin D1 and E genes, respectively, were generated by PCR using human genomic DNA as a template. Putative regulatory regions in the promoter DNAs of human cyclin D1 and E genes were deleted as indicated in Fig. 6 by using specific PCR primers. After the wild-type and mutant promoters of the human cyclin D1 and E genes were subcloned into a promoterless luciferase expression vector, pGL3 (Promega), the nucleotide sequence of each construct was verified by DNA sequencing. The pGL3 vector containing the promoter DNA was transfected into CAGE transfectant HeLa cells, and 2 days post-transfection cells were treated with or without doxycycline for 8 h. Luciferase activity in cell lysates was measured using the Promega luciferase assay system according to manufacturer's instructions. To normalize luciferase activity, each of the pGL3 vectors was co-transfected with a pRL-SV40ΔE, which expresses Renilla luciferase by an enhancerless SV40 promoter (15).
FIGURE 6.
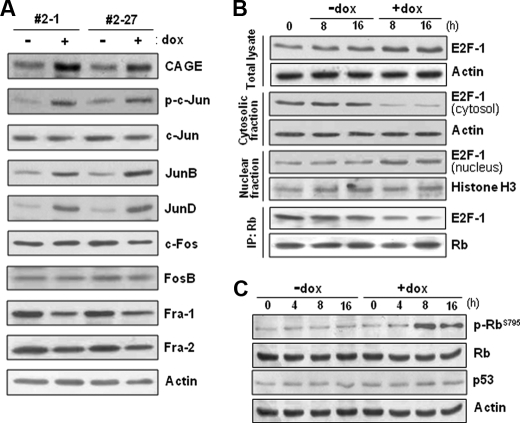
Levels of AP-1 proteins, Rb phosphorylation, and nuclear E2F in CAGE-overexpressing cells. A, protein levels of Jun and Fos subfamily members of AP-1 factors were examined in tetracycline-inducible CAGE transfectant HeLa cells by immunoblotting analysis after doxycycline (dox) treatment for 8 h. The phosphorylation level of c-Jun was evaluated by using an antibody specific to the phospho-Ser-63/Ser-73 residues of c-Jun. B, CAGE transfectant clonal cells (#2-27) were treated with or without doxycycline for the indicated time periods, and E2F-1 protein levels in the total cell lysate, the cytosolic and nuclear fractions, and the Rb immunoprecipitate (IP) were examined for by immunoblotting analysis. C, the phosphorylation level of Rb in cells (#2-27) treated with or without doxycycline was assessed by immunoblotting analysis using an antibody specific to Rb phosphorylated at the Ser-795 residue.
Electrophoretic Mobility Shift Assay
Double-stranded DNA probes corresponding to the putative AP-1 binding site (−943 to −920, 5′-AAAAAAAATGAGTCAGAATGGAGA-3′; the AP-1 recognition sequence is underlined) and the putative NF-κB site (−847 to −822, 5′-TGCAGTTGGGGACCCCCGCAAGGACC-3′; the NF-κB recognition sequence is underlined) in the proximal cyclin D1 promoter sequence were prepared by oligonucleotide synthesis. A double-stranded oligonucleotide containing the consensus binding motif of E2F (5′-ATTTAAGTTTCGCGCCCTTTCTCAA-3′; the E2F consensus recognition sequence is underlined) was obtained from Santa Cruz Biotechnology. The DNA probes were labeled with [γ-32P]ATP using T4 polynucleotide kinase and purified by a G-50 Sephadex column. The 32P-labeled probes (∼40,000 cpm) were then incubated with nuclear extracts (10 μg of protein) prepared from doxycycline-treated and untreated cells for 20 min at room temperature. For supershift experiments, 2–3 μg of antibody against c-Jun, JunB, JunD, E2F1, or p65 were preincubated with the nuclear extracts. Samples were resolved on native 5% polyacrylamide gel and then dried and exposed to x-ray film for autoradiography.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed using the chromatin immunoprecipitation assay kit from Upstate Chemicon according to the manufacturer's instructions. The PCR primers specific to the AP-1 binding region of the human cyclin D1 promoter were: 5′-ATTCAGTCCCAGGGCAAATTCTAAA-3′ and 5′-CCTTCATCTTGTCCTTCTAGCCTGG-3′. The sequences as a negative control in the cyclin D1 promoter were: 5′-TTTCGGAAGCGTTTTCCC-3′ and 5′-AGCGCGTTCATTCAGGAA-3′.
Cyclin-dependent Kinase (CDK) Assay
After cell lysis, immunoprecipitation was performed by using anti-CDK4 and anti-CDK2 antibodies. Immunoprecipitates were washed twice with kinase buffer (20 mm Tris-HCl, pH 7.5, 5 mm β-glycerolphosphate, 2 mm dithiothreitol, 0.1 mm Na3VO4, 1 mm NaF, 10 mm MgCl2) and then mixed with kinase reaction mixture containing 10 μg of histone H1 (Hoffmann-La Roche), 20 μm ATP, and 10 μCi of [γ-32P]ATP (Amersham Biosciences). After incubation for 30 min at 30 °C, the reactions were terminated by boiling with SDS sample buffer. Proteins were separated using 12% SDS-PAGE, and the dried gel was subjected to autoradiography.
Cell Proliferation Assay
In vitro growth of CAGE cDNA-transfected cells and CAGE siRNA-transfected cells was measured by direct cell counting and an MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2H-tetrazolium) assay, respectively. For the MTS assay, at 24 h following the transfection of control and CAGE siRNAs, 1 × 103 cells were seeded into the individual wells of a 96-well plate. At 16 h post-seeding, the MTS reagent (Promega) was added to each well and incubated for 30 min at 37 °C followed by measurement at 490 nm.
