Abstract
Huntington disease (HD) is caused by an expansion of more than 35–40 polyglutamine (polyQ) repeats in the huntingtin (htt) protein, resulting in accumulation of inclusion bodies containing fibrillar deposits of mutant htt fragments. Intriguingly, polyQ length is directly proportional to the propensity for htt to form fibrils and the severity of HD and is inversely correlated with age of onset. Although the structural basis for htt toxicity is unclear, the formation, abundance, and/or persistence of toxic conformers mediating neuronal dysfunction and degeneration in HD must also depend on polyQ length. Here we used atomic force microscopy to demonstrate mutant htt fragments and synthetic polyQ peptides form oligomers in a polyQ length-dependent manner. By time-lapse atomic force microscopy, oligomers form before fibrils, are transient in nature, and are occasionally direct precursors to fibrils. However, the vast majority of fibrils appear to form by monomer addition coinciding with the disappearance of oligomers. Thus, oligomers must undergo a major structural transition preceding fibril formation. In an immortalized striatal cell line and in brain homogenates from a mouse model of HD, a mutant htt fragment formed oligomers in a polyQ length-dependent manner that were similar in size to those formed in vitro, although these structures accumulated over time in vivo. Finally, using immunoelectron microscopy, we detected oligomeric-like structures in human HD brains. These results demonstrate that oligomer formation by a mutant htt fragment is strongly polyQ length-dependent in vitro and in vivo, consistent with a causative role for these structures, or subsets of these structures, in HD pathogenesis.
Keywords: Amyloid, Atomic Force Microscopy, Huntington Disease, Neurodegeneration, Polyglutamine Disease, Protein Folding, Fibril, Oligomer, Protein Aggregation
Introduction
Huntington disease (HD),3 a fatal neurodegenerative disorder, is caused by an expansion of a polyglutamine (polyQ) repeat in the protein huntingtin (htt) (1). This expanded polyQ domain mediates the deposition of cytoplasmic and intranuclear inclusion bodies that contain fibrillar material as determined by electron microscopy of HD brain tissues (2). PolyQ expansions are also responsible for a growing number of less common neurodegenerative disorders, such as the spinocerebellar ataxias (3). One of the most fascinating aspects of polyQ diseases is that the age of onset and severity of disease are tightly correlated with the length of the polyQ expansion. For example, HD repeat lengths of fewer than 35 do not result in disease, 35–39 repeats may or may not cause disease, 40–60 repeats elicits adult onset HD, and more than 60 repeats results in juvenile forms of HD (4–6).
Biochemical analyses of purified mutant htt fragments with polyQ repeats longer than 39Q demonstrated the polyQ length-dependent formation of detergent-insoluble protein aggregates characteristic of amyloid fibrils (7, 8). Synthetic polyQ peptides with expanded polyQ repeats also form amyloid-like fibrils, but surprisingly, in contrast to studies with htt exon 1, synthetic peptides with non-disease-causing polyQ lengths readily aggregated into fibrils (9). In cell culture, mutant htt fragments form intranuclear and cytoplasmic inclusion bodies in a polyQ length-dependent manner (10–12). Thus, polyQ length directly correlates with the kinetics of aggregation into fibrils and formation of inclusion bodies. Mutant htt fragments and other expanded polyQ proteins, as demonstrated with transmission electron microscopy and atomic force microscopy (AFM), also assemble into spherical and annular oligomeric structures similar in size and morphology to those formed by Aβ, α-synuclein, and other proteins that have been implicated in neurodegeneration (13–15). However, whether or not such structures form in a polyQ length-dependent manner has not been rigorously established.
Although inclusion bodies are a major neuropathological hallmark of HD (3), whether or not intraneuronal aggregates mediate HD pathogenesis is still not resolved. Considering the direct correlation between mutant htt aggregation and severity of pathogenesis with polyQ length (7), aggregation was thought to be causative in neurodegeneration. However, how the misfolded/aggregated form(s) mediates pathogenesis is unclear. Although, the onset of symptoms trails inclusion body formation in a transgenic mouse model of HD (16), it might also be a conserved cellular mechanism to protect against diffuse, soluble, toxic forms of mutant htt (11, 17, 18). Indeed, such soluble aggregates that precede symptoms may exist (19). In agreement with this hypothesis, a stable, toxic, monomeric β-sheet conformation of a thioredoxin-polyQ fusion protein was recently reported (20), suggesting that polyQ adopts multiple, distinct conformations that may have toxic consequences even in the absence of higher order aggregation. However, a mutant htt fragment may also misfold into distinct amyloid conformations and, depending on whether the polyQ domain was exposed or buried in a β-sheet, the amyloids can be either toxic or nontoxic, respectively (21). Our recent studies used AFM and other techniques to show that an expanded polyQ domain in a mutant htt fragment adopts a stable multitude of monomeric (22) and oligomeric4 conformations that can readily be discriminated by monoclonal antibodies in vitro and in situ (22).4
Because polyQ length only above a critical threshold (35–40Q) causes HD and is correlated to the onset and severity of the disease, the formation, abundance, and/or persistence of toxic conformers that mediate neuronal dysfunction and degeneration in HD must also be polyQ length-dependent. Here, we sought to systematically quantify the effects of polyQ length on the kinetics, morphologies, and abundance of aggregates formed by a mutant htt fragment in comparison to those formed by synthetic peptides. Because traditional light and electron microscopic approaches have important limitations for studying heterogeneous mixtures of htt aggregates, we used AFM to characterize the morphology of the assembly states. AFM is uniquely well suited to study amyloidogenic proteins. It yields three-dimensional surface maps with nanometer resolution in solution in the absence of artifacts from sample processing. Our aim was to determine the precise relationship between aggregate morphology, kinetics of formation, and polyQ length.
EXPERIMENTAL PROCEDURES
Protein Purification
Glutathione S-transferase (GST)-HD fusion proteins were purified as described (24). Cleavage of the GST moiety by a PreScission Protease (GE Healthcare) initiates aggregation. Fresh, unfrozen GST-HD fusion protein was used for each experiment. Solutions with all fusion proteins were centrifuged at 20,000 × g for 30 min at 4 °C to remove any preexisting aggregates before the addition of the PreScission protease.
Peptide Preparation
All peptides were custom synthesized and obtained from 21st Century Biochemicals (Marlboro, MA). The flanking lysines were included to ensure solubility. Disaggregation and solubilization of polyQ peptides were performed as described (25). Briefly, peptides were dissolved in a 50:50 mixture of trifluoroacetic acid and hexafluoroisopropanol. After vortexing, the solution was incubated at room temperature for 30 min with agitation. The solvent was then evaporated off. The remaining peptide residue was resuspended in water adjusted to pH 3 with trifluoroacetic acid and to a concentration of 0.5 mg/ml. This was in turn diluted into Buffer A (50 mm Tris-HCl, pH 7, 150 mm NaCl).
Atomic Force Microscopy
For experiments on monomeric preparations of mutant htt proteins, GST-HD fusion proteins were incubated at 30 and 2 μm with 1 mm dithiothreitol (DTT) or at 60 μm without DTT in buffer A (50 mm Tris-HCl, pH 7, 150 mm NaCl). PreScission protease (4 units of 100 μg of fusion protein) was added at time 0 to initiate GST cleavage and aggregation. Samples were incubated at 37 °C and 1400 rpm for the duration of the experiment. At 1, 3, 5, 8, and 24 h after cleavage of the GST, a sample (5 μl) of each incubation solution was deposited on freshly cleaved mica (Ted Pella Inc., Redding, CA) and allowed to sit for 30 s. Then the substrate was washed with 200 μl of ultrapure water and dried under a gentle steam of air. For experiments on preformed fibrils, 40 μm solutions of HD53Q were incubated without DTT for 5–6 h after the removal of the GST tag to allow the formation of fibrils.
Each sample was imaged ex situ using a MFP3D scanning probe microscope (Asylum Research, Santa Barbara, CA). Images were taken with silicon cantilever with a nominal spring constant of 40 newtons/m and resonance frequency of ∼ 300 kHz. Typical imaging parameters were drive amplitude, 150–500 kHz with set points of 0.7–0.8 V, scan frequencies of 2–4 Hz, image resolution 512 by 512 points, and scan size of 5 μm. All experiments were performed in triplicate.
For in situ AFM experiments tracking aggregation of monomeric preparations of HD53Q or synthetic peptide, bare substrate was imaged in buffer A, and the sample was injected directly into the fluid cell to reach the final concentration for the experiment. For in situ AFM experiments of preformed HD53Q fibrils, solutions containing preformed fibrils of HD53Q were allowed to rest on mica until several fibrils were present on the surface. Then the substrate was washed with buffer A to remove proteins remaining in solution, and then fresh monomeric preparations of HD53Q that had been incubated with protease on ice for 1 h was injected directly into the fluid cell. All in situ AFM images were taken with V-shaped oxide-sharpened silicon nitride cantilever with a nominal spring constant of 0.5 newtons/m. Scan rates were set at 1–2 Hz with cantilever drive frequencies ranging from ∼8 to 12 kHz. Images were taken with an image resolution of 1024 by 1024 points with scan sizes ranging from 10 to 20 μm. Image analysis of all AFM images was performed with Matlab with the image processing toolbox (Mathworks, Natick, MA) (for more details, see the supplemental material). Volume measurements were partially corrected for error associated with the finite size of the AFM probe based on geometric models (26).
