Abstract
To help understand the dynamic nature of membrane fusion induced by the human immunodeficiency virus-1 (HIV-1) envelope protein, we developed a new cell-based real-time assay system employing a pair of novel reporter proteins. The reporter proteins consist of a pair of split Renilla luciferase (spRL) fused to split green fluorescent protein (spGFP). The spGFP modules were chosen not only to compensate weak self-association of spRL but also to provide visual reporter signals during membrane fusion. Use of this reporter together with a membrane permeable substrate for Renilla luciferase achieved a simple real-time monitoring of membrane fusion using live cells. We analyzed the HIV-1 envelope mutants whose membrane-spanning domains were replaced with that of glycophorin A or vesicular stomatitis virus G-protein. These mutants showed a slower kinetics of membrane fusion. The analysis of membrane fusion in the presence of fusion inhibitors, soluble CD4 and C34, revealed that these replacements prolonged the period during which the mutants were sensitive to the inhibitors, as compared with the wild type. These results suggest that the mutations within the membrane-spanning domains exerted an allosteric effect on the HIV-1 envelope protein, probably affecting the receptor-induced conformational changes of the ectodomain of the protein.
Keywords: Diseases/AIDS, Membrane/Fusion, Protein/Chimeras, Viruses/Retrovirus, Enzymes, GFP, HIV-1, Renilla Luciferase, gp41, Membrane-spanning Domain
Introduction
Membrane fusion is involved in many biological events such as myogenesis, fertilization, and vesicular transport (1, 2). As the result of membrane fusion, two independent compartments, each enclosed in its own membrane, merge to form one compartment. Invasion of the host cell by an enveloped virus also relies on membrane fusion (2). HIV-12 uses the membrane fusion to infect a host cell. During viral membrane fusion, the HIV-1 envelope protein (Env) is known to undergo time-dependent substantial conformational rearrangements (3–5). For elucidating the mechanisms of viral membrane fusion with such a dynamic nature, a method for real-time and quantitative monitoring of membrane fusion is essential.
To achieve a real-time analysis of membrane fusion, an appropriate assay system that is quick enough to generate a reporter signal is needed. A so-called transcription factor transfer assay that monitors membrane fusion by an activation of a reporter gene in one compartment by the transcription factor transferred from another compartment is not quick enough because it requires the steps of transcription and translation. A dye transfer assay is quick enough for a real-time analysis. One drawback of this assay is that the color signals are not induced by membrane fusion but exist throughout the events of membrane fusion; therefore, it requires the detailed analysis of the mixing or relocation of fluorescent dyes or proteins (green fluorescent protein (GFP), red fluorescent protein, etc.) preloaded into each compartment to be fused (6, 7). One may require using a sophisticated device such as a video recorder (8) for a real-time analysis. Recently the transfer of β-lactamase during fusion was used as a reporter. The cleavage of the membrane-permeable substrate by the transferred β-lactamase results in the alteration of the fluorescent color that serves as a reporter of membrane fusion (9). The change of the color in this assay rather than the mixing of colors in the dye transfer assay seems to be a better marker. The β-lactamase assay is also quite sensitive, but it requires an optic device that can differentiate the change of the wavelength of the emitted light before and after the fusion.
We are interested in developing a simple, easy-to-perform cell-based membrane fusion assay system where a positive reporter signal is produced only when membrane fusion takes place. Recently we have reported such a system using split GFP (spGFP) (10). This simple cell-based assay produces a green signal only when membrane fusion actually takes place. One drawback of the system is a difficulty in quantification. Therefore, we explored the possibility of an enzymatic reporter. In selecting a candidate enzyme, we set the availability of a membrane-permeable substrate as a critical criterion because the lack of such a substrate demands membrane permeabilization or fixing before the assay (11, 12). This makes a live cell monitoring impossible.
Here we report the development of a cell-based membrane-fusion assay system that uses newly engineered split Renilla luciferase (spRL). The availability of membrane-permeable substrates of RL, coelenterazines, ensures continuous and quantitative real-time monitoring of membrane fusion with living cells. Furthermore, because RL has been used widely in molecular biological analyses recently, the reagents and devices for detection are easily accessible. We fused spGFP to spRL to overcome a problematic low self-association ability of spRL (13). The added spGFP portion (14) not only serves as a strong association module but also provides a visible green signal upon membrane fusion. We named this new reporter protein dual split protein (DSP) because it recovers dual functions of RL and GFP upon reassociation.
This dual functional reporter system was applied to analyzing mutant HIV-1 envelope protein (Env) whose membrane-spanning domain (MSD) was replaced with heterologous MSD derived from other membrane proteins such as glycophorin A (GpA) or vesicular stomatitis virus G protein (VSV-G) (15). The assay revealed a kinetic defect of the membrane fusion of mutant envelope proteins of HIV-1. Further analyses of the membrane fusion of the mutants in the presence of fusion inhibitors or the temperature arrest suggested that the mutations introduced into the MSD manifested the negative allosteric effect on the conformational changes of gp120-gp41 complex upon interaction with the receptor complex.
