Abstract
The human LPIN1 gene encodes the protein lipin 1, which possesses phosphatidate (PA) phosphatase (3-sn-phosphatidate phosphohydrolase; EC 3.1.3.4) activity (Han, G.-S., Wu, W.-I., and Carman, G. M. (2006) J. Biol. Chem. 281, 9210–9218). In this work, we characterized human lipin 1 α, β, and γ isoforms that were expressed in Escherichia coli and purified to near homogeneity. PA phosphatase activities of the α, β, and γ isoforms were dependent on Mg2+ or Mn2+ ions at pH 7.5 at 37 °C. The activities were inhibited by concentrations of Mg2+ and Mn2+ above their optimums and by Ca2+, Zn2+, N-ethylmaleimide, propranolol, and the sphingoid bases sphingosine and sphinganine. The activities were thermally labile at temperatures above 40 °C. The α, β, and γ activities followed saturation kinetics with respect to the molar concentration of PA (Km values of 0.35, 0.24, and 0.11 mm, respectively) but followed positive cooperative (Hill number ∼2) kinetics with respect to the surface concentration of PA (Km values of 4.2, 4.5, and 4.3 mol %, respectively) in Triton X-100/PA-mixed micelles. The turnover numbers (kcat) for the α, β, and γ isoforms were 68.8 ± 3.5, 42.8 ± 2.5, and 5.7 ± 0.2 s−1, respectively, whereas their energy of activation values were 14.2, 15.5, and 18.5 kcal/mol, respectively. The isoform activities were dependent on PA as a substrate and required at least one unsaturated fatty acyl moiety for maximum activity.
Keywords: Diacylglycerol, Phosphatase, Phosphatidic Acid, Phospholipid, Triacylglycerol, Lipin
Introduction
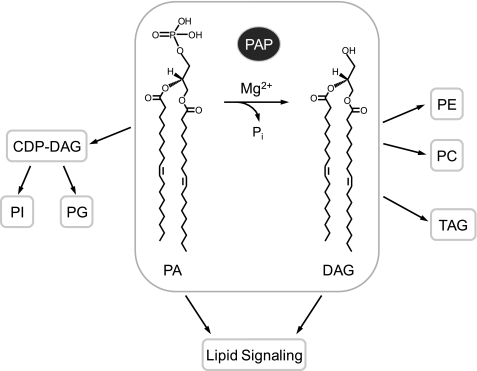
The PA2 phosphatase reaction (see Fig. 1) was first characterized in animal tissues by Kennedy and co-workers in 1957 (1). Subsequent studies during the last quarter of the 20th century implicated PA phosphatase as an important enzyme in lipid metabolism and signaling (2–5). The DAG generated in the PA phosphatase reaction may be used by a DAG acyltransferase enzyme to synthesize triacylglycerol and by ethanolaminephosphotransferase or cholinephosphotransferase enzymes to synthesize phosphatidylethanolamine or phosphatidylcholine, respectively (Fig. 1) (2, 3, 5, 6). The enzyme substrate PA is also used for phospholipid (e.g. phosphatidylinositol and phosphatidylglycerol) synthesis via CDP-DAG (Fig. 1) (2, 3, 6). With respect to lipid signaling, PA phosphatase may generate a pool of DAG used for protein kinase C activation, and by the nature of its reaction, it may attenuate the signaling functions of PA (7–11). Thus, it is expected that the regulation of PA phosphatase activity not only governs the pathways by which lipids are synthesized but also influences lipid signaling.
FIGURE 1.
PA phosphatase reaction and its roles in lipid synthesis and signaling. The reaction catalyzed by PA phosphatase (highlighted) is shown. The reaction product DAG is used for the synthesis of triacylglycerol (TAG) and the synthesis of phosphatidylethanolamine (PE) or phosphatidylcholine (PC). The reaction substrate PA is used for the synthesis of phosphatidylinositol (PI) or phosphatidylglycerol (PG) via CDP-DAG. The substrate and product of the PA phosphatase reaction play roles in lipid signaling.
Studies to establish the roles of PA phosphatase in lipid metabolism and signaling had been hampered by the lack of genetic and molecular information on the enzyme. However, this changed when yeast PAH1 was identified as the gene encoding PA phosphatase, which functions in lipid synthesis, PA signaling, and nuclear/endoplasmic reticulum membrane growth (6, 12–16). The homology between the yeast PA phosphatase protein and the lipin proteins encoded by the mouse Lpin1, Lpin2, and Lpin3 genes (17) led to the discovery that mammalian lipins are PA phosphatase enzymes (12, 18). That lipin 1 and lipin 2 complement phenotypes exhibited by the yeast pah1Δ mutant (19) indicates the evolutionarily conserved functions of the PA phosphatase enzymes. PA phosphatase (previously known as PA phosphatase 1) is specific for PA and catalyzes the Mg2+-dependent PA phosphatase reaction based on a DXDX(T/V) motif within a haloacid dehalogenase-like domain (6, 12, 14). The enzyme is distinguished from the family of lipid phosphate phosphatase enzymes (previously known as PA phosphatase 2) that dephosphorylate a broad spectrum of substrates (e.g. PA, lyso-PA, DAG pyrophosphate, sphingoid base phosphates, and isoprenoid phosphates) by a distinct catalytic mechanism that does not require divalent cations (5, 6, 20, 21). The functions of those lipid phosphate phosphatase enzymes are known to control the signaling properties of lipid phosphates, such as lyso-PA (5, 11, 22, 23).
Lpin1, identified by Reue and co-workers (17) through positional cloning, is the gene mutated in the fatty liver dystrophy (fld) mouse (24). In their seminal studies, Reue and co-workers (17, 25) showed that the loss of lipin 1 in mice prevents normal adipose tissue development, leading to lipodystrophy and insulin resistance, whereas its overexpression leads to obesity and insulin sensitivity. Thus, the molecular function of lipin 1 as a PA phosphatase enzyme provides a mechanistic basis for why the absence or overexpression of lipin 1 in mice has a major effect on lipid metabolism (12, 17, 25). In addition, mice lacking lipin 1 exhibit peripheral neuropathy that is characterized by myelin degradation, Schwann cell dedifferentiation and proliferation, and a reduction in nerve conduction velocity (24, 26, 27). These effects are mediated through the mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) signaling pathway that is activated by elevated levels of PA due to the loss of PA phosphatase activity (27). Moreover, the level and compartmentalization of lipin 1 exert a major impact on the assembly and secretion of hepatic very low density lipoprotein (28). With respect to humans, mutations in the LPIN1 gene are associated with metabolic syndrome, type 2 diabetes, and recurrent acute myoglobinuria in children, whereas mutations in the LPIN2 gene are the basis for the anemia and inflammatory disorders associated with the Majeed syndrome (29–31). Recent work has shown that a conserved serine residue that is mutated in lipin 2 of Majeed syndrome patients is essential for PA phosphatase activity (32).
In addition to its PA phosphatase activity, lipin 1 functions as a transcriptional coactivator in the regulation of lipid metabolism gene expression (33–36). Lipin 1 interacts with a complex containing peroxisome proliferator-activated receptor α (PPARα) and PPARγ coactivator-1α (PGC-1α) to regulate the expression of genes involved in fatty acid oxidation (33). This transcriptional coactivator function is dependent on an LXXIL motif found near the DXDX(T/V) motif but independent of PA phosphatase activity (33).
Lpin1 expression is required for adipocyte differentiation (19, 34), and it is stimulated by glucocorticoids (37, 38). Interestingly, Lpin2 expression is repressed during adipogenesis, indicating that lipin 1 and lipin 2 have distinct and non-redundant functions in adipocytes (19). In addition, alternative splicing of mouse Lpin1 mRNA gives rise to two lipin 1 isoforms (α and β) that are postulated to play distinct functions in adipogenesis (34). It is also known that human LPIN1 mRNA expression is induced by sterol depletion (39).
Lipin 1 is localized to the cytosol, endoplasmic reticulum, and nucleus (30, 35, 40). The cellular locations of lipin 1 are influenced by covalent modifications that include phosphorylation (19, 41, 42) and sumoylation (43) and by interaction with 14-3-3 proteins (44). Phosphorylation of lipin 1 is stimulated by insulin and mediated by the mTOR signaling pathway (41). Lipin 1 is also phosphorylated during the mitotic phase of the cell cycle (19). Phosphorylated forms of lipin 1 are enriched in the cytosolic fraction, whereas the dephosphorylated forms are enriched in the membrane fraction (19, 42). The translocation of lipin 1 from the cytosol to the membrane is essential to its function as a PA phosphatase enzyme. The association of lipin 1 with the nucleus, where it functions as a transcriptional coactivator, is facilitated by the sumoylation of the protein (43). However, the nuclear localization of lipin 1 is blocked by its interaction with 14-3-3 proteins in the cytosol (44).