Protein Analyses
Cell lysis, immunoblotting, and immunoprecipitation were performed as described previously (14). Protein concentration was determined using the BCA protein assay with bovine serum albumin as standard.
RESULTS
Foci Formed by CAGE in Fibroblast Cells
Because transformation of cultured cells provides a simple and rapid assay for the oncogenic potential of a certain gene, we first examined whether the C/T antigen CAGE could induce cell transformation in culture. Following transfection of the CAGE expression vector into NIH3T3 mouse fibroblast cells, 10-fold more foci were formed as compared with those transfected with empty vector alone (supplemental Fig. S1). Given that foci formation in a normal fibroblast cell monolayer represents a transformed cell phenotype, this result suggests the possibility that CAGE possesses the oncogenic ability to induce cell transformation.
Establishment of CAGE Stable Transfectant Clones and Anchorage-independent Growth
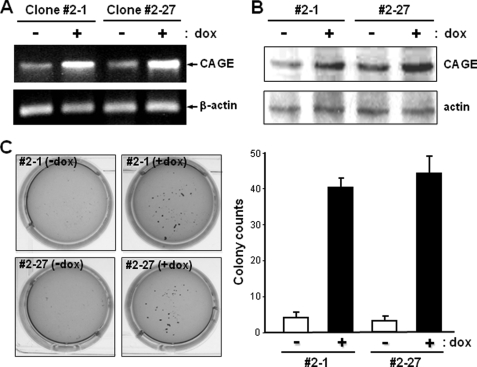
To generate human cancer cell clones with exogenous CAGE, we transfected a tetracycline-inducible expression vector containing the human CAGE cDNA into Tet-On sublines of HeLa human cervical cancer cells in which expression of the exogenous gene is induced by tetracycline treatment. Stable transfectant clones were analyzed for the tetracycline-responsive expression of CAGE. The CAGE expression level in transfectant clones 2-1 and 2-27 was significantly increased by doxycycline, a derivative of tetracycline (Fig. 1, A and B). Using these HeLa transfectant clones with the doxycycline-inducible CAGE gene, we further characterized the functional effect of CAGE overexpression on the phenotype of neoplastic transformation. Following equal plating of CAGE transfectant clones in soft agar suspension, doxycycline-treated cells formed ∼8-fold more colonies than untreated cells (Fig. 1C). As only transformed cells are able to form colonies in a semisolid medium through anchorage-independent growth, this result provides another line of evidence for the transforming ability of the CAGE gene.
FIGURE 1.
Generation of stable tetracycline-inducible CAGE transfectant clones of HeLa cells and their colony-forming ability in soft agar. A and B, Tet-On sublines of HeLa cells were transfected with CAGE cDNA subcloned into a pTRE2 tetracycline-inducible expression vector. The resultant stable transfectant clones were analyzed for CAGE expression in the absence or presence of doxycycline (dox) (1 μg/ml) by RT-PCR (A) and immunoblotting (B) analyses. C, stable tetracycline-inducible CAGE transfectant clones 2-1 and 2-27 suspended in 0.3% soft agar were cultured in the absence or presence of doxycycline for 21 days. After crystal violet staining, colonies with a diameter larger than 1 mm were counted directly. Values are the means ± S.D. of triplicate cultures.
Effects of CAGE Overexpression on HeLa Cell Growth in Vivo and in Vitro
We next examined whether CAGE plays a role in tumor formation and growth in vivo. Following subcutaneous inoculation of athymic nude mice with tetracycline-inducible CAGE transfectant clones of HeLa cells, tumor formation occurred at almost the same time regardless of doxycycline administration. However, doxycycline-administered mice exhibited faster tumor growth than control mice (Fig. 2, A and B). Quantitatively, administration of doxycycline resulted in a 2-fold greater increase in both tumor volume and tumor weight. Because CAGE-induced tumor growth may originate from increased cell proliferation as a consequence of CAGE overexpression, we next analyzed HeLa cell growth in culture in the presence of doxycycline. CAGE transfectant clones treated with doxycycline showed a significantly increased cell growth rate compared with untreated cells (Fig. 2C). In particular, the growth of CAGE transfectant cells cultured in the absence of doxycycline was dramatically increased without delay after doxycycline treatment. It thus appears that increased expression of CAGE indeed stimulates proliferation of transfected HeLa cells in vitro and in vivo.
FIGURE 2.
In vivo and in vitro growth rate of HeLa cells with tetracycline-inducible CAGE. A and B, stable tetracycline-inducible CAGE transfectant clones (106 cells) were inoculated subcutaneously into athymic nude mouse. Following inoculation 10 mice were divided into two groups, with one group devoid of treatment and the other group administered doxycycline (dox) (1 μg/ml). At the onset of tumor formation, tumor diameter was measured every other day with calipers (A). At 35 days after inoculation, the mice were euthanized, and tumor weight was measured (B). Values are the means ± S.D. of tumors from five mice. CAGE abundance in representative primary tumors (P) from each mouse was assessed by immunoblotting with an antibody to CAGE (inset in B). C, tetracycline-inducible CAGE transfectant clones were seeded at a low density and cultured in the absence or presence of doxycycline (1 μg/ml), and cell numbers were counted directly every 24 h over the course of 6 days. For delayed overexpression of CAGE, doxycycline was added to the culture medium of untreated cells 3 days after initial seeding of the cells (arrows above Δ+dox). *, p < 0.01 versus −dox at day 6 in culture.