EXPERIMENTAL PROCEDURES
Construction of the Plasmids for Split and Dual Split Protein Expression
The expression vectors were derivatives of phRL-CMV (Promega). The split point of RL was located between the 229th and 230th amino acid as reported previously (13). The N- or C-terminal fragment of spRL was named nRL or cRL, respectively, and introduced to phRL-CMV by replacing with the full-length RL. Several proteins to enhance the association between nRL and cRL were examined (Fig. 1). One is a pair of leucine zipper coiled-coil protein, called Velcros, the engineered leucine zipper fragments described previously (16). The basic or acidic residue-containing fragment (baseVel or acidVel, respectively) was fused to nRL or cRL as shown in Fig. 1. Between the spRL and Velcro, the long linker sequence with 36 amino acid residues, GLQGGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSVD, was introduced.
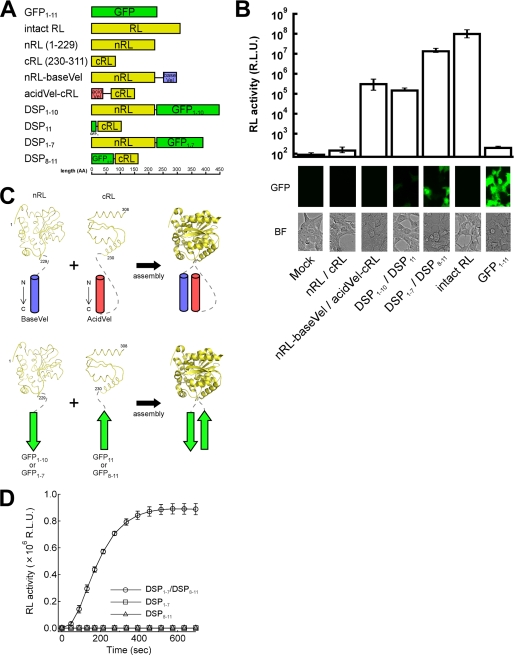
FIGURE 1.
Design and complementation efficiency of split proteins. A, shown are the constructs used in this study. GFP1–11 and intact RL, a full-length green fluorescent protein and Renilla luciferase; nRL and cRL, the N- or C-terminal fragment of spRL (13); baseVel and acidVel, a derivative of the heterodimeric leucine zipper containing basic or acidic residues (16); DSP, dual split protein composed of fragments of spGFP and spRL. The β-sheet contained within each spGFP is indicated by subscripts. The bottom bar indicates a scale in amino acids (AA). B, shown is the result of the complementation. The activities of RL (upper panel) and GFP (lower panel) were determined after 36 h of co-transfection in 293FT cells. R.L.U., relative light units. The bright field (BF) images are also shown. All the abbreviations are same as A. C, the schematic diagram of the arrangement of the association modules and target spRL is shown. The shown structures of spRL were generated based on the reported structure of RL (22). The number at the terminus of each chain indicates the position of the residue. The gray dashed line indicates the linker region. Top panel, the pattern of assembly of spRL with AcidVel (red) and BaseVel (blue) motives is shown. Bottom panel, the pattern of association of spRL with the self-assembling spGFPs (green) is shown. The spGFP-1 and -2 indicates the N-terminal fragment and C-terminal fragment of spGFP, respectively. D, shown are the kinetics of the complementation of DSP in a cell-free system. The cell lysates containing DSP1–7 or DSP8–11 prepared from the transfected 293FT cells were mixed (DSP1–7/DSP8–11), and the recovered RL activity was monitored in a time-dependent manner. Individual DSP alone (DSP1–7 or DSP8–11) showed no activities. The data are averages of three or more independent experiments.
To fuse spGFPs to spRL as SbfI-XbaI (GFP1–10 and GFP1–7) or NheI-SalI (GFP8–11) fragments, PCR was performed using KOD (+) polymerase (Toyobo) or Pfu turbo (Stratagene) with GFPopt1–11 (10) as a template. The number in the subscript of GFP indicates the β-sheets included in the fragment. Thus, for example, GFP1–10 contains, first, 10 β-sheets of 11 β-sheets present in an intact GFP. The amplicons were cloned and sequenced in pCR-4TOPOblunt. The GFP1–10 and GFP1–7 fragments were cloned downstream of nRL using SbfI and XbaI sites. The GFP8–11 fragment was ligated to the upstream of cRL using NheI and SalI sites. The GFP11 fragment with a 5′-NheI site and 3′-SalI site was made by annealing a pair of synthetic oligonucleotides and cloned similarly in the case of GFP8–11. In the spGFP-containing plasmids, two types of linkers were tested; that is, the long linker with the same 36 amino acid residues as that for Velcro and the short linker with 4 (GLQG) or 2 (VD) amino acid residues for nRL-spGFP or spGFP-cRL, respectively. The constructed gene with the linker was called DSP with the suffix reflecting the fused spGFP. The number in the subscript of DSP indicated the range of the GFP β-sheets included in the DSP. The amino acid sequences of DSP1–7 and DSP8–11 are shown in supplemental Fig. 1.
