Abstract
Moloney leukemia virus 10 (MOV10) protein is a superfamily-1 RNA helicase, and it is also a component of the RNA-induced silencing complex. Recent studies have shown that MOV10 plays an active role in the RNA interference pathway. Here, we report that MOV10 inhibits retrovirus replication. When it was overexpressed in viral producer cells, MOV10 was able to reduce the infectivity of human immunodeficiency virus type 1 (HIV-1), simian immunodeficiency virus, and murine leukemia virus. Conversely, when MOV10 expression was reduced by small interfering RNAs, HIV-1 infectivity was increased. Consistently, silencing of MOV10 expression in a human T cell line enhanced HIV-1 replication. Furthermore, we found that MOV10 interacts with HIV-1 nucleocapsid protein in an RNA-dependent manner and is packaged into virions. It blocks HIV-1 replication at a postentry step. In addition, we also found that HIV-1 could suppress MOV10 protein expression to counteract this cellular resistance. All of these results indicate that MOV10 has a broad antiretroviral activity that can target a wide range of retroviruses, and it could be actively involved in host defense against retroviral infection.
Keywords: HIV, Retrovirus, RNA Helicase, RNA Interference (RNAi), Viral Replication, Restriction Factor, Antiretroviral
Introduction
Helicases share seven conserved motifs (I, Ia, II, III, IV, V, and VI), and are classified into three superfamilies (SF-1 to SF-3) and two smaller families (F4 and F5) (1, 2). Most helicases belong to SF-2 and are known as DExH/D (DEAD, DEAH, DExH) box proteins that are named for a sequence fingerprint in the motif II.
The MOV10 gene was discovered from the MOV-10 mouse strain. The MOV mouse strains were derived from preimplantation embryos infected with Moloney murine leukemia virus (MLV).2 A total of nine strains that carry a single copy of the provirus at different chromosomal positions were created and designated MOV-5 to MOV-13. Unlike most of the other strains, no viremia was developed in the MOV-10 mice, which indicated the provirus was inactive (3). Later, it was found that this provirus is integrated into a gene locus that codes for a 110-kDa GTP-binding protein, which was named MOV10 (4). MOV10 was classified as a SF-1 RNA helicase because of its DEAG fingerprint (5). It is expressed in a variety of cell types, including embryonic stem cells, and high levels of MOV10 transcripts are found at different stages of development in different organs such as testes and thymus (4). The function of MOV10 remained unknown until its orthologs SDE3 in Arabidopsis (6) and Armitage in Drosophila (7) were found to play a critical role in the RNAi pathway.
Originally discovered from transgenic plants, RNAi is a highly conserved gene regulation mechanism that silences genes in a sequence-specific manner, mainly via microRNAs (miRNAs) and small interfering RNAs (8). miRNAs are from endogenous RNAs transcribed in the nucleus and are sequentially processed by the RNase III enzymes, Drosha and Dicer. Small interfering RNAs are from exogenous double strand RNAs that are directly processed by Dicer in the cytoplasm. These small RNAs serve as guide RNAs and direct RISC to sequence-complementary mRNAs (9). RISC, which either cleaves mRNAs or inhibits their translation to cause gene silencing, is a large molecular mass complex containing one of four Argonaute (Ago) proteins and several other proteins (10, 11). Notably, miRNA-mediated silencing normally does not require perfect sequence complementary to the target mRNA, and one miRNA could potentially target 100 discrete mRNAs (12).
SDE3 was isolated as the first non-RNA-dependent RNA polymerase factor in plants that amplifies RNAi by increasing the production of double strand RNA (6), whereas Armitage was found to be required for RISC assembly (7). The role of MOV10 in RNAi was further highlighted in human. It was found that MOV10 not only interacts with Ago2 (13), but MOV10 also is associated with a multiprotein complex containing the HIV-1 TAR RNA-binding protein (TRBP) and eIF6, which are two additional important components of RISC (14). A depletion of MOV10 from cells seriously compromised the activity of the RNAi pathway (13, 14). In addition, MOV10 has been found to co-localize with Ago1 and Ago2 in mRNA processing (P) bodies and stress granules (13). Interestingly, MOV10 also interacts with APOBEC3G (A3G) and APOBEC3F (A3F) in these cellular compartments (15–17). Thus, although the precise role is still unclear, MOV10 is an important player in the RNAi pathway. Here, we demonstrate that MOV10 inhibits retrovirus replication.
EXPERIMENTAL PROCEDURES
Plasmids
Proviral constructs pNL4-3Δvif, pNL-Luc, pNL-LucΔvif, SIVagm-Luc, SIVmac-Luc, pLNPX-Luc, and pCgp, and mammalian expression vectors for VSV-G, A3G, and A3G-GST fusion proteins were described before (18). pNL-LucΔenv and pNL-LucΔenvΔvif were created by NheI digestion of pNL-Luc and pNL-LucΔvif, followed by large Klenow fragment treatment before T4 ligation. To construct Gag-GST fusion expression vectors, Gag, MA-CA, MA, CA, NC-p6, and NC genes were first amplified from 3900 (see below) by PCR. They were then used to replace the A3G gene in pcDNA.3.1-A3G-GST vector by BamHI/NotI digestion.