SDS-Agarose Gel Electrophoresis (SDS-AGE) of Purified Mutant Htt Fragments
The analysis of mutant htt fragments by SDS-AGE with Western analysis was performed as described (19). Mutant htt oligomers were resolved by immunoreactions with the EM48 antibody (Chemicon, MAB5374). HD20Q and HD53Q proteins were purified as described (15, 24). Fresh, unfrozen HD53Q and HD20Q proteins were used for all experiments. Before each experiment, HD20Q or HD53Q was centrifuged (20,000 × g) for 30 min at 4 °C to remove pre-existing aggregates. HD20Q or HD53Q were incubated at 12 μm in 20 mm Tris-HCl, pH 8, 150 mm NaCl, 1 mm DTT at 37 °C with shaking at 800 rpm. Solutions of PreScission Protease (4 units/100 μg of fusion protein, Amersham Biosciences) were added at time 0 to initiate the aggregation of HD53Q. At the indicated time points after GST cleavage, equal amounts of protein (15 μg) were removed from the 12 μm aggregation reactions, diluted 1:1 into non-reducing Laemmli sample buffer (150 mm Tris-HCl, pH 6.8, 33% glycerol, 1.2% SDS, and bromphenol blue), and analyzed by SDS-AGE and Western analysis with EM48 (1:1000). Molecular mass standards used in SDS-AGE were ferritin (400 kDa), thyroglobulin (660 kDa), and IgM (1000 kDa).
SDS-AGE of ST14A Cell Homogenates
Complete Httex1p plasmids encoding either a normal range (25Q), intermediate (46Q), or expanded (97Q) polyQ tract with alternating CAG/CAA amino acid repeats were fused in-frame to GFP at the C terminus and subcloned into pcDNA3.1 (Invitrogen) as described (27, 28). All constructs were verified by sequencing and transient transfection into ST14A cells to observe expression using light microscopy (Zeiss AxioObserver.Z1).
Immortalized rat striatal neurons (ST14A cells) were grown in 6-well plates and transiently transfected with Httex1p plasmids (1 μg) with Lipofectamine 2000 (Invitrogen). ST14A cells were grown in Dulbecco's modified Eagle's medium and 10% fetal bovine serum at 33 °C (all reagents from Invitrogen). Each plasmid was transfected in duplicate per experiment, and three independent experiments were performed. At 24 and 48 h post-transfection, cells were lysed in radioimmune precipitation assay buffer (10 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA (from a concentrated stock at pH 8.0), 1% Nonidet P-40, 0.5% SDS) containing Complete Protease Inhibitor (Roche Diagnostics). A DC protein assay (Bio-Rad) was performed to determine protein concentration. Lysate (30 μg) was added in a 1:1 ratio to loading buffer (150 mm Tris, pH 6.8, 33% glycerol, 1.2% SDS) and loaded onto a 1% agarose gel containing 0.1% SDS and run until the dye front had migrated at least 12 cm to allow for maximum resolution of aggregates from the dye front. The proteins were then semidry-blotted (Owl HEP-1) onto a polyvinylidene difluoride membrane in transfer buffer (192 mm glycine, 2 5 mm Tris-base, 0.1% SDS, 15% MeOH). This blot was blocked for 1 h in 5% bovine serum albumin in Tris-buffered saline containing 0.1% Tween 20 at room temperature. The blot was then probed with either EM48 (Millipore, 1:1000) or anti-GFP antibody (Clontech, 1:1000). Peroxidase-conjugated AffiniPure goat anti-mouse secondary (Jackson ImmunoResearch Laboratories) was used at 1:50,000 for 1 h at room temperature. Blots were detected using PICO detection reagent (Pierce).
The same lysates used in the SDS-AGE assays were analyzed by cellulose acetate filter-retardation assays. Lysate (30 μg) was diluted to 200 μl with 2% SDS and filtered through cellulose acetate membrane (Schleicher & Schuell, 0.2-μm pore size) with a Bio-Rad dot blot filtration unit. The assay was performed as described (7). The blot was then probed with primary and secondary antibodies and developed as described above.
SDS-AGE of R6/2 Mouse Brain Homogenates
At 4, 7, and 14 weeks of age R6/2 and wild type cortex were homogenized with 10 volumes of ice-cold sample buffer (100 mm Tris-HCl, pH 7.4, 150 mm NaCl, and a protease inhibitor mixture) by a ball-bearing homogenizer and sonification with 10 pulses/min and 15% power. Samples were either analyzed without centrifugation (total homogenates) or were centrifuged at 20,000 × g for 45 min at 4 °C and were then diluted 1:1 into non-reducing Laemmli sample buffer (150 mm Tris-HCl, pH 6.8, 33% glycerol, 1.2% SDS, and bromphenol blue). Bradford assays were used to determine protein concentration in the homogenates before the addition of Laemmli sample buffer. Total protein (50 μg) was loaded per SDS-AGE well. Mutant htt oligomers were imaged after immunoreaction with EM48 antibody as described above.
Electron Microscopic Analysis of Inclusions
Immunoelectron microscopy (EM) analysis of human HD cortex and MCF-7 cells transiently transfected with FH969–100Q were performed as described (2, 29). Specifically, the cortex was dissected from a HD postmortem brain and fixed in 4% paraformaldehyde in phosphate-buffered saline overnight, immersed in 20–30% sucrose for 1 to 2 days, frozen on dry ice, and stored at −70 °C. The number of CAG repeats identified in the HD allele was 69. Immunoperoxidase labeling was performed by cutting frozen sections (40 μm thick), incubated in 5% normal goat serum (NGS), 1% bovine serum albumin, 0.2% Triton X-100, and 1% H2O2 in phosphate-buffered saline, and then incubated with a primary antibody specific to htt1–17 at a concentration of 1 μg/ml in 5% NGS and 1% bovine serum albumin for 40 h at 4 °C. Control experiments included omission of the primary antibody and preadsorption with 50 μg of N-terminal peptide. Both of these controls resulted in the absence of staining. For analysis of ultrastructure, some immunoperoxidase-labeled sections were embedded in Epon, and thin sections were cut on an ultramicrotome and mounted on Formvar-coated slot grids. MCF-7 cells were cultured onto poly-l-lysine-coated microslips and transfected with FH969–100. Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline 3 days after transfection. Cells were incubated with antibody M5 and horseradish peroxidase-conjugated anti-mouse IgG antibody. The htt bodies were visualized with diaminobenzidine. Cells were postfixed in 2.5% glutaraldehyde, incubated in 1% osmium tetroxide and 1% uranyl acetate, dehydrated in increasing grades of alcohol, and embedded in an ethanol-soluble resin (LX112, LADD). Embedded cells were sectioned (Ultracut E; Reichert-Jung). Both cortex and MCF-7 cells were examined with a JEOL 100CX electron microscope.
RESULTS
Formation of Spherical Oligomers in Vitro Depends on PolyQ Length and Is Transient
In these experiments we used mutant htt fragments with various polyQ repeat lengths (HD20Q, HD35Q, HD46Q, and HD53Q) that were purified from Escherichia coli as a fusion to GST (Fig. 1A) (24). After purification and centrifugation to remove small aggregates that might serve as seeds, GST-HD fusion proteins appeared to be non-aggregated by AFM (data not shown). Cleavage of the GST moiety with a site-specific protease (PreScission protease) released the intact mutant htt fragments, initiating aggregation in a time-dependent manner as reported (8, 15).
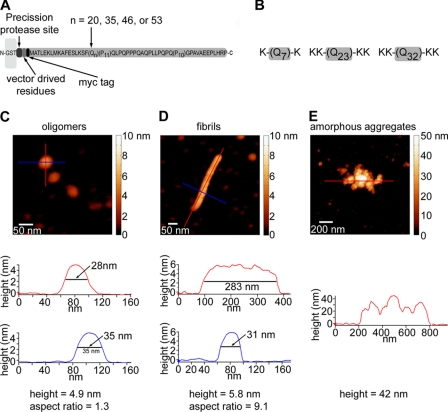
FIGURE 1.
Experimental systems. A, a schematic representation is shown of the GST-htt exon 1 fusion proteins used in this study (HD20Q, HD35Q, HD46Q, or HD53Q). These fusion proteins contain a PreScission protease site located between GST and the htt fragment (not drawn to scale). Cleavage of the GST initiates the aggregation reaction. B, schematics of the synthetic polyQ peptides used in this study are shown. Flanking lysine residues were added to increase solubility. Representative AFM images and aggregate profiles that compare basic dimensions of oligomers (C), fibrils (D), and amorphous aggregates of HD53Q (E) are shown. Height profiles under each image are indicated by colored lines. By using a combination of height and aspect ratio, relative populations of aggregates types can be distinguished and quantified in a heterogeneous mixture.