Effect of Increased CAGE Expression on Cell Cycle Progression
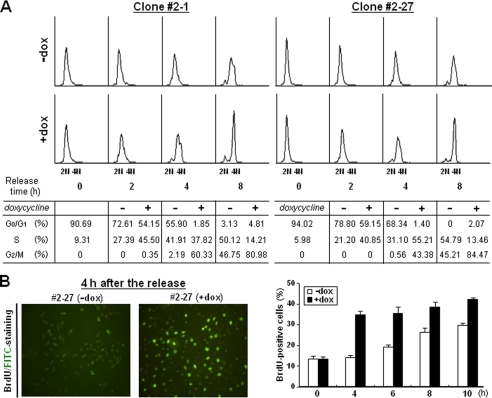
It was found previously that CAGE expression level is related with the cell cycle in a cervical cancer cell line, C33A (9). To examine the functional effect of CAGE overexpression on the cell cycle of HeLa cells, we synchronized CAGE transfectant cells at the G1/S phase boundary by double thymidine block. When cells were permitted to proceed to the next stage of the cell cycle, a larger cell population was found to progress to the S phase in the doxycycline-treated group as compared with the untreated group (Fig. 3A). Also, in cells synchronized at the M phase by thymidine-nocodazole block, doxycycline-treated cells exhibited a faster progression through G1 than untreated cells (supplemental Fig. S2). CAGE-promoted progression to the S phase was also demonstrated by BrdUrd staining (Fig. 3B), in which doxycycline-treated cells exhibited DNA synthesis earlier than untreated cells. Collectively, these results strongly suggest that CAGE functions as a positive regulator of cell cycle progression at the G1 phase and for entry into S phase.
FIGURE 3.
Cell cycle progression in CAGE-overexpressing HeLa cells. A, tetracycline-inducible CAGE transfectant clones were synchronized by double thymidine for G1/S block. After treatment with or without doxycycline for 2 h, synchronized cells were released in complete medium supplemented with or without doxycycline for the indicated time periods. After cells were labeled with propidium iodide, DNA content was analyzed by flow cytometry. Shown are representative histograms and the means of cell cycle phase distribution from three independent experiments. B, cells synchronized by double thymidine block were released in the absence or presence of doxycycline (dox) for the indicated time periods. After BrdUrd treatment, cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-BrdUrd antibody and viewed under a fluorescence microscope. Shown are representative images and the means ± S.D. of BrdUrd-positive cell counts in triplicate cultures.
Increased Expression of the G1 Cyclins in CAGE-overexpressing Cells
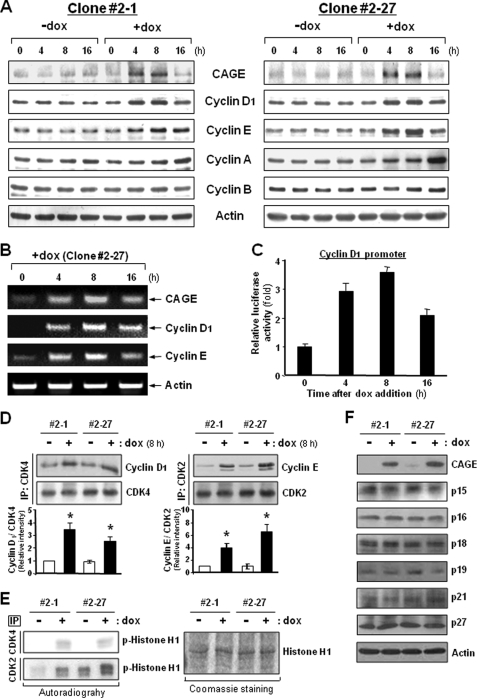
In an effort to identify molecules that mediate the cell cycle-promoting activity of CAGE, we compared the expression levels of cell cycle machinery components between doxycycline-treated and untreated CAGE transfectant HeLa cells. In asynchronized cells, no apparent alteration was observed in the expression levels of the four major cyclin types. However, doxycycline treatment enhanced the cellular levels of cyclins D1 and E expressed in the G1 phase, concomitantly with CAGE overexpression (Fig. 4A). CAGE-overexpressing cells also displayed increased levels of cyclin A, which mediates S phase progression. However, the increase in cyclin A levels occurred substantially later than CAGE induction, suggesting that CAGE is not linked directly to cyclin A expression. Increased mRNA levels of cyclins D1 and E following doxycycline treatment indicated that CAGE expression levels influence the transcription of cyclin D1 and E genes (Fig. 4B). Moreover, a cyclin D1 promoter assay further supported CAGE-induced transcription of the cyclin D1 gene (Fig. 4C). CAGE-induced expression of the G1 cyclins was also observed in HeLa cells in a nonproliferating state, such as cells in suspension or cells grown to 100% confluency (supplemental Fig. S3). Because cyclins D and E specifically interact with CDK4/6 and CDK2, respectively, we next examined the amount of cyclins D1 and E associated with these CDK partners in CAGE-overexpressing cells. As expected, higher amounts of cyclin D1 and E proteins were found to be associated with CDK4 and CDK2, respectively, in doxycycline-treated cells as compared with untreated cells (Fig. 4D). Furthermore, both CDK4 and CDK2 immunoprecipitates obtained from doxycycline-treated cells exhibited higher protein kinase activity than those from untreated cells (Fig. 4E), indicating that CAGE overexpression results in the activation of CDK4 and CDK2. Meanwhile, cellular levels of CDK inhibitors such as p15, p16, p18, p19, p21, and p27 were found to be minimally altered by CAGE overexpression (Fig. 4F). Taken together, these data suggest that CAGE-induced expression of the G1 cyclins leads directly to the activation of the CDK4 and CDK2, which play a central role in cell cycle progression from G1 to S phase.
