Abstract
BubR1 is essential for the mitotic checkpoint that prevents aneuploidy in cellular progeny by triggering anaphase delay in response to kinetochores incorrectly/not attached to the mitotic spindle. Here, we define the molecular architecture of the functionally significant N-terminal region of human BubR1 and present the 1.8 Å crystal structure of its tetratricopeptide repeat (TPR) domain. The structure reveals divergence from the classical TPR fold and is highly similar to the TPR domain of budding yeast Bub1. Shared distinctive features include a disordered loop insertion, a 310-helix, a tight turn involving glycine positive Φ angles, and noncanonical packing of and between the TPR motifs. We also define the molecular determinants of the interaction between BubR1 and kinetochore protein Blinkin. We identify a shallow groove on the concave surface of the BubR1 TPR domain that forms multiple discrete and potentially cooperative interactions with Blinkin. Finally, we present evidence for a direct interaction between BubR1 and Bub1 mediated by regions C-terminal to their TPR domains. This interaction provides a mechanism for Bub1-dependent kinetochore recruitment of BubR1. We thus present novel molecular insights into the structure of BubR1 and its interactions at the kinetochore-microtubule interface. Our studies pave the way for future structure-directed engineering aimed at dissecting the roles of kinetochore-bound and other pools of BubR1 in vivo.
Keywords: Cancer, Cell/Checkpoint, Cell/Cycle, Organisms/Human, Proteases/Ubiquitination, Protein/Protein-Protein Interactions, Protein/Protein-Protein Interactions, Protein/Structure
Introduction
The spindle assembly checkpoint (SAC2; also referred to as the mitotic checkpoint) performs the critical role of triggering anaphase delay in response to chromosomes incorrectly/not attached to the mitotic spindle (1). This prevents aneuploidy and associated genetic instability that result from anaphase commencing prior to the proper bi-orientation of replicated chromosomes. At the molecular level, the SAC inhibits the E3 ubiquitin ligase activity of the anaphase promoting complex/cyclosome (APC/C) (2). This prevents the ubiquitination and degradation of anaphase inhibitors, Securin and cyclin B1 (3). Securin inhibits the protease that would otherwise cleave the Cohesin complex holding sister chromatids together (4), whereas cyclin B1 regulates the activity of Cdk1 (cyclin-dependent kinase 1). The multidomain kinase BubR1 (budding uninhibited by benomyl related-1) is an essential component of the SAC of higher eukaryotes (5, 6). BubR1 mutations and/or mis-regulation have been detected in human cancers (7) and the related disease of mosaic variegated aneuploidy (8, 9). Reduced levels of BubR1 in mice also lead to increased tumor incidence and accelerated tumor progression (10–13). BubR1 has also recently been shown to be important for the segregation of acentric chromosome fragments that lack a kinetochore (14) and for oocyte meiotic transitions (15).
BubR1 inhibits APC/C activity during metaphase as part of the mitotic checkpoint complex (MCC) (Fig. 1A) (16). The MCC is composed of BubR1, Bub3, Mad2 (mitotic arrest deficient 2), and Cdc20 (cell division cycle 20). Cdc20 is an essential APC/C co-factor that mediates the recognition of substrates that harbor short sequence motifs such as KEN boxes (consecutive lysine, glutamate, and asparagine residues) (17). The formation of the MCC inhibits APC/C ubiquitin ligase activity by abrogating Cdc20 co-factor function (16). Within the MCC, Cdc20 directly binds a conformer of Mad2 and the constitutive BubR1-Bub3 complex. These interactions are independently capable of APC/C-Cdc20 inhibition, although highest potency inhibition is achieved through simultaneous binding, likely because of Mad2 promotion of the BubR1-Cdc20 interaction (18–20). Two complementary molecular mechanisms have been proposed for APC/C-Cdc20 inhibition by BubR1-Bub3 and Mad2. First is an inhibitory conformational shift of Cdc20, in which single particle electron microscopy reveals distinct orientations of Cdc20 when it is bound to the APC/C alone or as part of the MCC (21). It is possible that one of these orientations induces Cdc20 ubiquitin-mediated degradation (22). Second is competitive binding of BubR1 to substrate-binding sites of APC/C-Cdc20; BubR1 contains two conserved KEN boxes (human residues 26–28 and 304–306) that are essential for Cdc20 binding and cell viability and act to competitively block substrate binding to APC/C-Cdc20 (23, 24). This function of BubR1 is dependent upon PCAF (p300/CBP-associated factor)-dependent acetylation at Lys250, which prevents BubR1 ubiquitin-mediated degradation (25).
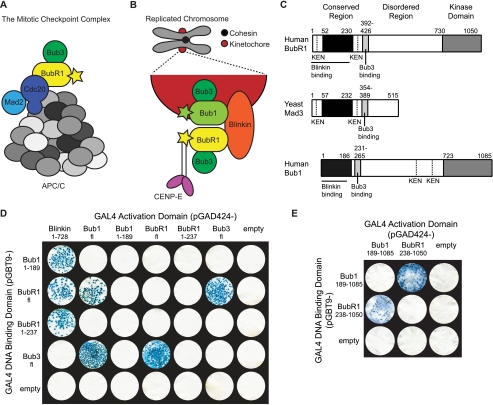
FIGURE 1.
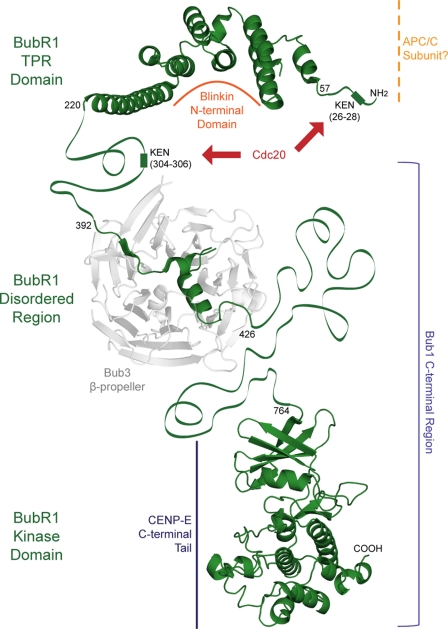
Architecture and interactions of Bub proteins. A and B, BubR1 interactions away from (A) and at (B) the kinetochore during SAC activation. The stars represent kinase domains, and the gray circles represent different APC/C subunits. C, domain organization of human BubR1, yeast Mad3, and human Bub1. The proteins contain a conserved N-terminal region (black), a central disordered region incorporating the Bub3-binding site (light gray), and numerous KEN boxes (dotted line). The C-terminal kinase domain (gray) and Blinkin-binding site (solid line) of the human proteins are also shown. D and E, BubR1 interacts with Blinkin, Bub1, and Bub3 in a yeast two-hybrid assay. Strains Y187(pGBT9-bait) and AH109(pGAD424-target)were mated and X-gal filter lift assays performed on colonies grown on SD/−Ade/−His/−Leu/−Trp plates. Colony growth requires the activation of ADE and HIS reporter genes, and the blue-producing X-gal reaction requires the activation of the lacZ reporter gene. Data for pGBT9-Blinkin(1–728) and pGBT9-Bub1fl vectors are not shown due to insurmountable technical difficulties of autoactivation and toxicity, respectively.