To determine the effects of polyQ length on the kinetics and morphologies of aggregates formed by a mutant htt fragment, preparations of soluble HD20Q, HD35Q, HD46Q, or HD53Q were incubated at concentrations of 2 and 30 μm and sampled for ex situ AFM analysis at 0, 1, 3, 5, 8, and 24 h after the addition of protease (Fig. 2). To quantify the relationship between oligomers, fibrils, and amorphous aggregates, the number of each aggregate type per μm2 was measured as a function of time at both concentrations (Fig. 3). Oligomers were defined as 2–10 nm in height with an aspect ratio (longest distance across to shortest distance across) less than 2.5, indicating a globular structure. Fibrils were defined as aggregates greater than 4 nm in height that had an aspect ratio greater than 2.5. Amorphous aggregates were defined as any aggregate greater than 10 nm in height that did not have any obvious fibrillar morphology. These criteria were based on measured characteristics of representative examples of each respective aggregate type (Fig. 1, C–E). The number of fibrils in a bundle was estimated by counting the number of fibrils visible in the bundle.
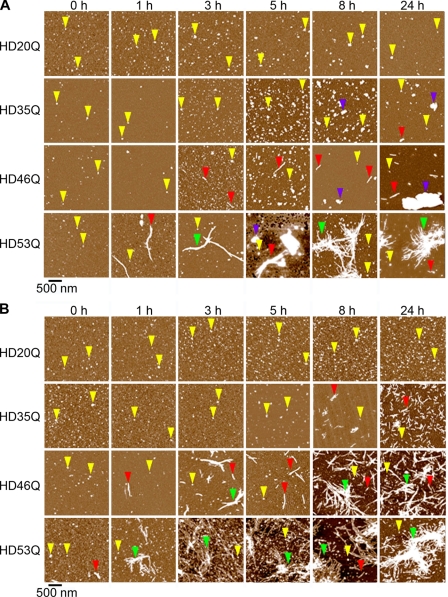
FIGURE 2.
Oligomers and fibrils formed by a mutant htt fragment depend on polyQ length and concentration. AFM images demonstrate the aggregation of mutant htt fragments with polyQ repeats of 20Q, 35Q, 46Q, or 53Q. Incubations with protein concentrations of 2 μm (A) and 30 μm (B) were imaged at different time points after the addition of protease. Examples of oligomers, fibrils, fibril clusters, and large amorphous aggregates are indicated by yellow, red, green, and blue arrows, respectively. Although oligomers were observed for HD20Q, fibrillar aggregates were not. For mutant htt fragments with longer polyQ repeats, heterogeneous mixtures of aggregate types appeared at later times. The appearance of fibrillar aggregates of HD35Q was concentration-dependent. At 2 μm, HD35Q began to form large amorphous aggregates but at 30 μm HD35Q formed a large number of short fibrils. At late times, fibrils and fibril bundles were the dominant morphology for HD46Q and HD53Q, with fibrils appearing earlier for HD53Q at both concentrations.
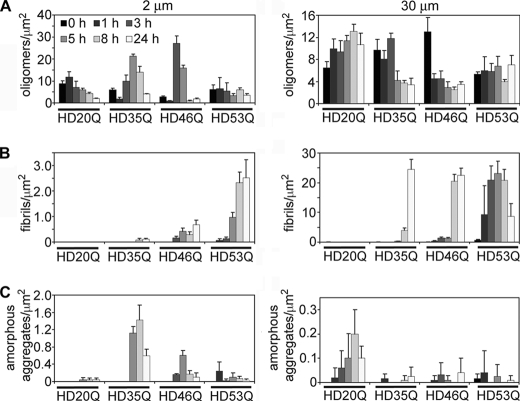
FIGURE 3.
Aggregate species of mutant htt fragments change temporally in a polyQ length- and concentration-dependent manner. Quantification of aggregate types observed in AFM images for incubations HD20Q, HD35Q, HD46Q, and HD53Q at 2 μm and 30 μm. The numbers of oligomers (A), fibrils (B), and amorphous (C) aggregates per μm2 were calculated for all images in Fig. 2. The appearance of fibrils occurred earlier for longer polyQ repeat lengths. Although HD20Q formed oligomers at both concentrations, fibrils were not observed. For HD35Q, a peak population of oligomers preceded the formation of amorphous aggregates (2 μm) or fibrils (30 μm). A peak population of oligomers also preceded the appearance of fibrils of HD46Q at both concentrations. As fibrils of HD53Q appeared early in the experiment, a peak oligomer population preceding the appearance of fibrils was only observed if the kinetics of aggregation reactions were decreased (see supplemental Fig. 2).
Oligomeric aggregates were observed for all htt fragments studied, including HD20Q. As reported (7, 8), the appearance of fibrils was polyQ length and concentration-dependent, with fibrils appearing earlier, growing longer, and forming bundles with longer polyQ lengths and higher concentrations. Quantification of distinct aggregate types per unit area (Fig. 3) demonstrated that aggregation results in a complex mixture of aggregate types at any given time for mutant htt fragments. Oligomers were detected at both concentrations of HD20Q but gradually dissipated (2 μm) or remained relatively stable (30 μm) in number, depending on concentration. Importantly, fibrils were not observed for HD20Q at any time, unlike those observed with longer polyQ lengths (see below). For both concentrations of HD20Q, low amounts (< 0.3 aggregates/μm2) of amorphous aggregates were observed relative to other aggregate types.
For HD35Q, a significant number of large amorphous aggregates (>1.0 aggregates/μm2) and very few (<0.1 fibrils/μm2) short putative fibrils formed at 2 μm after 8–24 h of incubation (Figs. 2 and 3). Short fibrillar species became the predominant aggregate type for HD35Q incubated at 30 μm after 8 h. The appearance of amorphous aggregates of HD35Q at 2 μm and fibrils at 30 μm was preceded by a peak population of oligomers, which quickly dissipated with time as the amorphous or fibrillar aggregates increased in number. The resultant distinct aggregate types of HD35Q (i.e. amorphous aggregates versus fibrils) (supplemental Fig. 1) that coincide with the dissipation of oligomers under these conditions suggest competing, concentration-dependent aggregation pathways for mutant htt fragments. Furthermore, the amorphous aggregates have a granular appearance, suggesting they may be composed of small oligomers that have accumulated together. This is consistent with the corresponding disappearance of the discrete oligomer population when these amorphous aggregates begin to appear.
For HD46Q, fibrils appeared after 3 and 1 h of incubation for 2 and 30 μm, respectively. However, the HD46Q fibrils were much longer and had a higher tendency to form bundles at 30 μm than at 2 μm incubations (Figs. 2 and 3). Similar to HD35Q, the appearance of HD46Q fibrils was preceded by a peak population in oligomers that dissipated with time coincident with the accumulation of fibrils.
Incubations of HD53Q resulted in fibrils and fibril bundles at both concentrations, with these structures appearing after 1 and 3 h, respectively, for 2 μm incubations and 0 and 1 h, respectively, for 30 μm incubations (Figs. 2 and 3). Surprisingly, an increase in the oligomer population before the appearance of fibrils was not observed for HD53Q under these experimental conditions. Rather, oligomer levels remained relatively stable over time even though the number of fibrils steadily increased. We hypothesized that the lack of a transient oligomer peak for HD53Q resulted from the rapid kinetics of fibril formation in this experiment and that the peak oligomer population may have already occurred by the time the reaction buffer was sampled by AFM.
To test this hypothesis, we changed conditions of the aggregation reactions. Our previous experiments were all performed with the reducing agent DTT in the reaction buffer. However, in independent experiments we serendipitously found that the kinetics of fibril formation by mutant htt fragments was much slower when DTT was omitted from the reaction buffer, apparently by reducing the efficiency of cleavage of GST from htt due to a single cysteine residue in vector-derived residues located between GST and htt (data not shown). Analysis of 60 μm HD53Q in the absence of DTT resulted in the formation of short fibrils only after 3 h of incubation (supplemental Fig. 2A). Quantification of the number of oligomers or fibrils per μm2 showed a gradual increase in oligomers (supplemental Fig. 2B) that peaked at 3 h, coinciding with the appearance of fibrils (supplemental Fig. 2C). After 3 h, the oligomer population steadily decreased, whereas the number of fibrils remained relatively constant. In agreement with the results with HD35Q and HD46Q, formation of HD53Q oligomers preceded fibril formation. Moreover, as fibrils appeared, oligomers gradually dissipated, indicating that oligomers were destabilized in the presence of fibrils, incorporated into fibrils, or were direct precursors of fibrils.
These results indicate that, although a mutant htt fragment with a non-pathological polyQ length (20Q) forms oligomers, these oligomers do not structurally transition to fibrils, as observed with mutant htt fragments that contain polyQ repeats in the pathological range (>35Q). These results suggest that these oligomers may have a different structure than those formed with disease-causing polyQ repeat lengths.
Oligomers Formed by a Mutant htt Fragment Are Highly Heterogeneous in Size and Subunit Composition
We next quantified height and volume distributions for oligomers formed by each mutant htt construct (2 μm) as a function of time (supplemental Fig. 3). Volume measurements were partially corrected for the contribution due to the finite size of the probe (26). Although the size distributions for HD20Q oligomers were relatively constant during aggregation (mode of height ∼2–4 nm and volume ∼400–750 nm3, respectively), distributions for oligomers, formed by mutant htt fragments with >35Q, became larger and more heterogeneous with increasing time (mode of height ∼6–9 nm and volume ∼900–1200 nm3). Using an independent analysis, we used the corrected volume distributions of oligomers to estimate the number of protein molecules per oligomer for HD20Q, HD35Q, HD46Q, or HD53Q, respectively (Fig. 4) (for details on this analysis see supplemental Experimental Procedures). Collectively, these analyses indicate that oligomeric species with a broad size range are observed only with longer polyQ repeats and that oligomers formed in vitro are highly heterogeneous and change in subunit composition over time.