Understanding the biochemical properties of the PA phosphatase activities of the lipin proteins is fundamental to elucidating the mode of action and control of these activities in vivo. Up to now, however, attempts to purify mammalian forms of PA phosphatase for defined biochemical studies have been unsuccessful (3, 5). In this work, we purified human lipin 1 isoforms (α, β, and γ) and characterized their PA phosphatase activities with respect to their enzymological and kinetic properties. These studies revealed that although the enzymological properties of the lipin 1 isoforms were generally similar, they differed significantly with respect to their kinetic properties. In addition, this work showed that the purified LPIN1-encoded PA phosphatase activities were potently inhibited by sphingoid bases, indicating a connection between human PA phosphatase and sphingolipid metabolism.
EXPERIMENTAL PROCEDURES
Materials
Growth medium components were purchased from Difco. Restriction endonucleases, modifying enzymes, and Vent DNA polymerase were from New England Biolabs. Human liver total RNA was from Clontech. DNA purification and one-step reverse transcription-PCR kits were from Qiagen. Nucleotides, oligonucleotides, Triton X-100, and ammonium molybdate were from Sigma. Malachite green was from Fisher. Protease inhibitor mixture tablets were from Roche Applied Science. Lipids were from Avanti Polar Lipids. Protein assay reagents, electrophoretic reagents, and DNA and protein size standards were from Bio-Rad. TLC glass plates (silica gel 60) were from EM Science. Radiochemicals were from PerkinElmer Life Sciences. Scintillation counting supplies were from National Diagnostics.
Strains and Plasmids
Escherichia coli strains and plasmids used in this study are listed in Table 1. Oligonucleotides used for plasmid construction are listed in Table 2. DH5α and Rosetta 2(DE3)pLysS were used for plasmid maintenance and protein expression, respectively.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR, recA1 endA1 hdR17(rnk−mk+) phoA supE44l−thi-1 gyrA96 relA1 | Ref. 65 |
| Rosetta2(DE3)pLysS | F−ompT hsdSB (rB−mB−) gal dcm (DE3) pLysSRARE2 (CamR) | Novagen |
| Plasmids | ||
| pET-28b(+) | E. coli expression vector for C-terminal His6-tag fusion (KanR) | Novagen |
| pGH321 | LPIN1γ coding sequence inserted into pET-28b(+) | This study |
| pGH322 | LPIN1α coding sequence inserted into pET-28b(+) | This study |
| pGH327 | LPIN1β coding sequence inserted into pET-28b(+) | This study |
TABLE 2.
Oligonucleotides used for PCR
| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| LPIN1-F4 | AATTACGTGGGGCAGTTAGC |
| LPIN1-R2670 | AACTCGAGCGCTGAGGCAGAATGAATGTCC |
| LPIN1-F1080 | GATCGAGGAGCTCAAACCCC |
| LPIN1-R1630 | CCCTCATGATAGATTCCACAGTGGCCTTTGGCAAAGGTTTCTGGAAGGCCTGC |
| LPIN1-F1578 | GCAGGCCTTCCAGAAACCTTTGCCAAAGGCCACTGTGGAATCTATCATGAGGG |
| LPIN1-R2130 | ATGGTACAGCTTAGCGATGC |
| LPIN1-F562 | TCGGATGAGGCCATGGAGCTGC |
| LPIN1-R1065 | CGCTGCTGCTCCTAAGGTCTCC |
Plasmid Constructions
The human LPIN1 plasmids were constructed on the E. coli expression vector pET-28b(+), which carries the C-terminal His6 tag. pGH321 was generated by insertion of the human LPIN1γ sequence into pET-28b(+) at the NcoI and XhoI sites. The LPIN1γ sequence (2,756 bp) was amplified from the human LPIN1 cDNA (12) by PCR with the LPIN1-F4 and LPIN1-R2670 primers, followed by digestion with XhoI. pET-28b(+) was digested with NcoI, filled in with Klenow, and digested with XhoI. The vector and insert with compatible ends were ligated to produce pGH321. pGH322 was derived from pGH321 by removal of the LPIN1γ-specific sequence using the overlap extension PCR method (45). The upstream (551 bp) and downstream (553 bp) flanking regions of the LPIN1γ-specific sequence were amplified from pGH321 by PCR with the LPIN1-F1080/LPIN1-R1630 and LPIN1-F1578/LPIN1-R2130 primer sets, respectively. The two PCR products (551 and 553 bp) containing the 53-bp overlapping ends were combined through denaturation, annealing, and extension. The combined 1051-bp DNA (LPIN1α) was then amplified with the LPIN1-F1080 and LPIN1-R2130 primers, followed by digestion with BamHI and HindIII. The resulting 693-bp LPIN1α DNA was substituted for the 771-bp fragment in pGH321 to produce pGH322. pGH327 was derived from pGH322 by insertion of the LPIN1β-specific sequence. The LPIN1 cDNA was produced from the human liver mRNA by reverse transcription-PCR with the LPIN1-F562 and LPIN1-R1065 primers. Of two LPIN1 DNA fragments (504 and 612 bp) amplified, the larger one corresponding to LPIN1β was purified from the gel. After digestion with NcoI and BsaI, the 575-bp LPIN1β fragment was substituted for the 467-bp fragment in pGH322 to produce pGH327.
Expression and Purification of the His6-tagged Human LPIN1-encoded PA Phosphatase Isoforms
A single colony of Rosetta 2(DE3)pLysS bearing a human LPIN1 plasmid was inoculated into 5 ml of LB medium containing kanamycin (30 μg/ml) and chloramphenicol (34 μg/ml), followed by incubation overnight at 37 °C. Two ml of the saturated culture was inoculated into 1 liter of fresh medium and was grown at 37 °C with shaking at 250 rpm until the absorbance at 600 nm reached 0.5. The culture was added with 1 mm isopropyl β-d-1-thiogalactopyranoside and was further incubated for 3 h at 37 °C. The induced culture for LPIN1 expression was harvested by centrifugation at 5,000 × g for 10 min at 4 °C, and the cell pellet was resuspended in 40 ml of 20 mm Tris-HCl (pH 8.0) buffer containing 500 mm NaCl, 5 mm imidazole, and one tablet of EDTA-free protease inhibitor mixture. The resuspended cells were disrupted by two passes through a French press at 20,000 p.s.i., and the lysate was centrifuged at 12,000 × g for 30 min at 4 °C. The supernatant was incubated with 2 ml of Ni2+-nitrilotriacetic acid-agarose (50% slurry) by gentle agitation for 2 h at 4 °C, and the mixture was packed in a 10-ml chromatography column. The packed column was washed with 50 ml of 20 mm Tris-HCl (pH 8.0) buffer containing 500 mm NaCl, 45 mm imidazole, 7 mm 2-mercaptoethanol, and 10% glycerol, and the His6-tagged enzymes were eluted from the column in 1-ml fractions with a total of 5 ml of 20 mm Tris-HCl (pH 8.0) buffer containing 500 mm NaCl, 250 mm imidazole, 7 mm 2-mercaptoethanol, and 10% glycerol. The protein concentration of the purified enzymes was determined by the method of Bradford (46) using bovine serum albumin as the standard. The enzyme preparations, which had a final protein concentration of about 0.5 mg/ml, were stored at −80 °C.
SDS-PAGE and Immunoblot Analysis
SDS-PAGE (47) was performed with 8% mini slab gels, and immunoblotting (48) was performed with polyvinylidene difluoride membranes. Proteins on the SDS-polyacrylamide gels were visualized by staining with Coomassie Blue R250. The His6-tagged proteins were detected with mouse monoclonal anti-His6 antibodies at a dilution of 1:1,000 and goat anti-mouse IgG-alkaline phosphatase conjugates at a dilution of 1:5,000. Chemifluorescence signals produced by the alkaline phosphatase reaction were subjected to FluorImaging analysis.
Preparation of [32P]PA and Malachite Green-Molybdate Reagent
[32P]PA was synthesized enzymatically from DAG and [γ-32P]ATP using E. coli DAG kinase as described by Carman and Lin (49). A color reagent to detect inorganic free phosphate was prepared by modification of the procedure described by Mahuren et al. (50) and was composed of 3 volumes of 0.1 mm malachite green and 1 volume of 34 mm ammonium molybdate in 5 m HCl.