Complementation Analysis of Split and Dual Split Protein by Co-transfection into Mammalian Cells
All cells were kept at 37 °C under 5% CO2 in a complete medium composed of Dulbecco's modified Eagle's medium (Sigma) with 10% fetal bovine serum (Hyclone). The 2 × 105 of 293FT cells in a 96-well plate (Packard View, PerkinElmer Life Sciences) were transfected with 100 ng of DNA with FuGENE HD (Roche Applied Science). The volume ratio of FuGENE HD to DNA (20 ng/ml in Dulbecco's modified Eagle's medium) was 1:17. Two days after transfection, images of the transfected cells were taken using an IN Cell Analyzer 1000 (GE Healthcare) (×10 objective lens, five sampling fields per well). Immediately after the image capture, the RL activities of the samples were measured using the Renilla luciferase assay kit (Promega) and Glomax (Promega).
Cell-free Complementation of Dual Split Proteins
After 48 h of seeding (6-cm-diameter dishes BD Falcon; 2 × 105 cells in 5 ml), the 293FT and the 293CD4 cells were transfected with the DSP1–7 gene or the DSP8–11 gene, respectively. After 36–48 h of the transfection, cells were scraped off the dish, pelleted down (20,000 × g, 4 °C, 10 min) in phosphate-buffered saline, and then lysed with 500 μl of the sample lysing buffer as suggested by the manufacturer (Renilla luciferase assay kit, Promega). Twenty μl of each of the lysates were mixed, and the time course measurement of the recovered RL activity was performed at room temperature using the Glomax instrument (Promega). Three or more independent experiments were performed.
Live-cell Monitoring of Membrane Fusion Using the Dual Split Proteins and a Membrane-permeable Substrate
The 293FT cells with more than 50% confluency in the 96-well plate were transfected with both the DSP1–7 and the HIV-1 Env expression vectors. The latter vector, pNHcRedEluc (10), contains the wild type or mutant envelope gene derived from the HXB2 strain. The envelope mutants used were those whose MSD was replaced with that of GpA or VSV-G protein as described previously (15). Because the pNHcRedEluc vector expresses nuclear-localizing red fluorescent proteins, HcRed, together with Env, it generated HIV-1 Env-expressing cells with red nuclei (10). Separately, 293CD4 cells in an UpCell dish (6-cm diameter, Nunc) were transfected with the vector to express DSP8–11. 48–60 h later, the culture medium for the 293FT cells was replaced with fresh medium. To normalize transfection efficiency, the firefly luciferase activity was measured using a portion of transfected 293FT cells with Dual-Glo reagent (Promega) before co-cultivation. The culture medium for the 293CD4 cells was also removed and replaced with 5 ml of fresh medium containing 60 μm membrane-permeable substrate, EnduRen (Promega). After incubation for 2 h at 37 °C, the EnduRen-loaded 293CD4 cells were detached from the UpCell plate by leaving them at room temperature. After gentle but thorough pipetting, the detached 293CD4 cells were layered over the 293FT cells to induce membrane fusion. During the co-culture, the RL activity was measured in a time-dependent manner with Glomax (Promega). The p values between the wild type and mutants at the indicated time points (n = 8) were calculated using the t test. To evaluate the necessity of the de novo protein synthesis during membrane fusion, the inhibitor of protein synthesis, cycloheximide (Sigma) or puromycin (Sigma), 100 or 10 μm, respectively, was added to the culture medium immediately before the co-cultivation. The RL activity was measured 200 min after co-cultivation. The green fluorescence of the same culture was monitored using an IN Cell Analyzer 1000 (GE Healthcare) in a time-dependent manner.
The effects of the fusion inhibitors were examined using either soluble CD4 (R&D Systems) or C34 (SciLight Biotech). Each inhibitor was added to the medium at 0, 20, 40, 60, 100, and 150 min after co-culturing as described above. The RL activity was measured at 200 min after co-culture. In the case of the mutants, GpA and VSV-G, the time of co-cultivation was extended to 20 and 32 h, respectively. The extent of fusion inhibition was calculated using the following equation: inhibition of fusion (%) = 100 − 100 × (RL activity in the presence of inhibitors)/(RL activity in the absence of inhibitor). The curve-fitting of data points was performed using the equation described previously (17). Each value was calculated from three or more independent experiments.
The experiment in the temperature-arrested state (TAS) was performed as follows. The fusion assay by the co-culturing was performed as described above, except that the culture was first incubated for 2 h at 25 °C with 5% CO2 to induce TAS. After this low temperature treatment, then the temperature was shifted to 37 °C, and the RL activity was measured in a time-dependent manner.