Codon-optimized HIV-1 gag and gag-pol expression vector 3900 and 4200 were obtained from G. Nabel (NIAID/National Institutes of Health). Δ8.9 was obtained from D. Trono (Ecole Polytechnique Fédérale de Lausanne). pNLΔPr was from E. O. Freed (NCI-Frederick). pMM310 was from M. D. Miller (Merck). pBS/HIV (78-340) was from P. Bieniasz (Aaron Diamond AIDS Research Center). pFLAG/HA-MOV10, pIRESneo-FLAG/HA Ago2, and pcDNA4/TO/cmycDrosha were from T. Tushchl (Rockefeller University) through a public repository (Addgene). A TRBP expression vector was obtained from K. T. Jeang (NIAID). A PACT expression vector was obtained from V. N. Kim (Seoul National University). An RHA expression vector was from L. Chen (McGill University). The cDNA for eIF6 was amplified from 293T cells by reverse transcription-PCR. The I.M.A.G.E. clones for human Ago1, DDX1, DDX3, and murine MOV10 were obtained from Open Biosystems. Ago1, Ago2, MOV10, TRBP, PACT, eIF6, RHA, DDX1, and DDX3 cDNAs were then subcloned into pcDNA3.1-V5-His vector, which contains a V5 tag to be expressed at the C terminus of the target proteins (Invitrogen). The pcDNA3.1–5′Myc-Dicer vector was obtained from P. Provost (CHUL Research Center).
The 5′-region of macMOV10 cDNA encoding amino acids 1–867 was generated by introducing two point mutations (A330T and T512M) into the human MOV10 gene, and the 3′-region of macMOV10 cDNA encoding amino acids 868–1003 was amplified from macaque peripheral blood mononuclear cells (PBMCs). These two fragments were then ligated into one gene via an EcoRI site and inserted into pcDNA3.1-V5-His. The seven human MOV10 helicase motif mutants, Mut-I, Mut-Ia, Mut-II, Mut-III, Mut-VI, Mut-V, and Mut-VI were created by site-directed mutagenesis in the pcDNA3.1-MOV10 vector and verified by DNA sequencing.
Retroviral vector (pSM2) expressing human MOV10-specific miRNA from miR-30 precursor (RHS1764-97189396) and a control miRNA (HMR4867) were purchased from Open Biosystems. They were used to produce recombinant viruses to create miR-MOV10- and miR-Ctrl-expressing CEM-SS cell lines. After replacing the two cloning sites XhoI and EcoRI with MluI and XbaI for cloning the stem-loop coding region in pSM2, the miR-30 precursor expression cassette was cloned into an HIV-1 proviral construct pNL-Luc by NotI/XhoI digestion, generating pNL-miR-30. After that, the MOV10, Dicer, and Ago2-specific sequences, as well as a control sequence, were inserted into pNL-miR-30 by MluI/XbaI digestion. These miRNA expression cassettes were cloned into pcDNA3.1/V5-His-TOPO (Invitrogen) by TOPO-cloning strategy. The sequences for these miRNAs are listed in Fig. S2.
Virus and Cell Culture
HIV-1, SIV, and MLV were produced by transfection of 293T cells as described previously (18). In particular, recombinant MLV-expressing miR-MOV10 and miR-Ctrl were produced by transfection of 293T cells with pSM2, pCgp, and VSV-G-expressing vectors.
Human PBMCs were isolated from buffy coats of healthy blood donors by a protocol approved by the Michigan State University Institutional Review Board. The human embryonic kidney cell line HEK293T was purchased from American Type Culture Collection. The human osteosarcoma cell line HOS-derived HIV GFP-reporter cell line GHOST (3) R3/X4/R5, human cervical cancer cell line HeLa-derived HIV-reporter cell line TZM-bl, and CD4+ T cell lines Jurkat, A3.01, CEM-T4, and CEM-SS were obtained from the NIH AIDS Research and Reference Reagent Program. Rhesus monkey PBMCs were from B. Jia and D. T. Evans (New England Primate Research Center). A detailed protocol was described previously for virions, cores, and purification of viral lysates (19).
Viral Entry Assay
HIV-1 particles carrying a β-lactamase reporter protein and MOV10 were produced by co-transfection of 293T cells with pNL4-3, pMM310, and pcDNA3.1-MOV10. pMM310 is a construct encoding β-lactamase (BlaM) fused to the N terminus of the virion protein Vpr (BlaM-Vpr), thereby targeting BlaM to the virion. TZM-bl cells were first incubated with the virus at 37 °C for 4 h to allow viral entry. 20 μm CCF2-AM (Invitrogen), a fluorescent substrate for BlaM, was added, and the cultures were incubated for 14 h at room temperature. Cells were pelleted and resuspended in phosphate-buffered saline, and the fluorescence was measured by flow cytometry at 447 and 520 nm to detect cell populations containing cleaved (blue) or uncleaved (green) CCF2.
Real-time PCR
To measure viral reverse transcripts, virus stocks from transfected 293T cells were first treated with 50 units/ml RQ1-DNase (Promega) for 2 h to remove any plasmid DNAs. A total of 1 × 107 CEM-SS cells were incubated at 37 °C for 1 h with these viruses, which were normalized to an input of 200 ng of p24CA. Cells were then washed with phosphate-buffered saline and cultured. One-fifth of infected cells were removed from these cultures at 0, 2, 6, and 18 h after infection. The total cellular DNAs were extracted from these cells by the DNeasy kit (Qiagen), and purified DNAs were further treated with DpnI at 37 °C for 2 h to remove any plasmid DNA contamination. Equal amounts of cellular DNAs were then used to measure early and late viral reverse transcripts by real-time quantitative PCR, as described before (20).
To measure MOV10 transcripts in HIV-infected cells, cells were similarly infected with HIV-1 and total RNAs were prepared by TRIzol reagent (Invitrogen). 100 ng of total RNA was subjected to reverse transcription using SuperScriptase II reverse transcriptase and oligo(dT)12–18 as a primer (Invitrogen). Synthesized cDNAs were then subjected to real-time PCR using a TaqMan® Master Mix gene expression kit (Applied Biosystems). The forward primer was MOV10.2670.S (5′-AATTCCAAGGCCAAGAACGA-3′), the reverse primer was MOV10.2748.A (5′-TCCAGATCCAGCTGCACAAA-3′), and the probe was MOV10.probe.1 (5′-VIC®-ATCTCCACCGTGCGAAG-MGBNFQ-3′). Reactions were performed in triplicate. After initial incubation at 95 °C for 10 min, 40 cycles of amplification were carried out for 15 s at 95 °C followed by 1 min at 60 °C. Reactions were analyzed using a 7900HT system (Applied Biosystems). Finally, MOV10 mRNA levels were normalized to the levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, which were determined using a SYBR Green® PCR Master Mix kit (Applied Biosystems) and primers GAPDH.S (5′-CGGAGTCAACGGATT-3′) and GAPDH.A (5′-AGCCTTCTCCATGGTGGTG-3′).