FIGURE 4.

Oligomers formed by mutant htt fragments are highly heterogeneous in size. Based on corrected volume measurements (see supplemental Fig. 3) and the molecular mass of each htt exon 1 fragment, the numbers of molecules per oligomer observed for 2 μm incubations of HD20Q, HD35Q, HD46Q, or HD53Q were calculated for each time point of aggregation from Fig. 2. The plots are color-coded such that darker colors represent a greater abundance of oligomers composed of that number of molecules. Black arrows indicate where ∼400 kDa oligomers would be observed, and the maximum size in kDa is represented in the plots as marked. Based on this analysis, htt exon 1 fragments form a mixture of oligomers ∼400 kDa and larger, similar to what was observed by SDS-AGE in vitro and in vivo (see Fig. 7).
Mutant htt Fragments with Expanded PolyQ Repeats Form Discrete Annular Aggregates
Although oligomeric and fibrillar structures account for the vast majority of aggregates in the incubation studies, other discrete annular structures were also occasionally observed (supplemental Fig. 4). These structures were exceedingly rare, were not observed in all incubations, and only appeared in incubations of HD46Q and HD53Q. Large annular structures (supplemental Fig. 4, A and B) with inner diameters of ∼30–130 nm were the most common. For HD46Q and HD53Q, they appeared at ∼1 per 500 μm2 when imaged by AFM. By contrast, spherical oligomers appeared at a rate of 10,000 per 500 μm2. Smaller annular aggregates (supplemental Fig. 4, C and D) observed for HD46Q and HD53Q with inner diameters of ∼10 nm were detected even more rarely. Each of their five globular subunits was comparable in size and morphology to spherical oligomeric aggregates, suggesting that some oligomers might be able to organize into these structures. Because of the limited number of observations of both annular structures, however, it was impossible to systematically quantify the effects of polyQ length on their formation, except that they were never observed for HD20Q or HD35Q.
Tracking the Fate of HD53Q Aggregates in Solution
To directly observe the fate of individual aggregates formed by a mutant htt fragment in solution, in situ AFM experiments were performed with HD53Q (Fig. 5). Fresh monomeric preparations of purified HD53Q were injected directly into the fluid cell of the AFM at a final concentration of 2 μm and imaged continuously (Fig. 5A). Within 9 min after injection, a diffuse film of soluble protein and several distinct oligomers (Fig. 5A, yellow arrows) appeared on the surface, entirely covering the mica surface. The number of oligomers gradually increased over time as more oligomers formed in solution and were deposited onto the surface (Fig. 5B). The oligomeric structures did not exhibit lateral mobility, as commonly observed for other amyloid forming peptides, such as Aβ (30, 31), perhaps due to their affinity for the protein film on the surface. The vast majority of oligomers were stable, neither desorbing from the surface nor seeding the appearance of fibrillar aggregates during the time course of the experiment. On rare occasions, oligomers appeared to seed the formation of a small fibril (Fig. 5A, red arrows). However, most fibrils appeared in the absence of oligomeric precursors (Fig. 5A, blue arrows). These fibrils may have formed from oligomers in solution before depositing onto the mica surface. The fibrils observed were quite short (<200 nm) and much less abundant than oligomers (Fig. 5B).
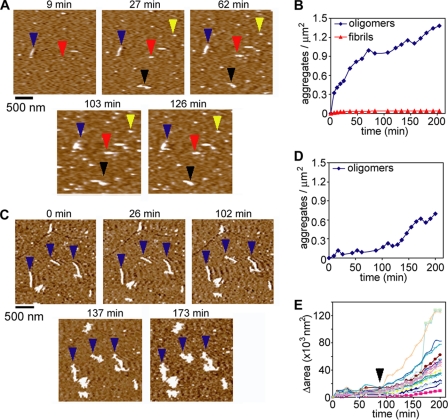
FIGURE 5.
A mutant htt fragment forms oligomers in solution. A, monomeric preparations of HD53Q were imaged in solution on mica at a concentration of 2 μm. The yellow arrows indicate oligomers that are stable throughout the experiment. The red arrows indicate an oligomer that coalesced into a fibril. The blue and black arrows indicate fibrils that appeared without a clear oligomeric precursor. B, the number of oligomers and fibrils per unit area was determined as a function of time. Oligomers appeared on the surface by 9 min with a steady increase in abundance over time. The vast majority of these oligomers did not further aggregate into fibrillar structures. Very short (<200 nm in length) fibrillar structures also appeared within 9 min. Although the number of fibrils did increase slightly, they were far less abundant than oligomers. C, HD53Q was aggregated to a fibrillar state, deposited on mica, and imaged in solution. Next, a monomeric preparation of HD53Q (2 μm) was injected into the fluid cell. D, the number of oligomers per unit area was determined as a function of time, illustrating that fewer oligomers formed in the presence of fibrils. E, the surface area occupied by individual fibrils was monitored as a function of time. Initially, fibrils appeared to elongate predominately at the fibril ends, resulting in a slight increase in occupied surface area. However, ∼88 min (marked by black arrowhead) after the addition of HD53Q monomer, branching appeared on several fibrils, increasing the rate of change in occupied surface area. The fibrils then appeared to accumulate soluble HD53Q as the fibrils expanded in all directions (also see supplemental Movie 1).
We next performed an order of addition experiment in which purified HD53Q monomers were added to preformed HD53Q fibrils previously immobilized on the mica surface (Fig. 5C and supplemental Movie 1). Control experiments demonstrated that conditions for these experiments did not appear to alter or damage pre-existing fibril morphology for up to 6 h of direct imaging (not shown). HD53Q was incubated at 37 °C until fibrils were formed. These fibrils were deposited on mica and washed with buffer to remove any residual HD53Q monomer and/or aggregates remaining in solution. Next, a freshly purified 2 μm HD53Q monomeric preparation, which had been incubated with protease on ice for 1 h, was added to the fluid cell. Initially, elongation of individual preformed fibrils occurred gradually from both fibril ends. This is indicated by tracking the change in surface area of individual fibrils that gradually increased due to this elongation (Fig. 5E). However, ∼80–120 min after the addition of HD53Q monomer, branching appeared along the fibrils both at the ends where elongation first appeared and from their periphery, resulting in an accelerated increase in fibril area (indicated by the black arrow in Fig. 5E). The fibrils then appeared to accumulate soluble HD53Q around their entire periphery both as fibrillar branches and some amorphous clumping, suggesting that large amorphous aggregates may have underlying fibrillar structure (supplemental Movie 1). In the presence of fibrils, the formation of oligomeric species was suppressed (Fig. 5, compare D and B), indicating that there was competition between soluble HD53Q-forming oligomers and incorporation into and/or accumulation around the preformed fibrils.
Synthetic PolyQ Peptides Form Oligomers and Fibrils in Solution with Faster Kinetics Than Observed with Mutant htt Exon 1
In studies by Wetzel and co-workers (9, 25, 32–35), synthetic polyQ peptides aggregate into fibrils in a polyQ length-dependent manner. Unlike htt exon 1, even non-pathological polyQ lengths readily formed fibrils. Surprisingly, oligomeric structures were apparently never observed in these studies. More recently, they added sequences that normally flank the polyQ domain in htt exon 1 to synthetic polyQ peptides (36) to partially mimic the htt exon 1. They stated that these flanking sequences imparted a novel aggregation pathway on the polyQ peptide in which it formed oligomers similar to htt exon 1 and that these sequences were in fact necessary for oligomer formation (i.e. that synthetic polyQ peptides alone could not form oligomers under any conditions).
We examined the effects of polyQ length on the real-time aggregation of synthetic peptides by in situ AFM to determine what structures these simple peptides could form and to compare these results directly with those we obtained using authentic htt exon 1. Freshly prepared solutions of completely solubilized synthetic peptides of K-Q7-K, KK-Q23-KK, and KK-Q32-KK were injected directly into the AFM fluid cell to a final concentration of 20 μm and imaged continuously on mica (supplemental Fig. 5A). A few oligomers of K-Q7-K appeared initially and steadily increased in abundance (supplemental Fig. 5B). Short, metastable fibrils appeared as early as 25–50 min after the addition of K-Q7-K. These early fibrils would often quickly disappear; however, stable fibrils eventually formed (beyond 150 min). Despite this dynamic fibril stability, the total number of fibrils detected at any given time increased steadily but at a slower rate than oligomers (supplemental Fig. 5B). For K-Q7-K, oligomers remained the dominant aggregate morphology. KK-Q23-KK initially aggregated into a large number of oligomers (supplemental Fig. 5, A and B), with a small population of short but stable fibrils appearing within 17 min. This fibril population steadily increased. By 44 min, large bundles of KK-Q23-KK fibrils appeared that continued to accumulate and grow in size, often branching in many directions. Fibrils eventually became the dominant morphology of KK-Q23-KK aggregates, with oligomers reaching a peak population at 71 min before gradually beginning to dissipate. When freshly solubilized preparations of the KK-Q32-KK peptide were imaged in solution (supplemental Fig. 5A and Movie 2), small oligomeric and short putative fibrillar structures appeared on the surface within a few min (supplemental Fig. 5B). For higher resolution AFM images of KK-Q32-KK oligomers and putative fibrils, see supplemental Fig. 6. With time, more aggregates of various types (i.e. oligomers, fibrils, and fibril bundles) appeared on the surface; however, fibrillar structures dominated the aggregate morphology by 35 min. For KK-Q32-KK, oligomers quickly reached a peak population before dissipating, with a much sharper transition than KK-Q23-KK (supplemental Fig. 5B), reminiscent of studies with a mutant htt fragment with longer polyQ lengths (Fig. 3). Once fibrils were established on the surface, most of the new peptide appearing on the surface was associated with these fibrillar structures (supplemental Fig. 5C), which was also similar to the observations for HD53Q monomer addition to preformed fibrils described above (Fig. 5, C–E). The total amount of aggregation was quantified for each peptide as the percent surface area occupied by aggregates as a function of time (supplemental Fig. 5C). Based on this analysis, the rate of total aggregation correlated with polyQ length. For KK-Q23-KK and KK-Q32-KK, an initial lag phase preceded acceleration in aggregation, and the acceleration was more pronounced for KK-Q32-KK.
