Abstract
Neutrophil gelatinase-associated lipocalin (NGAL) is a siderophore-binding antimicrobial protein that is up-regulated in epithelial tissues during inflammation. We demonstrated previously that the gene encoding NGAL (LCN2) is strongly up-regulated by interleukin (IL)-1β in an NF-κB-dependent manner but not by tumor necrosis factor (TNF)-α, another potent activator of NF-κB. This is due to an IL-1β-specific synthesis of the NF-κB-binding co-factor IκB-ζ, which is essential for NGAL induction. We demonstrate here that NGAL is strongly induced by stimulation with TNF-α in the presence of IL-17, a pro-inflammatory cytokine produced by the newly discovered subset of CD4+ T helper cells, TH-17. In contrast to the murine NGAL orthologue, 24p3/lipocalin 2, we found no requirement for C/EBP-β or C/EBP-δ for NGAL induction by IL-17 and TNF-α as neither small interfering RNAs against the two C/EBP mRNAs nor mutation of the C/EBP sites in the LCN2 promoter abolished IL-17- and TNF-α-induced up-regulation of NGAL. NGAL induction is governed solely by NF-κB and its co-factor IκB-ζ. This was demonstrated by a pronounced reduction in the amount of NGAL mRNA and NGAL protein synthesized in cells treated with small interfering RNA against IκB-ζ and a total lack of activation of an LCN2 promoter construct with a mutated NF-κB site. As IL-17 stimulation stabilizes the IκB-ζ transcript, we propose a model where TNF-α induces activation and binding of NF-κB to the promoters of both NFKBIZ and LCN2 genes but induce only transcription of IκB-ζ. Co-stimulation with IL-17 leads to accumulation of IκB-ζ mRNA and IκB-ζ protein, which can bind to NF-κB on the LCN2 promoter and thus induce NGAL expression.
Keywords: Cytokines/Interleukins, Cytokines/Tumor Necrosis Factor, Gene/Regulation, Inflammation, Innate Immunity, NF-κB, Transcription Factors
Introduction
The innate immune system has evolved in higher eukaryotes to combat microorganisms. It is activated rapidly and acts as the first line of defense against invading pathogens. Cells of the epithelial lining are among the initial players of the innate immune response. Receptors on epithelial cells, interstitial macrophages, and dendritic cells recognize pathogen-associated molecular patterns on microorganisms and respond by de novo synthesis of antimicrobial proteins and peptides as well as pro-inflammatory cytokines. Subsequently, specialized mobile phagocytes such as neutrophils and monocytes appear at the scene to muster a more full-blown counterattack against the microorganisms. Growing evidence indicates that the response of epithelial cells to the invading pathogen is fine tuned to eliminate the specific pathogen. As a consequence, expression of genes encoding the antimicrobial proteins is differentially regulated to match the specific stimuli encountered by the cells (1–4).
One of the genes found to be differentially regulated codes for the protein neutrophil gelatinase-associated lipocalin (NGAL)2 (1). NGAL is a 25-kDa glycoprotein originally identified as a matrix protein of specific granules of human neutrophils (5) and later also as a protein strongly up-regulated in epithelial cells during inflammation (1, 2, 6–8). NGAL belongs to the lipocalin superfamily whose members share a barrel-shaped tertiary structure with a hydrophobic pocket that can bind lipophilic molecules (9). The ligand of NGAL is bacterial ferric siderophores, which are used by bacteria for uptake of iron, an essential nutrient (10, 11). Binding of siderophores by NGAL deprives bacteria of iron. This bacteriostatic effect has been demonstrated both in vitro (10) and in a mouse model with a targeted disruption of the murine orthologue of NGAL (24p3/lipocalin 2) (11).
Expression of human NGAL in epithelial cells is induced by IL-1β in an NF-κB-dependent manner but not by TNF-α despite the well known ability of both cytokines to induce activation and binding of NF-κB to the LCN2 promoter (1, 2, 11, 12). The reason for this IL-1β specificity is a requirement for co-factor IκB-ζ (which itself is induced specifically by IL-1β) for activation of the LCN2 promoter through the NF-κB signaling pathway.
Recently, a new subgroup of T helper cells (TH-17) was characterized that act in the interface between the adaptive and innate immune system. When activated by pathogens and specific cytokines, naive CD4+ T cells develop into TH-17 cells that produce IL-17 and release it to the surroundings (13). IL-17 can stimulate epithelial cells to synthesize cytokines such as IL-6 and granulocyte macrophage-colony-stimulating factor as well as chemokines such as CXCL1 and CXCL8 (IL-8) (14, 15). It was recently demonstrated that IL-17 is also able to induce expression of antimicrobial proteins such as the murine orthologue of NGAL, 24p3/lipocalin 2, in mouse cells (16, 17), and hBD2 in some human cell systems (4). Furthermore, it was shown that TNF-α can potentiate the expression of IL-17-induced lipocalin 2 expression (16, 17). This prompted us to investigate whether IL-17 could also induce synthesis of human NGAL. Here we demonstrate that co-stimulation of A549 cells with IL-17 and TNF-α results in strong up-regulation of NGAL, whereas neither of the two cytokines alone have an effect on NGAL expression. Furthermore, we found that induction of NGAL expression is dependent on NF-κB and its co-activator IκB-ζ but not on C/EBP-β or C/EBP-δ.
EXPERIMENTAL PROCEDURES
Cell Culture
A549 (ATCC CCL-185) cells were obtained from the American Type Culture Collection and grown in Ham's F-12 (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen), 100 units/ml of penicillin, and 100 μg/ml of streptomycin (Invitrogen) at 37 °C in a humid atmosphere with 5% CO2. For transfection and/or induction with IL-17 and TNF-α (both eBioscience) or incubation with IL-1β or cycloheximide (both Sigma), 4 × 105 A549 cells were plated in a 10-cm dish and grown for 2 days before use. When stimulating with cytokines, the medium was changed to Ham's F-12 with 0.5% fetal calf serum and 100 units/ml of penicillin, and 100 μg/ml of streptomycin prior to the experiment.
RNA Isolation and Northern Blot
Total RNA was prepared with TRIzol (Invitrogen) according to the manufacturer's recommendations and the concentration determined by spectrophotometry. For Northern blotting, 5 μg of RNA was run on a 1% agarose gel, transferred to a Hybond-N membrane (GE Healthcare), and hybridized as described (1). The membranes were washed as described (1) and developed with a Fuji BAS2500 PhosphorImager. The size of the mRNAs was determined by reference to 18 S and 28 S ribosomal RNA. The membranes were stripped by boiling in 0.1% SDS before rehybridization. The NGAL and β-actin cDNA probes have been described earlier (1). The probes were radiolabeled with [α-32P]dCTP using the random Primers DNA Labeling System (Invitrogen). For quantitative assessments, the intensities of the NGAL signals were normalized to the hybridization intensity from a probe against β-actin.
Quantitative Real Time PCR
Reverse transcription of RNA to first strand cDNA was performed on 1 μl of RNA diluted in 10 μl of diethyl pyrocarbonate-treated water. 1 μl of 50 μm random hexamer primers (Invitrogen) were added. The solution was denatured at 70 °C for 10 min followed by addition of 4 μl of 5 × First Strand Buffer (Promega), 1 μl of 10 mm dNTP, 1 μl of RNAsin (Invitrogen), 2 μl of 0.1 m dithiothreitol (Promega), and 1 μl of SuperScript II Reverse Transcriptase (Invitrogen). The conditions for reverse transcription was incubation at 22 °C for 10 min, 45 °C for 45 min, and 98 °C for 5 min. Quantitative PCR analysis was performed on a MX 3000P Real Time PCR system (Stratagene) with the commercial gene expression assay Assay-on-demandTM (Applied Biosystems). The assays used were: LCN2 (NGAL), Hs00194353_m1; IL-8, Hs00174103_m1; IL-6, Hs00174131_m1; NFKBIZ (IκB-ζ), Hs00230071_m1; CEBPB (C/EBP-β), Hs00270923_s1; CEBPD (C/EBP-δ), Hs00270931_s1; and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) Hs99999905_m1.