In addition to a role in the MCC, BubR1 is enriched and exchanged at kinetochores incorrectly/not attached to the mitotic spindle during metaphase (Fig. 1B) (26). Kinetochores are large proteinaceous complexes that form on centromeric chromatin and mediate microtubule binding (27). BubR1 kinetochore enrichment is dependent on Bub1 and Bub3 (28, 29) and is required to sustain a SAC arrest, as well as for stable kinetochore-microtubule interactions (23). The mechanistic links between BubR1 kinetochore enrichment and these activities, however, have yet to be deciphered. Regulated cycling between kinetochore-bound and MCC pools of BubR1, similar to that observed for Mad2, is thought not to occur (19), whereas BubR1 interaction with the kinetochore protein Blinkin (Bub-linking) and microtubule binding, molecular motor CENP-E (centromeric protein E) may be important (30–32). Blinkin is vital for the formation of wild type kinetochore-microtubule interactions and interacts with an N-terminal α-helical region of BubR1 (32, 33); CENP-E regulates BubR1 kinase activity and localizes to the kinetochore in a BubR1-dependent manner (30, 31). Post-translational modification of BubR1 at the kinetochore is also likely to play a role (34). These links remain as conjecture as such mechanistic insight requires a detailed molecular understanding of BubR1. Molecular detail of BubR1 is not available as attempts to produce stable protein for high resolution structural studies have been hampered by BubR1 susceptibility to proteolysis, rendering such studies thus far unsuccessful (35, 36).
Insight into the structure of BubR1 can be gained by comparison with Bub1, which shows a similar overall domain organization and shares ligands Blinkin and Bub3 (Fig. 1C) (32, 36, 37). Both proteins contain an N-terminal α-helical region that mediates Blinkin binding (32), a central disordered region containing a short motif for Bub3 binding (36, 37), and a C-terminal serine/threonine kinase domain that is absent in lower eukaryotic BubR1 orthologues such as yeast Mad3 (38). The crystal structure of the N-terminal α-helical region of budding yeast Bub1 has been solved and reveals a divergent tetratricopeptide repeat (TPR) domain (39). Based on sequence analysis and biophysical studies, the N-terminal α-helical region of BubR1 is also expected to adopt a TPR fold (40). In this study, we define the molecular architecture of the N-terminal region of human BubR1 and present the 1.8 Å crystal structure of human BubR1 residues 48–237 (BubR1(48–237)). These residues include a triple tandem arrangement of the TPR motif with predicted similarity to that found in budding yeast Bub1. We thus define the structural platform that mediates BubR1 interaction with Cdc20 in the MCC and Blinkin at the kinetochore. We further examine the molecular determinants of the interaction between BubR1 and Blinkin, revealing the presence of multiple discrete and potentially cooperative interactions along a groove of BubR1. Finally, we identify an interaction between BubR1 and Bub1 mediated by regions C-terminal to their TPR domains. These data are invaluable for future studies aimed at defining the precise roles of kinetochore-bound and MCC pools of BubR1 in the SAC in vivo.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Analysis
Human gene fragments encoding for BubR1 full-length (BubR1fl), residues 1–237 (BubR1(1–237)) and residues 238–1050 (BubR1(238–1050)) (GenBankTM accession number AAD11941), Bub1 full-length (Bub1fl), residues 1–189 (Bub1(1–189)) and residues 189–1085 (Bub1(189–1085)) (GenBankTM accession number AAC06259), Blinkin residues 1–728 (Blinkin(1–728)) (GenBankTM accession number AAM45143), and Bub3 full-length (Bub3fl) (GenBankTM accession number AAC06258) were cloned into pGBT9 and pGAD424 (Clontech). BubR1fl point mutants (P119A, L126A, R130A, S157A, E161A, and R165A) were created by overlapping primer extension methods and cloned to pGBT9.
Yeast two-hybrid protocols were based on the Matchmaker 3 yeast two-hybrid system (Clontech), where the rate of yeast growth, the number of resultant colonies, and the intensity of X-gal reactions are directly related to the level of reporter gene activation (41). Using a polyethylene glycol/single-stranded DNA/LiAc procedure, pGBT9 vectors were transformed into the Y187 strain, and pGAD424 vectors were transformed into the AH109 strain. For mating, single colonies of Y187[pGTB9-bait] and AH109[pGAD424-target] were resuspended in 0.5 ml of 2× YPDA and incubated at 30 °C, 50 rpm for 24 h. Cultures were diluted 1 in 10 with 0.5× YPDA and 100 μl plated on SD/−Leu/−Trp (double dropout) and SD/−Ade/−His/−Leu/−Trp (quadruple dropout) plates. Plates were incubated at 30 °C for 2, 3, or 5 days. Matings with BubR1fl mutants were also grown with 0–15 mm 3-amino-1,2,4-triazole (3-AT; Sigma) on the quadruple dropout plates. Colonies were lifted onto filters (Whatman No. 5, 70 mm), lysed by two cycles of freeze-thaw and incubated in 100 mm sodium phosphate, pH 7.0, 10 mm KCl, 1 mm MgSO4, 0.3% (v/v) β-mercaptoethanol containing 1 mg/ml X-gal (Promega) at 30 °C for 5 h. Shown filters are representative of at least three replicates performed with independent yeast transformants.
For time course and 3-AT analysis, densitometry was performed by measuring mean pixel intensities of color-inverted scanned filters. The intensity of a relevant negative control, such as pGBT9-bait × pGAD424-empty, was subtracted from each value. “Relative intensity of interaction with Blinkin(1–728)” refers to (pGBT9-BubR1fl-mutant × pGAD424-Blinkin(1–728) at a particular day or 3-AT concentration) divided by ((pGBT9-BubR1fl-mutant × pGAD424-Blinkin(1–728)) at day 5 or with 0 mm 3-AT); likewise, “relative intensity of interaction with Bub3fl refers to ((pGBT9-BubR1fl-mutant × pGAD424-Bub3fl) at a particular day or 3-AT concentration) divided by ((pGBT9-BubR1fl-mutant × pGAD424-Bub3fl) at day 5 or with 0 mm 3-AT).
Protein Expression and Purification
Human gene fragments encoding for BubR1 residues 1–230 (BubR1(1–230)) and 1–280 (BubR1(1–280)) were cloned into pGEX-6P-1 (GE Healthcare) with a PCR-introduced C-terminal His6 tag, whereas BubR1 residues 39–230 (BubR1(39–230)) and 48–237 (BubR1(48–237)) were cloned into pHAT4 (42).
All proteins were expressed in Escherichia coli BL21(DE3) grown in 2× YT (Sigma) with 100 μg/ml ampicillin by induction with 0.4 mm isopropyl-d-thiogalactopyranoside for 3 h at 37 °C. Cells were suspended in 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1 mm 1,4-dithiothreitol with protease inhibitor mixture (Roche Applied Science) and lysed using an EmulsiFlex-C5 (Avestin). The soluble fraction was obtained by centrifugation at 15,000 × g at 4 °C for 30 min. Glutathione S-transferase fusions (BubR1(1–230) and BubR1(1–280)) and histidine tag fusions (BubR1(39–230) and BubR1(48–237)) were then purified by standard affinity chromatography and cleaved by overnight incubation with PreScissionTM (GE Healthcare) or tobacco etch virus protease, respectively. Protease, tag, and uncleaved fusion were removed by a second affinity chromatography step. BubR1(1–230) and BubR1(1–280) were further purified with Ni2+- nitrilotriacetic acid affinity chromatography, whereas BubR1(39–230) and BubR1(48–237) were further purified by ion exchange at pH 9.0 using a Resource Q column (GE Healthcare). All proteins were polished with size exclusion chromatography. Proteins were biochemically and biophysically characterized as described in the supplemental material.
Crystallization and Data Collection
BubR1(48–237) was concentrated to 9 mg/ml following size exclusion chromatography in 20 mm CHES, pH 9.0, 200 mm NaCl, 1 mm 1,4-dithiothreitol. Reducing agent tris(2-carboxyethyl)phosphine, pH 8.0, was then added at a final concentration of 1 mm and the sample used for crystallization. The best crystals were grown at 15 °C using the sitting drop vapor diffusion method with 140 μl of reservoir solution (22 g/100 ml of PEG 8000, 200 mm Tris-HCl, pH 8.0, 10 mm praseodymium(III) acetate) and a 200-nl drop (equal volume of protein and reservoir solution) in a 96-well format (Greiner). The crystals were cryoprotected by addition of 20% (v/v) ethylene glycol (soaked for ∼1 min) and frozen in liquid nitrogen for data collection at 100 K. Data were collected for two crystals at a wavelength of 1.06 Å at beam I03, Diamond Synchrotron Light Source, Oxford, UK, and processed using MOSFLM and SCALA from the CCP4 suite (43, 44).