Preparation of Triton X-100/Lipid-mixed Micelles
PA in chloroform was transferred to a glass test tube, and the solvent was eliminated in vacuo for 1 h. Triton X-100 was added to PA to prepare Triton X-100/PA-mixed micelles. Micelles containing other lipids were prepared in the same manner. The mol % of a lipid in a Triton X-100/lipid-mixed micelle was calculated using the following formula, mol %lipid = 100 × [lipid (molar)]/([lipid (molar)] + [Triton X-100 (molar)]). The total lipid concentration in the Triton X-100/lipid-mixed micelles was kept below 15 mol % to ensure that the structure of the lipid-mixed micelles was similar to that of pure Triton X-100 micelles (51, 52).
Enzyme Assays
PA phosphatase activity was measured by following the release of water-soluble Pi from chloroform-soluble PA. Unless otherwise specified, a reaction mixture contained 50 mm Tris-HCl (pH 7.5) buffer, 0.5 mm MgCl2, 10 mm 2-mercaptoethanol, 1 mm dioleoyl [32P]PA (5,000 cpm/nmol), 10 mm Triton X-100, and 50 ng of enzyme protein in a total volume of 0.1 ml. The reaction mixture was incubated at 37 °C for 20 min, after which the enzyme reaction was terminated by adding 0.5 ml of 0.1 m HCl in methanol, 1 ml of chloroform, and 1 ml of 1 m MgCl2. Following phase separation, 0.5 ml of the aqueous (upper) phase was measured for radioactivity. In the assay with non-radioactive PA, the enzyme reaction was terminated as described above except that 1 m MgCl2 was replaced with water. After phase separation, 1 volume of the upper phase was mixed with 2 volumes of malachite green-molybdate reagent, and the mixture was measured for absorbance at 650 nm. The same assay was used to examine the dephosphorylation of other nonradioactive lipid phosphate molecules. Enzyme assays were conducted in triplicate, and the average S.D. value of the assays was ± 5%. All enzyme reactions were linear with time and protein concentration. A unit of enzymatic activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of product/min.
Data Analyses
Kinetic data were analyzed according to the Michaelis-Menten and Hill equations using the SigmaPlot enzyme kinetics module. Student's t test (SigmaPlot software) was used to determine statistical significance, and p values of <0.05 were taken as a significant difference.
RESULTS
A Novel Splice Form of the Human LPIN1 Gene
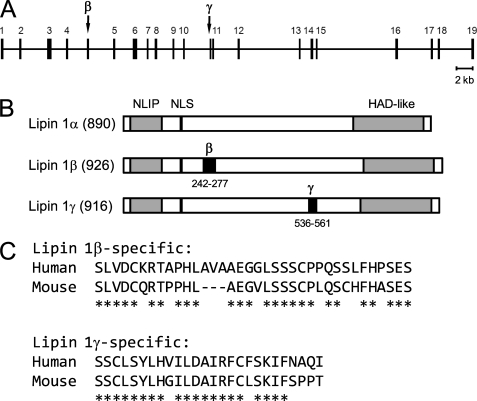
In our previous work, the LPIN1 cDNA (TC125492, OriGene) derived from the human fetal brain mRNA has been shown to encode a PA phosphatase enzyme (12). Sequence analysis of the LPIN1 cDNA revealed that its coding sequence was neither LPIN1α nor LPIN1β but corresponded to a new splice variant that consists of LPIN1α and an additional 78 nucleotides inserted at position 1606 of the sequence. Accordingly, the new human LPIN1 splice form that encodes a functional PA phosphatase was named LPIN1γ. In the exon-intron organization of the human LPIN1 gene, the LPIN1γ-specific exon (NC_000002.11: 11931833–11931910) is close to its downstream flanking exon with the intervening 128-nucleotide intron (Fig. 2A). In the primary structure of human lipin 1, the lipin 1γ-specific sequence consisting of 26 amino acids is located in a non-homologous region and does not alter the conserved NLIP and haloacid dehalogenase-like lipin domains (Fig. 2B). A homology search against the mouse genome identified a counterpart of the human LPIN1γ-specific exon, which is also composed of 78 nucleotides (NC_000078.5: c16565526–16565449) and flanked with a short (142 bp) downstream intron. When aligned, the human and mouse exon sequences showed 77% identity in deduced amino acids (Fig. 2C).
FIGURE 2.
Schematic representation of the human LPIN1 gene and its protein products. A, the exon-intron organization of the human LPIN1 gene is shown with respect to coding exons. The exons numbered 1–19 comprise LPIN1α, whereas those combined with an additional exon (indicated as β or γ) constitute LPIN1β and LPIN1γ, respectively. B, the human lipin 1 isoforms (α, β, and γ) are represented with the number of amino acids and the conserved lipin domains NLIP and haloacid dehalogenase-like (HAD-like). The specific sequences for lipin 1β and lipin 1γ are indicated as β and γ, respectively. NLS, nuclear localization signal. C, the human lipin 1β- and lipin 1γ-specific sequences are aligned with the mouse counterparts. *, conserved amino acid.
The Human LPIN1β-specific Sequence
The human LPIN1β-specific sequence, like the mouse Lpin1β-specific sequence, is known to contain 99 nucleotides (53). However, from sequence analysis of the LPIN1β cDNA derived from human liver mRNA, we found that 108 nucleotides coding for 36 amino acids comprise the LPIN1β-specific sequence (sequence not shown). This finding was confirmed by inspection of a reference human genome where the LPIN1β-specific exon (NC_000002.11: 11916211–11916318) flanked with canonical AG/GT splice sites is composed of the same 108 nucleotides. When compared with the mouse counterpart, the human LPIN1β-specific sequence shows 72% identity in deduced amino acids (Fig. 2C).
Heterologous Expression and Purification of the Human LPIN1-encoded PA Phosphatase Isoforms
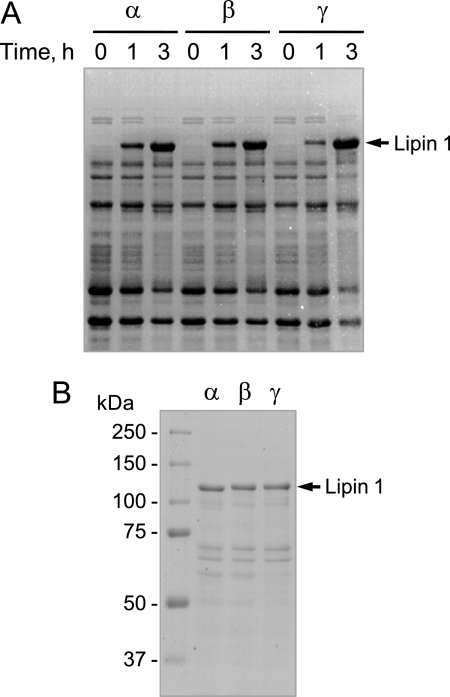
We expressed in E. coli the human LPIN1-encoded isoforms (α, β, and γ) fused with a His6 tag sequence at the C terminus. To enhance the heterologous expression of the human LPIN1 gene, the Rosetta 2 strain that overproduces seven rare tRNAs was used as an expression host. The isopropyl β-d-1-thiogalactopyranoside-induced expression of the His6-tagged PA phosphatase isoforms was confirmed by SDS-PAGE (Fig. 3A) as well as by immunoblot analysis using anti-His6 antibodies (data not shown). A significant amount of the expressed proteins was associated with an insoluble fraction containing inclusion bodies (data not shown). However, the recombinant proteins in the soluble fraction of cell lysates were purified by affinity chromatography using Ni2+-nitrilotriacetic acid-agarose. SDS-PAGE analysis showed that the procedure resulted in highly purified enzyme preparations (Fig. 3B). The PA phosphatase activities of the purified lipin 1 isoforms were relatively stable for a few cycles of freezing and thawing. The dilution of the enzyme preparations resulted in the loss of the PA phosphatase activities. The full-length recombinant proteins migrated more slowly upon electrophoresis to positions in the polyacrylamide gel that were ∼15 kDa greater than their predicted molecular masses of about 100 kDa (Fig. 3B). The faster migrating proteins below each of the full-length isoforms appeared to be breakdown products because their amounts increased during storage. The PA phosphatase activities of the purified lipin 1 isoforms were in the range of the activities of nearly homogenous preparations of the PAH1-encoded PA phosphatase expressed in Saccharomyces cerevisiae (15, 54) or expressed in E. coli (12).
FIGURE 3.