Northern Blotting
Levels of virion-associated genomic RNAs were determined by Northern blotting. Virions from transfected 293T cells were first enriched and purified by ultracentrifugation through a 20% sucrose cushion and then treated with RQ1-DNase. Viruses were normalized by the levels of p24Gag, and equal amounts of virions were used for RNA extraction by TRIzol (Invitrogen). To ensure the recovery of viral RNAs, virions were mixed with a fixed number of DF1 chicken embryo fibroblast cells before RNA extraction. In addition, total cellular RNAs were also extracted from the transfected 293T cells. A total of 4 μg of RNA from each preparation was resolved in 1% denatured agarose gel and transferred to a HybondTM-N nylon membrane (Amersham Biosciences). After UV cross-linking, the membrane was hybridized with a radioactive HIV-specific RNA probe in ULTRAhyb® Ultrasensitive Hybridization buffer (Ambion, Austin, TX). To prepare this probe, the plasmid pBS/HIV (78-340) containing the major 5′-splice donor sequence of HIV was digested by HindIII. It was then used as a template for in vitro transcription using MAXIscript® T7 kit (Ambion) in the presence of [32P]UTP (PerkinElmer Life Science). After TURBO DNase digestion, this 32P-labeled antisense RNA probe was used directly for overnight hybridization. To serve as an internal control for cellular RNAs, a fragment of the human GAPDH gene from nucleotide 17 to 324 was cloned into the pGEM®-T Easy vector (Promega), which contains a T7 promoter. A 32P-labeled antisense RNA probe was prepared in a similar manner to detect GAPDH mRNAs. After washing, the membrane was scanned by Typhoon 9200 (Amersham Biosciences).
Viral RNA RNase Protection Assay and Radiolabeling
A detailed protocol for a viral RNase protection assay was described in Ref. 21. A detailed protocol for 35S-pulse-chase radiolabeling was described in Ref. 22.
Western Blotting
Human MOV10 was detected by a rabbit polyclonal antibody (catalogue no. 10370-1-AP; Proteintech Group). Human Dicer and Ago2 proteins were detected by two mouse monoclonal antibodies (catalogue nos. ab14601 and ab57113; Abcam). HIV-1 p24Gag was detected by monoclonal antibody (183-H12-5C) from the National Institutes of Health AIDS Research and Reference Reagent Program. Actin was detected by a rabbit polyclonal antibody C-11 (Santa Cruz Biotechnology). Horseradish peroxidase-conjugated anti-V5 and anti-c-Myc antibodies were from Invitrogen; horseradish peroxidase-conjugated rabbit or mouse IgG secondary antibodies were from Pierce. Detection of the horseradish peroxidase-conjugated antibody was performed using Supersignal WestPico Chemiluminescence Substrate kit (Pierce).
RESULTS
Inhibition of HIV-1 Replication by Human MOV10 Protein
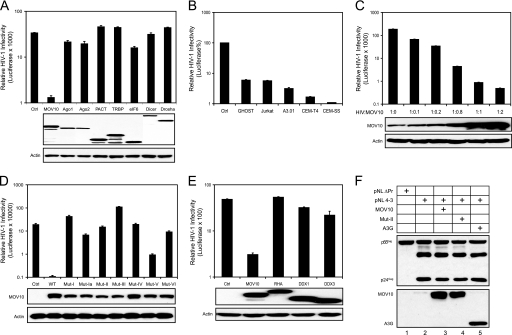
Based on the knowledge that the RNAi pathway participates in antiviral immunity in plants and invertebrate animals, our initial goal was to test whether the RISC has any antiretroviral activity. We ectopically expressed eight well known host proteins involved in this pathway in viral producer cells and measured HIV-1 infectivity by a single round replication assay. Drosha, Dicer, Ago2, and several other RISC-associated proteins, including Ago1, MOV10, TRBP, PACT, and eIF6, were selected for expression (11). HIV-1 luciferase reporter viruses were produced from 293T cells expressing one of these proteins, and viral infectivity was then determined in a GHOST cell line. All of these proteins were expressed well in 293T cells (Fig. 1A, lower panels). However, only MOV10 had anti-HIV activity in target cells, reducing viral infectivity by ∼25-fold (Fig. 1A, upper panel). Interestingly, the effect of MOV10 on reducing viral infectivity was much greater if the target cells were replaced with human T cell lines, as the reduction of viral infectivity observed in A3.01, CEM-T4, and CEM-SS reached 30, 60, or 100-fold, respectively (Fig. 1B). Importantly, the reduction of viral infectivity in CEM-SS cells correlated positively with those of MOV10 expression in 293T cells in a dose-dependent manner (Fig. 1C), indicating that this reduction was MOV10-specific.
FIGURE 1.