Plasmids and siRNA
The promoter constructs pNGP1695CAT, pNGP1695(NF-κB)CAT, pNGP1695(C/EBP-1)CAT, pNGP1695(C/EBP-2)CAT, pNGP1695(NF-Y)CAT, and pNGP1695(AP-1)CAT are described elsewhere (1). pNGP1695(C/EBP 1 and 2) was made by use of the QuikChange kit (Stratagene) using pNGP1695(C/EBP-2)CAT as template and primers 5′- ACTCTCCCCGTCCCTCTGTCccGCCCAATCCTGACCAGGTGC-3′ and 5′-GCACCTGGTCAGGATTGGGCggGACAGAGGGACGGGGAGAGT-3′ for site-directed mutagenesis (the altered bases in the mutant are shown as lowercase italic letters). The substitution mutants Sub C1 and Sub C2 are identical to pNGP1695(154/135) and pNGP1695(139/120) described in Ref. 1. The promoter construct Sub C1-C2 was made by PCR amplification with primers 5′-AGAATTCTTTCGTTCTTTGATCTTGCCAAGTGTTTCCGCAGG-3′ and 5′-AAGAATTCTCCGCTTCTCCGCTCGGACGGGGAGAGTGAGGG-3′, which anneal on pNGP1695CAT 33 bp apart on each side of C/EBP-1 and C/EBP-2, respectively. The entire plasmid was PCR amplified and the resulting PCR product digested with EcoRI (sequence shown in italics). Following re-ligation, a promoter construct was generated where the region of the LCN2 promoter with the two C/EBP sites was substituted by a nonsense sequence (underlined in the primers). The plasmid with 4 tandem repeats of the IL-8 NF-κB site (IL-8(NF-κB × 4)) has been described previously (1). The plasmid with 4 tandem repeats of the IL-6 NF-κB site (IL-6(NF-κB × 4)) was made by digestion of the pCAT3-promoter plasmid (Promega) with BglII and NheI and ligation of a double-stranded DNA fragment formed by annealing primers 5′-CTAGC(AATGTGGGATTTTCCCATGA)4A-3′ and 5′-GATCT(TCATGGGAAAATCCCACATT)4G-3′. By annealing the two oligoes, an overhang resembling DNA digested with BglII and NheI was formed. All promoter mutations were verified by sequencing. The 24p3 (Lcn2) promoter constructs 24p3–282-Luc, 24p3–282-κBm-Luc, and 24p3–282-C/EBPm-Luc were kindly provided by Dr. Sarah Gaffen, University of Pittsburg (17).
All siRNAs were purchased as pre-designed SilencerTM siRNAs from Ambion. For knockdown of IκB-ζ, an siRNA (siκB) (ID number 33380) was used, which we previously found very efficient against IκB-ζ mRNA (18). Two siRNAs against C/EBP-β (siB1 (ID number 42013) and siB2 (ID number 42089)) and two siRNAs against C/EBP-δ (siD1 (ID number 45017) and siD2 (ID number 15987)) were found to efficiently knock down their cognate transcripts. A Silencer Negative Control number 1 siRNA (Ambion) was included as control for any nonspecific effects of siRNA treatment.
Cell Transfection and Reporter Enzyme Assay
For experiments with siRNA, transfection with Lipofectamine 2000 (Invitrogen) was performed according to the manufacturer's instructions. Transfections involving only plasmid DNA were performed by use of Effectene (Qiagen, Hilden, Germany). For promoter studies, 0.8 μg of the CAT plasmid promoter construct was co-transfected with 0.2 μg of Rous sarcoma virus-Luc plasmid (19) (kindly provided by Dr. M. Schuster, Gene Therapy Laboratory, Rigshospitalet, Denmark) to compensate for differences in transfection efficiency. Expression of the reporter enzymes was quantitated by CAT ELISA (Roche Diagnostics) and Luciferase Reporter Assay (Promega) according to the manufacturer's recommendations. For each sample the CAT activity was normalized to firefly luciferase activity. For analysis of the 24p3 promoter 0.8 μg of the firefly luciferase reporter plasmid was co-transfected with 0.2 μg of the plasmid pGL4.74 (Promega) expressing Renilla luciferase for normalization of the transfection. Expression of the luciferase was determined by the Dual Luciferase Reporter Assay System (Promega).
Nuclear Extracts and EMSA
Nuclear proteins were prepared from A549 cells (either unstimulated or stimulated for 1½ h with IL-17 and TNF-α) as previously described (20). Double-stranded oligonucleotides used as probes were labeled with [γ-32P]ATP and incubated with nuclear extracts for 30 min at room temperature in 20 mm HEPES buffer (pH 7.9), containing 50 mm KCl, 1 mm EDTA, 2 mm dithiothreitol, 10% (w/v) glycerol, 2 μg of poly(dI-dC), 0.1% Nonidet P-40, 1 mg/ml of nuclease-free BSA, and 0.5 mm phenylmethylsulfonyl fluoride. If competitor oligos were included, the nuclear extract was added following a 20-min preincubation period. For supershift analysis 1 μl of antibody against C/EBP-β (sc-150X), C/EBP-δ (sc-151X), C/EBP-ϵ (sc-158X), NF-κB p50 (sc-114X), or NF-κB p65 (sc-109X) (all Santa Cruz) were added. All incubations were subjected to electrophoresis on 6% non-denaturing polyacrylamide gels and subsequently dried and autoradiographed. The oligoes used for EMSA were (only the sequence of the upper strand of the double-stranded probe/competitor is shown): NGAL-κB, 5′-CACTCCGGGAATGTCCCTCACT-3′; NGAL-κB*, 5′-CACTCCAATAATGTCCCTCACT-3′; C1, 5′-CTCTGTCTTGCCCAATCCTGAC-3′; C1*, 5′-CTCTGTCCCGCCCAATCCTGAC-3′; C2, 5′-TGACCAGGTGCAGAAATCTTGC-3′; C2*, 5′-TGACCAGGTGCAGGGATCTTGC-3′; cons. κB, 5′-GATCCGGGGACTTTCCATGGATGGGGACTTTCCATGG-3′; and CRP, 5′-TACATAGTGGCGCAAAGTGATAT-3′.
Quantitation of NGAL, IL-8, and IL-6 in Medium
NGAL was quantitated by ELISA as described previously (21). IL-8 was quantitated with the IL-8 opti-EIA ELISA kit (Pharmingen), and IL-6 was measured with the Ready-SET-Go ELISA (eBioscience) according to the manufacturer's recommendations.
SDS-PAGE and Immunoblotting
For immunodetection, the proteins were separated on a 4–12% NuPAGE BisTris gel (Invitrogen) and electrotransferred to a TransBlot nitrocellulose membrane (Bio-Rad) according to the manufacturer's instructions. The membrane was blocked for 1 h with 5% skimmed milk and washed four times for 5 min in phosphate-buffered saline with 0.5% BSA. The primary antibodies (C/EBP-β (sc-150), C/EBP-δ (sc-151), and glyceraldehyde-3-phosphate dehydrogenase (sc-20357), all from Santa Cruz) were incubated overnight at 4 °C in phosphate-buffered saline with 0.5% BSA and then washed four times for 5 min in phosphate-buffered saline with 0.5% BSA. The membranes were then incubated for 2 h with secondary antibody (peroxidase-conjugated goat anti-rabbit antibodies (Dako P0448)), washed four times for 5 min in phosphate-buffered saline with 0.5% BSA, and visualized by chemiluminescence (SuperSignal West Pico, Thermo Scientific). For incubation with anti-IκB-ζ (Cell Signaling number 9244), the membrane was blocked in Tris-buffered saline (TBS) with 0.1% Tween 20 (TBS/T) containing 5% skimmed milk according to the manufacturer's recommendations. The membranes were washed four times for 5 min with TBS/T, incubated overnight at 4 °C with the primary antibody in TBS/T plus 5% BSA, washed four times for 5 min with TBS/T, incubated for 1 h with the secondary antibody (peroxidase-conjugated goat anti-rabbit) in TBS/T with 5% skimmed milk, and finally washed four times for 5 min with TBS/T before detection of the proteins by chemiluminescence.
Statistical Analysis
The data are presented as the mean ± S.D. The significance of differences between two groups was determined by the Welch t test. Statistical significance was reported if p < 0.05 was achieved.
RESULTS
NGAL Synthesis Is Induced by Co-stimulation with IL-17 and TNF-α
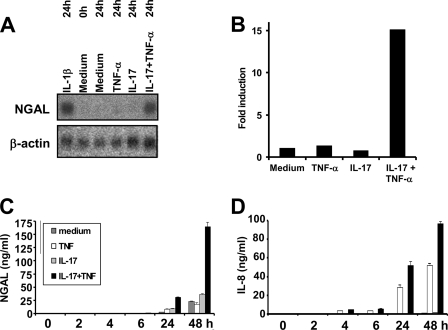
We previously demonstrated that transcription of the human LCN2 promoter is strongly up-regulated by the proinflammatory cytokine IL-1β in A549 cells, whereas no response is observed upon stimulation with TNF-α (1). This is in contrast to the gene encoding the murine orthologue of NGAL (24p3/lipocalin 2), as expression of this gene (Lcn2) can be induced by TNF-α (16, 22). Recently, it was shown that transcription of the murine Lcn2 gene was synergistically up-regulated in the osteoblast cell line MC3T3-E1 when stimulated with a combination of TNF-α and IL-17 (16). For this reason we speculated whether it would be possible to induce expression of the human LCN2 gene by stimulation of cells with TNF-α in the presence of IL-17. To investigate this, A549 cells were incubated with TNF-α and/or IL-17 for 24 h followed by RNA purification and Northern blot analysis. Cells stimulated with IL-1β were included as a positive control for NGAL induction. As seen in Fig. 1, strong induction of NGAL mRNA was observed in cells stimulated with IL-17 and TNF-α or IL-1β. A 15-fold increase in NGAL mRNA was measured in cells co-stimulated with IL-17 and TNF-α, whereas no effect was seen when the cells were stimulated with either of the two cytokines. A second experiment, where the amount of NGAL secreted from A549 cells to the medium was measured over a 48-h period, demonstrated than IL-17 combined with TNF-α also induced NGAL protein synthesis (Fig. 1C). IL-8 was included as control (Fig. 1D), as was found previously to be induced by TNF-α alone in these cells (1).