We observed two apparent mechanisms for the disappearance of oligomers; oligomers appeared to be nucleation sites for fibril growth or were incorporated into adjacent fibrillar structures. However, the relative stability of individual oligomers varied greatly. We tracked the fate of 66 oligomers of KK-Q32-KK that first appeared at 26 min (20 μm peptide concentration) to determine how long each remained in the oligomeric state (supplemental Fig. 5D). Half of the oligomers had nucleated fibril growth within 18 min and 71% within 50 min. However, nine oligomers were stable for longer than 150 min, with three lasting more than 200 min. Varied oligomer stability and dissipation mechanisms are further illustrated by time-lapse sequences of AFM images (supplemental Fig. 7). Although most oligomers nucleated fibril growth within minutes of appearing (supplemental Fig. 7, red arrows), other oligomers remained stable for extended periods (>200 min) (supplemental Fig. 7, black arrows). Some apparently stable oligomers were eventually assimilated by adjacent fibrillar structures that grew directly onto the stable oligomers (supplemental Fig. 7, blue arrow), incorporating them into their structure. The varied stability of individual oligomers suggests that these structures have to overcome an energy barrier to a structural rearrangement to initiate fibril growth, allowing some oligomers to remain relatively stable for extended times. These results are also consistent with the observed heterogeneity in the oligomer size for mutant htt fragments (Fig. 4), which may underlie their relative differences in stability.
We also tracked elongation and growth once individual fibrils appeared in the KK-Q32-KK experiment (supplemental Fig. 8). Although some fibrils arose from clear oligomeric precursors, most initially appeared as intact fibrils (supplemental Fig. 8, A and B). Fibrils grew in various directions, and individual fibril structures often exhibited different growth patterns and morphologies at different times. Elongation often occurred from fibril ends, but extensive branching, resulting in elongation in multiple directions, was common (supplemental Fig. 8C). Often a single elongated fibril would exhibit variable height/width along its contour. With time, the shorter/thinner regions of fibrils transitioned to resemble the taller/thicker regions. Fibrils grew together, forming a large network of intertwining fibers.
In situ AFM experiments with KK-Q32-KK were also carried out at a concentration of 5 μm and directly compared with the aggregation of HD53Q at the same concentration (Fig. 6A). At this concentration, the number of KK-Q32-KK oligomers and fibrils initially increased at similar rates (Fig. 6B), but as the number of fibrils continued to increase, oligomers again dissipated. The transition was not as sharp as in the 20 μm experiment and occurred at a later time (∼80 min). This was in stark contrast with HD53Q aggregation in which oligomer formation dominated the entire duration of the experiment with only a few putative fibrils forming (Fig. 6, A and B). Large amorphous aggregates of KK-Q32-KK appeared that were not seen at higher concentrations. Nevertheless, fibrils were still the dominant morphology observed. The excess solution of KK-Q32-KK (i.e. that was not injected into the fluid cell) was sampled after 5 h of preparation and imaged ex situ (Fig. 6C). Interestingly, the observed mixture of aggregate morphologies consisted of oligomers and two distinct types of fibrils: shorter (∼1–2 nm tall, red arrows) and taller (∼5–7 nm tall, blue arrows) fibrils. This was in contrast to experiments with the mutant htt fragment in which only a single type of fibril morphology was observed (Fig. 2). Thus, our studies show synthetic polyQ peptides can clearly form on oligomeric structures even in the absence of flanking htt exon 1-like sequences albeit with much faster kinetics than htt exon 1 that may have eluded their previous identification. However, no annular morphologies were ever observed in experiments with synthetic polyQ peptides.
FIGURE 6.
The synthetic polyQ peptide KK-Q32-KK forms oligomers with faster kinetics than a mutant htt fragment. A, in situ AFM experiments were performed with 5 μm HD53Q and KK-Q32-KK. B, the number of oligomeric and fibrillar aggregates formed by HD53Q and KK-Q32-KK was measured as a function of time. For KK-Q32-KK, the number of oligomers peaked (∼80 min) before dissipating, as the number of fibrils continued to increase. Large amorphous aggregates of KK-Q32-KK also appeared. Under these conditions, HD53Q only formed oligomers, which steadily increased in number as a function of time. C, ex situ AFM image of KK-Q32-KK sampled 5 h after incubation at 5 μm demonstrating that the peptide formed a mixture of aggregates consisting of oligomers (yellow arrows) and two distinct types of fibrils: narrower fibrils (∼1–2 nm tall, red arrows) and taller fibrils (∼5–7 nm tall, blue arrows).
A Mutant htt Fragment Forms Oligomers in a PolyQ Length-dependent Manner in Neurons and Brains
Although our analyses indicate mutant htt fragments form oligomers in a polyQ length-dependent manner under cell free conditions, we wanted to determine whether they exhibited similar properties in cultured neurons, brains from a transgenic mouse model of HD, and HD patient brains. Oligomers, however, in the absence of purification procedures that could potentially introduce artifacts cannot be readily identified in cell or brain homogenates by AFM due to the high background of other cellular contaminants. Therefore, we used SDS-AGE with Western analysis to directly compare oligomers formed by a mutant htt fragment under cell-free conditions to those in total homogenates from neurons and brains. Although this technique was used recently to resolve oligomers formed by mutant htt fragments from yeast (37) and mammalian systems (19), it has not been used under cell-free conditions nor to systematically analyze the effects of polyQ length dependence on oligomer formation.
We first incubated purified GST-HD20Q and GST-HD53Q in the absence and presence of protease at 12.5 μm for 72 h. SDS-AGE with Western analysis probed with an antibody raised to the N terminus of htt (EM48) (38) indicated that, only in the presence of protease, HD53Q, but not HD20Q, formed oligomeric species that fractionated as a smear above 400 kDa (Fig. 7A), similar in size to those described in cultured cells and brains from HD mouse models (19). The apparent molecular masses of these oligomers ranged from ∼400 kDa to several mDa (supplemental Fig. 9). HD53Q oligomers initially increased up to 8 h and were relatively stable from 8–48 h but decreased in abundance at 72 h of incubation.
FIGURE 7.
A mutant htt fragment forms oligomers in a polyQ length-dependent manner in neurons and brains. A, SDS-AGE with Western analysis indicates that a mutant htt fragment (HD53Q) forms oligomers that migrate as a smear above ∼400 kDa when detected with an anti-htt antibody (EM48). B and C, a mutant htt fragment forms oligomers in a polyQ length-dependent manner in ST14A cells similar in apparent size to those formed under cell-free conditions. Note that oligomers formed by HD97Q-GFP are less heterogeneous in size than those formed with shorter polyQ lengths but that the intensity of oligomers as detected by antibody reactivity increases as a function of time in ST14A cells. D, a filter-trap retardation assay shows that the total amount of SDS-insoluble aggregates increases as a function of polyQ length and time in ST14A cells that express a mutant htt fragment. E and F, a mutant htt fragment forms oligomers in total brain homogenates from R6/2 mice that are similar in apparent size to those formed under cell-free conditions. Note that oligomers formed by a mutant htt fragment with 220Q are less heterogeneous in size than those formed with shorter polyQ lengths and that the intensity of antibody reactivity also is significantly decreased in R6/2 mice with 220Q. WT, wild type. G and H, SDS-AGE with Western analysis on total brain homogenates show that a mutant htt fragment forms oligomers that significantly increase as a function of time in R6/2 mice with 110Q. I, SDS-AGE with Western analysis on the supernatant fraction of a total brain homogenate after centrifugation shows that a mutant htt fragment forms soluble oligomers that increase as a function of time in R6/2 mice with 110Q. *, p < 0.05; ***, p < 0.0001 (one-way analysis of variance).