SDS-PAGE analysis of the E. coli-expressed and purified human LPIN1-encoded isoforms. A, Rosetta 2(DE3)pLysS cells bearing a LPIN1 plasmid (pGH321, pGH322, or pGH327) were grown to A600 nm = 0.5. After induction of LPIN1 expression with 1 mm isopropyl β-d-1-thiogalactopyranoside for the indicated time intervals, an equal number of the induced cells were lysed with Laemmli sample buffer and resolved on an 8% SDS-polyacrylamide gel. B, the E. coli-expressed lipin 1 isoforms were subjected to affinity purification with Ni2+-nitrilotriacetic acid resin. The purified His6-tagged lipin 1 isoforms (5 μg of protein) were subjected to SDS-PAGE (8% slab gel). Proteins on the SDS-polyacrylamide gels were visualized with Coomassie Blue. The positions of the full-length lipin 1 isoforms and molecular mass markers are indicated.
Enzymological Properties of the Human LPIN1-encoded PA Phosphatase Isoforms
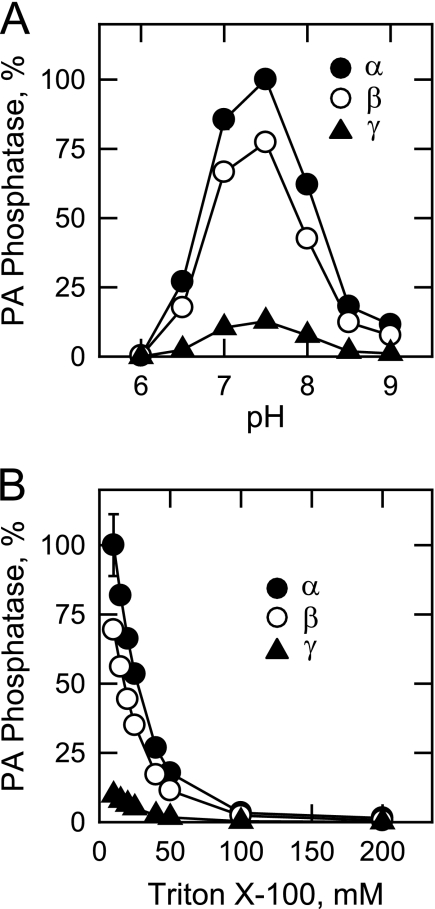
The human LPIN1-encoded isoforms (α, β, and γ) were specific for PA and did not catalyze the dephosphorylation of other lipid phosphate molecules, such as lyso-PA, DAG pyrophosphate, sphinganine-1-phosphate, sphingosine 1-phosphate, and ceramide 1-phosphate (data not shown). Under optimum assay conditions, the specific activities of the human LPIN1-encoded PA phosphatase α, β, and γ isoforms were 35, 25, and 3 μmol/min/mg, respectively. For ease of comparison, the data shown for the characterizations of the isoforms were presented relative (%) to the maximum activity of the α isoform. All three isoforms exhibited PA phosphatase activities in the range of pH 6.5–8.5, with optimum activities at pH 7.5 (Fig. 4A). Triton X-100 was used to solubilize the water-insoluble substrate PA by formation of Triton X-100/PA-mixed micelles (54, 55). Using 1 mm PA, the maximum activity for each of the isoforms was obtained with 10 mm Triton X-100 (10:1 molar ratio of Triton X-100 to PA) (Fig. 4B). The PA phosphatase activities of the isoforms were reduced in a dose-dependent manner at Triton X-100 concentrations that were higher than 10 mm (Fig. 4B). These results are characteristic of surface dilution kinetics, a property exhibited by many enzymes that utilize lipid substrates in detergent/lipid-mixed micelles (55).
FIGURE 4.
Effects of pH and Triton X-100 on the human LPIN1-encoded PA phosphatase isoform activities. A, PA phosphatase activity was measured at the indicated pH values with 50 mm Tris-maleate-glycine buffer. B, the enzyme activity was measured in 50 mm Tris-HCl (pH 7.5) buffer with the indicated concentrations of Triton X-100. The highest activity of the α isoform was set at 100%, and the activities of the β and γ isoforms were calculated relative to the activity of the α isoform. The data shown are means ± S.D. from triplicate enzyme determinations.
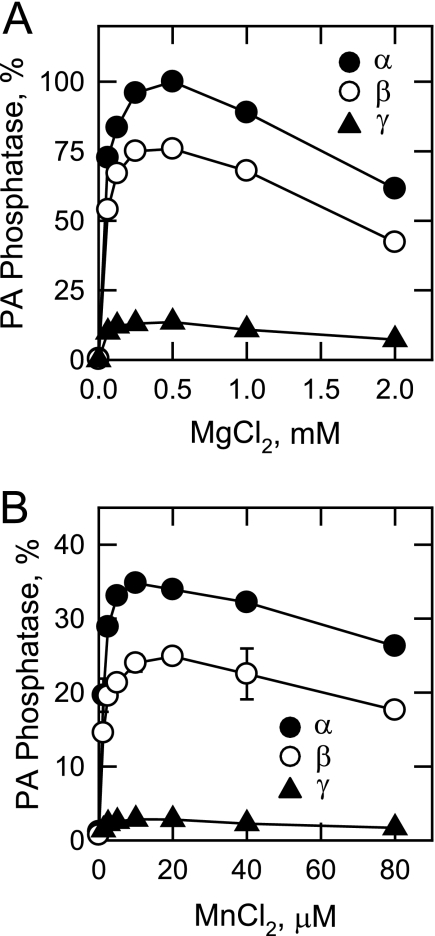
The PA phosphatase activities of the human LPIN1-encoded isoforms were absolutely dependent on either Mg2+ (Fig. 5A) or Mn2+ (Fig. 5B) ions, with Mg2+ being the preferred cofactor based on maximum activities (Fig. 5). For example, the activity of the α isoform with Mg2+ as the cofactor was 3-fold greater than the activity with Mn2+. However, a relatively high concentration (∼0.5 mm) was required for the maximum activities with Mg2+, whereas a relatively low concentration (∼10 μm) was required for the maximum activities with Mn2+ (Fig. 5). Concentrations of Mg2+ or Mn2+ above their optimums resulted in the inhibition of each of the LPIN1-encoded isoform activities (Fig. 5). The stimulatory effects of Mg2+ or Mn2+ on the PA phosphatase activities were abolished by chelation with EDTA (Table 3). With Mg2+ as the cofactor, the PA phosphatase activities were abolished by the addition of 1 mm concentrations of Mn2+ or Ca2+ (Table 3). Zn2+ was also inhibitory (33–50%) to the activities but to a lesser extent (Table 3).
FIGURE 5.
Effects of Mg2+ and Mn2+ on the human LPIN1-encoded PA phosphatase isoform activities. PA phosphatase activity was measured in the absence and presence of the indicated concentrations of MgCl2 (A) or MnCl2 (B). Relative activities were presented as described in the legend to Fig. 3. For B, the highest activity of the α isoform was calculated relative to its activity measured with MgCl2 in A. The data shown are means ± S.D. from triplicate enzyme determinations.
TABLE 3.
Effectors of the LPIN1-encoded PA phosphatase isoform activities
PA phosphatase activity was measured under standard assay conditions with 9.1 mol % PA in the presence of the indicated compounds. The data shown are means ± S.D. from triplicate enzyme determinations.