FIGURE 1.
Induction of NGAL synthesis by co-stimulation with IL-17 and TNF-α. A, cells were harvested at time “0” and at 24 h after addition of fresh medium without cytokines or supplemented with IL-1β (100 pg/ml), TNF-α (20 ng/ml), IL-17 (200 ng/ml), or IL17 and TNF-α (200 and 20 ng/ml). RNA was isolated and hybridized to 32P-labeled probes as indicated. B, schematic representation of the fold-induction of NGAL mRNA in A following normalization to β-actin intensities. The fold-induction is shown relative to the amount measured at time 0. C and D, the amount of NGAL and IL-8 in the medium of A549 cells was determined at the indicated time points after addition of fresh medium without cytokines or supplemented with IL-17 and/or TNF-α in the concentrations given. Data from one of two independent experiments showing essentially the same results are presented. Data are presented as the mean ± S.D.
C/EBP-β and C/EBP-δ mRNA Levels Increase following Stimulation with IL-17 and TNF-α
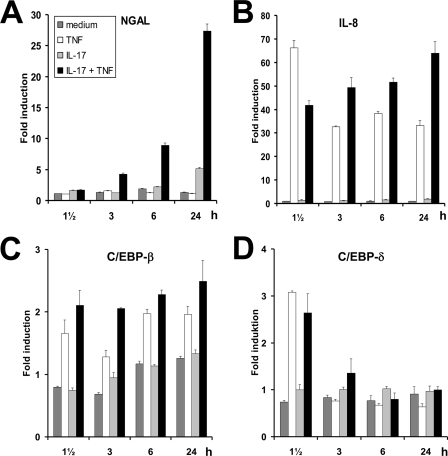
To further investigate the time course of NGAL induction following IL-17 and TNF-α-stimulation, we measured transcript levels by real-time PCR during a 24-h period. Accumulation of NGAL mRNA was observed during the entire period of stimulation (Fig. 2A) in concordance with the NGAL protein levels depicted in Fig. 1C. As seen previously (1), a rapid and sustained increase in IL-8 mRNA levels was found after stimulation with TNF-α (Fig. 2B). A 40–60-fold increase in the amount of IL-8 mRNA was also observed for cells administered IL-17 and TNF-α, whereas stimulation with IL-17 alone had no effect on IL-8 transcript levels.
FIGURE 2.
Time courses of NGAL, IL-8, C/EBP-β, and C/EBP-δ mRNAs following stimulation with IL-17 and/or TNF-α. The amount of NGAL (A), IL-8 (B), C/EBP-β (C), and C/EBP-δ (D) mRNA was determined by real time PCR at the indicated time points after addition of fresh medium without cytokines or supplemented with TNF-α, IL-17, or IL-17 and TNF-α at the concentrations indicated in the legend to Fig. 1. Expression of the transcripts is shown as fold-induction relative to expression measured in unstimulated cells at time 0. Data are presented as the mean ± S.D.
Recent data demonstrate that C/EBP-β and C/EBP-δ are required for IL-17- and TNF-α-induced expression of the murine Lcn2 gene (17) and for this reason we analyzed the mRNA profiles for these two transcription factors. A 2–3-fold increase of both C/EBP-β and C/EBP-δ mRNAs was observed for cells growing in the presence of TNF-α. The transcript for C/EBP-β remained elevated during the entire stimulation period, whereas the amount of C/EBP-δ mRNA peaked at 1½ h and then declined to the background level from 3 h and onwards (Fig. 2, C and D).
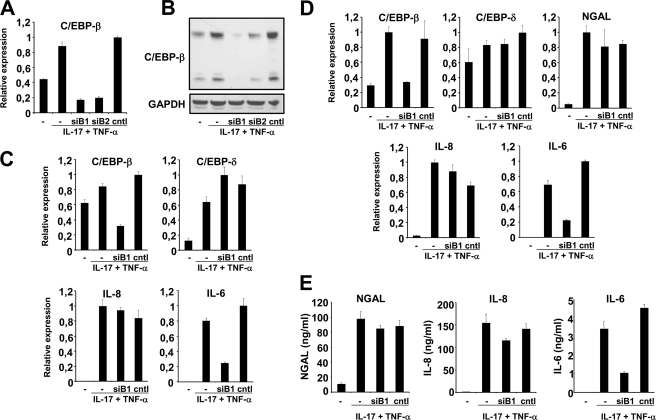
Knockdown of C/EBP-β mRNA Does Not Affect NGAL Production
Because C/EBP-β and C/EBP-δ are reported to be involved in IL-17- and TNF-α-induced expression of the murine Lcn2 promoter (17), we examined the effect of knocking down the mRNAs for these two transcription factors. Two different siRNAs against C/EBP-β (Fig. 3A, siB1 and siB2) were found to reduce the level of C/EBP-β mRNA significantly. Compared with unstimulated cells, a 2–2½-fold increase in the transcript level was observed for C/EBP-β mRNA in untreated and control siRNA-treated cells at 2 h post-induction (Fig. 3A). This is in agreement with the data in Fig. 2C. In contrast, the amount of C/EBP-β mRNA in stimulated cells that had received siB1 or siB2 was about one-third of that found in unstimulated cells and consequently less than 20% of that seen for stimulated cells that were untransfected or transfected with control siRNA. The amount of C/EBP-β protein was significantly reduced in the cells treated with siB1 and siB2 (Fig. 3B) and the treatment affected both the 35- and 20-kDa forms of C/EBP-β (23). In the following experiments only data for siB1 are shown but similar results were obtained with siB2. At 2 h post-induction, the amount of C/EBP-β in cells receiving siB1 was about one-third of that seen in untransfected and Cntl siRNA-treated cells, whereas the amounts of C/EBP-δ and IL-8 mRNAs were similar in all three stimulated cell populations (Fig. 3C). As expression of IL-6 is induced by C/EBP-β and C/EBP-δ (24) we included a measurement of the IL-6 mRNA level in our study to determine whether an effect of knocking down C/EBP-β could be measured. This was found to be the case as the level of IL-6 mRNA was severely diminished in cells treated with siB1 for 2 h (Fig. 3C). When examining the transcript profiles 24 h after co-stimulation with TNF-α + IL-17 the amount of C/EBP-β in siB1-treated cells was also found to be about one-third of that seen in the two other cytokine-treated cell populations that had not been treated with siB1. Also the amount of IL-6 mRNA was around one-third in the siB1-treated cells compared with the other two cell populations stimulated with cytokines. In contrast, the amount of C/EBP-δ, IL-8, and NGAL mRNA was unaffected by siB1 treatment as the amount of transcript was similar in all three stimulated cell populations. When measuring the amount of NGAL, IL-6, and IL-8 in the medium of siRNA-treated cells, the same pattern was observed as for the mRNAs (Fig. 3, D and E). Transfection with siB1 did not diminish the level of either NGAL or IL-8 in the medium, whereas the amount of IL-6 was one-third of that observed for untreated and Cntl siRNA-transfected cells following cytokine stimulation (Fig. 3E).
FIGURE 3.