FIGURE 4.
Expression levels of cyclins and kinase activities of CDKs in CAGE-overexpressing HeLa cells. A and B, asynchronized HeLa cells transfected with the tetracycline-inducible CAGE were cultured in the absence or presence of doxycycline (dox) for the indicated time periods. Expression levels of cyclins D1, E, A, and B were examined by immunoblotting (A) and RT-PCR (B) analyses. C, following transfection with a control pGL3 reporter vector or vector containing cyclin D1 promoter DNA, CAGE transfectant cells were treated with doxycycline for the indicated time periods. Promoter activity was assessed by a luciferase assay. D, the amount of cyclins D1 and E associated with CDK4 and CDK2, respectively, were assessed by immunoprecipitation (IP) followed by immunoblotting analysis. *, p < 0.05 versus −dox. E, immunoprecipitates with anti-CDK4 and anti-CDK2 antibodies were subjected to an in vitro kinase assay using histone H1 as a substrate. F, CDK inhibitor proteins were examined for their cellular levels by immunoblotting analysis.
Functional Involvement of AP-1 and E2F Factors in CAGE-induced Cyclin D1 and E Gene Transcription
We next investigated the transcriptional regulation of the cyclin D1 and E genes by using several mutants of their 5′-proximal promoter regions (16, 17). When a reporter vector containing a wild-type promoter of the cyclin D1 gene was transiently transfected into CAGE transfectant HeLa cells, doxycycline-treated cells exhibited about a 3-fold higher promoter activity than untreated cells (Fig. 5A). However, promoters with deletions at the E2F binding sites exhibited significantly decreased activities for reporter gene expression and responded to doxycycline to a lesser extent than the wild-type promoter. The effect of doxycycline on cyclin D1 promoter activity was further decreased by the AP-1 binding site deletion. In contrast, little effect of the NF-κB binding site deletion was observed on doxycycline-induced activity of the cyclin D1 promoter. These results suggest that the E2F and AP-1 binding sites in the cyclin D1 gene promoter are functionally involved in CAGE-mediated regulation of cyclin D1 gene transcription. An essential role of the E2F binding sites in CAGE-induced promoter activity was more evident in the cyclin E gene promoter. As shown in Fig. 5B, more E2F sites in the cyclin E promoter were removed, and a significantly lower stimulatory effect of doxycycline on promoter activity was observed. We next compared the level of nuclear proteins bound to the oligonucleotides corresponding to the recognition sites of E2F and AP-1 factors between doxycycline-treated and untreated cells. Doxycycline-treated CAGE transfectant cells displayed significantly more nuclear proteins bound to the E2F and AP-1 recognition oligonucleotides than did the untreated cells (Fig. 5C). Moreover, incubation with anti-E2F-1 antibody resulted in a partial supershift of the nuclear protein-DNA complex in the gel mobility shift assay. Also, among the antibodies against Jun subfamily members of AP-1 factors, anti-c-Jun and anti-JunD antibodies were shown to bind to the nuclear proteins complexed with the AP-1 recognition oligonucleotide. Conversely, a minimal difference in the amount of nuclear proteins bound to the NF-κB recognition oligonucleotide was found between doxycycline-treated and untreated cells. In a further attempt to support the involvement of c-Jun and JunD AP-1 factors in CAGE-mediated activation of cyclin D1 promoter, a ChIP assay was performed (Fig. 5D). Antibodies targeting c-Jun and JunD, but not JunB, were able to precipitate more AP-1 binding site-containing cyclin D1 promoter region upon CAGE overexpression, indicating that the amount of c-Jun and JunD bound to the cyclin D1 promoter was increased by CAGE. It thus appears that CAGE overexpression results in increased binding of the AP-1, including c-Jun and JunD, and E2F transcription factors to the promoters of cyclin D1 and E genes, possibly leading to the up-regulation in transcription of these G1 cyclin genes.
FIGURE 5.
Functional involvement of AP-1 and E2F binding sites within cyclin D1 and E gene promoters in CAGE-induced G1 cyclin expression. A and B, human wild-type (WT) promoters of the cyclin D1 (A) and E (B) genes and the mutant promoters for E2F, AP-1, NF-κB, and Sp1 binding sites were fused to a luciferase reporter gene of the pGL3 vector. Following transient transfection with pGL3 vector containing wild-type or mutant promoters of cyclin D1 and E genes, the tetracycline-inducible CAGE transfectant clone 2-27 was treated with or without doxycycline (dox) for 8 h, and a dual luciferase assay was performed. Normalized luciferase activity ± S.D. of triplicate experiments are plotted. C, double-stranded oligonucleotides containing AP-1, E2F, or NF-κB recognition sequences were 32P-labeled and incubated with nuclear extracts of CAGE transfectant cells treated with or without doxycycline. Specific DNA binding activities of AP-1, E2F, and NF-κB factors were determined by an electrophoretic mobility shift assay in the absence or presence of an 8-fold excess of cold competitors. Anti-JunB, anti-c-Jun, anti-JunD, anti-E2F-1 and anti-NF-κB antibodies were used for supershift analysis. D, CAGE transfectant cells treated with or without doxycycline were fixed with formaldehyde and then harvested and sonicated to shear DNA. Following incubation with anti-JunB, anti-c-Jun, or anti-JunD antibodies or normal mouse IgG, the immunocomplex was subjected to 25 PCR cycles using primers specific to the AP-1 binding region of the human cyclin D1 promoter.