Structure Determination and Refinement
The structure of BubR1(48–237) was solved by single wavelength anomalous dispersion using praseodymium ions from the crystallization solution, found stably in the crystal lattice. Using data collected from crystal 1 (anomalous completeness of 100% and anomalous redundancy of 11.1), two praseodymium ions were found and refined using autoSHARP (45). The final figure of merit was 0.82; the phasing power was 1.75, and the Rcullis was 0.66. Following density modification with 43.2% solvent content, these phases were used to calculate an initial map used for automated model building with ARP/wARP (46). Iterative cycles of model building and refinement were then carried out in COOT (47) and REFMAC 5 (48) using data collected from crystal 2, including a single cycle of Translation, Liberation, and Screw refinement with two groups (residues 57–114 and 115–220) identified by the TLMSD server (49). During refinement, 5% of reflections were kept aside for evaluation of Rfree. Residues 48–56, 92–95, and 221–237 are absent in the final model. PyMOL (DeLano Scientific) was used to create images.
RESULTS
BubR1 Directly Interacts with Blinkin and Bub1
A major challenge in developing a molecular understanding of the SAC is establishing the structural basis of BubR1 kinetochore recruitment and APC/C-Cdc20 inhibition. We first set out to confirm the reported interaction between the N-terminal region of BubR1 and Blinkin and to investigate the interactions between BubR1 and functionally related proteins Bub1 and Bub3. Human sequences of full-length BubR1 (BubR1fl), BubR1(1–237), BubR1(238–1050), Bub1fl, Bub1(1–189), Bub1(189–1085), Blinkin(1–728), and Bub3fl were fused to GAL4 DNA-binding domains and GAL4 activator domains. Interactions were tested through the highly stringent Matchmaker 3 yeast two-hybrid system in which positive interactions were determined by independent activation of three reporter genes (HIS, ADE, and lacZ). In this system, the rate of yeast growth, the number of resultant colonies, and the intensity of X-gal reactions are directly related to the underlying level of reporter gene activation (41). BubR1(1–237) was found to interact with Blinkin(1–728) in a manner equivalent to that of full-length protein (Fig. 1D), confirming that Blinkin(1–728) forms contacts with the N-terminal region of BubR1. In contrast, interaction with Bub3fl was detected only for full-length BubR1 (Fig. 1D), consistent with the known Bub3-binding site downstream of the N-terminal region (36, 37). Related protein Bub1 showed a similar pattern of binding to Blinkin(1–728) through its N-terminal region and Bub3fl through a downstream region (Fig. 1D). We were regrettably unable to test interactions using pGBT9-Blinkin(1–728) and pGBT9-Bub1fl vectors due to insurmountable technical difficulties of autoactivation and toxicity, respectively. We thus conclude that the BubR1 N-terminal region interacts with Blinkin, whereas a more C-terminal region interacts with Bub3.
The yeast two-hybrid experiments also indicate an interaction between BubR1 and Bub1 (Fig. 1, D and E). The interaction does not rely on the N-terminal region of Bub1 or BubR1, is retained upon deletion of the N-terminal regions, and specifically allows formation of a BubR1-Bub1 heterodimer but not a BubR1-BubR1 homodimer. Failure to detect a heterodimeric complex of BubR1(1–237) and Bub1(1–189) is consistent with in vitro analytical size exclusion chromatography and chemical cross-linking analysis with recombinant proteins (data not shown). We thus conclude that BubR1 and Bub1 undergo heterodimerization through the interaction of distinct sequences in their C-terminal regions (Fig. 1, D and E). Our findings conflict with previously published yeast two-hybrid data suggesting both homo- and heterodimerization of BubR1 and Bub1 (32). We account for this through our observation that the BubR1fl GAL4 DNA-binding domain fusion exhibits a low level of autoactivation that hampers investigations using only the lacZ reporter gene (supplemental Fig. S1) but is completely eliminated by the three independent reporter genes (HIS, ADE, and lacZ) employed in this study. The formation of a BubR1-Bub1 complex is consistent with reports that BubR1 kinetochore recruitment is dependent on Bub1 when the capacity of Blinkin to bind BubR1 is abrogated in vivo (32).
Molecular Architecture of the BubR1 N-terminal Region
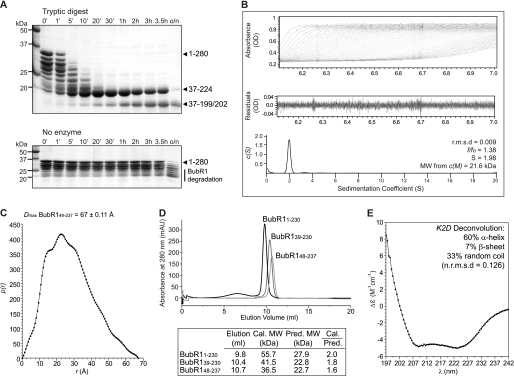
We next investigated the molecular architecture of the BubR1 N-terminal region. To explore the presence of domain structure, recombinant human BubR1(1–280) was subjected to limited proteolysis using trypsin, which specifically cleaves peptide chains C-terminal to Lys and Arg residues. Despite Lys and Arg residues being distributed throughout BubR1(1–280), this identified protease-resistant fragments of ∼18 and 20 kDa (Fig. 2A). Edman degradation and tandem mass spectrometry peptide fragmentation analyses subsequently showed that these fragments contain residues Ile37–Arg199 or Arg202 and Ile37–Arg224, respectively, suggesting the presence of a folded domain within these limits. The trypsin cleavage site immediately downstream of Arg36 is Lys59, indicating that the N-terminal boundary of this folded domain lies between Ile37 and Lys59. This is supported by our observation that degradation products of BubR1(1–230) formed during purification are N-terminally truncated at residues Arg33, Ser39, Glu42, Gly43, Ala44, and Ala46 (shown by Edman degradation; data not shown). Interestingly, the N-terminal fragment cleaved from BubR1 is conserved in BubR1 but not Bub1 orthologues and contains one of the conserved KEN box motifs essential for interaction with Cdc20 (Fig. 3A) (23). Similarly, the presence of protease-resistant fragments that undergo C-terminal cleavage at Arg199/202 or Arg224 suggests a mobile C-terminal region. One frequently sampled conformation of this region must constrain residues Arg199–Arg224 as cleavage at the intervening trypsin site Lys210 was not observed. Our analysis thus predicts a molecular architecture for the N-terminal region of BubR1 in which a folded domain is preceded by an N-terminal extension of 36–58 amino acids that is connected by a flexible linker.
FIGURE 2.
Biochemical and biophysical characterization of the BubR1 N-terminal region. A, limited proteolysis of BubR1(1–280) identified protease-resistant fragments of ∼18 and 20 kDa. These fragments encompass residues Ile37–Arg199 or Arg202 and Ile37–Arg224, respectively. B, sedimentation velocity analysis of BubR1(48–237) reveals it to be an asymmetrical monomer. Using absorbance data (top), the sedimentation coefficient distribution c(S) (bottom) was calculated using SEDFIT to minimize residuals (middle) and with a root mean square deviation (r.m.s.d.) of less than 0.01. The resultant frictional coefficient (f/f0), sedimentation coefficient (S), and molecular weight (MW) are shown. C, small angle x-ray scattering of BubR1(48–237) measures a Dmax of 67 ± 0.11 Å, indicating an elongated shape. The intra-particle distance distribution function p(r) was calculated using GNOM. D, analytical size exclusion chromatography of BubR1 proteins shows that the calculated (cal.) to predicted (pred.) molecular weight ratio increases as more of the N-terminal of BubR1 is included. mAU, milli-absorbance units. E, far-UV circular dichroism spectra of BubR1(48–237) reveal a predominantly α-helical structure.