Knockdown of C/EBP-β expression does not affect NGAL expression. Untransfected A549 cells (−) or cells transfected with 40 nmol of two different siRNAs against C/EBP-β (siB1 and siB2) or control siRNA (cntl) and stimulated with IL-17 (200 ng/ml) and TNF-α (20 ng/ml) for 2 h were analyzed for mRNA (A) and protein expression (B). A, the amount of C/EBP-β mRNA, as determined by real time PCR, showed a 2–2½-fold induction for untransfected and Cntl siRNA-transfected cells relative to unstimulated cells. In contrast, cells treated with siB1 and siB2 showed a C/EBP-β mRNA level about 20% of that found in cells with Cntl siRNA after stimulation with the two cytokines. B, analysis of total cell lysates showed a similar strong reduction in the amount of C/EBP-β protein in siB1- and siB2-transfected cells. Both the 35- and 20-kDa forms of C/EBP-β was affected. C, a new series of experiments again showed a significant reduction of C/EBP-β mRNA as well as of the IL-6 mRNA in cells treated with siB1 and stimulated for 2 h with IL-17 and TNF-α, whereas the amounts of C/EBP-δ and IL-8 mRNAs were unaffected. D, at 24 h post-stimulation the levels of C/EBP-β and IL-6 mRNAs was still reduced in siB1-transfected cells, whereas the transcripts for C/EBP-δ, IL-8, and NGAL were unaffected by the treatment. E, this observation also holds true for NGAL, IL-6, and IL-8 also when measuring the amount of these three proteins in the medium of the cytokine-stimulated cells, as comparable levels were found for NGAL and IL-8 irrespective of whether the cells were transfected with siRNA or not, whereas the amount of secreted IL-6 was strongly diminished from siB1-treated cells. The real time PCR data are shown relative to the most highly expressed transcript in each experiment. Data are presented as the mean ± S.D. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
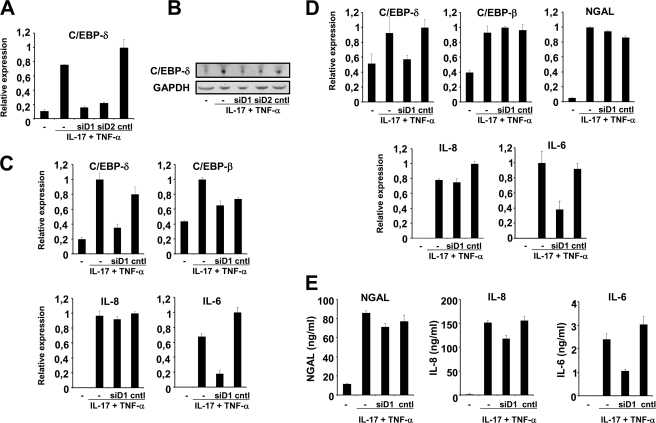
Knockdown of C/EBP-δ mRNA Does Not Affect NGAL Production
A similar series of experiments was performed with siRNAs against C/EBP-δ mRNA. A reduction in mRNA levels to 20- 25% of that observed for control-transfected and untransfected cells was observed for the two siRNAs, siD1 and siD2, both of which target C/EBP-δ mRNA (Fig. 4A). A similar effect on the C/EBP-δ protein levels was observed as demonstrated by Western blot (Fig. 4B). Results for siD1-treated cells are shown in the following, but comparable data were found when transfecting with siD2. When measured at 2 h post-induction, siD1-treated cells showed a significant reduction in the amount of C/EBP-δ and IL-6 mRNAs, whereas the amounts of the C/EBP-β and IL-8 mRNAs were at a level similar to those seen in stimulated cells with or without control siRNA (Fig. 4C). At 24 h post-stimulation again both C/EBP-δ and IL-6 mRNA levels were affected by siD1 transfection and comparable levels of C/EBP-β, IL-8, and NGAL mRNAs were found in the stimulated cells irrespective of whether they had received siD1, Cntl siRNA, or neither of the two (Fig. 4D). The same was true for NGAL and IL-8 protein in the medium as no difference was observed between stimulated cells without siRNA and cells transfected with siD1 or Cntl siRNA (Fig. 4E). In contrast, less than half the amount of IL-6 was secreted from cells treated with siD1 compared with the other two stimulated cell populations (Fig. 4E).
FIGURE 4.
Knockdown of C/EBP-δ expression does not affect NGAL expression. Untransfected A549 cells (−) or cells transfected with 40 nmol of two different siRNAs against C/EBP-δ (siD1 and siD2) or control siRNA (cntl) and stimulated with IL-17 (200 ng/ml) and TNF-α (20 ng/ml) for 2 h were analyzed for mRNA (A) and protein expression (B). A, the amount of C/EBP-δ mRNA was strongly induced after 2 h stimulation with IL-17 and TNF-α in untransfected and Cntl siRNA-transfected cells relative to unstimulated cells (as seen also in Fig. 2). However, if the stimulated cells were transfected with siD1 and siD2, the level was 4 to 5 times lower. B, transfection with siD1 or siD2 in the same manner affected the amount of C/EBP-δ protein measured in total cell lysates after a 2-h stimulation. C, transfection with siD1 and stimulation with the two cytokines for 2 h reduced the levels of C/EBP-δ and IL-6 mRNAs without having any effect on the levels of the C/EBP-β and IL-8 trancripts. D, when analyzing the cells after 24 h of stimulation with IL-17 and TNF-α, the amount of C/EBP-δ and IL-6 mRNAs was still lower in siD1-treated cells, whereas the levels of C/EBP-β, IL-8, and NGAL mRNAs were very similar in all cytokine-treated cells. E, the amount of secreted NGAL, IL-6, and IL-8 from the cells were affected in the same manner as their cognate transcripts by siD1 treatment compared with Cntl siRNA-transfected and untransfected cells. The real time PCR data are shown relative to the most highly expressed transcript in each experiment. Data are presented as the mean ± S.D. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
An Intact NF-κB Binding Site Is Required for IL-17- and TNF-α-induced Transcription of the LCN2 Promoter
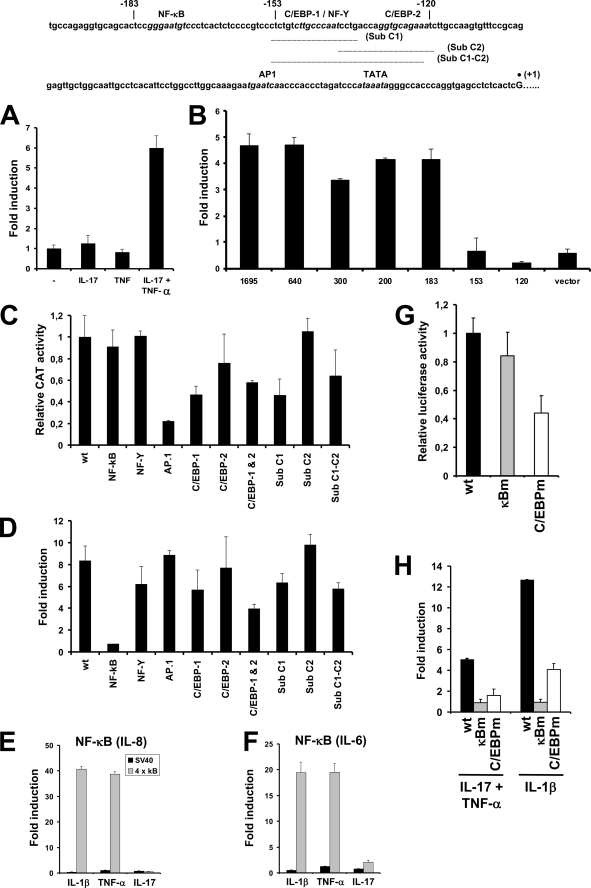
To further investigate the regulatory mechanisms involved in up-regulation of NGAL we examined whether transcription of a 1695-bp LCN2 promoter fragment could be induced by IL-17 and TNF-α. As seen in Fig. 5A transcription was only induced following co-stimulation with IL-17 and TNF-α, whereas stimulation with the two cytokines alone did not have any effect. We next analyzed a number of 5′-deletions of the LCN2 promoter. A comparable level of activation was observed for deletions ranging from −1695 to −183 bp upstream of the transcriptional start site (Fig. 5B). Deletion of a further 30 bp, however, completely abolished the induction of the LCN2 promoter activity. An NF-κB site situated at position −180 to −171 has previously been shown to be essential for IL-1β-induced up-regulation of LCN2 transcription (1). For this reason, we decided to investigate the importance for IL-17- and TNF-α-induced transcription of the NF-κB binding site and four other potential transcription factor binding sites in the LCN2 promoter. As the −183 deletion showed the same level of up-regulation as the −1695 deletion, we focused on transcription factor binding sites in the promoter downstream of −183. Point mutations of the −1695 deletion mutant in NF-κB, AP.1, NF-Y, and two C/EBP sites were tested (see sequence in Fig. 5). Mutation of the AP.1 site reduced basal (uninduced) expression of the LCN2 promoter to 20% of that measured for the wild type sequence and mutation of the distal C/EBP site (C/EBP-1) caused a 50% reduction in activity (Fig. 5C). However, these two mutations did not affect the level of induction in response to stimulation with IL-17 and TNF-α as a 6–8-fold up-regulation was still observed for all the mutated promoter constructs except for the NF-κB mutant, which had expression levels similar to unstimulated cells (Fig. 5D). This indicates that binding of NF-κB is essential for up-regulation of the NGAL promoter, whereas binding to the two C/EBP sites does not appear to be required for this process. This notion is further supported by the observation that mutation of the NF-Y site, which potentially could affect binding of C/EBP to the adjacent (and partly overlapping) C/EBP-1 site, did not influence the ability of this promoter construct to be activated. However, because the LCN2 promoter contains two potential C/EBP-binding sites it is possible that mutation of only one C/EBP site still allows C/EBP to regulate cytokine induction by binding to the other intact C/EBP site. To investigate this, we therefore made a promoter construct with mutations in both C/EBP sites (C/EBP-1 and C/EBP-2). As seen in Fig. 5D, a 6-fold induction was observed also for this construct.
FIGURE 5.