MOV10 inhibits HIV-1 replication. A, single round HIV-1 infection assays. Expression vectors of the indicated proteins, as well as an empty vector (control, Ctrl), were co-transfected with an HIV-1 luciferase reporter construct (pNL-Luc) into 293T cells using a 1:1 ratio. Protein expression in 293T cells was determined by Western blotting with an anti-V5 (MOV10, Ago1, Ago2, PACT, TRBP, eIF6) or anti-myc (Dicer and Drosha) antibody. After normalization of viral input by p24Gag enzyme-linked immunosorbent assay, equal amounts of virus were used to infect the GHOST (3) R3/X4/R5 cell line. 24 h later, viral infectivity was determined by measuring cellular luciferase activity. B, cell type dependence of MOV10 anti-HIV-1 activity. Reporter viruses were produced from 293T cells by transfection of pNL-LucΔenvΔvif in the presence of VSV-G and MOV10 expression vectors. The indicated cell lines were then infected using equal amounts of each virus based on p24Gag. Viral infectivity is presented as relative values (%) between viruses with and without MOV10 in each cell line. C, MOV10 dose-dependent anti-HIV activity in CEM-SS cells. MOV10 expression in 293T cells was determined by a polyclonal anti-MOV10 antibody. D, anti-HIV-1 activity of seven MOV10 mutants in CEM-SS cells. Expression of these mutants in 293T cells was determined by Western blotting with an anti-V5 antibody. E, expression of RHA, DDX1, and DDX3 did not reduce HIV-1 infectivity. F, HIV-1 Gag protein expression in the presence of MOV10, its mutant (Mut-II), or APOBEC3G (A3G) in 293T cells.
To confirm its specificity further, we mutated all seven helicase motifs (I, Ia, II, III, IV, V, and VI, as described in Fig. 2A) (1) to determine whether the activity of MOV10 in reducing viral infectivity could be disrupted. The key residues in each motif were changed to alanines, thereby creating seven mutants (designated Mut-I to VI) that were tested for anti-HIV-1 activity. All mutants lost almost all anti-HIV activity, except Mut-V, which retained some activity (Fig. 1D). This result not only confirms the specificity of MOV10, but also indicates that helicase motif V is the least critical for this activity. In addition, we compared MOV10 activity with three other RNA helicases involved in the HIV-1 life cycle (2), including RHA (23), DDX1 (24), and DDX3 (25). Notably, RHA is also required for the RNAi pathway (26). We found that none of these other RNA helicases could reduce viral infectivity as MOV10 did (Fig. 1E). Moreover, we determined whether MOV10 could affect Gag processing, resulting in a loss of viral infectivity. Gag proteins were nicely processed regardless of whether functional MOV10 proteins were expressed during viral production (Fig. 1F).
FIGURE 2.
Antiretroviral activity of MOV10 from different species. A, amino acid sequence alignment of MOV10 from human, rhesus macaque, and mouse. Dots indicate identical sequence, and residues different from this sequence are directly indicated. Seven RNA helicase motifs are underlined and marked. The GenBankTM accession numbers of human, rhesus macaque, and mouse MOV10 are NM_020963, XM_001108588.1, and NM_008619, respectively. B, antiretroviral activity of MOV10 from different species. HIV-1, SIVagm, SIVmac, or MLV luciferase reporter viruses were produced from 293T cells in the presence of human (hu), macaque (mac), or murine (mu) MOV10 expression vectors. Proviral constructs for SIVagm, SIVmac, and MLV did not express a functional viral envelope so these viruses were pseudotyped with VSV-G during viral production. Viruses were normalized by either p24Gag for HIV-1 or reverse transcriptase activity for SIVs and MLV; viral infectivity was determined in GHOST cells.
Finally, we addressed whether MOV10 could inhibit the infectivity of incoming viruses. When MOV10 or rhesus monkey Trim5α proteins were expressed in target cells instead of viral producer cells, rhesus monkey Trim5α reduced HIV-1 infectivity by 3-fold, whereas MOV10 did not have any effect (supplemental Fig. S1). Taken together, we conclude that MOV10 present in virus-producing cells restricts HIV-1 replication by reducing the infectivity of virions released from these cells.
Activity of MOV10 from Other Species
We compared MOV10 amino acid sequences from human, rhesus macaque, and mouse. The MOV10 sequence is well conserved in all these species (Fig. 2A). For example, human shares 99% similarity with macaque and 91% similarity with mouse. In particular, all seven RNA helicase motifs are completely identical. Thus, MOV10 is evolutionarily conserved, implying an important function during development. We then compared the activity of these three MOV10 proteins against four different retroviruses: HIV-1, SIVagm, SIVmac, and MLV. Human, macaque, and murine MOV10 had very similar anti-HIV-1 and SIV activities, reducing viral infectivity by 30–150-fold (Fig. 2B). However, murine MOV10 had much less activity against MLV compared with human or macaque MOV10 (Fig. 2B, lanes 15 and 16). Nevertheless, we conclude that MOV10 from all species has antiretroviral activity.