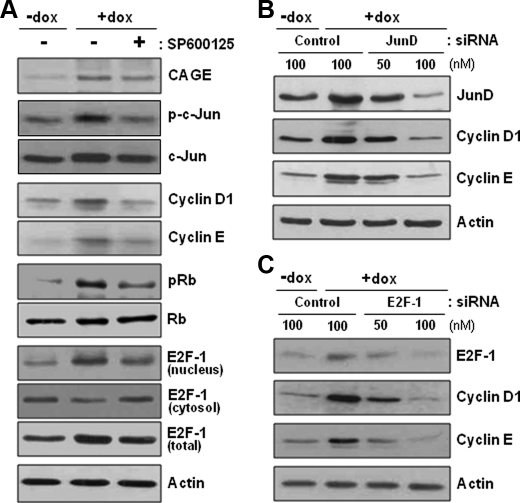
CAGE-induced Expression/Activation of Jun AP-1 Proteins and Nuclear Translocation of E2F-1
We next examined the levels of AP-1 proteins in CAGE-overexpressing HeLa cells. Among the Jun subfamily AP-1 members, the protein levels of JunB and JunD in doxycycline-treated cells were higher than those in untreated cells (Fig. 6A). The phosphorylation level of c-Jun was also increased by doxycycline treatment, although the c-Jun protein level was not altered. Conversely, none of the cellular levels of the Fos subfamily AP-1 members were shown to be altered by doxycycline treatment. We also investigated the effect of CAGE on the expression levels and subcellular localization of the E2F-1 transcription factor. The total amount of E2F-1 was found to be increased by doxycycline treatment to some extent as compared with untreated cells (Fig. 6B). Importantly, the E2F-1 level in the nuclear fraction was significantly increased by CAGE overexpression, whereas its cytosolic level was decreased. Because the association of E2F with retinoblastoma (Rb) in the cytosol prevents E2F from nuclear translocation, the amount of E2F-1 complexed with Rb was compared between doxycycline-treated and untreated cells. Doxycycline-treated cells exhibited significantly less E2F-1 protein bound to Rb than did the untreated cells (Fig. 6B). Moreover, doxycycline treatment resulted in significantly increased phosphorylation levels of Rb, especially at the Ser-795 residue, without affecting protein levels (Fig. 6C). Because phosphorylated Rb does not form a complex with E2F, it is likely that CAGE overexpression induces nuclear translocation of E2F by stimulating Rb phosphorylation and subsequent E2F release from the Rb-E2F complex in the cytosol. Taken together, the data in this study illustrate the potential contribution of CAGE to AP-1- and E2F-mediated gene transcription.
Ablation of CAGE-induced Cyclin D1 and E Expression by an Inhibitor of JNK and siRNAs for JunD and E2F-1
Because an increase in the phosphorylation level of c-Jun, but not the protein level, was observed in CAGE-overexpressing cells, we treated CAGE transfectant cells with an inhibitor of c-Jun N-terminal kinase (JNK), SP600125, to clarify the participation of phosphorylated c-Jun in CAGE-induced G1 cyclin expression. As shown in Fig. 7A, inhibition of c-Jun N-terminal phosphorylation abolished the positive effect of CAGE overexpression not only on cyclin D1 and E expression but also on Rb phosphorylation and E2F-1 nuclear translocation. These results indicate a critical role of JNK-mediated c-Jun phosphorylation in CAGE-promoted cell cycle progression. Next, CAGE-overexpressing cells were transfected with siRNAs targeted to JunD and E2F-1. CAGE-induced expression of cyclins D1 and E was abrogated by siRNA knockdown of JunD and E2F-1 (Fig. 7, B and C). Along with the gel mobility shift assay, these data indicate that CAGE induces expression of the G1 cyclins in a c-Jun-, JunD-, and E2F-1-dependent manner.
FIGURE 7.
Ablation of CAGE-induced G1 cyclin expression by an inhibitor of JNK and siRNAs for JunD and E2F-1. A, the tetracycline-inducible CAGE transfectant clone (#2-27) of HeLa cells was pretreated with a JNK inhibitor, SP600125, for 4 h and further incubated in the absence or presence of doxycycline (dox) for 8 h. The levels of cyclin D1 and E proteins, phospho-c-Jun, phospho-Rb, and cytosolic and nuclear E2F-1 proteins were assessed by immunoblotting analysis. B and C, the clonal cells (#2-27) were transfected with control and JunD (B) or E2F-1 (C) siRNAs. At 48 hours after transfection, cells were treated with or without doxycycline for 8 h and analyzed for protein levels of cyclins D1 and E.
Effect of CAGE Knockdown on Cell Proliferation and Expression/Activation of G1 Cyclins, Jun AP-1 Proteins, and E2F-1
Next, we knocked down CAGE using siRNAs in HeLa human cervical cancer cells and Malme-3M human melanoma cells, both of which normally express CAGE. CAGE knockdown reduced the levels of cyclins D1 and E and JunD and the phosphorylation level of c-Jun in both cancer cell lines (Fig. 8A). Rb phosphorylation and nuclear localization of E2F-1 were also diminished upon CAGE knockdown (Fig. 8, B and C). Additionally, CAGE knockdown retarded the cell proliferation of these two cancer cell lines (Fig. 8, D and E). These data strongly suggest that CAGE plays a role in regulating the normal cell cycle progression of these CAGE-expressing human cancer cells.
FIGURE 8.