We next used SDS-AGE with Western analysis to analyze total homogenates from an immortalized cell line (ST14a) derived from rat striatal neurons transiently transfected with plasmids that express GFP alone or mutant htt exon 1 with 25Q, 46Q, or 97Q fused at the C terminus to GFP. Homogenates from cells that expressed HD46Q-GFP and HD97Q-GFP, but not GFP or HD25Q-GFP, formed oligomers that migrated as smears at an apparent molecular mass above 400 kDa (Fig. 7, B and C), similar to those formed by HD53Q in vitro, although the apparent size distribution was broader in cell homogenates (supplemental Fig. 9). Interestingly, oligomers from cell homogenates that expressed HD97Q-GFP showed a smaller and less heterogeneous apparent size distribution than from HD46Q-GFP homogenates. As HD97Q-GFP-expressing cells form significantly more inclusion bodies than HD46Q-GFP cells (data not shown), we hypothesize that the narrower size distribution observed in HD97Q-GFP-expressing cells is due to a greater propensity for mutant htt exon 1 with this polyQ repeat length to form fibrils, as observed in our in vitro studies. However, oligomers formed by HD46Q-GFP and HD97Q-GFP continued to accumulate with incubation time (Fig. 7, B and C). A similar result was observed in a filter retardation assay that detects all SDS-insoluble aggregated material greater or equal to the pore size (0.2 μm) of the membrane (Fig. 7D).
We next performed SDS-AGE and Western analysis using total brain homogenates from R6/2 mice, a transgenic mouse model of HD that expresses mutant htt exon 1 (16, 39), and non-transgenic littermate controls. We analyzed homogenates from three independent transgenic lines of R6/2 mice that expressed mutant htt exon 1 with 110Q, 160Q, or 220Q repeats. As observed in vitro and in ST14a cells, mutant htt exon 1 formed oligomers in R6/2 mouse brains that migrated above 400 kDa as detected by the EM48 antibody (Fig. 7E). Homogenates from 220Q mice had the narrowest apparent size distribution and a significantly lower level of antibody staining (Fig. 7, E and F, and supplemental Fig. 9) when compared with R6/2 mice with 110Q or 160Q despite the rapid and widespread distribution of inclusion bodies in these mice (40). As described (19), oligomers in R6/2 brain homogenates appeared to increase in a time-dependent manner (Fig. 7, G and H). Because the oligomeric species we detected in SDS-AGE could also be derived in part from fibrillar species, we performed SDS-AGE with Western analysis with the supernatant fractions of brain homogenates after centrifugation at 20,000 × g, which sediments all fibrillar material (15). SDS-AGE of these samples indicated mutant htt exon 1 forms soluble, non-sedimentable oligomers in a time-dependent manner (Fig. 7I). Similar results were obtained after centrifugation of recombinant HD53Q (data not shown). Finally, a direct comparison of recombinant HD53Q oligomers with those from neurons and R6/2 brain homogenates demonstrated that oligomers fractionate in SDS-AGE to similar apparent molecular masses ranging from 400 kDa to several mDa in size (supplemental Fig. 9).
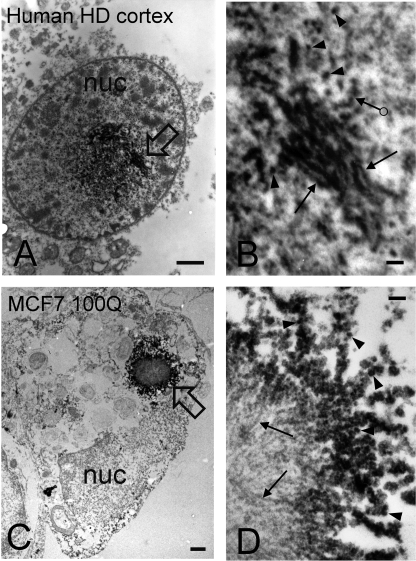
We next used EM to examine the size and morphology of mutant htt inclusions formed in cortical neurons from a postmortem HD brain (69Q in the HD allele) (Fig. 8, A and B) and in MCF7 cells (a human breast adenocarcinoma cell line) transfected with a mutant htt fragment containing 100Q (Fig. 8, C and D). After immunoperoxidase labeling with a primary antibody to htt1–17 (2), mutant htt inclusions occupying large areas within nuclei of cortical neurons from the HD brain were readily observed (Fig. 8A, black arrow). Such structures were never observed in control, age-matched brains (data not shown). At higher resolution (Fig. 8B), these inclusions appeared to be comprised of a heterogeneous mixture of aggregates consisting of granular/oligomeric (arrowheads) and fibrillar (arrows) morphologies, with similar dimensions to oligomers and fibrils observed by AFM with mutant htt exon 1 under cell-free conditions (Fig. 1). In a MCF7 cell transfected with Flag-htt1–969-100Q and incubated with antibody M5, a large cytoplasmic htt inclusion appeared adjacent to the nucleus (Fig. 8C). This htt inclusion was composed of a core of unlabeled fibrils (Fig. 8D, arrows) surrounded by a periphery of immunoreactive granular/oligomeric species (Fig. 8D, arrowheads). Importantly, the granular/oligomeric structures observed by EM in both cortical neurons and transfected MCF7 cells were similar in lateral dimension to the oligomers observed by AFM. Many of the granular/oligomeric structures observed by EM in both the cortical neurons (Fig. 8B, ringed arrow) and in MCF7 cells (Fig. 8D, arrowheads) appeared to be laterally associated, similar to beads on a string.
FIGURE 8.
Mutant htt forms oligomers in HD brains. Mutant htt was detected with EM using the immunoperoxidase method and a primary antibody to htt1–17. Analysis was performed with a transmission electron microscope (JEOL 100CX). The peroxidase reaction product appears as electron dense label. A and B, electron micrographs are from a cortical neuron in human postmortem HD brain. The CAG repeats were 17 and 69. Note that the labeled inclusion occupies a large area at the center of the nucleus (nuc). The composition of the inclusion is highly heterogeneous consisting of granular/oligomeric and fibrillar structures. Stacks of labeled fibrils are present at the upper part of the inclusion and along the right at the open arrow, which is magnified in B. In B, note the presence of small, labeled granular/oligomeric structures of different sizes (arrowheads) that are similar in lateral dimensions to those observed by AFM analysis of mutant htt fragments in vitro. Fibrils (arrows) and structures that resemble beads on a string (ringed arrow) are also indicated. C and D, electron micrographs of an MCF7 cell transfected with Flag-htt1–969-100Q. In C, a large cytoplasmic inclusion appears adjacent to the nucleus (nuc) at the open arrow. A portion of the inclusion is shown at higher magnification in D. The inclusion consists of a core of unlabeled fibrils (arrows) and a periphery of immunoreactive granular/oligomeric structures (arrowheads). Some of the granules/oligomers form linear arrays (“beads on a string”) similar to that observed in the human HD brain (B, ringed arrow). Scale bars on left panels = 1 μm and on right panels = 100 nm.
DISCUSSION
In this study we present several findings. First, we provide direct evidence that a mutant htt fragment and synthetic polyQ peptides readily form spherical oligomers. Second, we show that oligomer formation precedes fibril formation, that oligomers quickly disappear as fibrils form, and that this transition appears to be polyQ length- and concentration-dependent. Third, we show that mutant htt fragments with expanded polyQ domains, but not synthetic polyQ peptides, can form a variety of discrete annular structures. Fourth, we show that on rare occasions spherical oligomers can be direct precursors of fibrils; however, the individual fate, stability, and size of oligomers vary widely, suggesting different underlying structures of these aggregates. Fifth, we show a mutant htt fragment forms oligomers in a polyQ length-dependent manner in an immortalized striatal cell line and in brain homogenates from a mouse model of HD that are similar in apparent size to those formed in vitro. Finally, we show by immunoelectron microscopy that oligomeric-like structures are readily detected in HD brains. Collectively, these results underscore the complex nature of polyQ aggregation and the importance of polyQ length and amino acid context on the kinetics and morphologies of mutant htt aggregation. As the length of the polyQ expansion in htt is tightly correlated with disease onset and severity in HD, the results we present are consistent with the hypothesis that oligomers or subsets of oligomers may play a causative role in HD pathogenesis.
Based on these and previous studies, we propose a model of mutant htt aggregation (Fig. 9). In this model, aggregation initiates from monomers that are continuously sampling multiple, distinct conformations. By analogy to a freely jointed homopolymeric chain, one can speculate that the relative number and stability of these conformers would depend on polyQ length and that a subset of these conformers might seed higher order aggregation pathways. Although our previous studies with monoclonal antibodies strongly implicate multiple monomeric polyQ conformations in a mutant htt fragment (22), Kim et al. (41) recently provided direct structural evidence of this at near-atomic resolution. These results suggest that, at least in principle, different monomeric conformations might lead to oligomers that also possess different structures, perhaps mediated by different hydrogen-bonded networks between polyQ residues (42). Fibril formation appears to proceed via two distinct pathways; 1) direct nucleation from a monomer or small multimer and 2) one with a higher order, spherical, oligomeric intermediate. Although it is difficult to determine whether either pathway is dominant, these pathways do not appear to be mutually exclusive, and at least at high protein concentrations, monomer addition to preformed fibrils appears to predominate.
FIGURE 9.