| Compound | Relative PA phosphatase activity of isoform |
||
|---|---|---|---|
| α | β | γ | |
| % | % | % | |
| Control | 100 | 100 | 100 |
| Control + 1 mm EDTA | 2 ± 0.1 | 2 ± 0.1 | 1 ± 0.1 |
| Control + 50 mm NaCl | 96 ± 0.9 | 92 ± 2.7 | 93 ± 4.4 |
| Control + 50 mm KCl | 98 ± 1.9 | 97 ± 3.0 | 99 ± 2.2 |
| Control + 1 mm MnCl2 | 0 ± 0.0 | 0 ± 0.0 | 0 ± 0.0 |
| Control + 1 mm CaCl2 | 1 ± 0.0 | 1 ± 0.2 | 1 ± 0.1 |
| Control + 1 mm ZnCl2 | 67 ± 1.3 | 63 ± 1.2 | 50 ± 2.3 |
| Control + 1 mm Na3VO4 | 50 ± 0.7 | 47 ± 1.0 | 53 ± 2.3 |
| Control + 10 mm NaF | 95 ± 5.9 | 97 ± 6.0 | 103 ± 3.5 |
| Control + 50 mm potassium phosphate (pH 7.5) | 67 ± 1.4 | 70 ± 1.3 | 62 ± 2.6 |
| Control + 10 mm glycerophosphate | 94 ± 3.0 | 94 ± 2.4 | 87 ± 2.1 |
| Control + 25 mm imidazole | 79 ± 2.6 | 71 ± 0.9 | 86 ± 6.2 |
| Control + 1 mm propranolol | 32 ± 1.4 | 36 ± 0.9 | 28 ± 1.1 |
| Control + 5 mm phenylglyoxal | 86 ± 0.9 | 100 ± 6.0 | 93 ± 4.3 |
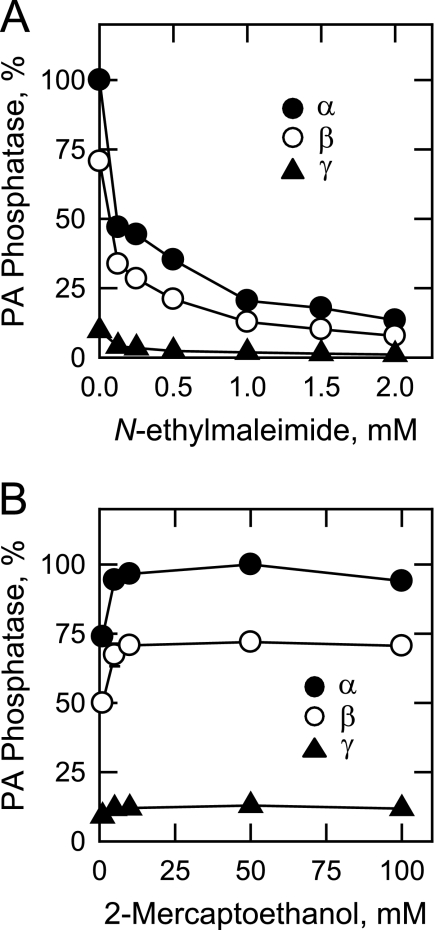
A characteristic property of PA phosphatase enzymes from mammalian cells is their sensitivities to the alkylating reagent N-ethylmaleimide (2, 3). For each of the isoforms, N-ethylmaleimide inhibited PA phosphatase activity in a dose-dependent manner (IC50 = 0.1 mm; Fig. 6A). The inhibitory effect of this reagent on the PA phosphatase activities could be prevented by preincubation with 10 mm 2-mercaptoethanol. In addition, the enzyme activities were stimulated (25%) by the addition of 10 mm 2-mercaptoethanol to the assay systems that did not contain N-ethylmaleimide (Fig. 6B).
FIGURE 6.
Effects of N-ethylmaleimide and 2-mercaptoethanol on the human LPIN1-encoded PA phosphatase isoform activities. PA phosphatase activity was measured in the presence of the indicated concentrations of N-ethylmaleimide (A) or 2-mercaptoethanol (B). In A, 2-mercaptoethanol was not present. Relative activities were presented as described in the legend to Fig. 3. The data shown are means ± S.D. from triplicate enzyme determinations.
The effects of general phosphatase inhibitors on the LPIN1-encoded PA phosphatase isoform activities were examined (Table 3). Na3VO4 was the only compound that had a significant inhibitory effect on the isoform activities (50% inhibition at 1 mm). At a concentration of 1 mm, propranolol (a non-selective β-blocker) inhibited the activities by about 70%, whereas a 5 mm concentration of phenylglyoxal (an arginine-reactive compound) did not have an inhibitory effect on the activities (Table 3).
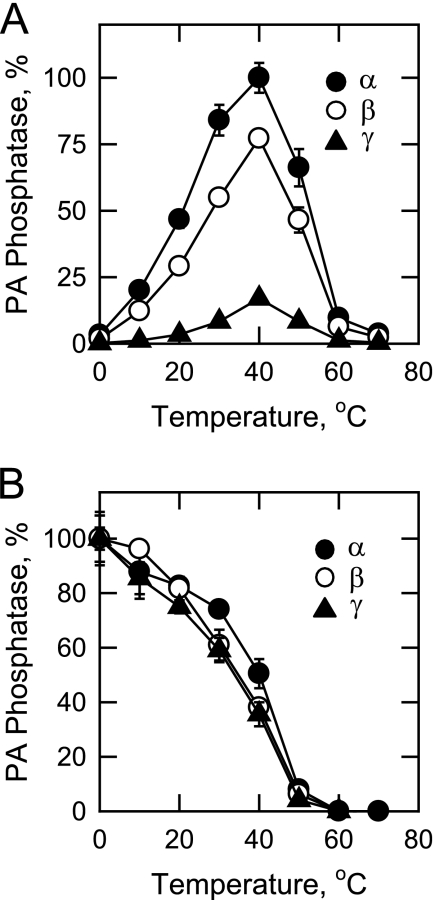
Effect of Temperature on the LPIN1-encoded PA Phosphatase Isoforms
The effect of temperature on the PA phosphatase activities of the LPIN1-encoded isoforms was examined (Fig. 7A). For each of the isoforms, maximum activity was observed at 40 °C. However, the enzymes were thermally labile above 40 °C and were essentially inactive at 60 °C. Arrhenius plots of the data from 0 to 40 °C were constructed for each of the isoforms. The energy of activation for the α, β, and γ isoforms was calculated to be 14.2, 15.5, and 18.5 kcal/mol, respectively. The purified PA phosphatase isoforms were examined for their thermal stabilities by preincubation at temperatures ranging from 0 to 70 °C for 20 min (Fig. 7B). Following the preincubation, the enzyme samples were cooled on ice for renaturation and then measured for PA phosphatase activity at 37 °C. About 50% of the PA phosphatase activities of the LPIN1-encoded isoforms were lost after preincubation at 40 °C, with a 90% loss at 50 °C.
FIGURE 7.
Effect of temperature on the activities and stabilities of the human LPIN1-encoded PA phosphatase isoforms. A, PA phosphatase activity was measured at the indicated temperatures for 20 min in a temperature-controlled water bath. Relative activities were presented as described in the legend to Fig. 3. B, the enzyme samples were first incubated for 20 min at the indicated temperatures. After incubation, the samples were cooled in an ice bath for 10 min to allow for enzyme renaturation, and PA phosphatase activity was then measured for 20 min at 37 °C. The highest activity of each isoform was set at 100%. The data shown are means ± S.D. from triplicate enzyme determinations.
Kinetic Properties of the Human LPIN1-encoded PA Phosphatase Isoforms
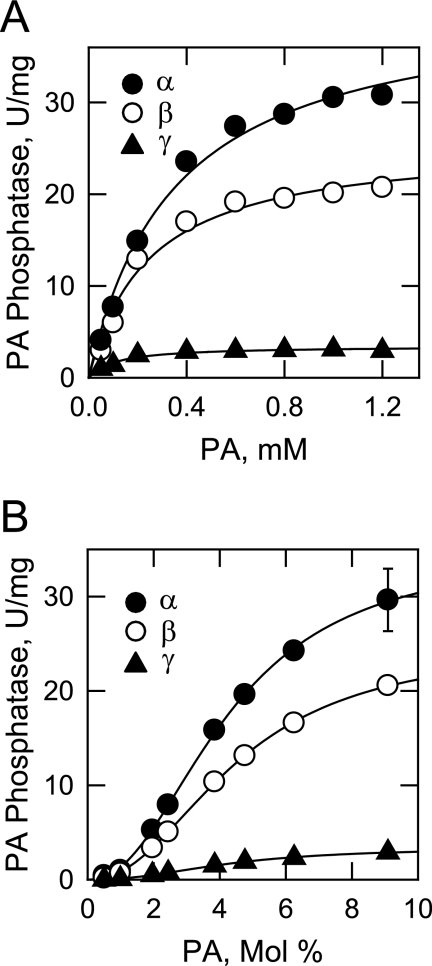
The kinetic analyses of the LPIN1-encoded isoforms were performed using Triton X-100/PA-mixed micelles. This micelle system permitted the analysis of the PA phosphatase activities in an environment that mimics the surface of the cellular membrane (55, 56). In the first set of kinetic experiments, the activities were measured as a function of the molar concentration of PA by maintaining the molar ratio of Triton X-100 to PA in the micelle at 10:1 (i.e. 9.1 mol % PA). This surface concentration of PA was chosen because it afforded the maximum PA phosphatase activities of the isoforms (Fig. 4B). Under these conditions, each of the isoforms followed Michaelis-Menten kinetics (Fig. 8A). Analysis of the data according to the Michaelis-Menten equation showed that the α isoform had the highest turnover number (kcat), followed by the β and γ isoforms (Table 4). The kcat values for the α and β isoforms were 12- and 7.5-fold greater, respectively, when compared with the γ isoform. On the other hand, the γ isoform had a better binding affinity for PA (as reflected in the lowest Km value), followed by the β and α isoforms. The Km value for the γ isoform was 3- and 2-fold lower, respectively, when compared with those of the α and β isoforms, yet the catalytic efficiencies (kcat/Km) of the PA phosphatase isoforms were in the order α > β > γ.