RESULTS
Design and Complementation Analyses of Fusion Proteins
Our assay system of membrane fusion relies on the recovery of the activity of the split enzyme, spRL, via reassociation induced by membrane fusion. A problematic aspect of spRL to be used as a reporter for membrane fusion is that spRL itself has a very weak ability to self-associate (Fig. 1, A and B, nRL/cRL). Therefore, a better association of spRL was aimed by fusing a pair of association modules. The split point of spRL was the same as the one used in a previous study (13). To evaluate the efficiency of the different association modules to augment the association of the spRL, co-transfection experiments using spRLs fused with the various different association modules were performed. The previous study showed that the fusion of spRL to a parallel homodimeric leucine zipper (coiled-coil) motif only recovered a low activity of spRL (13). This may be due to the possible interfering homodimeric interaction between nRL and nRL or between cRL and cRL against the desirable heterodimeric nRL and cRL interaction. Therefore, we tried the heterodimeric parallel coiled-coil motif called Velcro (16) instead of the homodimeric coiled-coil motif. This is supposed to preferentially induce a proper pairing between nRL and cRL. This combination, however, resulted in the recovery of only 0.1% of the intact RL enzymatic activity (Fig. 1, A and B, nRL-baseVel/acidVel-cRL), which was similar to the result of the case of the homodimeric coiled coil motif (13). Therefore, we sought for another self-association motif and chose spGFP whose capacity to self-associate has been dramatically augmented by molecular evolution (14). As schematically shown in the top panel of Fig. 1C, the use of the parallel coiled-coil motif (acidVel/baseVel) requires at least one long linker sequence between the association motif and spRL (see AcidVel-cRL in Fig. 1C, top panel). On the other hand, because the two split ends of spGFPs are expected to be located near each other after reassociation, the spRLs can be brought closer without the need of a long linker sequence (Fig. 1C, bottom panel). Furthermore, the inclusion of spGFP, as shown in our recent study (10), is expected to provide the visual monitoring of membrane fusion as well. We called these fusion proteins between spRL and spGFP dual split proteins, DSPs, because they would manifest dual functions of RL and GFP upon successful reassociation.
We first tried the previously described pair of spGFP, GFP1–10 and GFP11(10,14), as a fusion partner of spRL. We tested two types of linker sequences between spGFP and spRL; one is long and rich in glycine and serine residues, and the other is a short sequence derived from the employed restriction sequences (see “Experimental Procedures”). There was no significant difference in recovered activities between the two different linkers, and the level of recovery was similar to that of Velcro. The data for the shorter linker sequence is shown (Fig. 1, A and B, DSP1–10/DSP11). We hypothesized that this low complementation achieved by DSP1–10 and DSP11 might be due to weaker reassociation via their small interaction regions between GFP1–10 and GFP11. Therefore, we employed a new pair, GFP1–7 and GFP8–11, by shifting the split point of GFP to after the 157th amino acid residue (Fig. 1, A and B, DSP1–7 and DSP8–11). This new spGFP pair is expected to have larger interaction regions than the pair of GFP1–10 and GFP11. We used similar linker sequences as in the case of GFP1–10 and GFP11. The shift of the split point combined with a short linker sequence dramatically increased the RL activity by 2 orders of magnitude higher than that produced with GFP1–10 and GFP11 or Velcro (Fig. 1B). The constructs with the long linker showed about 20% activity that observed for the short linker (data not shown). In addition to the higher recovery of the RL activity, the new pair recovered a brighter GFP signal than did DSP1–10/DSP11 (Fig. 1B). The individual fusion proteins produced neither a GFP signal nor RL activity (supplemental Fig. 2). This is not due to a failure of stable expression of the individual DSP, because the expression of DSP1–7 and DSP8–11 was verified by immunoblotting (supplemental Fig. 3). Both DSP1–7 and DSP8–11 were shown to distribute homogenously in the cell by immunofluorescence analysis (supplemental Fig. 3). A quick complementation of DSP1–7 and DSP8–11, reaching its plateau after 400 s, was observed when cell extracts containing either DSP1–7 or DSP8–11 were mixed in vitro (Fig. 1D). This time scale of the recovery is useful to analyze the early phase of virus Env-mediated membrane fusion.
Real-time Analysis of Membrane Fusion Using Dual Split Proteins
We monitored HIV-1 Env-mediated membrane fusion with the DSP1–7/DSP8–11 system (Fig. 2). Similar to a simple spGFP-based assay (10), the progress of the membrane fusion at an individual cell level was easily monitored by the time-dependent increase in the number of green fluorescent cells (Fig. 3A). The population-based fusion kinetics for the wild type (WT) Env was monitored with the RL activities using the membrane-permeable substrate EnduRen. This showed a sigmoidal profile similar to that reported previously (17) (Fig. 3B, open square). The time required to achieve 50% that of the maximal fusion after co-culture, t½MAX, was 94 ± 9 min (Fig. 3B, open square), slightly longer than that obtained in a previous study with the dye transfer assay (17). This difference could be due to the different experimental settings or reflect the lag time that associates with the recovery of DSP activity.