CAGE siRNA-mediated suppression of G1 cyclin expression, Jun AP-1 protein expression/activation, E2F-1 nuclear translocation, and cell proliferation in human cancer cell lines. A and B, protein levels of CAGE, cyclins D1 and E, and JunD and the phosphorylation level of c-Jun (A) and Rb (B) were determined in CAGE siRNA-transfected HeLa and Malme-3M cells. Cont, control. C, the level of cytosolic and nuclear E2F-1 was assessed by immunoblotting analysis after subcellular fractionation. D and E, cell proliferation was measured by MTS assay upon siRNA knockdown of CAGE in HeLa and Malme-3M cells as described under “Experimental Procedures.” *, p < 0.01 versus −doxycycline at day 4 in culture.
DISCUSSION
Many studies have analyzed C/T antigens for their potential use in cancer immunotherapy (2, 3). Cytotoxic T lymphocyte (CTL) reactions have been found to be induced by C/T antigens such as BAGE, GAGE, and MAGE (6, 18–20). We have also demonstrated herein that CAGE is capable of inducing CTL activity (11). Cancer-specific expression coupled with recognition by CTL makes these C/T antigens ideal targets for cancer-specific immune therapy. However, the functional role or involvement of C/T antigens in cancer development and progression remains to be elucidated. In this study, we have demonstrated the oncogenic potential of the C/T antigen CAGE through commonly used assays of transformation in culture. Transfection of the CAGE-expressing gene construct into NIH3T3 fibroblast cells resulted in the formation of foci on a cultured cell monolayer (supplemental Fig. S1), indicating that CAGE expression leads to a transformed cell phenotype demonstrated by the loss of contact inhibition of movement and loss of density-dependent inhibition of growth. Additionally, overexpression of CAGE in cells suspended in soft agar rendered HeLa cells able to form more colonies (Fig. 1C), indicating that CAGE contributes to anchorage-independent growth of transformed cells. CAGE overexpression in HeLa cells was also found to stimulate xenograft tumor growth in nude mice and anchorage-dependent in vitro cell growth (Fig. 2). Although we did not directly evaluate the role of CAGE in tumor pathogenesis through a transgenic mouse model, these experiments illustrate the potential contribution of the C/T antigen CAGE in tumor development.
The positive role of CAGE in the regulation of cancer cell proliferation was further supported by cell cycle analysis. CAGE overexpression resulted in a rapid transition from G1 to S phase upon release into the cell cycle following cell cycle synchronization at the G1/S boundary or at the M phase (Fig. 3 and supplemental Fig. S2). CAGE-induced cell cycle progression from G1 to S phase was shown to be correlated with an increase in the expression levels of D1-type G1 phase cyclin and E-type G1/S phase cyclin, along with increased protein kinase activity of CDK4-cyclin D1 and CDK2-cyclin E complexes (Fig. 4). CAGE knockdown in cell lines that normally express CAGE, HeLa, and Malme-3M resulted in retarded cell proliferation and reduced levels of cyclins D1 and E (Fig. 8). These results raised the possibility that CAGE overexpression might be critical for the process of tumorigenesis in these cancer cell lines. Taken together, it thus appears that up-regulation of cyclins D1 and E, and abnormalities in cell proliferation, are likely attributable to the action of CAGE.
Cyclin D is a mitogenic growth factor-inducible member of the cyclin family, and its expression persists as long as growth factors are present. Numerous studies have demonstrated that cyclin D plays a key role in mediating the functional effect of mitogenic growth factors on the cell cycle machinery of stimulated cells (21). The importance of cyclin D in controlling cell proliferation has been further supported by its identification as an oncogene-encoded protein. In particular, cyclin D1 is encoded by the PRAD-1 oncogene, which is frequently found to be activated by chromosomal translocation in parathyroid adenomas and some B-cell lymphomas and by amplification in breast and squamous cell carcinomas (22, 23). Subsequently, elevated expression of cyclin D1 has been found in other human cancers such as gastric, colon, prostate, and esophageal carcinomas (24). The oncogenic potential of cyclin D1 has been further evidenced by its ability to cooperate with myc and ras to induce neoplastic transformation in mice (25, 26). It thus appears that abnormal expression of the D-type cyclins, including cyclin D1, provides a direct link between perturbations of the cell cycle regulatory machinery and cell transformation.
The present study revealed the functional involvement of AP-1 and E2F transcription factors in CAGE-induced transcription of cyclin D1 and E genes. The 5′-proximal promoter region of the cyclin D1 gene contains one AP-1 and three E2F binding sites (16). We found here that all of these sites contribute to CAGE-induced cyclin D1 gene transcription (Fig. 5A). Promoter analysis of the cyclin E gene also clearly indicated the requirement of E2F binding sites for CAGE-induced cyclin E expression (Fig. 5B). Further evidence for the participation of AP-1 and E2F transcription factors in CAGE-mediated regulation of cyclin D1 and E gene transcription were obtained from the gel mobility shift assay (Fig. 5C). Gel supershift and ChIP analyses indicated that CAGE increases the amount of c-Jun, JunD, and E2F-1 bound to the cyclin D1 promoter (Fig. 5, C and D). CAGE thus appears to induce transcription of the D- and E-type cyclin genes in both AP-1- and E2F-dependent manner.