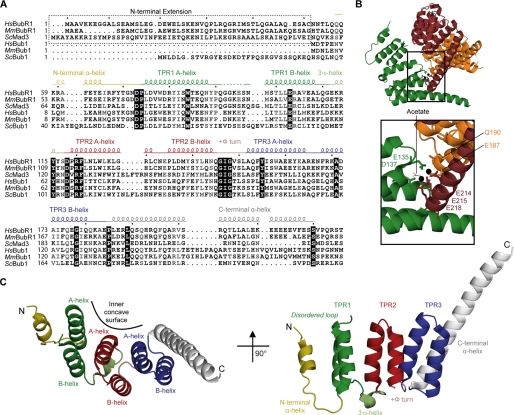
FIGURE 3.
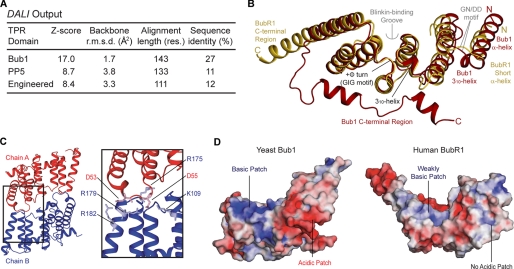
1.8 Å crystal structure of the BubR1 N-terminal domain reveals a TPR fold. A, sequence alignment of the N-terminal region of Bub proteins. The alignment was created with ClustalX2 and visualized with ESPRIPT. Sequences shown are from Homo sapiens (Hs), Mus musculus (Mm), and Saccharomyces cerevisiae (Sc). Secondary structure elements are shown and colored as in C. Many residues are conserved (black box) or semi-conserved/similar (gray). B, coordination of Pr(III) in the BubR1 crystals. The crystal contains two Pr(III) (black spheres) per BubR1 protomer coordinated by two waters (small spheres), an acetate (black), and seven residues from three protomers (sticks). C, orthogonal views of the BubR1 TPR domain.
To probe further the molecular architecture of the BubR1 N-terminal region, biophysical analysis was performed on a construct in which the putative N-terminal extension was deleted, BubR1(48–237). The N-terminal boundary of this construct was based on secondary structure prediction and the Ile37–Lys59 range defined by limited proteolysis. Similarly, the C-terminal boundary was selected to be outside of the folded domain between predicted α-helices such that the fold is expected be retained. Sedimentation velocity analysis of BubR1(48–237) revealed the presence of a single species of molecular mass 21.6 kDa (theoretical molecular mass of 22.4 kDa) with a frictional coefficient of 1.38 (Fig. 2B), suggesting a monomer with an elongated, asymmetrical shape. The monomeric status was consistent with chemical cross-linking (data not shown) and is in keeping with the yeast two-hybrid finding that BubR1 is incapable of homodimerization (Fig. 1D). An extended conformation is also supported by the large Dmax of 67 ± 0.11 Å measured with small angle x-ray scattering (Fig. 2C) and analytical size exclusion chromatography in which BubR1(48–237) eluted as a single species at a volume corresponding to 1.6 times the mass predicted for a compact, globular monomer (Fig. 2D). Interestingly, on the same Superdex column, constructs in which the N-terminal extension was progressively re-introduced, BubR1(39–230) and BubR1(1–230), showed elution volumes corresponding to 1.8 and 2.0 times the mass predicted for compact, globular monomers, respectively. Circular dichroism of BubR1(48–237) further revealed a predominantly α-helical content, with spectra minima at 208 and 222 nm (Fig. 2E). Together, these biophysical analyses lead to a model in which the BubR1 N-terminal region contains an α-helical domain with a protruding N-terminal extension that is connected by a flexible linker. Importantly, these data also demonstrate that removal of the N-terminal extension does not compromise the fold, thus allowing the production of a stable construct of the BubR1 N-terminal region suitable for structural analysis in vitro.
Overall Structure of the BubR1 N-terminal Region
The BubR1 N-terminal region 1.8 Å crystal structure was solved using single wavelength anomalous dispersion of BubR1(48–237) crystals grown in the presence of 10 mm praseodymium(III) acetate (Table 1). High quality electron density was present for residues 57–220 (supplemental Fig. S2) excluding residues 92–95, which likely constitute a disordered loop. The structure is composed of eight α-helices arranged in an anti-parallel fashion reminiscent of a TPR domain (Fig. 3, A and C) (50). The central six α-helices fold into three sequential helix-loop-helix units (TPR1–3), with their α-helices forming concave inner and outer surfaces. Each of the helix-loop-helix units resembles a single TPR motif, a well known motif in which the consensus sequence contains small hydrophobic residues at positions 8, 20, and 27, large hydrophobic residues at positions 4, 7, 11, and 24, and an α-helix-breaking residue, typically a proline, at position 32 (Fig. 5A) (50–52). The three TPR units are packed together such that “A” α-helices form the inner surface and “B” α-helices form the outer surface, and the entire structure has a subtle right-handed super-helical twist. Important to this arrangement is a tight turn involving positive Φ angles of Gly146 and Gly148 in the conserved GIG motif connecting TPR2 and TPR3.
TABLE 1.
Data collection and refinement statistics
Crystal 1 is for SAD, and Crystal 2 is for refinement.
| Crystal 1 | Crystal 2 | |
|---|---|---|
| Data collection | ||
| Space group | P3221 | P3221 |
| Cell dimensions | ||
| a, b, c | 62.67, 62.76, 90.29 Å | 62.67, 62.76, 90.29 Å |
| α, β, γ | 90, 90, 120° | 90, 90, 120° |
| Resolution | 2.0 Å (2.1-2.0 Å)a | 1.8 Å (1.9-1.8 Å)a |
| Rmerge | 11.2% (42.0%) | 10.7% (51.0%) |
| I/σI | 23.9 (6.6) | 21.0 (4.0) |
| Completeness (%) | 100 (100) | 99.9 (100) |
| Redundancy | 20.8 (21.3) | 15.6 (15.8) |
| Refinement | ||
| Resolution (Å) | 54.4-1.8 | |
| No. reflections (Rfree) | 18643 (1005) | |
| Rwork/Rfree | 0.222/0.259 | |
| Protein atoms | 1358 | |
| Water atoms | 117 | |
| Ligand/Ion atoms | 14 | |
| (2 Pr, 1 acetate, 2 ethylene glycol) | ||
| B-factors (Å2) | ||
| Protein | 36.8 | |
| Ligand/Ion | 32.5 | |
| Water | 41.2 | |
| Root mean square deviations | ||
| Bond lengths | 0.016 Å | |
| Bond angles | 1.685° | |
| Residues in structure | Gln57–Gln91, Ser96–Ser220 | |
| Residues in preferred/allowed Φ-Ψ region of Ramachandran plot | 96.15/3.85% | |
| Protein Data Bank code | 2WVI | |
a Values in parentheses are for highest resolution shell.
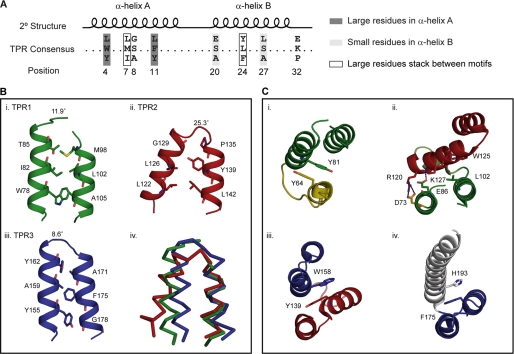
FIGURE 5.
BubR1 TPR domain exhibits both canonical and noncanonical packing interactions. A, TPR motif consensus sequence. B, interactions within BubR1 TPR motifs (panels i–iii) involving TPR consensus positions 4, 11, 8, 20, 24, and 27 (sticks). The angle between α-helices is noted. Superposition of the TPR motifs (panel iv) highlights the noncanonical packing of TPR2. C, packing between BubR1 TPR motifs (panels i–iv). Between TPR1 and TPR2, the canonical hydrophobic stacking of TPR consensus positions 7 and 24 (sticks) is replaced by ionic interactions (dotted lines). For all images, the N-terminal α-helix is yellow; TPR1 is green; TPR2 is red; TPR3 is blue, and the C-terminal α-helix is white.