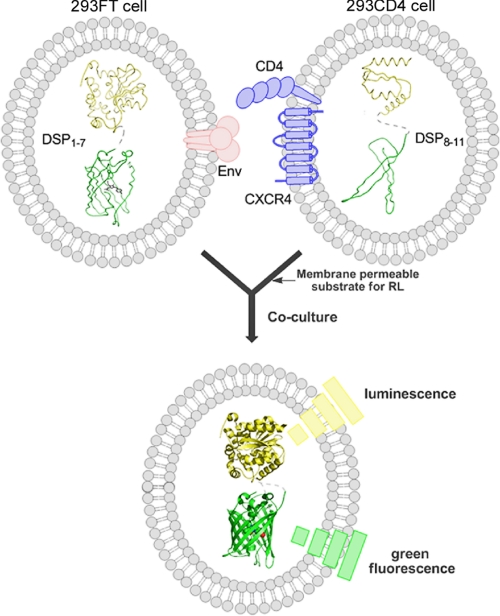
FIGURE 2.
Schema of the cell-based membrane fusion assay with DSPs. The 293FT cells, expressing HIV-1 Env and DSP1–7, and 293CD4 cells that harbor receptors (CD4 and CXCR4) and DSP8–11 were co-cultured to induce membrane fusion. The degree of membrane fusion was monitored in a time-dependent manner using RL and GFP activities. The DSP1–7 and DSP8–11 are schematically shown with structures of the RL (yellow, based on PDB code 2pse) and superfolder GFP (green, based on PDB code 2b3p) represented in a ribbon diagram.
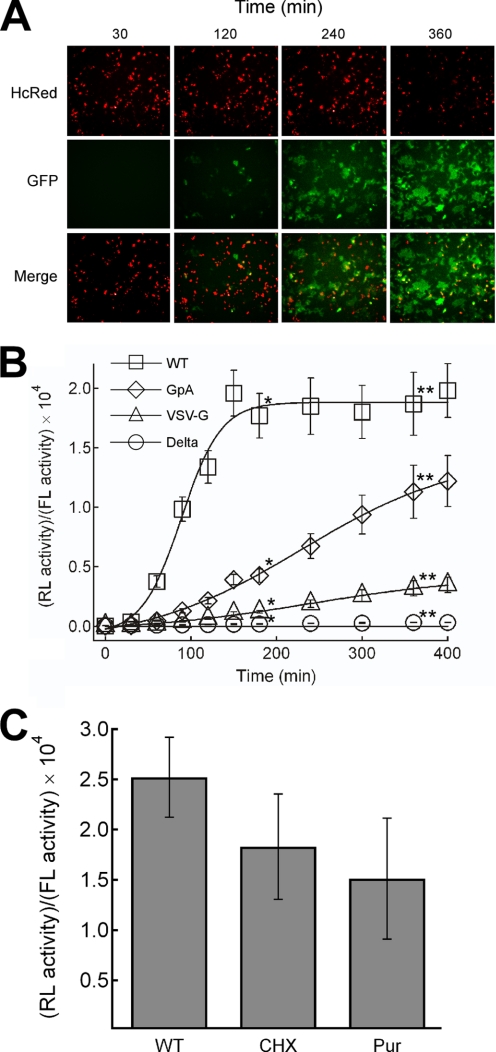
FIGURE 3.
Real-time monitoring of HIV-1 Env-mediated fusion. A, monitoring of GFP signal after co-culture of 293FT cell harboring wild type Env and DSP1–7 with 293CD4 cells harboring DSP8–11. Images were obtained with IN Cell Analyzer 1000. The Env-expressing cells bear red fluorescence in nuclei derived from the HcRed included in the Env expression vector (HcRed) (10). The views of the green signal (GFP) and merged images (Merge) are also shown. B, monitoring of the RL activity is shown. The recovered RL activity was divided by the firefly luciferase (FL) activity to normalize the transfection efficiency of the Env expression vector as described previously (15). Open squares, WT Env of HIV-1; open diamonds, GpA mutant; open triangles, the VSV-G mutant. Delta depicts the vector with the deleted envelope gene (open circle). * and **, statistically significant by independent t test (p < 0.0001). C, shown is the effect of protein synthesis inhibitor on the efficiency of fusion in DSP-mediated system. Cycloheximide (CHX, 100 μm) and puromycin (Pur, 10 μm) were added immediately before co-cultivation. The RL activities were measured 200 min after co-cultivation. The values were normalized by dividing them with the firefly luciferase activities and compared with that in the absence of the inhibitors (WT).
One of the advantages of the DSP system is its rather quick generation of the reporter signal after the co-cultivation. This is different from the transcription factor transfer assay system that relies on the synthesis of the reporter proteins induced by the transfer of the transcription factor, such as T7 RNA polymerase (15), during membrane fusion. To assess the effect of an on-going protein synthesis during co-culture in the DSP system, the inhibitors of protein synthesis, cycloheximide (CHX) or puromycin (Pur), were added before the co-culturing. The presence of the inhibitor resulted in a slightly lower reporter activity (Fig. 3C). This result suggests that the pre-existing DSPs before co-cultivation can account for the majority of the signal. Probably on-going synthesis of DSP is not required for the recovery of the DSP activity, as shown in the results of in vitro mixing of DSPs in cell lysates (Fig. 1D). The possibility of the enhancement of complementation of DSPs by the active protein synthesis or the involvement of labile proteins other than DSPs in membrane fusion, however, could not be ruled out completely.