A model for the misfolding and aggregation of a mutant htt fragment. A native monomer can continuously misfold to sample distinct monomeric conformers. The relative number and stability of these conformers may be polyQ length-dependent. Some of these misfolded monomers potentially can lead to off-pathway aggregates, such as large annular aggregates. Others can form globular oligomeric aggregates. The size and stability of oligomeric aggregates can vary widely and are also polyQ length-dependent. These oligomers can further self-assemble or accumulate into other aggregate morphologies, such as small annular aggregates composed of oligomeric subunits, large amorphous aggregates, and fibrils. In this model a major structural transition must occur within an oligomer to initiate fibril elongation. Once this transition occurs, oligomers and monomers can directly accumulate on the fibril, causing fibril elongation. Many of these higher order aggregates can accumulate together to form large aggregates that make up inclusions.
Our AFM and biochemical analyses indicate that oligomers are highly heterogeneous in size. The stability of oligomeric aggregates is also highly variable, and the transition from oligomeric to fibrillar aggregates occurs with faster kinetics at longer polyQ lengths. Interestingly, it appears that a thermodynamically driven structural transition is necessary for an oligomer to nucleate fibril formation. Our data also suggest oligomers form and may be direct components of other higher order assemblies, such as annular structures and large amorphous aggregates. Once formed, fibrils often appear to branch while simultaneously accumulating monomer, oligomers, and amorphous structures. Overall, our results demonstrate clearly that a heterogeneous mixture of aggregate morphologies can co-exist in a dynamic equilibrium at any given time during an aggregation reaction. We speculate that the propensity for the polyQ domain to adopt multiple conformations in monomeric and higher order structures may in fact underlie mutant htt toxicity, as the number of toxic, gain-of-function interactions that mutant htt participates in may be proportional to the number of toxic conformers that it samples. However, it is also conceivable that perhaps only some subsets of these structures are most chemically reactive.
Our results provide further evidence to support the hypothesis that flanking amino acids modulate the conformation and aggregation of polyQ tracts and likely their biological activities. The polyQ repeat in mutant htt exon 1 is flanked by an N-terminal 17-amino acid α-helix and a C-terminal polyproline helix (41). These ordered flanking sequences might exert influence over the polyQ tract, perhaps restraining its ability to adopt multiple conformers (43) and by strongly inhibiting aggregation (32, 36, 43). The addition of oligoprolines to polyQ peptides decreases the rate of formation and stability of amyloid aggregates (32). The importance of amino acid context on polyQ aggregation has been shown in several studies and systems (44–48). Sequences that flank the polyQ repeat in mutant htt fragments also modulate toxicity in yeast models in cis and trans during aggregation (49, 50). In this study the most striking difference between synthetic polyQ peptides and a mutant htt fragment was the length of the polyQ tract necessary to form fibrils. Although synthetic peptides with even 7Q readily formed fibrils, only polyQ lengths in the disease-causing range (≥35Q) did so in a mutant htt fragment.
Although previous studies suggested synthetic polyQ peptides do not form oligomers and aggregate slower than peptides designed to partially mimic htt exon 1 (9, 33, 34, 36), our in situ AFM experiments where we directly compared these peptides with bona fide mutant htt exon 1 under identical experimental conditions provide unequivocal evidence that polyQ peptides indeed form oligomers albeit with much faster kinetics than htt exon 1. We believe this faster kinetics may be partly explained by the influence of amino acid context. Although experiments by Wetzel and co-workers (36) used “htt exon 1-like” polyQ peptides that lack the ∼40 amino acids C-terminal to the polyproline repeat in htt exon 1, our studies were performed with full-length, purified mutant htt exon 1. More importantly, our in situ AFM experiments allowed us to sample synthetic polyQ peptides continuously within minutes of initiating aggregation, whereas previous studies that used transmission electron microscopy were sampled only at later time points of aggregation when fibrils predominate (36). Differences between our results and those described previously might also be explained by differences in buffer conditions, as all of our studies were performed using a Tris-based buffer, whereas those by Wetzel and co-workers (36) were performed in phosphate-buffered saline. Although the precise fragments of mutant htt that accumulate in HD is unknown, fragments slightly larger than exon 1 have been detected, providing a strong rationale for the use of exon 1 as a model system to study htt aggregation (10, 51). However, it will also be interesting to determine whether longer mutant htt fragments and intact mutant htt itself exhibit a similar polyQ length dependence of oligomer formation.
Here, we used SDS-AGE with Western analysis to directly compare the effects of polyQ length on mutant htt oligomerization with purified proteins to that in an immortalized cell line derived from striatal neurons and in the brains of a mouse model for HD. We observed several important similarities and differences between the behavior of mutant htt fragments in vitro and in vivo. Although purified mutant htt fragments form oligomeric species in the same apparent size range to that observed in cell and brain homogenates, the latter appear to be even more heterogeneous in size. In contrast to that observed in vitro, where mutant htt oligomers are transient structures that dissipate coincident with fibril accumulation (at least at higher protein concentrations), oligomers formed in cell and brain homogenates accumulate as a function of time. One parsimonious interpretation of these results is that oligomers persist in vivo due to the constant supply of newly synthesized monomeric mutant htt that leads to steady-state equilibrium, in contrast to in vitro aggregation reactions, when monomeric mutant htt is limiting and a chemical equilibrium exists. This is not to say that fibrils do not form in vivo, as these structures have been described in a mouse model of HD (16, 39) and in HD brains (2) as well as in this study (Fig. 8). It is noteworthy that at lower protein concentrations in vitro, oligomers appear to predominate. Although we and others (2, 52–54) showed that oligomers exist in vivo, we hypothesize that the high local concentration of mutant htt fragments in an inclusion body in neurons is likely sufficient for fibril formation to predominate. This model would be consistent with cellular studies that suggest inclusion body formation is a conserved, microtubule-dependent homeostatic mechanism to detoxify mutant htt, where increasing the local concentration of mutant htt would presumably lead to an increase in fibril formation (17, 18). As electrophysiological abnormalities have been described even in embryonic and early postnatal neurons cultured from mouse models of HD (55, 56), well before inclusion bodies are deposited, these results suggest that oligomer formation likely predominates during early stages of disease, whereas fibril formation may only occur at later stages and may in fact represent a coping response. We speculate that the striking susceptibility of medium spiny striatal neurons to the effects of mutant htt and even the cell-to-cell variability observed among these cells may simply be explained by their differential ability to degrade mutant htt or shuttle it into an inclusion body and concomitant effects on the types of toxic conformers that may accumulate at different protein concentrations in cellular microenvironments.
Understanding if different aggregate species relate to polyQ length in some manner could help in identifying the conformers that mediate pathogenesis in HD. A systematic analysis of the heterogeneous mix of mutant htt aggregates and the elucidation of the dependence of polyQ length on this process in vivo may ultimately elicit the design of potential therapeutics (such as intrabodies) to neutralize toxic mutant htt assemblies (57). However, without precise knowledge of what constitutes a toxic conformer in vivo, steps to alter aggregation or deplete specific conformers could also potentially result in detrimental effects. For example, an intrabody specific for the N terminus of htt (VL12.3) had varying effects in different mouse models of HD ranging from being beneficial in a lentiviral model of HD to exacerbating phenotypes in the R6/2 HD model (57). In contrast, treatment with an intrabody specific for the polyproline and proline-rich domains of Htt (Happ1) conferred beneficial effects on several mouse models (57). As cellular dysfunction and toxicity could lie in the balance between fibrillar and diffuse forms of mutant htt, the risk/benefit ratio of therapies designed to modulate aggregation might depend critically on the extent to which the relative ratio of monomeric/oligomeric/fibrillar aggregates is altered. Clearly, the polyQ repeat in mutant htt can adopt multiple monomeric (41) and higher order conformations (21), of which only a subset may mediate pathogenesis in HD. Although we found that mixtures of aggregates are quite heterogeneous even with purified proteins, our data also indicates that this complexity is even greater in the crowded environment of the cytosol and nucleus of vulnerable neurons. To understand which mutant htt species are toxic entities, the major challenge will be to develop novel experimental tools capable of identifying and quantifying specific conformers in vivo in a highly sensitive manner. To this end, conformational specific antibodies (22, 58, 23) appear to have great potential in probing the complex diffuse fraction of mutant htt.
Supplementary Material
Acknowledgments
We acknowledge Thomas Möller for providing R6/2 brains, Carl Johnson, Steve Finkbeiner, Jason Miller, and Earl Rutenber for insightful discussions, and G. Howard and S. Ordway for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS054753 and R01NS047237.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. 1–9, and Movies 1 and 2.
E. J. Mitchell, G. Lotz, B. Apostol, A. Weiss, G. P. Bates, P. Paganetti, P. J. Muchowski, C. Glabe, and L. M. Thompson, manuscript in preparation.
- HD
- Huntington disease
- polyQ
- polyglutamine
- AFM
- atomic force microscopy
- DTT
- dithiothreitol
- HD20Q
- mutant htt fragment with 20 glutamine repeats
- HD35Q
- mutant htt fragment with 35 glutamine repeats
- HD46Q
- mutant htt fragment with 46 glutamine repeats
- HD53Q
- mutant htt fragment with 53 glutamine repeats
- GST
- glutathione S-transferase
- AGE
- agarose gel electrophoresis
- EM
- immunoelectron microscopy
- GFP
- green fluorescent protein.