The BubR1 structure also contains a short N-terminal α-helix connected by a kinked loop and an extended C-terminal α-helix that mimics an A-type helix but protrudes greater than 20 Å from the molecule (Fig. 3, A and C). These α-helices are unlikely to form additional TPR motifs in the full-length protein given the hydrophilic nature of their outer edges. The extended C-terminal α-helix resembles a capping α-helix as observed in numerous TPR domains (50). The protrusion of the C-terminal α-helix, however, may be an artifact of crystal packing as glutamate residues in this region (Glu212, Glu214, Glu215, and Glu218) coordinate two praseodymium(III) ions in the crystal (Fig. 3B). The lower average B-factor of the protruding region (30.8 for residues 203–220 compared with 37.5 for residues 57–202) reinforces its constraint in the crystal lattice. In solution or in the context of the full-length protein, this sequence may become disordered or may contribute to novel structure. Such a scenario has been reported for protein phosphatase 5 (PP5), a TPR domain protein in which an extended C-terminal α-helix unwinds to form a disordered loop in the full-length protein (53, 54). As also suggested by the aforementioned limited proteolysis data, the extended conformation of the C-terminal α-helix is likely to be one of many conformations sampled in solution. It is not known if this extended conformation is relevant to a physiological interaction.
Comparison of BubR1 and Bub1 TPR Domains
To confirm the TPR nature of the BubR1 N-terminal region, the DALI server was employed (55). Such confirmation is important as differences between all α-helical repeat domains such as HEAT repeats, Armadillo repeats, and clathrin heavy chain repeats can be subtle and difficult to predict (56). The analysis revealed the most similar structure to be the budding yeast Bub1 TPR domain, with moderate similarity to canonical TPR domains such as defining member PP5 and an engineered three-repeat TPR domain (Fig. 4A). Similar to the BubR1 structure, the Bub1, PP5, and engineered TPR domains each contain three TPR motifs and, in the case of PP5 and the engineered TPR domains, a C-terminal capping α-helix. As such, these results attest to a TPR fold with noncanonical features shared between the BubR1 and Bub1 TPR domains (Fig. 4B). A TPR fold similar to the Bub1 TPR domain was predicted for the BubR1 N-terminal region. Key features common to BubR1 and Bub1 but not other TPR domains include a disordered loop within TPR1, a short, approximately perpendicular 310-helix connecting TPR1 and TPR2, a tight turn involving positive Φ angles between TPR2 and TPR3, and noncanonical packing of and between TPR motifs (see below). These features define the “Bub protein” TPR domain that shows diversification of the classical TPR fold. The conservation of these defining features further suggests that they are important for the structural integrity and/or specific functionality.
FIGURE 4.
Comparison of the human BubR1 and yeast Bub1 TPR domains. A, summary of the output from the DALI server. Protein Data Bank codes are 3ESL (Bub1), 1A17 (PP5), and 1NA0 (engineered three-repeat TPR). Res., residues; r.m.s.d., root mean square deviation. B, superposition of the BubR1 (yellow) and Bub1 (red) TPR domains. Shared features are labeled black, and different features are labeled yellow/red for BubR1/Bub1. The Blinkin-binding groove and G(N/D)D motif are labeled gray. C, dimerization interface of the yeast Bub1 TPR domain. Residues involved in salt bridge formation important for dimer stability are shown as sticks and are lightly colored. D, electrostatic surface of the yeast Bub1 TPR domain dimerization interface (left) and the equivalent interface in the human BubR1 TPR domain (right). The electrostatic surface (positive shown in blue, negative shown in red) was calculated using APBS in PyMOL.
The BubR1 and Bub1 TPR domains, however, do adopt distinct conformations peripheral to the three TPR motifs (Fig. 4B). Namely, a disordered region, a short α-helix, and a kinked loop at the N terminus of the BubR1 TPR domain are replaced by an α-helix, a 310-helix, and a short loop at the N terminus of the Bub1 TPR domain. Although potentially due to crystal packing, these distinct conformations may reflect species-specific differences, i.e. the Bub1 conformation may be unique to yeast Bub1 as sequence conservation between Bub1 orthologues is moderate in this region (Fig. 3A). Alternatively, the comparatively flexible conformation of BubR1 may be required for the BubR1-specific function of presenting the upstream KEN box to APC/C-Cdc20. The BubR1 and Bub1 TPR domains also have different C-terminal regions (Fig. 4B). The protruding segment of the C-terminal α-helix and immediately adjacent disordered region of the BubR1 TPR domain are replaced by an unfolded segment that connects to two short α-helices that lie across the outer concave surface of the TPR motifs of the Bub1 TPR domain. As discussed previously, these distinct conformations may be an artifact of crystal packing and/or protein truncation. The moderate sequence conservation of this region also raises the possibility of species-specific differences (Fig. 3A).
A final distinction between the BubR1 and Bub1 TPR domains relates to quaternary structure, with BubR1 being a monomer (Fig. 2B) and Bub1 a dimer (Fig. 4C) in both solution and crystalline states. This difference occurs as Bub1 dimerization is mediated by salt bridges involving residues whose homologous partners in BubR1 are mostly neutral (Fig. 3A and Fig. 4, C and D). This lack of conservation, combined with our yeast two-hybrid data showing that human Bub1fl does not interact with Bub1(1–189) (Fig. 1D), suggests yeast Bub1 dimerization to be species-specific and/or a result of protein truncation. Notably, the TPR domain of yeast Mad3 is monomeric. Thus, although the native oligomeric state of yeast Bub1 remains uncertain, dimerization of the Bub protein TPR domain seems to be the exception rather than the rule.
Noncanonical Packing of the Bub Protein TPR Domain
To further define the noncanonical features of the Bub protein TPR domain, we looked at interactions occurring within a TPR motif. Typical interactions of classical TPR domains involve large hydrophobic residues packing against small hydrophobic residues to create an angle of 6–20° between α-helices (Fig. 5A) (51). In the BubR1 TPR domain, such interactions are present in TPR1 and TPR3 but not TPR2 (Fig. 5B). TPR1 has an angle of 11.9° between α-helices as Trp78 and Thr85 pack against Ala105 and Met98 correspondingly, whereas TPR3 is somewhat extreme and has an angle of 8.6° between α-helices as Tyr155 and Tyr162 pack against Gly178 and Ala171 correspondingly. TPR2 is distinct as pairing of medium sized Leu122 and Leu142, and the presence of helix-truncating Pro135, induce an angle of 25.3° between α-helices. This atypical packing is reflected in the high backbone root mean square deviations between TPR2 and TPR1 (2.67 Å2) or TPR3 (2.92 Å2) compared with that between TPR1 and TPR3 (1.28 Å2). Like many TPR motifs, TPR1 and TPR3 adopt similar conformations despite having less than 4% sequence identity (Fig. 3A). This unique packing within TPR2 is not likely a result of crystal packing as it is conserved in the yeast Bub1 TPR domain structure where pairing of medium sized Leu108 and Met133 similarly causes a >20° angle between the α-helices of TPR2 (Fig. 4B). Ultimately, this unique packing within TPR2 causes it to have noncanonical interactions with neighboring repeats.