Analysis of Membrane Fusion in the Presence of Fusion Inhibitors
We next examined the effects of representative fusion inhibitors of HIV-1 Env. One is soluble CD4 (sCD4), which is a competitive inhibitor of the binding between gp120 and the CD4 receptor. Another is a peptide called C34, which corresponds to the C-terminal heptad repeat sequence of gp41 that forms a coiled coil structure, a six-helix bundle, with an upstream N-terminal heptad repeat sequence of gp41 (18, 19). It is generally known that gp120 first binds to CD4 receptor and then undergoes the conformational changes that lead to a six-helix bundle formation of gp41 during membrane fusion. Each inhibitor was added to the culture medium at different time points after the co-cultivation to determine when 50% inhibition of fusion occurred (t½INH). The t½INH for sCD4 was about 51 ± 12 min, and the t½INH for C34, a six-helix bundle-formation inhibitor, was 93 ± 13 min (Fig. 4). These values are quite consistent with the expected order of inhibition, sCD4 then C34, and similar to those described before (17). This specific time-dependent inhibition of fusion validated our membrane fusion assay system using DSPs. With our DSP assay, the action point of an inhibitor can be determined more easily because lysing or fixing the cell samples before the measurement was not necessary.
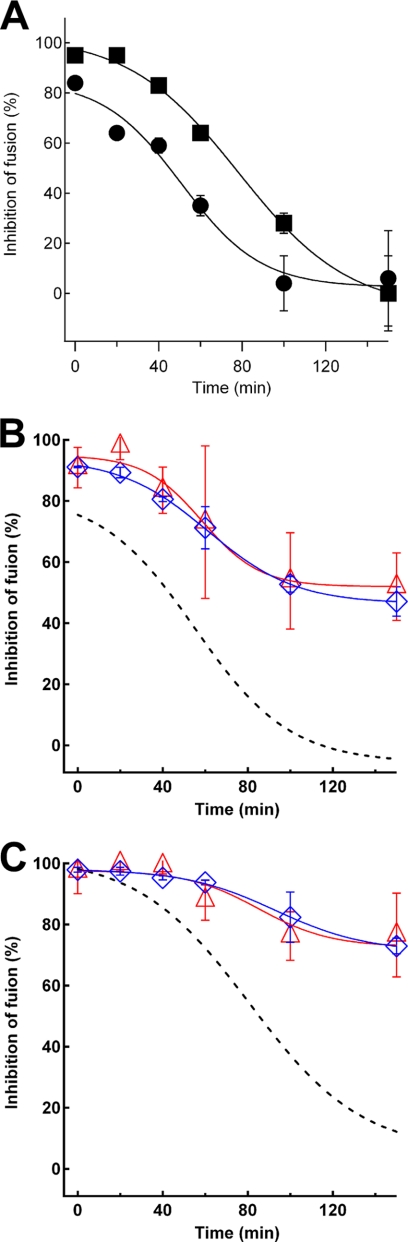
FIGURE 4.
Effect of the specific inhibitors of HIV-1 Env-mediated membrane fusion on the progress of membrane fusion. Soluble CD4 (0.1 μm) or C34 (2 μm) was added at the different time points during co-culture. A certain time after co-cultivation, the RL activity was determined. The degree of inhibition is shown in percentages. A representative data of four independent experiments is shown. A, shown is the inhibition profile of the WT Env of HIV-1. The filled circle and square indicate the results of soluble CD4 and C34, respectively. B and C, shown is the effect of soluble CD4 (panel B) and C34 (panel C) for mutants. The results for GpA (blue open diamond) and VSV-G (red open triangle) replacement mutants are shown together with those for WT. The profile of WT is shown with a broken line.
Analysis of Membrane Fusion of HIV-1 Mutant Env
We used the DSP system to examine the fusion kinetics of previously described mutants of HIV-1 Env that have their MSD replaced with that derived from GpA or VSV-G, respectively (10, 15). These MSD mutants showed incompetent fusion due to a defect(s) after the receptor binding step, but the mechanism of their fusion defect was unknown (15). The VSV-G and GpA mutants showed slower kinetics of membrane fusion than that of WT (Fig. 3B). Given the similar surface expression level and the CD4 binding activity among the tested mutants (15), these slower kinetics observed for these MSD mutants suggest the presence of processes that progress slower than those of WT after the interaction between Env and the CD4 receptor.
To obtain a further insight into the mechanism of the defect of fusion with the MSD mutants, we analyzed membrane fusion in the presence of fusion inhibitors, sCD4 and C34, and found that the period in which both mutants were sensitive to the inhibitors was extended, as compared with WT (Fig. 4, B and C). These results suggest that these mutants of Env have slower conformational changes of the extracellular domain of Env.