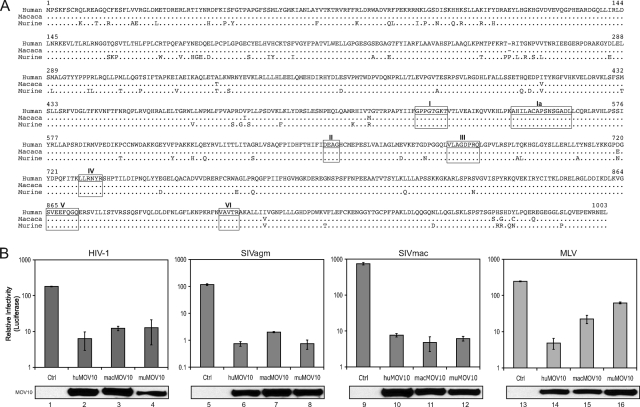
Association of MOV10 with Virions
Because MOV10 reduced the infectivity of HIV-1 released from producer cells, we hypothesized that MOV10 is packaged into HIV-1 virions. To test this hypothesis, viruses were produced from 293T cells co-transfected with the HIV-1 proviral clone pNL4-3 and a MOV10 expression vector and purified by spinning through a 16–65% continuous sucrose gradient via ultracentrifugation. As a control for exosome contamination, 293T cells were transfected separately with only the MOV10 expression vector, and cellular exosomes were purified similarly. Virions were mostly found in fractions 6, 7, and 8, where the viral antigen p24Gag proteins were detected (Fig. 3A, virion, upper panel). Notably, MOV10 was also enriched in these fractions (Fig. 3A, virion, lower panel). Next, we purified viral cores from the same virion samples by including a very brief detergent treatment as described before (19). Unlike virions, cores were enriched in fraction 12 because of a higher density (Fig. 3A, core, top panel). Similarly, the majority of MOV10 was found in fraction 12, indicating an association of MOV10 with cores (Fig. 3A, core, middle panel). However, some MOV10 proteins still remained in fractions 6, 7, and 8, even after detergent treatment. To understand the origin of these MOV10 proteins, the exosome samples were also subjected to the same core-preparation procedure. It was found that MOV10 proteins were also found in fractions 6, 7, and 8 from exosome samples (Fig. 3A, core, bottom panel). This result indicates that the MOV10 proteins found in fractions 6, 7, and 8 are from exosomes that are resistant to detergent treatment. To confirm this conclusion further, we disassembled the cores by a more extensive detergent treatment as described previously (19). Our results show that the cores were completely disrupted as evidenced by a shift of the p24Gag from the high density fraction 12 to the low density fraction 2 (Fig. 3A, viral lysates, top panel). Under these conditions, we found that the location of the majority of MOV10 also shifted to fraction 2 (Fig. 3A, viral lysates, middle panel). However, some MOV10 proteins were still found in fractions 6 and 7, which indicates that the MOV10 found in these fractions is likely from exosomes that remained stable even by this more extensive detergent treatment (Fig. 3A, viral lysates, bottom panel). Taken together, we conclude that MOV10 proteins are encapsidated by HIV-1 during viral assembly.
FIGURE 3.
Interaction of MOV10 with HIV-1 Gag. A, MOV10 in HIV-1 virions. Viruses were prepared from the supernatant of 293T cells co-transfected with MOV10 and HIV-1 expression vectors, and exosomes were prepared from supernatants of 293T cells transfected with only the MOV10 expression vector. After spinning through a 20% sucrose cushion, virus and exosome pellets were collected. For virion purification, pellets were loaded on the top of a 16–65% sucrose gradient. For core preparation, a layer of 1% Triton X-100 was laid on the top of the gradient. Pellets were also pretreated with 1% Triton X-100 at 37 °C for 3.5 h before loading on the gradient for viral lysate analysis. After spinning for 17 h at 100,000 × g, 12 fractions were collected from each preparation. Proteins in each fraction were analyzed by Western blotting with an anti-p24Gag antibody or anti-V5 antibody for MOV10. B, MOV10 incorporation into VLPs. 293T cells were transfected with MOV10 expression vector plus pNL4-3 (lanes 1 and 5), 3900 (lanes 2 and 6), 4200 (lanes 3 and 7), or Δ8.9 (lanes 4 and 8). Particles were collected by ultracentrifugation. MOV10 and Gag protein expression in cells and viral particles was determine by Western blotting. C, schematic representation of the full-length and truncated Gag-GST fusion proteins. MA, matrix protein; CA, capsid protein. D, MOV10 interaction with HIV-1 NC.
To determine how MOV10 proteins are encapsidated, we first tested whether MOV10 was encapsidated by virus-like particles (VLPs). Three different viral constructs (3900, 4200, and Δ8.9) were used to produce VLPs, and pNL4-3 was used to produce wild-type viral particles. Constructs 3900 and 4200 express codon-optimized gag and gag-pol, and Δ8.9 expresses the original gag-pol in the presence of Rev. When these constructs were co-transfected with MOV10 into 293T cells, the expression of Gag, as well as MOV10 proteins, was clearly detected (Fig. 3B, lane 1, pNL4-3; lane 2, 3900; lane 3, 4200; lane 4, Δ8.9). Supernatants from these cultures were then subjected to ultracentrifugation to isolate particles. Constructs of pNL4-3 (lane 5), 4200 (lane 7), and Δ8.9 (lane 8) produced mature viral particles or VLPs because the majority of p55Gag proteins were processed into p24Gag, whereas construct 3900 (lane 6) only produced immature VLPs because this construct does not express viral protease. Notably, MOV10 proteins were detected in all of these particles (Fig. 3B, right lower panel). This result demonstrates that gag alone is sufficient for MOV10 encapsidation, suggesting an interaction between MOV10 and Gag.
To determine how MOV10 interacts with Gag, a GST fusion protein co-precipitation assay was used. A series of Gag and Gag deletion mutants that were C-terminally fused to a GST tag were created: Gag-GST, MACA-GST, MA-GST, NCp6-GST, and NC-GST (Fig. 3C). These fusion proteins were co-expressed with MOV10 in 293T cells, and their expression was confirmed by Western blotting (Fig. 3D, left panels). Thereafter, cell lysates were incubated with glutathione-Sepharose beads that were subsequently washed, and bound proteins were analyzed by Western blotting with an anti-V5 antibody. Gag or its fragments containing the NC domain (NCp6-GST and NC-GST) bound to MOV10, whereas other Gag fragments did not. Notably, when samples were pretreated with RNase A, these interactions were no longer detectable. Taken together, we conclude that MOV10 proteins are packaged into virions in an RNA-dependent manner via an interaction with the NC domain of Gag.