The dominant interaction between neighboring repeats of a TPR domain involves the stacking of large hydrophobic residues at TPR consensus positions 7 and 24 (51) (Fig. 5A). In the BubR1 TPR domain, this interaction is observed between the short N-terminal α-helix and TPR1 (Tyr64–Tyr81), TPR2 and TPR3 (Tyr139–Trp158), TPR3, and the C-terminal α-helix (Phe175–His193) but not between TPR1 and TPR2 (Fig. 5C). Between TPR1 and TPR2, the canonical stacking of Leu102 and Trp125 is replaced by salt bridges (Asp73–Arg120 and Glu86–Lys127) and other hydrophobic packing interactions. The residues involved in these ionic interactions are either strictly conserved (Asp73 and Arg120) or conserved in higher eukaryotic Bub proteins (Glu86 and Lys127) (Fig. 3A). These noncanonical interactions are of note as they cause α-helix A of TPR2 to lie across both α-helices of TPR1 (Fig. 5C, panel ii). This ultimately reduces the regularity of the super-helical twist and flattens the concave surface which in a TPR domain is frequently a site of protein interaction (57). Such peculiar packing may be required to accommodate the 310-helix between TPR1 and TPR2 as it is also observed in the yeast Bub1 TPR domain, even though it is achieved through alternative interactions (Fig. 4B). Importantly, a flatter concave surface likely impacts the character of this putative protein interaction site.
Identification of a Blinkin-binding Groove in the BubR1 TPR Domain
A previous analysis of the interaction between human Bub1(1–189) and Blinkin(1–728) implicated the GIG and G(N/D)D motifs of the Bub1 TPR domain (Fig. 4B) (39). The conservation of these motifs in the BubR1 TPR domain prompted us to test whether they are similarly involved in the interaction between human BubR1(1–237) and Blinkin(1–728). The GIG motif superimposes very well between the two structures, although the conformation of the G(N/D)D motif varies (Fig. 4B). Yeast two-hybrid analysis showed that the BubR1-Blinkin interaction was unaffected by mutation of the G(N/D)D motif (N72A/D73A), as well as by most mutations of the GIG motif (G146N, G148N, and G146N/G148N).3 However, the BubR1-Blinkin interaction was abrogated by the GIG mutant G146N/G148V. This is likely explained by the ability of asparagine, but not valine, to adopt positive Φ angles observed for the native glycine residues at these positions that appear important for formation of the TPR domain structure (58). Thus, the G(N/D)D and GIG motifs of BubR1 do not seem to establish direct contacts with Blinkin. Nevertheless, the latter motif is important for the structural integrity of the Blinkin-binding site. Its location at the end of a groove on the surface of BubR1 (Fig. 6C), formed by TPR2 and TPR3, raises the possibility that this constitutes the Blinkin-binding region.
FIGURE 6.
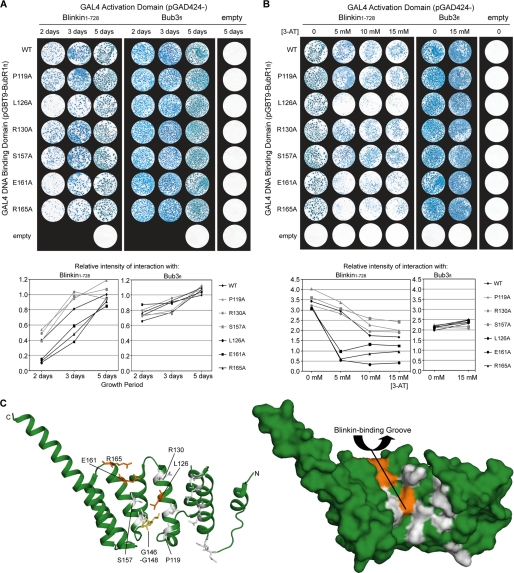
Blinkin binds a groove formed by TPR2 and TPR3 of the BubR1 TPR domain. Time course (A) and high stringency (B) analysis of the interaction between BubR1fl point mutants (wild type (WT), P119A, L126A, R130A, S157A, E161A, and E165A) and Blinkin(1–728) or Bub3fl is shown. Strains Y187(pGBT9-BubR1fl) and AH109(pGAD424-Blinkin(1–728)) or Bub3fl were mated and X-gal filter lift assays performed on colonies grown on SD/−Ade/−His/−Leu/−Trp plates for 2, 3, or 5 days in the presence of 0, 5, 10, or 15 mm 3-AT. Densitometry graphs represent the intensity of each mating condition relative to the intensity of wild type BubR1fl following 5 days of growth with 0 mm 3-AT. C, schematic (left) and surface (right) representation of the BubR1 TPR domain highlighting the residues involved in Blinkin binding (orange, residues not involved are white). The GIG motif (yellow) is not surface-exposed.
To test this, the importance of numerous residues exposed in the BubR1 groove formed by TPR2 and TPR3 was examined. Yeast two-hybrid analysis was performed using a series of BubR1fl alanine mutants (P119A, L126A, R130A, S157A, E161A, and R165A) fused to the GAL4 DNA-binding domain and Blinkin(1–728) or Bub3fl fused to the GAL4 activator domain (Fig. 6C). Although all BubR1fl mutants retained an ability to bind Blinkin(1–728), BubR1fl mutants L126A, E161A, and R165A experienced a highly reproducible reduction in the rate of growth and number of colonies on SD/−Ade/−His/−Leu/−Trp plates (selecting for the presence of both plasmids, along with HIS and ADE reported gene activation) and β-galactosidase activity (lacZ reporter gene activation) (Fig. 6A). This was particularly noticeable when mated yeast were incubated on selection plates for 2 or 3 days rather than 5 days. The rate of yeast growth, the number of resultant colonies, and the rate and intensity of X-gal reaction are directly related to the underlying level of reporter gene activation, which for constant GAL4 DNA-binding domain bait and GAL4 activation domain target fusion proteins (modified by point mutagenesis) provide “semi-quantitative” assessment of binding affinities (41). Thus, our observations indicate reduced levels of reporter gene activation, and hence reduced binding affinity, for BubR1fl mutants L126A, E161A, and R165A. Importantly, for all BubR1fl mutants, the growth of mated yeast on SD/−Leu/−Trp plates (selecting for the presence of both plasmids) was unaffected, and a wild type level of interaction with Bub3fl was detected at all time points (Fig. 6A). These observations exclude the possibility of the mutants affecting the growth characteristics of the yeast, fusion protein expression levels, or fusion protein stability. Instead, these data are consistent with the L126A, E161A, and R165A BubR1 mutants reducing the affinity of the interaction between BubR1fl and Blinkin(1–728).
To confirm the involvement of BubR1 residues Leu126, Glu161, and Arg165 in Blinkin(1–728) binding, mated yeast were selected on SD/−Ade/−His/−Leu/−Trp plates containing 3-AT, a competitive inhibitor of the HIS3 reporter gene product frequently used to eliminate background levels of reporter gene activation. Upon testing the interaction with Blinkin(1–728), the growth of colonies for BubR1fl mutants L126A, E161A, and R165A was almost completely eliminated by 5 mm 3-AT (Fig. 6B). This is in contrast to the growth of colonies for wild type BubR1fl and mutants P119A, R130A, and S157A, which were only mildly affected. Increasing 3-AT to 10 or 15 mm had little additional effect on BubR1fl mutants L126A, E151A, and R165A, while furthering the mild inhibition of wild type BubR1fl and mutants P119A, R130A, and S157A (Fig. 6B). The interactions between all BubR1fl mutants and Bub3fl were equally unaffected by the presence of 5–15 mm 3-AT, consistent with a Bub3-binding site downstream of the BubR1 TPR domain (36, 37). We therefore conclude that the affinity of the BubR1fl-Blinkin(1–728) interaction is substantially reduced by BubR1 mutations L126A, E161A, and R165A and is unaffected by the BubR1 mutations P119A, R130A, and S157A. The yeast two-hybrid data presented here thus demonstrate that side chains of Leu126, Glu161, and Arg165 contribute to the Blinkin-binding interface of the BubR1 TPR domain. These residues belong to a shallow groove on the inner concave surface of the BubR1 domain defined by TPR2 and TPR3 (Fig. 6C). Although each individual residue is not essential for the interaction, mutation results in a reduction in binding affinity. This could be characteristic of a “Velcro-like” mode of interaction where multiple discrete and potentially cooperative interfaces contribute to the overall protein-protein interaction.