Activity of the LCN2 promoter in stimulated A549 cells. Top, DNA sequence and putative regulatory consensus elements of the −200 to 1 region of the LCN2 gene (6). The underlined sequences denote putative transcription factor recognition sites and the numbers above the sequence show the end points of the −183, −153, and −120 deletion mutants. The transcriptional start site is indicated with a dot. The extent of the substituted sequences Sub C1, C2, and C1-C2 are shown with bars under the sequence. A, fold-induction of the 1695-bp LCN2 promoter following stimulation with IL-17, TNF-α, and IL-17 + TNF-α shown relative to the activity from unstimulated cells (−). B, LCN2 promoter with deletions ranging from −1695 to −120, as denoted under the columns, was transfected into A549 cells and stimulated with IL-17 (200 ng/ml) and TNF-α (20 ng/ml) for 24 h. The fold-induction following stimulation is shown. Deleting the region below −183 abolished the ability of the promoter to be induced by the cytokines. C, relative CAT activities of promoter constructs in unstimulated cells with point mutations or substitutions of different transcription factor binding sites. The activities are shown relative to the wild type −1695 promoter, which was given the value “1.” D, fold-induction of the point mutations and substitutions following stimulation with IL-17 and TNF-α for 24 h. Fold-induction in CAT activity of pCAT3-promoter (SV40) and the same construct with four tandem repeats of the NF-κB sequence (4 × κB) from the IL8 promoter (E) or IL6 promoter (F) inserted upstream of the SV40 basal promoter. The fold-induction was determined for IL-1β, TNF-α, and IL-17-stimulated cells relative to the promoter activities of the unstimulated SV40 and “4 × κB” constructs, respectively. G, relative firefly luciferase activities of the 24p3–282-luc (wt), 24p3–282-κBm-luc (κBm), and 24p3–282-C/EBPm-luc (C/EBPm) promoter constructs in unstimulated cells. The activities are shown relative to the wild type Lcn2 (24p3) promoter, which was given the value 1. H, fold-induction in firefly luciferase activity of the wt, κBm, and C/EBPm promoter constructs following stimulation with IL-17 and TNF-α or IL-1β relative to the promoter activities of the unstimulated promoter constructs. All results are the mean ± S.D. of three independent transfections. The CAT activities were normalized to the firefly luciferase activity from the co-transfected vector Rous sarcoma virus-Luc. The firefly luciferase activities of the Lcn2 promoter constructs were normalized to the Renilla luciferase activity of the co-transfected vector pGL4.74. Data are presented as the mean ± S.D.
To further evaluate the consequence of inactivating the two C/EBP sites, and to rule out that the observed effect of the point mutations was due to residual binding capacity of the mutated transcription factor binding sites, we analyzed a series of mutations where the entire sequence of either C/EBP-1 and/or C/EBP-2 was substituted by a nonsense sequence. The individual substitutions (sub C1 and sub C2) as well as a substitution covering both C/EBP binding sites (sub C1-C2) all showed a level of induction in response to IL-17 and TNF-α stimulation comparable with that observed for the equivalent point mutations of C/EBP-1 and/or C/EBP-2 (Fig. 5, C and D).
In some cells stimulation with IL-17 alone has been found to activate the NF-κB pathway (4, 25). This does not appear to be the case in our model system as IL-8 transcription, which is induced by numerous stimuli causing NF-κB activation (1, 26), did not increase as a result of IL-17 administration (Fig. 2B). To further evaluate whether IL-17 could induce NF-κB-mediated transcription, we tested two heterologous reporter constructs where four tandem repeats of the NF-κB element from the IL8 and IL6 promoters, respectively, were inserted upstream of a minimal SV40 promoter. Strong induction of the CAT-reporter was seen following stimulation with TNF-α or IL-1β, whereas no transcription or transcription only slightly above the background level was observed when stimulating with IL-17 (Fig. 5, E and F).
The murine Lcn2 promoter is dependent on both an intact NF-κB and C/EBP-binding element for induction by IL-17 and TNF-α in murine bone marrow stromal cells (17). To investigate whether this was also the case in a human cell system we transfected A549 cells with Lcn2 promoter constructs carrying a mutated NF-κB or C/EBP binding site. The wild type promoter and the NF-κB mutant showed comparable luciferase activities in unstimulated cells, whereas the activity of the C/EBP mutant was about half of that measured for the wild type promoter (Fig. 5G). This is analogous to the activities measured for the NF-κB and C/EBP-1 mutants of the LCN2 promoter where mutation of the C/EBP site also decreased basal expression by about 50% (Fig. 5C). A 5-fold increase in Lcn2 promoter activity was seen following stimulation with IL-17 and TNF-α, whereas no induction was observed for the promoter with a mutated NF-κB site and a less than 2-fold induction was found for the C/EBP mutant (Fig. 5H). We also analyzed the Lcn2 promoter after stimulation with IL-1β as this cytokine mimics the effect of stimulating with IL-17 and TNF-α with regard to NGAL induction and as the C/EBP sites of the LCN2 promoter are not needed for IL-1β-induced up-regulation (1). The data in Fig. 5H show no induction of the 24p3-NF-κB mutant and a more that two-thirds reduction in the promoter activity of the 24p3-C/EBP mutant compared with the wild type promoter following IL-1β stimulation. This indicates that there also are differences in the requirement for C/EBP binding for these two orthologous LCN2/Lcn2 promoters following stimulation with IL-1β.
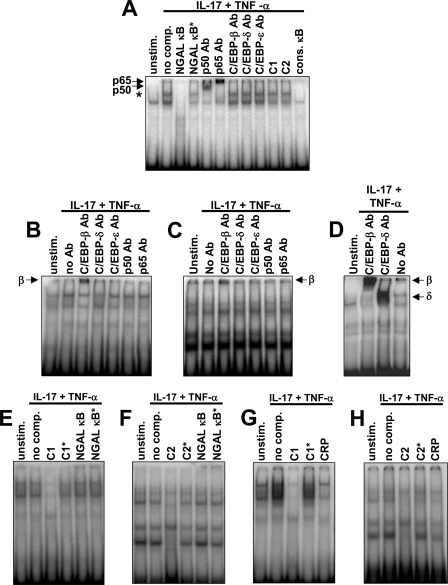
NF-κB and C/EBP Do Not Form a Complex on the κB Element of the LCN2 Promoter
It has been demonstrated that a multiprotein complex consisting of both C/EBP and NF-κB in some instances can be formed on a κB element of a promoter that lacks a recognizable adjacent C/EBP-binding element (27). In these cases, the C/EBP protein is believed to bind to the NF-κB complex sitting on the DNA without any direct contact between the C/EBP protein and the DNA. Although mutation of the two C/EBP sites of the LCN2 promoter did not affect the level of induction of the LCN2 promoter following stimulation with IL-17 and TNF-α (Fig. 5) we cannot rule out that a complex between C/EBP and NF-κB is formed on the κB element of the LCN2 promoter and thereby compensates for the lack of direct binding of the C/EBPs to the promoter. To evaluate whether such a complex might be formed we performed an EMSA using an oligo with the NF-κB element of the LCN2 promoter (Fig. 6A). Binding of NF-κB to the LCN2 κB site was observed when incubating with the nuclear extract from cytokine-stimulated cells (the band is indicated with an asterisk). As expected, this complex was not observed when a competitor oligo with an intact LCN2 NF-κB element (NGAL κB) or two tandem repeats of a consensus κB element (cons. κB) was added, whereas the complex could still be seen when competing with an oligo carrying a mutated κB site (NGAL κB*). Supershift was observed when incubating with antibodies against the p50 and p65 subunits of the NF-κB heterodimer, as seen previously (1). An analysis with antibodies against C/EBP-β, C/EBP-δ, and the myeloid-specific factor C/EBP-ϵ (included as a negative control) was also performed. No supershift was seen for either of these reactions indicating that the C/EBPs did not constitute a part of the complex formed on the NGAL κB element. Furthermore, an excess of C/EBP-1 (C1) or C/EBP-2 (C2) oligo did not alter the band pattern of the EMSA. An EMSA with oligoes containing the C/EBP-1 or C/EBP-2 binding sites of the LCN2 promoter as probes was also performed. No differences in the band pattern were observed between nuclear extracts from unstimulated and stimulated cells (Fig. 6, B–H) in accordance with the finding that both C/EBP-β and C/EBP-δ were present in the cells prior to stimulation (Figs. 2–4). A supershift of the C/EBP-1 probe was only detected with an antibody against C/EBP-β (Fig. 6B) indicating that neither C/EBP-δ nor NF-κB were part of the nuclear complexes formed on this probe. A similar, although less pronounced, supershift, was also seen for the C/EBP-2 probe with anti-C/EBP-β (Fig. 3C). We also made a series of competition experiments with the C1 and C2 probes. In this case binding of nuclear proteins to the C1 probe was found to be inhibited by addition of an excess of unlabeled C1 probe but not by a C1 probe with a 2-base pair mutation (C1*) of the C/EBP-recognition element (Fig. 6E). We, furthermore, tested whether the NGAL-κB oligo could affect the band pattern. The rationale behind this experiment is that if C/EBPs do form a complex with NF-κB bound to the κB element of the LCN2 promoter then competition with the NGAL-κB oligo should diminish the binding of C/EBPs to the C1 probe. No differences in band intensity were observed between a C1 probe without added competitor and a C1 probe competed with an excess of NF-κB oligo nor between the band intensities when the C1 probe was competed with NF-κB and NF-κB* (Fig. 6E). Similar results were found when testing the C2 probe (Fig. 6F). Finally, we made an EMSA with the C1 and C2 probes where we also included competition with an oligo (CRP) that has previously been shown to bind C/EBPs (27). Competition of the nuclear complexes by the CRP oligo was observed for both the C1 and C2 probes demonstrating that C/EBPs do bind to both these probes (Fig. 6, G and H). Together, these data indicate that the C/EBPs do not bind to the NF-κB protein complex when the C/EBP-binding regions of the LCN2 promoter are lacking but rather directly to the C/EBP-binding elements, C/EBP-1 and C/EBP-2, of the LCN2 promoter.