Mechanism of MOV10 Antiviral Activity
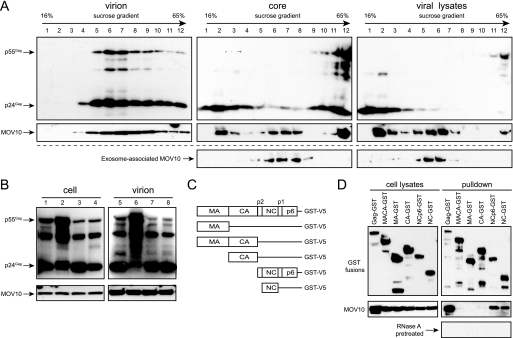
First, we determined whether MOV10 blocks viral entry by a previously described assay (27). HIV-1 particles were produced from 293T cells by co-transfection of HIV-1 and MOV10 expression vectors plus a construct (pMM310) encoding Escherichia coli BlaM fused to the N terminus of HIV-1 Vpr. BlaM-Vpr fusions are encapsidated via Vpr-Gag interaction and can cleave the cell-permeable fluorescence BlaM substrate CCF2 in target cells, resulting in a shift of emission spectrum from green to blue, which can be detected by flow cytometry. The intensity of the blue fluorescence signal correlates positively with the level of viral entry. Indeed, regardless of whether MOV10 was present, cells infected by virions containing BlaM-Vpr produced an almost equal intensity of blue fluorescence signal (Fig. 4A). This result indicates that MOV10 does not block viral entry.
FIGURE 4.
Mechanism of HIV-1 inhibition by MOV10. A, levels of viral entry. Representative FACS dot plots are shown for the detection of CCF2 substrate cleavage in TZM-bl cells that were either mock-infected or infected with virions carrying BlaM-Vpr fusion proteins in the presence or absence of exogenous MOV10. B, top, levels of viral genomic RNAs (9 kb) in viral producer cells determined by Northern blotting. B, bottom, levels of viral subgenomic RNAs determined by RNA splicing assay. C, levels of accumulation of viral reverse transcripts. vif-deficient virions carrying MOV10, Mut-II, or A3G were first treated with DNase and then used to infect CEM-SS cells. 2, 6, and 18 h later, cellular DNAs were extracted from these infected cells, and viral early and late reverse transcripts were quantified by real-time PCR. Values represent the mean from three replicates, and error bars represent the S.D. Results shown are from one of three independent experiments.
Second, we determined whether MOV10 affects viral RNA expression from producer cells. When full-length viral genomic RNAs were measured by Northern blotting, it was found that similar levels of unspliced genomic RNAs (∼9 kb) were detected regardless of MOV10 expression (Fig. 4B, top panel). We next determined whether MOV10 could affect viral RNA splicing, and it was found that all viral RNAs were spliced with a very similar efficiency (Fig. 4B, bottom panel). These results indicate that MOV10 does not affect viral RNA expression or splicing in viral producer cells.
Third, we determined how MOV10-containing viruses replicate immediately after entry by measuring viral reverse transcripts. Because A3G could reduce viral reverse transcripts of vif-defective HIV-1, we chose to use vif-defective HIV-1 for this determination so A3G could be included as a positive control. We confirmed that MOV10 inhibited both wild-type and vif-defective HIV-1 replication equally well (supplemental Fig. S3). The Mut-II of MOV10, which lost anti-HIV activity (Fig. 1D), was included as a negative control. Viruses were produced from 293T cells in the presence of MOV10, Mut-II, or A3G proteins and used to infect CEM-SS cells. 2, 6, and 18 h later, cellular DNAs were extracted, and viral early or late reverse transcripts were quantified by real-time PCR. As expected, it was found that A3G significantly reduced both early and late reverse transcription products (Fig. 4C). Similarly, the wild-type MOV10 also effectively reduced early and particularly the late reverse transcripts. In sharp contrast, Mut-II did not show any effect on reverse transcripts. These results indicate that MOV10 inhibits viral replication at a postentry step.
Activity of Endogenous MOV10 Proteins
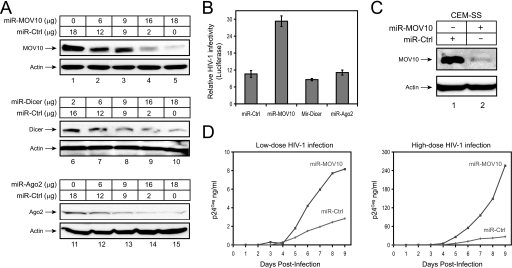
We have shown that viral replication was inhibited by ectopically expressed MOV10 in viral producer cells (Figs. 1 and 2). Next, we attempted to silence the MOV10 gene and determine whether HIV-1 replication could be enhanced. A pcDNA3.1 vector containing a miR-30-derived miRNA expression cassette was constructed to express Dicer, Ago2, and MOV10-specific miRNAs (miR-Dicer, miR-Ago2, miR-MOV10), as well as a control miRNA (miR-Ctrl) (supplemental Fig. S2). Previously, this expression cassette was shown to provide 12 times more efficiency than simple hairpin designs in producing the mature synthetic miRNAs (28). When these vectors were expressed in 293T cells, they all silenced their target genes in a sequence-specific manner (Fig. 5A). Next, we determined how HIV-1 infectivity is affected when these genes are silenced in viral producer cells. It was found that although silencing of Dicer and Ago2 had no effect, silencing of MOV10 increased HIV-1 infectivity by 3-fold in a single round of replication cycle (Fig. 5B).
FIGURE 5.
Endogenous MOV10 anti-HIV-1 activity. A, gene silencing by miRNAs expressed from pcDNA3.1. 293T cells transfected with 18 μg of decreasing amounts of vector-expressing miRNAs specific for MOV10, Dicer, or Ago2 plus increasing amounts of a control miRNA. The expression of endogenous MOV10, Dicer, and Ago2 was analyzed by Western blotting with actin as an internal control. B, effect of MOV10 silencing on HIV-1 infectivity. 20 μg of miR-Ctrl, miR-MOV10, miR-Dicer, or miR-Ago2 expression vectors were co-transfected with 4 μg of HIV-1 reporter construct (pNL-Luc) plus 1 μg of VSV-G expression vector into 293T cells, and viral infectivity was determined as in Fig. 1A. C, silencing of MOV10 gene in human T cells. CEM-SS cells were transduced with an MLV vector (pSM2) expressing miR-Ctrl or miR-MOV10, and stably transduced cells were selected by puromycin treatment. The expression of endogenous MOV10 proteins was determined by Western blotting. D, HIV-1 replication in T cells with reduced MOV10 expression. The above two cell lines were infected with 5 ng (low dose) or 50 ng (high dose) p24Gag of HIV-1 produced from 293T cells, and viral growth curves were monitored for 9 days by p24Gag enzyme-linked immunosorbent assay.