DISCUSSION
Distinct roles have been described for BubR1 in its enrichment at kinetochores incorrectly/not attached to the mitotic spindle and as a constituent of the MCC, which inhibits APC/C-Cdc20 ubiquitin ligase activity. The mechanistic links between these cellular pools of BubR1 and their contribution to the SAC, however, remain unknown. Such understanding requires detailed molecular information regarding BubR1 and the basis of its interactions both at the kinetochore and within the MCC. Thus far, this has not been possible as recombinant protein instability has hampered high resolution structural analysis of BubR1 or its paralogue Mad3 (35, 36). Here, we have overcome this difficulty and, through detailed mapping of the molecular architecture of the functionally significant N-terminal portion of human BubR1, have solved the crystal structure of its TPR domain at 1.8 Å resolution. The structure reveals subtle but significant deviations from a canonical TPR domain that likely provide protein-specific functionality. Such deviations are shared with the yeast Bub1 TPR domain and thus define a common structural scaffold of Bub proteins. Key distinctive features include the disordered loop insertion of TPR1, the insertion of a 310-helix between TPR2 and TPR3, the tight turn involving positive Φ angles between TPR2 and TPR3, and the noncanonical packing of and between TPR motifs.
Analysis based on the BubR1 structure also facilitated novel molecular insight into BubR1 interactions at the kinetochore. We confirmed the interaction between BubR1(1–237) and Blinkin(1–728) using a yeast two-hybrid assay and defined the molecular determinants of this interaction through structure-directed mutagenesis. This revealed a series of residues, Leu126, Glu161, and Arg165, located within a shallow groove on the inner concave surface of the BubR1 TPR domain that each contribute to the Blinkin interaction. The significant reduction, but not elimination, of the Blinkin interaction upon individual mutation of these residues, and the discrete distribution of these residues within the groove, suggests the interaction is mediated by a number of potentially cooperative interfaces. This binding groove is subtly distinct from that of other TPR domains as the unusual packing of TPR2 means it is formed by the A α-helices of TPR2 and TPR3 but not TPR1.3 Canonical TPR domains typically contain a binding pocket formed by all three A α-helices on the inner concave surface (57). The overall polarity of this groove and the importance of residues Glu161 and Arg165 suggest a role for ionic contacts in mediating the Blinkin interaction, possibly compensating for relatively weak hydrophobic packing associated with the shallowness of the groove. The Blinkin-binding groove of the BubR1 TPR domain thus differs from previous characterized TPR domains and likely provides a unique functionality.
A complementary aspect of BubR1 function is Bub1-dependent kinetochore enrichment (29, 32). Our data reveal a physical interaction between BubR1 and Bub1 that is mediated by regions downstream of their TPR domains. Both BubR1 and Bub1 undergo direct interactions with Blinkin via their TPR domains, with their respective “Blinkin-binding” grooves showing gross structural similarity. However, specific differences are predicted as Glu161 and Arg165 are not conserved in Bub1, and the G(N/D)D motif may be differentially required (39), suggesting that the BubR1 and Bub1 TPR domains may bind to distinct regions within Blinkin. This is supported by the finding that although BubR1 binding is restricted to the N-terminal region of Blinkin, Bub1 also interacts with the central region of Blinkin (32). Future work is clearly required to define the precise nature of these interactions. Nevertheless, this combination raises the possibility of a ternary complex between BubR1, Bub1, and Blinkin at kinetochores; their orderly assembly may be essential for SAC activation.
The molecular architecture of the larger BubR1 N-terminal region provides further insight into its role in APC/C-Cdc20 inhibition as part of the MCC. Specifically, the first KEN box of BubR1 is located within the flexible N-terminal extension that protrudes from the TPR domain. APC/C-Cdc20 recognition of the KEN box in this location may thus be affected by a recently reported phosphorylation event at Thr54, which lies at the boundary between the N-terminal extension and the TPR domain (59). In addition to BubR1 KEN boxes, APC/C-Cdc20 inhibition may also depend on interactions between the BubR1 TPR domain and other APC/C components. This is supported by a recent electron microscopy structure of the human APC-C-MCC complex, which reveals that the MCC, and in particular BubR1, is located in close proximity to APC/C TPR protein APC5 (21).
On the basis of findings reported here and in previous studies, we present a model for BubR1 SAC function (Fig. 7). First, BubR1 is recruited to kinetochores incorrectly/not attached to the mitotic spindle through a direct interaction with Bub1. A ternary complex of BubR1, Bub1, and Blinkin is thus formed (32). The TPR domains of BubR1 and Bub1 each bind Blinkin, although their C-terminal regions mediate interactions with each other and with Bub3 (36, 37). This complex is predicted to sustain the SAC, with the kinase activity and/or phosphorylation of BubR1 triggering prolonged mitotic arrest (23). Second, BubR1 inhibits APC/C-Cdc20 ubiquitin ligase activity as part of the MCC (16). This is achieved through direct interactions between BubR1 KEN boxes and APC/C-Cdc20 (24), and possibly additional interactions between the BubR1 TPR domain and APC/C subunits. A potential mechanism by which BubR1 kinetochore enrichment induces prolonged APC/C-Cdc20 inhibition is a signal cascade in which downstream targets include enzymes that post-translationally modify BubR1, other MCC components, or APC/C-Cdc20, thus affecting APC/C-Cdc20 ubiquitin ligase activity. Although details of SAC silencing remain to be elucidated (60), one redundant mechanism may involve inhibition of BubR1 kinase activity by CENP-E, upon CENP-E microtubule capture (31). However, these models have not yet been tested within the cellular context. The detailed molecular understanding of the BubR1 N-terminal domain provided by this study will now permit such an analysis to be performed; structure-directed engineering of BubR1 will for the first time allow its distinct roles in kinetochore recruitment and APC/C-Cdc20 inhibition to be dissected in vivo.
FIGURE 7.
BubR1 exerts a SAC function through interaction with numerous protein ligands. The N-terminal region of BubR1 mediates interactions with Blinkin, Cdc20, and possibly the APC/C, although the C-terminal region forms contacts with Bub3, Bub1, and CENP-E.
Supplementary Material
Acknowledgments
We thank L. Packman for mass spectrometry analysis; M. Weldon for Edman degradation; M. Hyvönen for reagents and advice; G. Grossman for assistance with small angle x-ray scattering; M. Moncrieffe for assistance with analytical ultracentrifugation; B. Luisi for advice on phasing; T. Kiyomitsu and M. Yanagida for initial yeast two-hybrid experiments; and S. Lowell for support.
This work was supported by Wellcome Trust Programme Grant RG44650 (to T. L. B.) and funding from the Royal Commission for the Exhibition of 1851 (to O. R. D.), the University of Sydney, Kings College London, and the Cambridge Commonwealth Trust (to S. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods,” Figs. S1 and S2, and additional references.
The atomic coordinates and structure factors (code 2WVI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
S. D'Arcy, T. Kiyomitsu, and V. Bolanos-Garcia, unpublished data.
- SAC
- spindle assembly checkpoint
- MCC
- mitotic checkpoint complex
- TPR
- tetratricopeptide repeat
- fl
- full-length
- 3-AT
- 3-amino-1,2,3-triazole
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- APC/C
- anaphase promoting complex/cyclosome.