The role of Jun family proteins in the regulation of cell proliferation and cyclin D1 gene transcription has been demonstrated in several studies (27, 28). Using c-jun knock-out mouse models, c-Jun has been shown to induce cyclin D1 gene transcription and cell proliferation. c-jun−/− mouse embryo fibroblasts (MEFs) exhibit severe proliferation defects and reduced cyclin D1 expression (29, 30). Also, fibroblasts of knock-in mouse expressing a phosphorylation-defective mutant form of c-Jun display reduced proliferation (31). Furthermore, it has been found that c-jun cooperates with activated oncogenes, such as H-ras, to induce transformation, and its oncogenic cooperating ability partially requires JNK-mediated phosphorylation (32, 33). JunD is also suggested to be a positive regulator of cell proliferation, as junD−/− MEFs have been shown to have a proliferation defect (34). However, JunD does not exert a pro-proliferative function as potent as c-Jun. Meanwhile, junB−/− MEFs do not exhibit a slower proliferation (35), and MEFs derived from junB-overexpressing transgenic mouse display limited proliferative capacity and decreased expression of cyclin D1 (36). Collectively, c-Jun and, to a lesser extent, JunD are thought to positively regulate cell proliferation, whereas JunB has an opposite effect. In the present study, we found that CAGE with cell proliferation-promoting ability induces expression of both JunB and JunD and phosphorylation of c-Jun (Fig. 6A). Importantly, both the inhibition of c-Jun N-terminal phosphorylation and siRNA knockdown of JunD suppressed CAGE-induced cyclin D1 expression (Fig. 7, A and B). Because the AP-1-DNA complex enhanced by CAGE was shown to contain c-Jun and JunD but not JunB (Fig. 5, C and D), an increase in c-Jun phosphorylation and JunD expression provides an important route for CAGE to stimulate cyclin D1 expression and cell proliferation.
The Rb protein normally functions as a cell cycle control checkpoint at the G1/S phase boundary and has been shown to be inactivated via G1 CDK-mediated phosphorylation at multiple Ser/Thr residues. We found here that CAGE overexpression increases the phosphorylation level of Rb, preferentially at the Ser-795 residue (Fig. 6C), which is known to be phosphorylated specifically by G1 CDKs, such as CDK4 and CDK2, leading to the dissociation of the E2F-Rb complex (37–39). As expected, the level of E2F-1 in the nucleus was increased by CAGE overexpression, whereas the cytosolic level was decreased (Fig. 6B). siRNA knockdown of E2F-1 abolished the positive effect of CAGE overexpression on both cyclin D1 and E expression (Fig. 7C). Because cyclin E transcription is not responsive to AP-1 factors, we postulated that CAGE induces cyclin D1 expression first through AP-1 activation, to some extent, and subsequently phosphorylation of Rb by the CDK4-cyclin D1 complex results in the nuclear translocation of E2F. This results not only in the induction of cyclin E gene transcription but also in further stimulation of cyclin D1 transcription. CAGE-induced cyclin D1 expression thus seems to be under a positive feedback control between AP-1- and E2F-dependent manners.
In conclusion, CAGE could be considered an oncogene in the following aspects, although its product is completely unknown for biochemical function/activity: 1) its frequent expression in various human cancers of different histological origins but no expression in normal counterparts; 2) the ability to transform cells in culture; and 3) the ability to accelerate tumor growth in vivo. Taken together, the data from the present study suggest that the oncogenic ability of CAGE is likely to be based on its function to stimulate cell proliferation by promoting cell cycle progression through the up-regulation of cyclin D1 and E expression.
Supplementary Material
This work was supported by Grants KRF-2006-312-C00677 from the Korea Research Foundation and R01-2007-000-11397-0 from the Korea Science and Engineering Foundation. This work was also supported by a grant from the Korean Ministry of Education, Science, and Technology (Regional Core Research Program/Medical & Bio-Materials Research Center).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and “Experimental Procedures.”
- C/T
- cancer/testis
- BrdUrd
- bromodeoxyuridine
- CDK
- cyclin-dependent kinase
- ChIP
- chromatin immunoprecipitation
- CTL
- cytotoxic T lymphocyte
- DMEM
- Dulbecco's modified Eagle's medium
- JNK
- c-Jun N-terminal kinase
- MEF
- mouse embryo fibroblast
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2H-tetrazolium
- Rb
- retinoblastoma
- RT
- reverse transcriptase
- siRNA
- small interfering RNA.