Moreover, we attempted to silence the MOV10 gene from a human T cell line. The miR-Ctrl and miR-MOV10 expression cassettes were expressed from the MLV-derived vector pSM2, and recombinant viruses were created to infect CEM-SS cells. After that, stable cell lines were created by puromycin selection. Indeed, the MOV10 gene was effectively silenced in this cell line (Fig. 5C, compare lanes 1 and 2). Importantly, the replication of HIV-1 was greatly enhanced when MOV10 expression was suppressed (Fig. 5D). Thus, we conclude that HIV-1 replication is enhanced when MOV10 expression is decreased, indicating that the endogenous MOV10 protein has antiretroviral activity.
Virus Counteraction
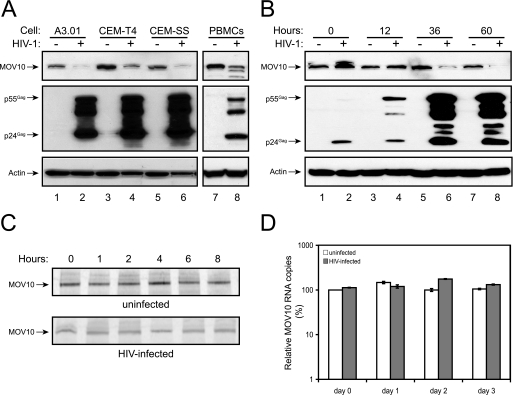
Because HIV-1 can establish a productive infection in human tissues, we hypothesized that the virus might be able to counteract the inhibitory effects of MOV10. To test this hypothesis, we determined MOV10 protein expression levels during viral replication. Three human T cell lines (A3.01, CEM-T4, and CEM-SS) were infected with HIV-1, and 3 days later, expression of MOV10 was determined by Western blotting. Interestingly, the levels of MOV10 expression were significantly reduced upon the establishment of viral infection in all these cell lines (Fig. 6A, lanes 2, 4, and 6). A similar observation was also made in human PBMCs (lane 8). We further determined the time course of MOV10 reduction in CEM-SS cells. Accompanying the detection of increasing levels of Gag expression, we saw a time-dependent decrease of MOV10 expression after 36 h (Fig. 6B).
FIGURE 6.
Effect of virus replication on MOV10 expression. A, suppression of MOV10 expression during HIV-1 infection. Human T cell lines A3.01, CEM-T4, and CEM-SS, as well as phytohemagglutinin-activated human PBMCs, were infected with HIV-1. 3 days later, expression of MOV10 and viral p24Gag proteins were determined by Western blotting. B, time course of MOV10 reduction during HIV-1 infection in CEM-SS cells. C, MOV10 protein stability in uninfected and HIV-infected CEM-SS cells. CEM-SS cells were infected with HIV-1. 60 h later, infected or uninfected cells were pulse-labeled with [35S]methionine/cysteine and incubated for the indicated time points. After immunoprecipitation of lysates with anti-MOV10 antibody, proteins were separated by SDS-PAGE, and the gels were subjected to autoradiography. D, levels of MOV10 transcripts in HIV-infected or uninfected CEM-SS cells, as determined by real-time PCR.
To understand how HIV-1 decreases MOV10 protein expression, we first measured MOV10 protein stability by pulse-chase assay. HIV-infected or uninfected CEM-SS cells were pulse-labeled with [35S]methionine/cysteine and chased for 1–8 h. MOV10 proteins were immunoprecipitated, separated by SDS-PAGE, and analyzed by autoradiography. Although the MOV10 protein levels were lower in infected compared with uninfected cells, all proteins were similarly stable (Fig. 6C). Thus, HIV-1 infection does not change MOV10 protein stability. Next, we compared MOV10 mRNA levels in HIV-infected and uninfected CEM-SS cells by real-time PCR. However, there was no significant decrease of MOV10 mRNA levels in infected cells (Fig. 6D). Thus, we could not completely understand the mechanism of how the virus suppresses MOV10 protein expression.
DISCUSSION
In this report, we have outlined several lines of evidence to show that MOV10 has antiretroviral activity: (i) overexpression of MOV10 reduced viral infectivity, and this activity required functional helicase motifs; (ii) reduction of MOV10 expression increased viral infectivity and enhanced viral replication; (iii) MOV10 was specifically encapsidated by the virus; (iv) MOV10 expression was suppressed when the virus established a productive infection. The fact that MOV10 from different species inhibited the replication of HIV-1, SIVs, and MLV indicates that this protein targets a wide range of retroviruses.
Moreover, we have shown that MOV10 blocks virus replication at a postentry step. It specifically interacts with the NC domain of Gag in an RNA-dependent manner and is packaged into virions during assembly. MOV10 does not change viral RNA expression in viral producer cells. This is in sharp contrast to ZAP, which inhibits MLV replication by degrading viral mRNAs in the cytoplasm of viral producer cells. Interestingly, the action of ZAP is dependent on a DEAD box RNA helicase p72 (29). It is thought that p72 helps recruit the RNA-processing exosome for degradation. In addition, another helicase, RHA, that increases viral production, was also found to bind to NC, and it was speculated that RHA may increase viral RNA packaging by restructuring these RNAs (23). We have found that MOV10 does not affect viral RNA packaging or viral RNA dimerization, nor does it change the thermostability of the virion RNA dimers (supplemental Fig. S4). Thus, the specific mechanism by which MOV10 inhibits viral replication after entry into a target cell is still unclear. Also, it should be noted that the inactivation of MLV provirus in the MOV-10 mice should not have a direct link to MOV10 antiretroviral activity because later it was found that the provirus in these mice carries a mutation that prevents synthesis of infectious viruses (30).