REFERENCES
- 1.Zich J., Hardwick K. G. (2010) Trends Biochem. Sci. 35, 18–27 [DOI] [PubMed] [Google Scholar]
- 2.Kops G. J. (2008) Front. Biosci. 13, 3606–3620 [DOI] [PubMed] [Google Scholar]
- 3.Thornton B. R., Toczyski D. P. (2003) Nat. Cell Biol. 5, 1090–1094 [DOI] [PubMed] [Google Scholar]
- 4.Uhlmann F., Wernic D., Poupart M. A., Koonin E. V., Nasmyth K. (2000) Cell 103, 375–386 [DOI] [PubMed] [Google Scholar]
- 5.Cahill D. P., Lengauer C., Yu J., Riggins G. J., Willson J. K., Markowitz S. D., Kinzler K. W., Vogelstein B. (1998) Nature 392, 300–303 [DOI] [PubMed] [Google Scholar]
- 6.Logarinho E., Bousbaa H. (2008) Cell Cycle 7, 1763–1768 [DOI] [PubMed] [Google Scholar]
- 7.Weaver B. A., Cleveland D. W. (2006) Curr. Opin. Cell Biol. 18, 658–667 [DOI] [PubMed] [Google Scholar]
- 8.Hanks S., Coleman K., Reid S., Plaja A., Firth H., Fitzpatrick D., Kidd A., Méhes K., Nash R., Robin N., Shannon N., Tolmie J., Swansbury J., Irrthum A., Douglas J., Rahman N. (2004) Nat. Genet. 36, 1159–1161 [DOI] [PubMed] [Google Scholar]
- 9.Matsuura S., Matsumoto Y., Morishima K., Izumi H., Matsumoto H., Ito E., Tsutsui K., Kobayashi J., Tauchi H., Kajiwara Y., Hama S., Kurisu K., Tahara H., Oshimura M., Komatsu K., Ikeuchi T., Kajii T. (2006) Am. J. Med. Genet. A 140, 358–367 [DOI] [PubMed] [Google Scholar]
- 10.Babu J. R., Jeganathan K. B., Baker D. J., Wu X., Kang-Decker N., van Deursen J. M. (2003) J. Cell Biol. 160, 341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker D. J., Jeganathan K. B., Cameron J. D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R. B., de Groen P. C., Roche P., van Deursen J. M. (2004) Nat. Genet. 36, 744–749 [DOI] [PubMed] [Google Scholar]
- 12.Dai W., Wang Q., Liu T., Swamy M., Fang Y., Xie S., Mahmood R., Yang Y. M., Xu M., Rao C. V. (2004) Cancer Res. 64, 440–445 [DOI] [PubMed] [Google Scholar]
- 13.Rao C. V., Yang Y. M., Swamy M. V., Liu T., Fang Y., Mahmood R., Jhanwar-Uniyal M., Dai W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4365–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Royou A., Gagou M. E., Karess R., Sullivan W. (2010) Cell 140, 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Homer H., Gui L., Carroll J. (2009) Science 326, 991–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sudakin V., Chan G. K., Yen T. J. (2001) J. Cell Biol. 154, 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfleger C. M., Kirschner M. W. (2000) Genes Dev. 14, 655–665 [PMC free article] [PubMed] [Google Scholar]
- 18.Fang G. (2002) Mol. Biol. Cell 13, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulukian A., Han J. S., Cleveland D. W. (2009) Dev. Cell 16, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davenport J., Harris L. D., Goorha R. (2006) Exp. Cell Res. 312, 1831–1842 [DOI] [PubMed] [Google Scholar]
- 21.Herzog F., Primorac I., Dube P., Lenart P., Sander B., Mechtler K., Stark H., Peters J. M. (2009) Science 323, 1477–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson J., Yekezare M., Minshull J., Pines J. (2008) Nat. Cell Biol. 10, 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malureanu L. A., Jeganathan K. B., Hamada M., Wasilewski L., Davenport J., van Deursen J. M. (2009) Dev. Cell 16, 118–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burton J. L., Solomon M. J. (2007) Genes Dev. 21, 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi E., Choe H., Min J., Choi J. Y., Kim J., Lee H. (2009) EMBO J. 28, 2077–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howell B. J., Moree B., Farrar E. M., Stewart S., Fang G., Salmon E. D. (2004) Curr. Biol. 14, 953–964 [DOI] [PubMed] [Google Scholar]
- 27.Cheeseman I. M., Desai A. (2008) Nat. Rev. Mol. Cell Biol. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 28.Millband D. N., Hardwick K. G. (2002) Mol. Cell. Biol. 22, 2728–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson V. L., Scott M. I., Holt S. V., Hussein D., Taylor S. S. (2004) J. Cell Sci. 117, 1577–1589 [DOI] [PubMed] [Google Scholar]
- 30.Mao Y., Desai A., Cleveland D. W. (2005) J. Cell Biol. 170, 873–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao Y., Abrieu A., Cleveland D. W. (2003) Cell 114, 87–98 [DOI] [PubMed] [Google Scholar]
- 32.Kiyomitsu T., Obuse C., Yanagida M. (2007) Dev. Cell 13, 663–676 [DOI] [PubMed] [Google Scholar]
- 33.Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. (2006) Cell 127, 983–997 [DOI] [PubMed] [Google Scholar]
- 34.Elowe S., Dulla K., Uldschmid A., Li X., Dou Z., Nigg E. A. (2010) J. Cell Sci. 123, 84–94 [DOI] [PubMed] [Google Scholar]
- 35.Yoon J., Kang Y., Kim K., Park J., Kim Y. (2005) Protein Expr. Purif. 44, 1–9 [DOI] [PubMed] [Google Scholar]
- 36.Larsen N. A., Al-Bassam J., Wei R. R., Harrison S. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1201–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Babu J. R., Harden J. M., Jablonski S. A., Gazi M. H., Lingle W. L., de Groen P. C., Yen T. J., van Deursen J. M. (2001) J. Biol. Chem. 276, 26559–26567 [DOI] [PubMed] [Google Scholar]
- 38.Hardwick K. G., Johnston R. C., Smith D. L., Murray A. W. (2000) J. Cell Biol. 148, 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolanos-Garcia V. M., Kiyomitsu T., D'Arcy S., Chirgadze D. Y., Grossmann J. G., Matak-Vinkovic D., Venkitaraman A. R., Yanagida M., Robinson C. V., Blundell T. L. (2009) Structure 17, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolanos-Garcia V. M., Beaufils S., Renault A., Grossmann J. G., Brewerton S., Lee M., Venkitaraman A., Blundell T. L. (2005) Biophys. J. 89, 2640–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacDonald P. N. (2001) Methods Mol. Biol. 177, v–viii [PubMed] [Google Scholar]
- 42.Peränen J., Rikkonen M., Hyvönen M., Kääriäinen L. (1996) Anal. Biochem. 236, 371–373 [DOI] [PubMed] [Google Scholar]
- 43.Collaborative Computational Project No. 4 (CCP4) (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 44.Leslie A. G. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 45.Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 46.Perrakis A., Sixma T. K., Wilson K. S., Lamzin V. S. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 448–455 [DOI] [PubMed] [Google Scholar]
- 47.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 48.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 49.Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 50.D'Andrea L. D., Regan L. (2003) Trends Biochem. Sci. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 51.Main E. R., Xiong Y., Cocco M. J., D'Andrea L., Regan L. (2003) Structure 11, 497–508 [DOI] [PubMed] [Google Scholar]
- 52.Sikorski R. S., Boguski M. S., Goebl M., Hieter P. (1990) Cell 60, 307–317 [DOI] [PubMed] [Google Scholar]
- 53.Das A. K., Cohen P. W., Barford D. (1998) EMBO J. 17, 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J., Roe S. M., Cliff M. J., Williams M. A., Ladbury J. E., Cohen P. T., Barford D. (2005) EMBO J. 24, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rice L. M., Brunger A. T. (1999) Mol. Cell 4, 85–95 [DOI] [PubMed] [Google Scholar]
- 57.Cliff M. J., Harris R., Barford D., Ladbury J. E., Williams M. A. (2006) Structure 14, 415–426 [DOI] [PubMed] [Google Scholar]
- 58.Deane C. M., Allen F. H., Taylor R., Blundell T. L. (1999) Protein Eng. 12, 1025–1028 [DOI] [PubMed] [Google Scholar]
- 59.Imami K., Sugiyama N., Kyono Y., Tomita M., Ishihama Y. (2008) Anal. Sci. 24, 161–166 [DOI] [PubMed] [Google Scholar]
- 60.Vanoosthuyse V., Hardwick K. G. (2009) Genes Dev. 23, 2799–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.