FIGURE 6.
Binding of nuclear complexes to the κB and C/EBP elements of the LCN2 promoter. Nuclear extracts from A549 cells that were either not stimulated (unstim.) or stimulated with IL-17 (200 ng/ml) and TNF-α (20 ng/ml) for 1½ h (IL-17 + TNF-α) were used for EMSA. A, a 32P-labeled probe containing the κB element of the LCN2 promoter was incubated with either no competitor (no comp.) or a 250-fold excess of an unlabeled oligo, which was identical in sequence to the probe (NGAL κB), only differed by a mutated κB element (NGAL κB*), contained sequences of the C/EBP-1 (C1) or C/EBP-2 (C2) elements of the LCN2 promoter, or carried a consensus κB sequence (cons. κB). Antibodies against p50, p65, C/EBP-β, C/EBP-δ, and C/EBP-ϵ were tested for their ability to cause a supershift of the nuclear complex associated with the probe. Supershifts with the p50 and p65 antibodies are shown with an arrow. The specific band formed on the NGAL-κB probe with the nuclear extract from cytokine-stimulated cells is indicated with an asterisk. B, a probe containing the C/EBP-1 element of the LCN2 promoter (C1) was incubated without antibody (no Ab) or with antibodies against C/EBP-β, C/EBP-δ, C/EBP-ϵ, p50, or p65. Supershift with anti-C/EBP-β is indicated with an arrow. C, an experiment similar to that in B was performed with a probe containing the C/EBP-2 element of the LCN2 promoter (C2). A supershift band was also in this case seen with anti-C/EBP-β (arrow). D, a control for supershift using the CRP oligo as probe. Supershift was observed for anti-C/EBP-β and anti-C/EBP-δ. E, the C1 probe was incubated with either no competitor (no comp.) or a 250-fold excess of an unlabeled oligo, which was identical in sequence to the probe (C1), had a 2-base pair mutation in the C/EBP-binding element (C1*), or contained the wild-type (NGAL κB) or mutated (NGAL κB*) NGAL-κB sequence. F, identical to the experiment in E except for C1 being exchanged by C2. G and H, identical to the experiments in E and F except for the NGAL-κB oligoes being substituted by the C/EBP-containing oligo CPR.
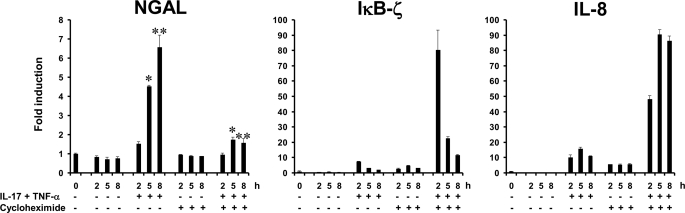
De Novo Protein Synthesis Is Required for Induction of NGAL Expression by TNF-α and IL-17
Transcription of the LCN2 gene following IL-1β stimulation requires de novo synthesis of IκB-ζ (18). To test whether induction of new protein synthesis is also required for induction of NGAL transcription with IL-17 and TNF-α, we measured the amount of NGAL mRNA generated in A549 cells co-stimulated with the two cytokines in the presence or absence of cycloheximide, a potent inhibitor of protein synthesis. As seen in Fig. 7, NGAL transcription was significantly inhibited in the presence of cycloheximide, whereas induction of IL-8 and IκB-ζ mRNAs still occurred. As reported also for IL-1β stimulation (18), the levels of IL-8 and IκB-ζ transcripts were found to be higher when cycloheximide was present. This probably reflects either a stabilization of these mRNAs due to a labile post-transcriptional regulatory protein not being synthesized (28) or the removal of a transcriptional repressor (29). Induction of the latter two mRNAs demonstrates that the NF-κB pathway was not disturbed by cycloheximide treatment because both mRNAs are encoded by genes, which require NF-κB for transcriptional activation. The failure of NGAL mRNA induction therefore must be due to lack of an essential transcriptional regulator, the production of which is blocked by cycloheximide.
FIGURE 7.
Induction of NGAL mRNA synthesis by co-stimulation with IL-17 and TNF-α requires de novo protein synthesis. Cells were harvested at the indicated time points after addition of fresh medium without cytokines or supplemented with IL-17 (200 ng/ml) and TNF-α (20 ng/ml) in the presence or absence of 10 μg/ml of cycloheximide to abolish synthesis of protein. Cycloheximide was added 30 min prior to stimulation with the cytokines. RNA was isolated and analyzed by real time PCR for transcripts of NGAL, IκB-ζ, and IL-8. A significantly higher level of NGAL mRNA was observed in cytokine-stimulated cells without cycloheximide at 5 and 8 h post-stimulation than in cytokine-stimulated cells also receiving cycloheximide; *, p = 0.047 and **, p = 0.031. Errors bars show the S.D. for each experiment.
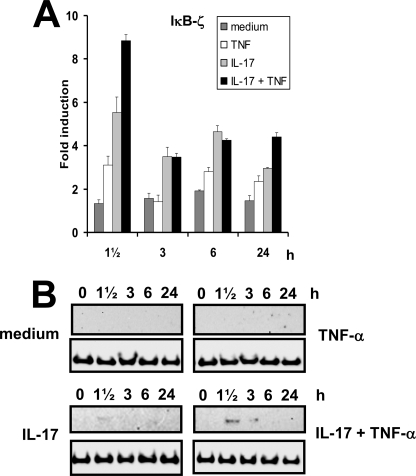
IκB-ζ Is Induced following Stimulation with Both IL-17 and TNF-α
As IκB-ζ has previously been shown to be essential for IL-1β-induced up-regulation of LCN2 transcription through binding to NF-κB (18), we looked at the mRNA profile for this factor in our experimental set-up. At 1½ h post-stimulation, IκB-ζ mRNA levels were slightly increased after TNF-α administration. Stimulation with IL-17 resulted in a 6-fold increase and an 8–9-fold increase was observed after co-stimulation with IL-17 and TNF-α (Fig. 8A). At later time points the amount of IκB-ζ mRNA in TNF-α-stimulated cells fell to almost the same levels seen in unstimulated cells, whereas a 2–3-fold higher expression was observed when IL-17 was present in the medium. The IκB-ζ protein was only detected in cells treated with IL-17 or IL-17 and TNF-α and was significantly increased above that in IL-17-stimulated cells in cells co-stimulated with IL-17 and TNF-α. The peak level of the IκB-ζ protein was found at 1½ h after stimulation with IL-17 and TNF-α as observed for the mRNA profile (Fig. 8B). A similar, but weaker, transient increase of IκB-ζ protein was found when stimulation was done with IL-17 alone.
FIGURE 8.
Time courses of IκB-ζ mRNA and protein levels following stimulation with IL-17 and/or TNF-α. The amount of IκB-ζ mRNA (A) and protein (B) was determined by real time PCR (A) or Western blot (B) at the indicated time points after addition of fresh medium without cytokines or supplemented with TNF-α, IL-17, or IL-17 and TNF-α in the concentrations indicated in the legend to Fig. 1. The expression of the transcripts is shown as fold-induction relative to the expression measured in unstimulated cells at time 0. A control (a lysate from cells stimulated 1½ h with IL-17 + TNF-α) was included on the blot for unstimulated and TNF-α-stimulated cells to ensure that the IκB-ζ protein could be detected under the experimental conditions employed (data not shown). Real time data are presented as the mean ± S.D.
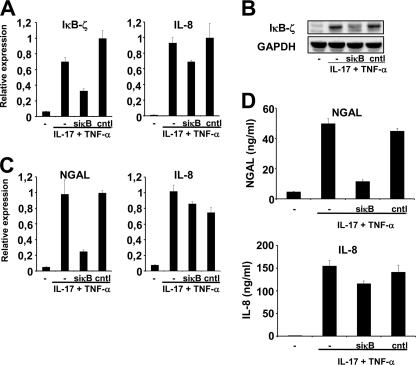
Inhibition of IκB-ζ Expression Results in Decreased NGAL Production
The nuclear factor IκB-ζ is essential for up-regulation of LCN2 transcription in response to IL-1β stimulation (18). To examine if this is also the case following IL-17 and TNF-α stimulation, we tested whether an siRNA against IκB-ζ mRNA would affect the amount of NGAL mRNA synthesized. As demonstrated in Fig. 9 an siRNA against IκBζ (siκB) reduced the amount of IκB-ζ mRNA in the cytokine-stimulated cells significantly compared with untransfected cells and to cells transfected with a control siRNA (Fig. 9A). The same effect was observed on the amount of IκB-ζ protein induced by the cytokines (Fig. 9B). The siRNA against IκB-ζ also reduced the amount of NGAL mRNA drastically in the cells, whereas no effect on IL-8 mRNA induction was observed (Fig. 9C). The same holds true with regard to the quantity of NGAL and IL-8 protein measured in the medium of siRNA-treated cells. The amount of NGAL protein measured in small interfering κB-treated cells was less than 25% of that found in untreated and control siRNA-treated cells, whereas the level of IL-8 protein found in the medium was unaffected by transfection with siRNAs (Fig. 9D).