FIGURE 8.
Dependence of the human LPIN1-encoded PA phosphatase isoform activities on the molar and surface concentrations of PA. PA phosphatase activity was measured as a function of the indicated molar concentrations of PA (A) and as a function of the indicated surface concentrations of PA (B). For the experiment shown in A, the molar ratio of Triton X-100 to PA was maintained at 10:1 (9.1 mol % PA). For the experiment shown in B, the molar concentration of PA was held constant at 1 mm, and the Triton X-100 concentration was varied to obtain the indicated surface concentrations. The data shown are means ± S.D. from triplicate enzyme determinations. The best fit curves were derived from the kinetic analysis of the data.
TABLE 4.
Kinetic constants for the LPIN1-encoded PA phosphatase isoform activities
Kinetic constants were calculated from the data in Fig. 8.
| Isoform | Kinetic constant based on PA molar concentration |
Kinetic constant based on PA surface concentration |
|||||
|---|---|---|---|---|---|---|---|
| kcat | Km | kcat/Km | kcat | Km | kcat/Km | Hill number | |
| s−1 | mm | mm−1s−1 | s−1 | mol % | mol %−1s−1 | ||
| α | 68.8 ± 3.5 | 0.35 ± 0.05 | 196.6 | 58.2 ± 1.3 | 4.2 ± 0.11 | 13.8 | 2.2 |
| β | 42.8 ± 2.5 | 0.24 ± 0.05 | 178.3 | 41.5 ± 0.5 | 4.5 ± 0.07 | 9.2 | 2.2 |
| γ | 5.7 ± 0.2 | 0.11 ± 0.02 | 51.8 | 5.7 ± 0.2 | 4.3 ± 0.14 | 1.3 | 2.3 |
In the second set of kinetic experiments, the LPIN1-encoded PA phosphatase activities were measured as a function of the surface concentration (mol %) of PA in the micelle by maintaining the molar concentration of PA at 1 mm (Fig. 8B). Under these conditions, the activities of the isoforms were independent of the PA molar concentration, and they followed positive cooperative kinetics with respect to the surface concentration of PA. The maximum activities of the isoforms were observed at 9.1 mol %. Analysis of the data according to the Hill equation showed that the α, β, and γ isoforms had similar Km and Hill values for PA surface concentration (Table 4). The turnover numbers for the isoforms were in the order α > β > γ, and thus the catalytic efficiencies of the isoforms were also in the same order.
Fatty Acyl Specificities of Human LPIN1-encoded PA Phosphatase Isoforms
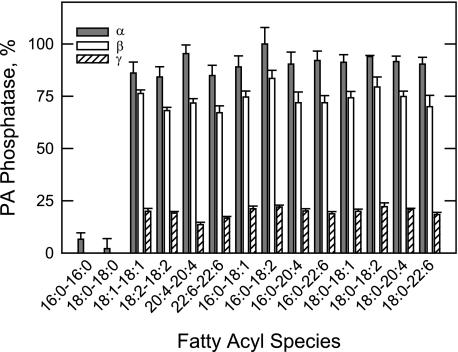
The fatty acyl specificities of the isoforms were examined using a saturating concentration of PA (9.1 mol %). PA substrates containing 1,2-diunsaturated and 1-saturated-2-unsaturated fatty acyl groups were equally good substrates (Fig. 9). Among the substrates with unsaturated fatty acyl groups, the chain length (from 16 to 22) and the degree of unsaturation (from 1 to 6 double bonds) had little effect on the isoform activities. PA molecules with saturated fatty acyl groups only were not good substrates for the three isoforms (Fig. 9).
FIGURE 9.
Fatty acyl specificity of the human LPIN1-encoded PA phosphatase isoform activities. PA phosphatase activity was measured with 9.1 mol % PA with the indicated fatty acyl moieties. The amount of phosphate released from PA in the enzyme reactions was determined by the colorimetric assay using the malachite green-molybdate reagent. Relative activities were presented as described in Fig. 3. The data shown are means ± S.D. from triplicate enzyme determinations.
Effects of Lipids on the LPIN1-encoded PA Phosphatase Isoforms
The effects of phospholipids, neutral lipids, and sphingolipids on the activities of the LPIN1-encoded isoforms were examined (Table 5). To better observe the stimulatory and inhibitory effects of the lipids on the isoforms, the activities were measured with a subsaturating concentration of PA and an equal concentration of the lipid effector. Of the phospholipids and neutral lipids tested, DAG pyrophosphate and dolichol, respectively, had a small inhibitory effect on the three isoforms. Sphingolipids and, in particular, the sphingoid bases sphinganine and sphingosine (by 80–90%) and ceramide-1-phosphate (by 60–80%) were potent inhibitors of the isoform activities.
TABLE 5.
Effect of lipids on the LPIN1-encoded PA phosphatase isoform activities
PA phosphatase activity was measured at a subsaturating concentration of PA (4.5 mol %) in the presence of the indicated lipids (4.5 mol %). The data shown are means ± S.D. from triplicate enzyme determinations.
| Lipid | Relative PA phosphatase activity of isoform |
||
|---|---|---|---|
| α | β | γ | |
| % | % | % | |
| Control | 100 | 100 | 100 |
| Control + phosphatidylcholine | 93 ± 1.4 | 85 ± 0.8 | 90 ± 1.1 |
| Control + phosphatidylethanolamine | 126 ± 1.6 | 115 ± 5.3 | 118 ± 3.9 |
| Control + phosphatidylserine | 118 ± 0.6 | 104 ± 1.0 | 103 ± 2.3 |
| Control + phosphatidylinositol | 117 ± 2.6 | 112 ± 0.3 | 102 ± 2.1 |
| Control + phosphatidylglycerol | 124 ± 3.7 | 121 ± 4.8 | 106 ± 5.3 |
| Control + cardiolipin | 108 ± 2.1 | 92 ± 2.5 | 99 ± 8.2 |
| Control + CDP-DAG | 136 ± 1.1 | 128 ± 3.5 | 97 ± 4.0 |
| Control + DAG pyrophosphate | 88 ± 1.9 | 76 ± 5.0 | 69 ± 5.3 |
| Control + lyso-PA | 109 ± 5.3 | 103 ± 3.6 | 92 ± 6.4 |
| Control + monoacylglycerol | 103 ± 3.6 | 95 ± 3.7 | 94 ± 3.5 |
| Control + DAG | 92 ± 6.8 | 94 ± 6.0 | 103 ± 4.8 |
| Control + triacylglycerol | 99 ± 3.1 | 93 ± 3.1 | 90 ± 2.0 |
| Control + dolichol | 55 ± 2.6 | 44 ± 2.8 | 32 ± 0.3 |
| Control + sphinganine | 8 ± 0.5 | 7 ± 0.7 | 7 ± 1.1 |
| Control + sphingosine | 16 ± 0.4 | 17 ± 0.8 | 17 ± 1.9 |
| Control + sphinganine 1-phosphate | 59 ± 6.9 | 38 ± 1.5 | 30 ± 0.8 |
| Control + sphingosine 1-phosphate | 93 ± 3.4 | 117 ± 2.6 | 87 ± 3.7 |
| Control + ceramide | 79 ± 0.3 | 69 ± 3.5 | 53 ± 1.5 |
| Control + ceramide 1-phosphate | 28 ± 6.5 | 37 ± 1.2 | 16 ± 0.9 |
DISCUSSION
PA phosphatase has emerged as a crucial enzyme in lipid metabolism that has a significant impact on obesity, lipodystrophy, and the metabolic syndrome (29–31). Previous attempts to purify PA phosphatase from mammalian tissues have been unsuccessful because the enzyme is thermally unstable and highly susceptible to proteolytic degradation (5, 57–59). The association of PA phosphatase with other proteins also complicates its purification (59). In addition, prior to the studies of Jamal et al. (60), lipid phosphate phosphatase enzymes that utilize PA as a substrate had been unknown (5, 20, 42), making it difficult to know which phosphatase enzyme was being isolated. Moreover, posttranslational modification may lead to enzyme degradation and/or inactivation and specific interactions with other proteins (15, 19). The identification of LPIN1 as the gene encoding PA phosphatase (12) allowed us to selectively express, purify, and characterize the human lipin 1 α, β, and γ isoforms.