REFERENCES
- 1.Boon T., Old L. J. (1997) Curr. Opin. Immunol. 9, 681–683 [DOI] [PubMed] [Google Scholar]
- 2.Scanlan M. J., Gure A. O., Jungbluth A. A., Old L. J., Chen Y. T. (2002) Immunol. Rev. 188, 22–32 [DOI] [PubMed] [Google Scholar]
- 3.Scanlan M. J., Simpson A. J., Old L. J. (2004) Cancer Immun. 4, 1. [PubMed] [Google Scholar]
- 4.Zendman A. J., Ruiter D. J., Van Muijen G. N. (2003) J. Cell. Physiol. 194, 272–288 [DOI] [PubMed] [Google Scholar]
- 5.van der Bruggen P., Traversari C., Chomez P., Lurquin C., De Plaen E., Van den Eynde B., Knuth A., Boon T. (1991) Science 254, 1643–1647 [DOI] [PubMed] [Google Scholar]
- 6.Boël P., Wildmann C., Sensi M. L., Brasseur R., Renauld J. C., Coulie P., Boon T., van der Bruggen P. (1995) Immunity 2, 167–175 [DOI] [PubMed] [Google Scholar]
- 7.Van den Eynde B., Peeters O., De Backer O., Gaugler B., Lucas S., Boon T. (1995) J. Exp. Med. 182, 689–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y. T., Boyer A. D., Viars C. S., Tsang S., Old L. J., Arden K. C. (1997) Cytogenet. Cell Genet. 79, 237–240 [DOI] [PubMed] [Google Scholar]
- 9.Cho B., Lim Y., Lee D. Y., Park S. Y., Lee H., Kim W. H., Yang H., Bang Y. J., Jeoung D. I. (2002) Biochem. Biophys. Res. Commun. 292, 715–726 [DOI] [PubMed] [Google Scholar]
- 10.Cho B., Lee H., Jeong S., Bang Y. J., Lee H. J., Hwang K. S., Kim H. Y., Lee Y. S., Kang G. H., Jeoung D. I. (2003) Biochem. Biophys. Res. Commun. 307, 52–63 [DOI] [PubMed] [Google Scholar]
- 11.Shim E., Shim H., Bae J., Lee H., Jeoung D. (2006) Biotechnol. Lett. 28, 515–522 [DOI] [PubMed] [Google Scholar]
- 12.Surjit M., Liu B., Chow V. T., Lal S. K. (2006) J. Biol. Chem. 281, 10669–10681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duxbury M. S., Ito H., Zinner M. J., Ashley S. W., Whang E. E. (2004) Oncogene 23, 465–473 [DOI] [PubMed] [Google Scholar]
- 14.Hong I. K., Jin Y. J., Byun H. J., Jeoung D. I., Kim Y. M., Lee H. (2006) J. Biol. Chem. 281, 24279–24292 [DOI] [PubMed] [Google Scholar]
- 15.Hah N., Lee S. T. (2003) Biochem. Biophys. Res. Commun. 305, 428–433 [DOI] [PubMed] [Google Scholar]
- 16.Herber B., Truss M., Beato M., Müller R. (1994) Oncogene 9, 1295–1304 [PubMed] [Google Scholar]
- 17.Geng Y., Eaton E. N. M., Roberts J. M., Lundberg A. S., Gifford A. (1996) Oncogene 12, 1173–1180 [PubMed] [Google Scholar]
- 18.De Backer O., Arden K. C., Boretti M., Vantomme V., De Smet C., Czekay S., Viars C. S., De Plaen E., Brasseur F., Chomez P., Van den Eynde B., Boon T., van der Bruggen P. (1999) Cancer Res. 59, 3157–3165 [PubMed] [Google Scholar]
- 19.Lucas S., De Smet C., Arden K. C., Viars C. S. B., Lurquin C., Boon T. (1998) Cancer Res. 58, 743–752 [PubMed] [Google Scholar]
- 20.Sadanaga N., Nagashima H., Mashino K., Tahara K., Yamaguchi H., Ohta M., Fujie T., Tanaka F., Inoue H., Takesako K., Akiyoshi T., Mori M. (2001) Clin. Cancer Res. 7, 2277–2284 [PubMed] [Google Scholar]
- 21.Musgrove E. A. (2006) Growth Factors 24, 13–19 [DOI] [PubMed] [Google Scholar]
- 22.Hunter T., Pines J. (1994) Cell 79, 573–582 [DOI] [PubMed] [Google Scholar]
- 23.King R. W., Jackson P. K., Kirschner M. W. (1994) Cell 79, 563–571 [DOI] [PubMed] [Google Scholar]
- 24.Sgambato A., Flamini G., Cittadini A., Weinstein I. B. (1998) Tumori 84, 421–433 [DOI] [PubMed] [Google Scholar]
- 25.Bodrug S. E., Warner B. J., Bath M. L., Lindeman G. J., Harris A. W., Adams J. M. (1994) EMBO J. 13, 2124–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez-Puebla M. L., Robles A. I., Conti C. J. (1999) Mol. Carcinog. 24, 1–6 [PubMed] [Google Scholar]
- 27.Shaulian E., Karin M. (2001) Oncogene 20, 2390–2400 [DOI] [PubMed] [Google Scholar]
- 28.Shaulian E., Karin M. (2002) Nat. Cell Biol. 4, E131–E136 [DOI] [PubMed] [Google Scholar]
- 29.Schreiber M., Kolbus A., Piu F., Szabowski A., Möhle-Steinlein U., Tian J., Karin M., Angel P., Wagner E. F. (1999) Genes Dev. 13, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wisdom R., Johnson R. S., Moore C. (1999) EMBO J. 18, 188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behrens A., Sibilia M., Wagner E. F. (1999) Nat. Genet. 21, 326–329 [DOI] [PubMed] [Google Scholar]
- 32.Vandel L., Montreau N., Vial E., Pfarr C. M., Binetruy B., Castellazzi M. (1996) Mol. Cell. Biol. 16, 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behrens A., Jochum W., Sibilia M., Wagner E. F. (2000) Oncogene 19, 2657–2663 [DOI] [PubMed] [Google Scholar]
- 34.Weitzman J. B., Fiette L., Matsuo K., Yaniv M. (2000) Mol. Cell 6, 1109–1119 [DOI] [PubMed] [Google Scholar]
- 35.Schorpp-Kistner M., Wang Z. Q., Angel P., Wagner E. F. (1999) EMBO J. 18, 934–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakiri L., Lallemand D., Bossy-Wetzel E., Yaniv M. (2000) EMBO J. 19, 2056–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garnovskaya M. N., Mukhin Y. V., Vlasova T. M., Grewal J. S., Ullian M. E., Tholanikunnel B. G., Raymond J. R. (2004) J. Biol. Chem. 279, 24899–24905 [DOI] [PubMed] [Google Scholar]
- 38.Keenan S. M., Lents N. H., Baldassare J. J. (2004) J. Biol. Chem. 279, 5387–5396 [DOI] [PubMed] [Google Scholar]
- 39.Giacinti C., Giordano A. (2006) Oncogene 25, 5220–5227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.