FIGURE 9.
Knockdown of IκB-ζ expression causes a decrease in NGAL expression. Untransfected A549 cells (−) or cells transfected with 40 nmol of IκB-ζ-siRNA (siκB) or control siRNA (cntl) were grown in medium with or without IL-17 (200 ng/ml) and TNF-α (20 ng/ml) for 2 (A and B) or 24 h (C and D) and then harvested for total RNA and protein isolation. The siRNA against IκB-ζ has been used previously (18). A, the amount of IκB-ζ and IL-8 mRNA was determined by real time PCR and demonstrated a reduction in IκB-ζ in siκB-treated cells. B, protein (whole cell lysates) from cells treated as above was analyzed by Western blot with antibodies against IκB-ζ and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and demonstrated a significant knockdown of IκB-ζ in siκB-treated cells. C, the amount of NGAL and IL-8 mRNA in cells stimulated for 24 h was determined by real time PCR and demonstrated a strong reduction of NGAL mRNA in cells that had received siκB. D, analogous to the mRNA profiles a significant drop of NGAL was measured in the medium of stimulated cells containing siκB. Treatment with siκB did not affect the amount of IL-8 secreted to the medium. All real time PCR data are shown relative to the most highly expressed transcript in each experiment. Data are presented as the mean ± S.D.
DISCUSSION
NGAL has been found to be up-regulated by IL-1β in an NF-κB-dependent manner in human epithelial cell cultures (1). In contrast, no activation of NGAL occurs by stimulating with TNF-α, another activator of the NF-κB pathway. This IL-1β specificity has been demonstrated in human epithelial cells originating from the lung, bronchus, and the skin (1, 2), as well as in a hepatocyte cell line (22). This is in contrast to the murine orthologue, 24p3/lipocalin 2, which can be induced by both IL-1β and TNF-α (16, 22). It was recently demonstrated that the TNF-α-induced expression of murine lipocalin 2 can be further boosted by co-stimulation with IL-17 (16). For this reason, we decided to examine whether it would be possible to induce NGAL expression in the epithelial cell by stimulation of combined TNF-α and IL-17.
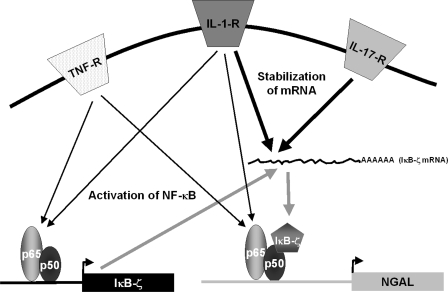
Neither IL-17 nor TNF-α had any stimulating effect on NGAL expression in our model system, whereas co-stimulation with both cytokines resulted in a 15-fold induction over the background level at 24 h. De novo protein synthesis was required for LCN2 transcription to occur by stimulation with IL-17 and TNF-α. This is analogous to the induction of NGAL transcription following IL-1β stimulation where new synthesis of the IκB-ζ protein is required for LCN2 gene transcription (18). The same appears to be true when stimulating with IL-17 and TNF-α as both the level of NGAL mRNA and amount of protein synthesized were severely reduced if the cells were treated with an siRNA against IκB-ζ prior to stimulation. The lack of NGAL induction following stimulation with TNF-α or IL-17 alone indicates that the cellular response to each of these cytokines alone does not activate all the components required for LCN2 transcription. Induction of LCN2 transcription with IL-1β requires both binding of NF-κB to the LCN2 promoter as well as synthesis of IκB-ζ, which acts as a co-activator with NF-κB on the promoter (18). Stimulation with TNF-α induces binding of NF-κB to the LCN2 promoter but does not result in NGAL protein synthesis (1). In contrast, IL-17 does not appear to activate NF-κB, as stimulation with IL-17 alone does not induce IL8 transcription (Fig. 2). This notion is further strengthened by the observation that IL-17 was unable to induce transcription from a reporter plasmid with 4 tandem repeats of an NF-κB binding site positioned directly upstream of a minimal SV40 promoter, whereas a strong induction was seen when stimulating with TNF-α or IL-1β (Fig. 5). A slight induction of IκB-ζ mRNA (and protein) was observed when stimulating with IL-17 alone (Fig. 7), which can be explained by stabilization of IκB-ζ mRNA, which is transcribed at a low level in unstimulated cells, or the removal of a transcriptional repressor sitting on the promoter of the NFKBIZ gene. In support of the former notion is the observation that IL-17 can induce stabilization of a number of transcripts (15) including the NFKBIZ mRNA (30). IκB-ζ does not bind DNA directly but functions through interaction with the p50 subunit of DNA-bound NF-κB (31). This explains why an increased level of IκB-ζ following IL-17 stimulation does not result in NGAL induction by itself. Based on these results and the knowledge that transcription of IκB-ζ is induced through NF-κB binding to the NFKBIZ promoter (30), we propose a model for LCN2 induction by co-stimulation with IL-17 and TNF-α as shown in Fig. 10. Stimulation with TNF-α induces activation of NF-κB and results in its binding to the promoters of both the NFKBIZ and LCN2 genes. This leads to transcription of IκB-ζ mRNA. The IκB-ζ mRNA is then stabilized by a signal emanating from the activated IL-17 receptor. This enables the IκB-ζ protein to be synthesized and act as a co-activator of the NF-κB complex bound to the LCN2 promoter, which finally allows transcription of the LCN2 gene to take place. This model is supported by our previous finding that LCN2 transcription can be induced by TNF-α alone if the cells are transfected with a plasmid that constitutively expresses IκB-ζ (18). Induction of LCN2 by IL-1β is possible because this cytokine is able to induce both NF-κB binding and stabilization of the IκB-ζ transcript (1, 18, 30).
FIGURE 10.
Model for up-regulation of NGAL by co-stimulation with IL-17 and TNF-α. Stimulation of the TNF-α receptor (TNF-R) induces activation and translocation of NF-κB to the promoters of the NFKBIZ (IκB-ζ) and LCN2 (NGAL) genes. Binding of NF-κB to the NFKBIZ promoter induces IκB-ζ transcription but the IκB-ζ mRNA is unstable and rapidly degraded. Binding of NF-κB to the LCN2 promoter is not sufficient to initiate transcription. Stimulation of the IL-17 receptor (IL-17-R) generates an intracellular signal that stabilizes the IκB-ζ transcript and allows translation of IκB-ζ protein. The newly synthesized IκB-ζ is then able to translocate and bind to NF-κB on the LCN2 promoter and thereby initiate transcription of the gene. Ligation of the IL-1 receptor (IL-1-R) generates signals that both activate NF-κB and stabilize the IκB-ζ mRNA. This explains why NGAL can be induced by stimulation with IL-1β alone.
Gaffen and co-workers (17) have previously shown that an intact C/EBP-binding element in the mouse Lcn2 promoter is essential for IL-17- and TNF-α-induced transcription of a luciferase reporter in murine cells. Furthermore, they demonstrated that C/EBP-β and C/EBP-δ can bind in vitro to the C/EBP site of the Lcn2 promoter in a gel shift assay (17). This is in contrast to the data presented here where neither siRNAs against C/EBP-β and C/EBP-δ nor mutations and substitutions of the two C/EBP sites in the human LCN2 promoter abolished transcriptional activation in response to IL-17 and TNF-α. Although a 20–30% lower degree of activation was found when the C/EBP-1 site was inactivated by a point mutation or substitution, these promoter constructs still gave rise to a 6-fold induction compared with unstimulated cells. This is in accordance with our previous characterization of the LCN2 promoter in response to IL-1β stimulation where the C/EBP-1 mutation reduced basal (uninduced) promoter activity by 50%, but did not affect the extent of induction (1). These data are also corroborated by the findings of Matsuo et al. (32) that the absolute expression level of an LCN2 promoter with a C/EBP-1 mutation is about one-third of that of the wild type promoter after IL-1β stimulation. As the basal expression level of the mutated promoter is also lowered by the same magnitude compared with the basal expression from the wild type construct this means that the degree of up-regulation in response to the cytokine is the same for both promoters. Although a direct interaction between C/EBP and a NF-κB complex bound to the κB element of a promoter, which lacks a C/EBP-binding DNA sequence, has been reported (27) we were not able to detect such an association between NF-κB and C/EBP on the κB element of the LCN2 promoter using short oligos in EMSAs. Binding of C/EBPs directly to C/EBP elements of the LCN2 promoter was, on the other hand, detected by EMSA. The supershift complex with anti-C/EBP-β formed on the C2 probe, although reproducibly observed with nuclear extracts from different stimulation experiments, appeared to be less pronounced than the complex formed on the C1 probe. That C2 does bind C/EBPs was shown by the disappearance of nuclear bands formed on the C2 probe when competing with the heterologous C/EBP-binding oligo CRP. Together, this indicates that the binding affinity of C/EBP-β to the C/EBP-2 element is lower than to the C/EBP-1 element. This may explain why mutation or substitution of the C/EBP-2 element of the LCN2 promoter only has a weak effect on the level of transcription (Fig. 5). The same pattern is also observed when examining the LCN2 promoter with a mutated C/EBP-2 element following stimulation with IL-1β (1). In conclusion, these data indicate that binding of C/EBPs to the C/EBP-1 site affects the absolute level of expression of the human LCN2 promoter, but does not play a role in the regulatory mechanism governing the up-regulation of LCN2 expression in response to stimulation with IL-17 and TNF-α or IL-1β.