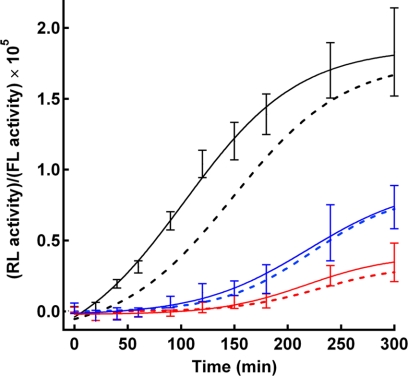
The HIV-1 Env is known to undergo sequential conformational changes during membrane fusion. One of these stages is operationally defined as a TAS (17, 20). This stage was induced by keeping the Env at the low temperature after its engagement with the CD4-co-receptor complex. At the TAS conformation, the Env is expected to have gone through a rate-limiting slow conformational change and is arrested at the state just before the productive lipid mixing (20). After the release from the TAS by raising the temperature, the rapid progress of membrane fusion was observed in the case of WT (Fig. 5, black lines). On the other hand, this rapid induction of membrane fusion was not observed with both GpA and VSV-G MSD mutants (Fig. 5). This result indicates the presence of the defect in the conformational changes of the Env of the MSD mutants after they interact with the CD4-co-receptor complex. Alternatively, it is also possible that these mutants have an additional defect after the TAS stage as well. In conclusion, mutations within the MSD manifest an allosteric effect on the conformation of Env.
FIGURE 5.
Analysis of membrane fusion by temperature shift. After the induction of TAS at 25 °C, the incubation temperature of the co-culture was shifted to 37 °C. The progress of membrane fusion was followed by the RL activity of the DSP system as described under “Experimental procedures.” The black, blue, and red lines are used to indicate the results of WT, GpA, and VSV-G mutants, respectively. The results in the presence (solid lines) or absence (broken lines) of the induction of TAS are shown. FL, firefly luciferase.
DISCUSSION
In this study we have utilized a newly developed real-time monitoring system of membrane fusion to analyze the membrane fusion induced by HIV-1 Env. The new method employed spRL, whose weak reassociation capacity was augmented by fusing them to spGFP. Between two spGFP pairs we tested, the pair of spGFP1–7 and spGFP8–11, which has broader contact regions than the pair of spGFP1–10 and spGFP11(10), induced the higher recovery of Renilla luciferase activity. Our data showed the usefulness of spGFP as a new association module to bring a pair of weakly interacting proteins. The anti-parallel nature of spGFP association may widen the catalogue of reassociation modules other than the widely used parallel coiled-coil-based association motif. In the case of the spGFP motif, we could avoid the use of a longer linker sequence to connect spRL and spGFP (Fig. 1C). This resulted in higher efficiency of the recovery of the RL activity. A longer linker sequence may interfere with the desired association or even provide a target of a proteolytic cleavage. The availability of the wide range of variants of GFP makes spGFP a versatile candidate association motif.
We applied DSPs to the analysis of the HIV-1 Env-mediated membrane fusion in the cell-based assay. Besides the estimation of the overall fusion efficiency from the RL activity or the GFP signal at an arbitrary end point, the combination of DSP with the membrane-permeable substrate enabled us to conduct a simple, continuous, and quantitative kinetic evaluation of the membrane fusion. The fact that the signal in the DSP system becomes detectable only after the actual fusion event is a great advantage. It makes a false positive rate very low. Other methods such as a dye transfer assay, on the other hand, require the careful determination of the redistribution of the dyes to distinguish the real fusion from mere aggregation of the cells (21). An assay based on the α-complementation of β-galactosidase is similar to our DSP in principle (12), but fluorogenic substrate of β-galactosidase, β-d-galactopyranoside, is not readily membrane-permeable (11), and a simple continuous live cell monitoring like ours is difficult.
Using our new method, HIV-1 Env-mediated membrane fusion was analyzed. The kinetic parameters of the membrane fusion induced by WT Env were similar to those described previously. We then analyzed the mutant Env in which its MSD was replaced with heterologous MSD derived from GpA and VSV-G. Our previous study indicated the potential defect(s) after the CD4 binding step, but the exact nature of the defect was unknown (15). The kinetic analysis by the DSP system showed an overall slower kinetics of membrane fusion in these mutants. The assay in the presence of the fusion inhibitors further revealed the prolonged period of vulnerability to sCD4 and C34. These data suggest that mutant Env has the altered structure in the ectodomain of the envelope protein, and there is the defect in conformational changes of gp120-gp41 complex after the binding of CD4 and co-receptors. The prolonged exposure to the lower temperature to promote the slow conformational changes (induction of TAS) still failed to facilitate the membrane fusion. This indicates the severe kinetic trap during the early conformational changes in these mutants. These results suggest that the mutations in MSD resulted in an altered structure in the extracellular portion of the gp120-gp41 complex by an allosteric effect. The exact nature of this altered conformation remains to be elucidated.