The human lipin 1 isoforms are predicted to have subunit molecular masses of ∼100 kDa, but they were estimated to be ∼115 kDa by SDS-PAGE. Although phosphorylation is known to decrease the electrophoretic mobility of lipin 1 in mammalian cells (41, 42), the anomalous electrophoretic behavior of the purified recombinant lipin 1 isoforms was not due to posttranslational modifications. A similar electrophoretic behavior has also been observed for recombinant yeast PAH1-encoded PA phosphatase (12), and like lipin 1, the electrophoretic mobility of the yeast enzyme decreases further upon its phosphorylation (15).
Early studies have provided useful information (e.g. pH optimum, Mg2+ ion requirement, and sensitivity to modifying chemicals like N-ethylmaleimide) about the basic enzymological properties of PA phosphatase from mammalian tissues (2, 5, 18, 57–60). However, the data derived from this work are difficult to interpret in a definitive way because they have been derived from studies using impure enzymes. The work reported here on the LPIN1-encoded PA phosphatase isoforms did not contain competing and/or modifying enzymes or molecules that inhibit or stimulate activity. Accordingly, the concentration of Mg2+ needed to stimulate the PA phosphatase activities and the concentrations of other effector molecules needed to inhibit the activities were found to be lower than those previously reported. Moreover, for assays with impure enzyme, EDTA is commonly added to inhibit endogenous protease activities, resulting in an increase in the amount of divalent cations needed to affect activity. This work also uncovered the fact that Mn2+ could partially substitute for the Mg2+ dependences of the isoform activities. However, Mg2+ and especially Mn2+ were inhibitory to the PA phosphatase activities at concentrations higher than their optimum levels. In addition, the purified isoforms were potently inhibited by Ca2+ and Zn2+. The inhibition of the activities by Ca2+ has been attributed to its association with the substrate PA to prevent the dephosphorylation reaction (2, 3).
Previous work on the kinetics of mammalian forms of PA phosphatase has been difficult to interpret because of undefined components in the enzyme preparations. Moreover, the PA phosphatase assays have been performed with PA-containing phospholipid vesicles along with the detergent Tween 20 (2, 18, 60). The availability of the purified LPIN1-encoded isoforms permitted defined kinetic studies on human PA phosphatase using Triton X-100/PA-mixed micelles. Although previous studies have indicated that Triton X-100 inhibits PA phosphatase activity (2, 3), the apparent inhibitory effect of the detergent was due to the dilution of PA within the Triton X-100/PA-mixed micelle (55). Indeed, the LPIN1-encoded isoforms followed surface dilution kinetics (55) because their PA phosphatase activities were dependent on both the molar (e.g. number of micelles containing PA) and the surface (e.g. number of PA molecules on a micelle surface) concentrations of PA. The Km values for the molar concentration of PA reflected the interactions of the enzymes with the micelle surface, whereas the Km values for the surface concentration of PA reflected the interactions with PA within the surface (55). These kinetic properties may reflect the in vivo condition, where soluble PA phosphatase must first interact with PA at the endoplasmic reticulum surface, and once bound, the enzyme must scoot along the membrane surface for interaction with other PA molecules for the catalytic step in the reaction. The α isoform displayed the highest turnover number, whereas the γ isoform had the lowest turnover number. The additional 36 amino acids near the NLIP domain of the β isoform reduced the activity by 30%, whereas the additional 26 amino acids near the haloacid dehalogenase-like domain of the γ isoform reduced activity by 90%. The rank orders for the isoform catalytic efficiencies were consistent with their respective energies of activation; the α isoform had the lowest energy of activation, whereas the γ isoform had the highest energy of activation. That the α isoform was more active than the β isoform differs from a recent finding that showed the opposite result (5, 18). The reason for this difference was not clear, but it might be attributable to the crude nature of the enzyme preparations used in that work or due to the effects of posttranslational modifications. Interestingly, the γ isoform displayed the lowest Km value for PA molar concentration, which may suggest better binding to the surface when compared with the other isoforms. However, once bound to the micelle surface, all three isoforms displayed similar affinities for PA within the micelle surface. The cooperative nature (Hill number ∼2) of the activities with respect to the PA surface concentration may reflect cooperative interactions with the PA substrate, suggesting that the enzyme interacts with one PA molecule before it dephosphorylates another PA molecule (55). That the isoform activities exhibited saturation kinetics with respect to the molar concentration of PA argues against the notion that the cooperativity with respect to PA surface concentration was due to enzyme oligomerization.
The early work of Mullmann et al. (61) demonstrated that PA phosphatase activity in human neutrophil homogenates is inhibited by the sphingoid base sphingosine. These workers proposed that sphingosine, which is a breakdown product of sphingomyelin by the sequential actions of sphingomyelinase and ceramidase, regulates the cellular levels and signaling functions of PA by activating phospholipase D and inhibiting PA phosphatase (61). This regulation would also govern the levels of DAG that activates protein kinase C activity (7). However, definitive conclusions about the regulation of PA phosphatase activity by sphingosine could not be made from the early studies because they were conducted with a crude enzyme preparation (61). The work performed in this study clearly showed that the human LPIN1-encoded PA phosphatase activity was potently inhibited by sphingosine as well as by sphinganine (dihydrosphingosine). Sphingosine is known to inhibit other enzymes of mammalian lipid metabolism, including monoacylglycerol acyltransferase (62) and phosphocholine cytidylyltransferase (63) but in a less potent manner when compared with the human lipin 1 isoform activities. That the human PA phosphatase activities were regulated by sphingoid bases provides a firm foundation for examining the interrelationship between PA metabolism and sphingolipid signaling in human cells.
The human LPIN1 gene, like the mouse Lpin1 gene, is expressed in two isoforms (α and β) by alternative mRNA splicing (34, 53). In this work, a third spliced variant (γ) was identified from LPIN1 cDNA derived from human fetal brain mRNA. The kinetic properties of the γ isoform suggested that it might play a distinct enzymological role when compared with the α and β isoforms. It is unclear whether the γ isoform is the major form in the brain and whether it is expressed along with the α and β isoforms. Moreover, the complete tissue distributions of all three isoforms have not been determined, and thus it unclear whether the γ isoform has a specialized role as a PA phosphatase in a particular tissue. It is tempting to speculate that the differential expression of γ isoform is a mechanism by which PA phosphatase activity is attenuated in a particular tissue or under some physiological condition.
The only other PA phosphatase enzyme for which defined enzymological and kinetic information is available is from the model eukaryote S. cerevisiae. The human and yeast enzymes share similar properties. For example, the yeast PAP also exhibits surface dilution kinetics where its activity is dependent on both the molar and surface concentrations of PA (12, 54). In addition, the yeast enzyme is also potently inhibited by sphingoid bases (64). Although both the human and yeast enzymes share similar domain structures involved in catalysis, they differ in other regions that may confer specialized regulatory functions (16). For example, both enzymes are regulated by phosphorylation, but the sites of phosphorylation are different (16). Whereas the work with yeast PA phosphatase has paved the way for studying mammalian PA phosphatase, the studies reported here advance understanding of the human LPIN1-encoded PA phosphatase and provide a foundation for elucidating its biochemical regulation.
Acknowledgment
We thank Florencia Pascual for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-28140 (to G. M. C.).
- PA
- phosphatidate
- DAG
- diacylglycerol.