REFERENCES
- 1.The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 2.DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 3.Zoghbi H. Y., Orr H. T. (2000) Annu Rev. Neurosci. 23, 217–247 [DOI] [PubMed] [Google Scholar]
- 4.Penney J. B., Jr., Vonsattel J. P., MacDonald M. E., Gusella J. F., Myers R. H. (1997) Ann. Neurol. 41, 689–692 [DOI] [PubMed] [Google Scholar]
- 5.Snell R. G., MacMillan J. C., Cheadle J. P., Fenton I., Lazarou L. P., Davies P., MacDonald M. E., Gusella J. F., Harper P. S., Shaw D. J. (1993) Nat. Genet. 4, 393–397 [DOI] [PubMed] [Google Scholar]
- 6.Tobin A. J., Signer E. R. (2000) Trends Cell Biol. 10, 531–536 [DOI] [PubMed] [Google Scholar]
- 7.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. (1997) Cell 90, 549–558 [DOI] [PubMed] [Google Scholar]
- 8.Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G. P., Lehrach H., Wanker E. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4604–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S., Berthelier V., Hamilton J. B., O'Nuallain B., Wetzel R. (2002) Biochemistry 41, 7391–7399 [DOI] [PubMed] [Google Scholar]
- 10.Lunkes A., Mandel J. L. (1998) Hum. Mol. Genet. 7, 1355–1361 [DOI] [PubMed] [Google Scholar]
- 11.Saudou F., Finkbeiner S., Devys D., Greenberg M. E. (1998) Cell 95, 55–66 [DOI] [PubMed] [Google Scholar]
- 12.Hackam A. S., Singaraja R., Wellington C. L., Metzler M., McCutcheon K., Zhang T., Kalchman M., Hayden M. R. (1998) J. Cell Biol. 141, 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirier M. A., Li H., Macosko J., Cai S., Amzel M., Ross C. A. (2002) J. Biol. Chem. 277, 41032–41037 [DOI] [PubMed] [Google Scholar]
- 14.Tanaka M., Morishima I., Akagi T., Hashikawa T., Nukina N. (2001) J. Biol. Chem. 276, 45470–45475 [DOI] [PubMed] [Google Scholar]
- 15.Wacker J. L., Zareie M. H., Fong H., Sarikaya M., Muchowski P. J. (2004) Nat. Struct. Mol. Biol. 11, 1215–1222 [DOI] [PubMed] [Google Scholar]
- 16.Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., Scherzinger E., Wanker E. E., Mangiarini L., Bates G. P. (1997) Cell 90, 537–548 [DOI] [PubMed] [Google Scholar]
- 17.Muchowski P. J., Ning K., D'Souza-Schorey C., Fields S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 19.Weiss A., Klein C., Woodman B., Sathasivam K., Bibel M., Régulier E., Bates G. P., Paganetti P. (2008) J. Neurochem. 104, 846–858 [DOI] [PubMed] [Google Scholar]
- 20.Nagai Y., Inui T., Popiel H. A., Fujikake N., Hasegawa K., Urade Y., Goto Y., Naiki H., Toda T. (2007) Nat. Struct. Mol. Biol. 14, 332–340 [DOI] [PubMed] [Google Scholar]
- 21.Nekooki-Machida Y., Kurosawa M., Nukina N., Ito K., Oda T., Tanaka M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9679–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legleiter J., Lotz G. P., Miller J., Ko J., Ng C., Williams G. L., Finkbeiner S., Patterson P. H., Muchowski P. J. (2009) J. Biol. Chem. 284, 21647–21658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trottier Y., Lutz Y., Stevanin G., Imbert G., Devys D., Cancel G., Saudou F., Weber C., David G., Tora L. (1995) Nature 378, 403–406 [DOI] [PubMed] [Google Scholar]
- 24.Muchowski P. J., Schaffar G., Sittler A., Wanker E. E., Hayer-Hartl M. K., Hartl F. U. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S., Berthelier V., Yang W., Wetzel R. (2001) J. Mol. Biol. 311, 173–182 [DOI] [PubMed] [Google Scholar]
- 26.Legleiter J., DeMattos R. B., Holtzman D. M., Kowalewski T. (2004) J. Colloid Interface Sci. 278, 96–106 [DOI] [PubMed] [Google Scholar]
- 27.Steffan J. S., Agrawal N., Pallos J., Rockabrand E., Trotman L. C., Slepko N., Illes K., Lukacsovich T., Zhu Y. Z., Cattaneo E., Pandolfi P. P., Thompson L. M., Marsh J. L. (2004) Science 304, 100–104 [DOI] [PubMed] [Google Scholar]
- 28.Kazantsev A., Preisinger E., Dranovsky A., Goldgaber D., Housman D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11404–11409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin Z. H., Wang Y., Sapp E., Cuiffo B., Wanker E., Hayden M. R., Kegel K. B., Aronin N., DiFiglia M. (2004) J. Neurosci. 24, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackley H. K., Sanders G. H., Davies M. C., Roberts C. J., Tendler S. J., Wilkinson M. J. (2000) J. Mol. Biol. 298, 833–840 [DOI] [PubMed] [Google Scholar]
- 31.Kowalewski T., Holtzman D. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharyya A., Thakur A. K., Chellgren V. M., Thiagarajan G., Williams A. D., Chellgren B. W., Creamer T. P., Wetzel R. (2006) J. Mol. Biol. 355, 524–535 [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharyya A. M., Thakur A. K., Wetzel R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15400–15405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S., Ferrone F. A., Wetzel R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11884–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thakur A. K., Wetzel R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 17014–17019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thakur A. K., Jayaraman M., Mishra R., Thakur M., Chellgren V. M., Byeon I. J., Anjum D. H., Kodali R., Creamer T. P., Conway J. F., Gronenborn A. M., Wetzel R. (2009) Nat. Struct. Mol. Biol. 16, 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meriin A. B., Zhang X., Alexandrov I. M., Salnikova A. B., Ter-Avanesian M. D., Chernoff Y. O., Sherman M. Y. (2007) FASEB J. 21, 1915–1925 [DOI] [PubMed] [Google Scholar]
- 38.Gutekunst C. A., Li S. H., Yi H., Mulroy J. S., Kuemmerle S., Jones R., Rye D., Ferrante R. J., Hersch S. M., Li X. J. (1999) J. Neurosci. 19, 2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S. W., Bates G. P. (1996) Cell 87, 493–506 [DOI] [PubMed] [Google Scholar]
- 40.Cummings D. M., Yim M. M., Alaghband Y., Malvar J. S., Tsuruyama K., Joshi P. R., Cepeda C., Levine M. S. (2008) Soc. Neurosci. Abstr. 34, 433.411 [Google Scholar]
- 41.Kim M. W., Chelliah Y., Kim S. W., Otwinowski Z., Bezprozvanny I. (2009) Structure 17, 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller J., Rutenber E., Muchowski P. J. (2009) Structure 17, 1151–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darnell G., Orgel J. P., Pahl R., Meredith S. C. (2007) J. Mol. Biol. 374, 688–704 [DOI] [PubMed] [Google Scholar]
- 44.Bulone D., Masino L., Thomas D. J., San Biagio P. L., Pastore A. (2006) PLoS ONE 1, e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ignatova Z., Gierasch L. M. (2006) J. Biol. Chem. 281, 12959–12967 [DOI] [PubMed] [Google Scholar]
- 46.La Spada A. R., Taylor J. P. (2003) Neuron 38, 681–684 [DOI] [PubMed] [Google Scholar]
- 47.Menon R. P., Pastore A. (2006) Cell. Mol. Life Sci. 63, 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nozaki K., Onodera O., Takano H., Tsuji S. (2001) Neuroreport 12, 3357–3364 [DOI] [PubMed] [Google Scholar]
- 49.Duennwald M. L., Jagadish S., Giorgini F., Muchowski P. J., Lindquist S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11051–11056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duennwald M. L., Jagadish S., Muchowski P. J., Lindquist S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11045–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ratovitski T., Gucek M., Jiang H., Chighladze E., Waldron E., D'Ambola J., Hou Z., Liang Y., Poirier M. A., Hirschhorn R. R., Graham R., Hayden M. R., Cole R. N., Ross C. A. (2009) J. Biol. Chem. 284, 10855–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi T., Kikuchi S., Katada S., Nagai Y., Nishizawa M., Onodera O. (2008) Hum. Mol. Genet. 17, 345–356 [DOI] [PubMed] [Google Scholar]
- 53.Takahashi Y., Okamoto Y., Popiel H. A., Fujikake N., Toda T., Kinjo M., Nagai Y. (2007) J. Biol. Chem. 282, 24039–24048 [DOI] [PubMed] [Google Scholar]
- 54.Wong S. L., Chan W. M., Chan H. Y. (2008) FASEB J. 22, 3348–3357 [DOI] [PubMed] [Google Scholar]
- 55.Zeron M. M., Hansson O., Chen N., Wellington C. L., Leavitt B. R., Brundin P., Hayden M. R., Raymond L. A. (2002) Neuron 33, 849–860 [DOI] [PubMed] [Google Scholar]
- 56.Starling A. J., André V. M., Cepeda C., de Lima M., Chandler S. H., Levine M. S. (2005) J. Neurosci. Res. 82, 377–386 [DOI] [PubMed] [Google Scholar]
- 57.Southwell A. L., Ko J., Patterson P. H. (2009) J. Neurosci. 29, 13589–13602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ko J., Ou S., Patterson P. H. (2001) Brain Res. Bull 56, 319–329 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.