Our new real-time fusion assay system has wide applications. It may be useful for a screening of potential fusion inhibitors of virus Env, identification of an unidentified receptor for a virus, or a tropism analysis of a viral Env. In addition to the analysis of viral membrane fusion, our DSP may be applied to a quantitative analysis of cell biological events involving membrane fusion such as vesicular transport. As DSP is a dual functional reporter protein, there is a choice of two modes of monitoring depending on the purpose of the analysis. One can employ the GFP mode that realizes a simple monitoring without an extra exogenous substrate. This can provide temporal and spatial information as a visible signal. One can also use the RL mode combined with a membrane-permeable substrate, which will provide a simple continuous quantitative monitoring. Both modes do not require very sophisticated laboratory equipment. Therefore, high throughput analysis is possible.
Supplementary Material
Acknowledgments
We thank Heng Ru and Yuxing Cheng for excellent technical assistance. We thank Drs. Kunito Yoshiike and Ai Kawana-Tachikawa for critical reading of the manuscript. We thank A. M. Menting, an editorial consultant, for help in the preparation of the manuscript.
This work was supported by a contract grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan for the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- HIV-1
- human immunodeficiency virus, type 1
- GFP
- green fluorescent protein
- spGFP
- split GFP
- spRL
- split Renilla luciferase
- DSP
- dual split protein
- MSD
- membrane-spanning domain
- GpA
- glycophorin A
- VSV-G
- vesicular stomatitis virus G protein
- TAS
- temperature-arrested state
- Env
- envelope protein
- WT
- wild type
- sCD4
- soluble CD4.
REFERENCES
- 1.Mayer A. (2002) Annu. Rev. Cell Dev. Biol. 18, 289–314 [DOI] [PubMed] [Google Scholar]
- 2.Söllner T. H. (2004) Curr. Opin. Cell Biol. 16, 429–435 [DOI] [PubMed] [Google Scholar]
- 3.Melikyan G. B. (2008) Retrovirology 5, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roux K. H., Taylor K. A. (2007) Curr. Opin. Struct. Biol. 17, 244–252 [DOI] [PubMed] [Google Scholar]
- 5.Harrison S. C. (2008) Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumenthal R., Gallo S. A., Viard M., Raviv Y., Puri A. (2002) Chem. Phys. Lipids 116, 39–55 [DOI] [PubMed] [Google Scholar]
- 7.Dimitrov D. S., Willey R. L., Martin M. A., Blumenthal R. (1992) Virology 187, 398–406 [DOI] [PubMed] [Google Scholar]
- 8.Markosyan R. M., Cohen F. S., Melikyan G. B. (2005) Mol. Biol. Cell 16, 5502–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavrois M., De Noronha C., Greene W. C. (2002) Nat. Biotechnol. 20, 1151–1154 [DOI] [PubMed] [Google Scholar]
- 10.Wang J., Kondo N., Long Y., Iwamoto A., Matsuda Z. (2009) J. Virol. Methods 161, 216–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell R. E. (2004) Trends Biotechnol. 22, 208–211 [DOI] [PubMed] [Google Scholar]
- 12.Holland A. U., Munk C., Lucero G. R., Nguyen L. D., Landau N. R. (2004) Virology 319, 343–352 [DOI] [PubMed] [Google Scholar]
- 13.Paulmurugan R., Gambhir S. S. (2003) Anal. Chem. 75, 1584–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabantous S., Terwilliger T. C., Waldo G. S. (2005) Nat. Biotechnol. 23, 102–107 [DOI] [PubMed] [Google Scholar]
- 15.Miyauchi K., Komano J., Yokomaku Y., Sugiura W., Yamamoto N., Matsuda Z. (2005) J. Virol. 79, 4720–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Shea E. K., Lumb K. J., Kim P. S. (1993) Curr. Biol. 3, 658–667 [DOI] [PubMed] [Google Scholar]
- 17.Gallo S. A., Puri A., Blumenthal R. (2001) Biochemistry 40, 12231–12236 [DOI] [PubMed] [Google Scholar]
- 18.Chan D. C., Fass D., Berger J. M., Kim P. S. (1997) Cell 89, 263–273 [DOI] [PubMed] [Google Scholar]
- 19.Hussey R. E., Richardson N. E., Kowalski M., Brown N. R., Chang H. C., Siliciano R. F., Dorfman T., Walker B., Sodroski J., Reinherz E. L. (1988) Nature 331, 78–81 [DOI] [PubMed] [Google Scholar]
- 20.Melikyan G. B., Markosyan R. M., Hemmati H., Delmedico M. K., Lambert D. M., Cohen F. S. (2000) J. Cell Biol. 151, 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabrijel M., Repnik U., Kreft M., Grilc S., Jeras M., Zorec R. (2004) Biochem. Biophys. Res. Commun. 314, 717–723 [DOI] [PubMed] [Google Scholar]
- 22.Loening A. M., Fenn T. D., Gambhir S. S. (2007) J. Mol. Biol. 374, 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.