The reason why the C/EBP element is required for full induction of the murine Lcn2 promoter, but not of the human LCN2 promoter, following stimulation with IL-17 and TNF-α, is not known. One explanation could be that differences in the nucleotide sequences of the C/EBP-binding elements of the two promoters (17) might cause the C/EBPs to bind with higher affinity to the murine C/EBP element and thus have a greater effect on Lcn2 promoter activity. Another explanation could be that the different spacing between the NF-κB and the C/EBP-binding elements of the murine and human promoters (17) may allow a better physical contact between the NF-κB and C/EBP transcriptions factors on the murine Lcn2 promoter than on the human LCN2 promoter.
The role of C/EBP sites for IL-17- and TNF-α-induced expression of the LCN2 gene is in contrast to the findings for the murine Lcn2 promoter, as mentioned above. This, however, is only one of several differences registered between the two orthologous genes in mouse and man. Another notable difference is that murine Lcn2 can be up-regulated in response to stimulation with TNF-α alone (16, 22). Furthermore, lipocalin 2 has been found to be an acute-phase protein in a turpentine injection model and to be synthesized in the liver of the mouse (33), whereas NGAL is neither expressed in human liver (6, 22) nor can be measured as an acute-phase protein in a human sepsis model (22). The involvement of C/EBP sites for up-regulation of NGAL in the mouse may explain the effect of TNF-α in mice if stimulation by this cytokine leads to C/EBP activation. A difference between man and mouse in the transcriptional regulation of an antimicrobial protein is not unique to LCN2 as exemplified by the gene for CAMP3 (hCAP-18) that is strongly induced by vitamin D3 in humans, whereas no induction is observed for the murine gene (34). These data underscore that data regarding gene regulation obtained in a mouse system may not always apply to humans.
Acknowledgments
The expert technical assistance of Inge Kobbernagel and Tesse Hornsyld is greatly appreciated. Dr. Sara Rørvig is thanked for help with statistical analysis. Dr. Sarah Gaffen, University of Pittsburg and the University of Buffalo, is thanked for providing the 24p3-Luc promoter reporter plasmids.
This work was supported by grants from the Danish Medical Research Council, the Novo Nordisk Foundation, the Lundbeck Foundation, and the A. P. Møller Foundation for the Advancement of Medical Science.
- NGAL
- neutrophil gelatinase-associated lipocalin
- IL
- interleukin
- TNF
- tumor necrosis factor
- siRNA
- small interfering RNA
- ELISA
- enzyme-linked immunosorbent assay
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TBS
- Tris-buffered saline
- CAT
- chloramphenicol acetyltransferase
- EMSA
- electrophoretic mobility shift assay
- BSA
- bovine serum albumin
- Cntl
- control
- CRP
- C-reactive protein.
REFERENCES
- 1.Cowland J. B., Sørensen O. E., Sehested M., Borregaard N. (2003) J. Immunol. 171, 6630–6639 [DOI] [PubMed] [Google Scholar]
- 2.Sørensen O. E., Cowland J. B., Theilgaard-Mönch K., Liu L., Ganz T., Borregaard N. (2003) J. Immunol. 170, 5583–5589 [DOI] [PubMed] [Google Scholar]
- 3.Sørensen O. E., Thapa D. R., Rosenthal A., Liu L., Roberts A. A., Ganz T. (2005) J. Immunol. 174, 4870–4879 [DOI] [PubMed] [Google Scholar]
- 4.Kao C. Y., Chen Y., Thai P., Wachi S., Huang F., Kim C., Harper R. W., Wu R. (2004) J. Immunol. 173, 3482–3491 [DOI] [PubMed] [Google Scholar]
- 5.Kjeldsen L., Bainton D. F., Sengeløv H., Borregaard N. (1994) Blood 83, 799–807 [PubMed] [Google Scholar]
- 6.Cowland J. B., Borregaard N. (1997) Genomics 45, 17–23 [DOI] [PubMed] [Google Scholar]
- 7.Friedl A., Stoesz S. P., Buckley P., Gould M. N. (1999) Histochem. J. 31, 433–441 [DOI] [PubMed] [Google Scholar]
- 8.Nielsen B. S., Borregaard N., Bundgaard J. R., Timshel S., Sehested M., Kjeldsen L. (1996) Gut 38, 414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flower D. R. (1996) Biochem. J. 318, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goetz D. H., Holmes M. A., Borregaard N., Bluhm M. E., Raymond K. N., Strong R. K. (2002) Mol. Cell 10, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 11.Flo T. H., Smith K. D., Sato S., Rodriguez D. J., Holmes M. A., Strong R. K., Akira S., Aderem A. (2004) Nature 432, 917–921 [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto M., Yamazaki S., Uematsu S., Sato S., Hemmi H., Hoshino K., Kaisho T., Kuwata H., Takeuchi O., Takeshige K., Saitoh T., Yamaoka S., Yamamoto N., Yamamoto S., Muta T., Takeda K., Akira S. (2004) Nature 430, 218–222 [DOI] [PubMed] [Google Scholar]
- 13.Bettelli E., Korn T., Oukka M., Kuchroo V. K. (2008) Nature 453, 1051–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fossiez F., Djossou O., Chomarat P., Flores-Romo L., Ait-Yahia S., Maat C., Pin J. J., Garrone P., Garcia E., Saeland S., Blanchard D., Gaillard C., Das Mahapatra B., Rouvier E., Golstein P., Banchereau J., Lebecque S. (1996) J. Exp. Med. 183, 2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartupee J., Liu C., Novotny M., Li X., Hamilton T. (2007) J. Immunol. 179, 4135–4141 [DOI] [PubMed] [Google Scholar]
- 16.Shen F., Ruddy M. J., Plamondon P., Gaffen S. L. (2005) J. Leukocyte Biol. 77, 388–399 [DOI] [PubMed] [Google Scholar]
- 17.Shen F., Hu Z., Goswami J., Gaffen S. L. (2006) J. Biol. Chem. 281, 24138–24148 [DOI] [PubMed] [Google Scholar]
- 18.Cowland J. B., Muta T., Borregaard N. (2006) J. Immunol. 176, 5559–5566 [DOI] [PubMed] [Google Scholar]
- 19.de Wet J. R., Wood K. V., DeLuca M., Helinski D. R., Subramani S. (1987) Mol. Cell. Biol. 7, 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjeldsen L., Koch C., Arnljots K., Borregaard N. (1996) J. Immunol. Methods 198, 155–164 [DOI] [PubMed] [Google Scholar]
- 22.Klausen P., Niemann C. U., Cowland J. B., Krabbe K., Borregaard N. (2005) Eur. J. Haematol. 75, 332–340 [DOI] [PubMed] [Google Scholar]
- 23.Descombes P., Schibler U. (1991) Cell 67, 569–579 [DOI] [PubMed] [Google Scholar]
- 24.Hu H. M., Baer M., Williams S. C., Johnson P. F., Schwartz R. C. (1998) J. Immunol. 160, 2334–2342 [PubMed] [Google Scholar]
- 25.Patel D. N., King C. A., Bailey S. R., Holt J. W., Venkatachalam K., Agrawal A., Valente A. J., Chandrasekar B. (2007) J. Biol. Chem. 282, 27229–27238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann E., Dittrich-Breiholz O., Holtmann H., Kracht M. (2002) J. Leukocyte Biol. 72, 847–855 [PubMed] [Google Scholar]
- 27.Agrawal A., Samols D., Kushner I. (2003) Mol. Immunol. 40, 373–380 [DOI] [PubMed] [Google Scholar]
- 28.Roger T., Out T., Mukaida N., Matsushima K., Jansen H., Lutter R. (1998) Biochem. J. 330, 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nourbakhsh M., Kalble S., Dorrie A., Hauser H., Resch K., Kracht M. (2001) J. Biol. Chem. 276, 4501–4508 [DOI] [PubMed] [Google Scholar]
- 30.Yamazaki S., Muta T., Matsuo S., Takeshige K. (2005) J. Biol. Chem. 280, 1678–1687 [DOI] [PubMed] [Google Scholar]
- 31.Motoyama M., Yamazaki S., Eto-Kimura A., Takeshige K., Muta T. (2005) J. Biol. Chem. 280, 7444–7451 [DOI] [PubMed] [Google Scholar]
- 32.Matsuo S., Yamazaki S., Takeshige K., Muta T. (2007) Biochem. J. 405, 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q., Nilsen-Hamilton M. (1995) J. Biol. Chem. 270, 22565–22570 [DOI] [PubMed] [Google Scholar]
- 34.Gombart A. F., Borregaard N., Koeffler H. P. (2005) FASEB J. 19, 1067–1077 [DOI] [PubMed] [Google Scholar]