REFERENCES
- 1.Smith S. W., Weiss S. B., Kennedy E. P. (1957) J. Biol. Chem. 228, 915–922 [PubMed] [Google Scholar]
- 2.Brindley D. N. (1984) Prog. Lipid Res. 23, 115–133 [DOI] [PubMed] [Google Scholar]
- 3.Nanjundan M., Possmayer F. (2003) Am. J. Physiol. Lung Cell Mol. Physiol. 284, L1–L23 [DOI] [PubMed] [Google Scholar]
- 4.Carman G. M. (1997) Biochim. Biophys. Acta 1348, 45–55 [DOI] [PubMed] [Google Scholar]
- 5.Brindley D. N., Pilquil C., Sariahmetoglu M., Reue K. (2009) Biochim. Biophys. Acta 1791, 956–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carman G. M., Han G. S. (2006) Trends Biochem. Sci. 31, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Exton J. H. (1990) J. Biol. Chem. 265, 1–4 [PubMed] [Google Scholar]
- 8.Exton J. H. (1994) Biochim. Biophys. Acta 1212, 26–42 [DOI] [PubMed] [Google Scholar]
- 9.Testerink C., Munnik T. (2005) Trends Plant Sci. 10, 368–375 [DOI] [PubMed] [Google Scholar]
- 10.Waggoner D. W., Xu J., Singh I., Jasinska R., Zhang Q. X., Brindley D. N. (1999) Biochim. Biophys. Acta 1439, 299–316 [DOI] [PubMed] [Google Scholar]
- 11.Sciorra V. A., Morris A. J. (2002) Biochim. Biophys. Acta 1582, 45–51 [DOI] [PubMed] [Google Scholar]
- 12.Han G. S., Wu W. I., Carman G. M. (2006) J. Biol. Chem. 281, 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. (2005) EMBO J. 24, 1931–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han G. S., Siniossoglou S., Carman G. M. (2007) J. Biol. Chem. 282, 37026–37035 [DOI] [PubMed] [Google Scholar]
- 15.O'Hara L., Han G. S., Peak-Chew S., Grimsey N., Carman G. M., Siniossoglou S. (2006) J. Biol. Chem. 281, 34537–34548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carman G. M., Han G. S. (2009) J. Biol. Chem. 284, 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Péterfy M., Phan J., Xu P., Reue K. (2001) Nat. Genet. 27, 121–124 [DOI] [PubMed] [Google Scholar]
- 18.Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. (2007) J. Biol. Chem. 282, 3450–3457 [DOI] [PubMed] [Google Scholar]
- 19.Grimsey N., Han G. S., O'Hara L., Rochford J. J., Carman G. M., Siniossoglou S. (2008) J. Biol. Chem. 283, 29166–29174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brindley D. N., Waggoner D. W. (1998) J. Biol. Chem. 273, 24281–24284 [DOI] [PubMed] [Google Scholar]
- 21.Brindley D. N., English D., Pilquil C., Buri K., Ling Z. C. (2002) Biochim. Biophys. Acta 1582, 33–44 [DOI] [PubMed] [Google Scholar]
- 22.Smyth S. S., Sciorra V. A., Sigal Y. J., Pamuklar Z., Wang Z., Xu Y., Prestwich G. D., Morris A. J. (2003) J. Biol. Chem. 278, 43214–43223 [DOI] [PubMed] [Google Scholar]
- 23.Tanyi J. L., Morris A. J., Wolf J. K., Fang X., Hasegawa Y., Lapushin R., Auersperg N., Sigal Y. J., Newman R. A., Felix E. A., Atkinson E. N., Mills G. B. (2003) Cancer Res. 63, 1073–1082 [PubMed] [Google Scholar]
- 24.Langner C. A., Birkenmeier E. H., Ben-Zeev O., Schotz M. C., Sweet H. O., Davisson M. T., Gordon J. I. (1989) J. Biol. Chem. 264, 7994–8003 [PubMed] [Google Scholar]
- 25.Phan J., Reue K. (2005) Cell Metab. 1, 73–83 [DOI] [PubMed] [Google Scholar]
- 26.Langner C. A., Birkenmeier E. H., Roth K. A., Bronson R. T., Gordon J. I. (1991) J. Biol. Chem. 266, 11955–11964 [PubMed] [Google Scholar]
- 27.Nadra K., de Preux Charles A. S., Médard J. J., Hendriks W. T., Han G. S., Grès S., Carman G. M., Saulnier-Blache J. S., Verheijen M. H., Chrast R. (2008) Genes Dev. 22, 1647–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bou Khalil M., Sundaram M., Zhang H. Y., Links P. H., Raven J. F., Manmontri B., Sariahmetoglu M., Tran K., Reue K., Brindley D. N., Yao Z. (2009) J. Lipid Res. 50, 47–58 [DOI] [PubMed] [Google Scholar]
- 29.Reue K., Donkor J. (2007) Diabetes 56, 2842–2843 [DOI] [PubMed] [Google Scholar]
- 30.Reue K., Brindley D. N. (2008) J. Lipid Res. 49, 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reue K., Dwyer J. R. (2009) J. Lipid Res. 50, (suppl.) S109–S114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donkor J., Zhang P., Wong S., O'Loughlin L., Dewald J., Kok B. P., Brindley D. N., Reue K. (2009) J. Biol. Chem. 284, 29968–29978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., Jr., Kelly D. P. (2006) Cell Metab. 4, 199–210 [DOI] [PubMed] [Google Scholar]
- 34.Péterfy M., Phan J., Reue K. (2005) J. Biol. Chem. 280, 32883–32889 [DOI] [PubMed] [Google Scholar]
- 35.Reue K., Zhang P. (2008) FEBS Lett. 582, 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koh Y. K., Lee M. Y., Kim J. W., Kim M., Moon J. S., Lee Y. J., Ahn Y. H., Kim K. S. (2008) J. Biol. Chem. 283, 34896–34906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manmontri B., Sariahmetoglu M., Donkor J., Bou Khalil M., Sundaram M., Yao Z., Reue K., Lehner R., Brindley D. N. (2008) J. Lipid Res. 49, 1056–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P., O'Loughlin L., Brindley D. N., Reue K. (2008) J. Lipid Res. 49, 1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishimoto K., Nakamura H., Tachibana K., Yamasaki D., Ota A., Hirano K., Tanaka T., Hamakubo T., Sakai J., Kodama T., Doi T. (2009) J. Biol. Chem. 284, 22195–22205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi K., Reue K. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E1195–E1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huffman T. A., Mothe-Satney I., Lawrence J. C., Jr. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., Lawrence J. C., Jr. (2007) J. Biol. Chem. 282, 277–286 [DOI] [PubMed] [Google Scholar]
- 43.Liu G. H., Gerace L. (2009) PLoS One 4, e7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Péterfy M., Harris T. E., Fujita N., Reue K. (2010) J. Biol. Chem. 285, 3857–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 46.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 47.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 48.Haid A., Suissa M. (1983) Methods Enzymol. 96, 192–205 [DOI] [PubMed] [Google Scholar]
- 49.Carman G. M., Lin Y. P. (1991) Methods Enzymol. 197, 548–553 [DOI] [PubMed] [Google Scholar]
- 50.Noguchi K., Kokubu A., Kitanaka C., Ichijo H., Kuchino Y. (2001) Biochem. Biophys. Res. Commun. 281, 1313–1320 [DOI] [PubMed] [Google Scholar]
- 51.Robson R. J., Dennis E. A. (1977) J. Phys. Chem. 81, 1075–1078 [Google Scholar]
- 52.Lichtenberg D., Robson R. J., Dennis E. A. (1983) Biochim. Biophys. Acta 737, 285–304 [DOI] [PubMed] [Google Scholar]
- 53.Croce M. A., Eagon J. C., LaRiviere L. L., Korenblat K. M., Klein S., Finck B. N. (2007) Diabetes 56, 2395–2399 [DOI] [PubMed] [Google Scholar]
- 54.Lin Y. P., Carman G. M. (1989) J. Biol. Chem. 264, 8641–8645 [PubMed] [Google Scholar]
- 55.Carman G. M., Deems R. A., Dennis E. A. (1995) J. Biol. Chem. 270, 18711–18714 [DOI] [PubMed] [Google Scholar]
- 56.Dennis E. A. (1983) in The Enzymes (Boyer P. D. ed) pp. 307–353, Academic Press, Inc., New York [Google Scholar]
- 57.Hosaka K., Yamashita S., Numa S. (1975) J. Biochem. 77, 501–509 [DOI] [PubMed] [Google Scholar]
- 58.Butterwith S. C., Hopewell R., Brindley D. N. (1984) Biochem. J. 220, 825–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siess E. A., Hofstetter M. M. (2005) Biol. Chem. 386, 1197–1201 [DOI] [PubMed] [Google Scholar]
- 60.Jamal Z., Martin A., Gomez-Muñoz A., Brindley D. N. (1991) J. Biol. Chem. 266, 2988–2996 [PubMed] [Google Scholar]
- 61.Mullmann T. J., Siegel M. I., Egan R. W., Billah M. M. (1991) J. Biol. Chem. 266, 2013–2016 [PubMed] [Google Scholar]
- 62.Bhat B. G., Wang P., Coleman R. A. (1995) Biochemistry 34, 11237–11244 [DOI] [PubMed] [Google Scholar]
- 63.Sohal P. S., Cornell R. B. (1990) J. Biol. Chem. 265, 11746–11750 [PubMed] [Google Scholar]
- 64.Wu W. I., Lin Y. P., Wang E., Merrill A. H., Jr., Carman G. M. (1993) J. Biol. Chem. 268, 13830–13837 [PubMed] [Google Scholar]
- 65.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]