Although MOV10 inhibits HIV-1 replication, the virus is able to counteract the effects of this host protein by suppressing its protein expression. The anti-HIV activity of MOV10 is reminiscent of that from A3G/A3F (31–33) and Tetherin (34). A3G and A3F are two cellular cytidine deaminases that restrict viral replication at the step of reverse transcription. Tetherin is an integral membrane protein that restricts viral replication at the step of viral release. However, HIV-1 produces two accessory proteins, Vif and Vpu, to counteract the effects of these antiviral proteins. Both Vif and Vpu can function as adaptor proteins that interact with a Cullin-5-based E3 ubiquitin ligase complex (35), or a β-transducin repeat-containing protein (βTrCP)-based E3 ubiquitin ligase complex (36), respectively. Subsequently, Vif and Vpu target the A3G/A3F and BST-2 proteins to the proteasomal pathway for degradation. In the case of MOV10, HIV-1 was able to suppress its protein expression, and thereby its antiviral activity, via an unknown mechanism that does not seem to involve the proteasomal pathway. Thus, the antiviral activity of MOV10 and the suppression of MOV10 by HIV-1 are another arm wrestling match between host and virus, and over the course of evolution the virus again appear to have established a winning balance, which allows it to productively infect its host. Without using ΔVif and ΔVpu mutant viruses, the discovery of A3G/A3F and Tetherin activities would not have been possible. Similarly, without overexpressing MOV10 in viral producer cells to disrupt the established balance between HIV-1 and MOV10, it would have been impossible to detect the antiretroviral activity of MOV10. Thus, therapeutic approaches that increase MOV10 expression or inhibit HIV-1 suppression of MOV10 may become useful strategies to amplify this innate intracellular antiretroviral immunity and fight HIV infection.
Supplementary Material
Acknowledgments
We thank N. Landau, P. Cannon, G. Nabel, D. Trono, E. O. Freed, M. D. Miller, P. Bieniasz, T. Tuschl, K. T. Jeang, V. N. Kim, L. Chen, P. Provost, D. T. Evans, and the National Institutes of Health AIDS Research and Reference Reagent program for reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants AI063944 and AI080225 (to Y.-H. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- MLV
- murine leukemia virus
- MOV10
- Moloney leukemia virus 10 protein
- RNAi
- RNA interference
- miRNA
- microRNA
- RISC
- RNA-induced silencing complex
- Ago
- Argonaute
- HIV-1
- human immunodeficiency virus type 1
- TRBP
- TAR RNA-binding protein
- A3G
- APOBEC3G
- A3F
- APOBEC3F
- Luc
- luciferase
- VSV
- vesicular stomatitis virus
- GST
- glutathione S-transferase
- mac
- rhesus macaque
- PBMC
- peripheral blood mononuclear cell
- SIV
- simian immunodeficiency virus
- BlaM
- β-lactamase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- VLP
- virus-like particle
- NC
- nucleocapsid.
REFERENCES
- 1.Caruthers J. M., McKay D. B. (2002) Curr. Opin. Struct. Biol. 12, 123–133 [DOI] [PubMed] [Google Scholar]
- 2.Jeang K. T., Yedavalli V. (2006) Nucleic Acids Res. 34, 4198–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaenisch R., Jähner D., Nobis P., Simon I., Löhler J., Harbers K., Grotkopp D. (1981) Cell 24, 519–529 [DOI] [PubMed] [Google Scholar]
- 4.Mooslehner K., Müller U., Karls U., Hamann L., Harbers K. (1991) Mol. Cell. Biol. 11, 886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koonin E. V. (1992) Trends Biochem. Sci. 17, 495–497 [DOI] [PubMed] [Google Scholar]
- 6.Dalmay T., Horsefield R., Braunstein T. H., Baulcombe D. C. (2001) EMBO J. 20, 2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook H. A., Koppetsch B. S., Wu J., Theurkauf W. E. (2004) Cell 116, 817–829 [DOI] [PubMed] [Google Scholar]
- 8.Lindbo J. A., Dougherty W. G. (2005) Annu. Rev. Phytopathol. 43, 191–204 [DOI] [PubMed] [Google Scholar]
- 9.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 10.Meister G. (2007) Cell 131, 25–28 [DOI] [PubMed] [Google Scholar]
- 11.Peters L., Meister G. (2007) Mol. Cell 26, 611–623 [DOI] [PubMed] [Google Scholar]
- 12.Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 13.Meister G., Landthaler M., Peters L., Chen P. Y., Urlaub H., Lührmann R., Tuschl T. (2005) Curr. Biol. 15, 2149–2155 [DOI] [PubMed] [Google Scholar]
- 14.Chendrimada T. P., Finn K. J., Ji X., Baillat D., Gregory R. I., Liebhaber S. A., Pasquinelli A. E., Shiekhattar R. (2007) Nature 447, 823–828 [DOI] [PubMed] [Google Scholar]
- 15.Gallois-Montbrun S., Holmes R. K., Swanson C. M., Fernández-Ocaña M., Byers H. L., Ward M. A., Malim M. H. (2008) J. Virol. 82, 5636–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallois-Montbrun S., Kramer B., Swanson C. M., Byers H., Lynham S., Ward M., Malim M. H. (2007) J. Virol. 81, 2165–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak S. L., Marin M., Rose K. M., Bystrom C., Kabat D. (2006) J. Biol. Chem. 281, 29105–29119 [DOI] [PubMed] [Google Scholar]
- 18.Dang Y., Wang X., Esselman W. J., Zheng Y. H. (2006) J. Virol. 80, 10522–10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Dolan P. T., Dang Y., Zheng Y. H. (2007) J. Biol. Chem. 282, 1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y., Wang X., Dang Y., Zheng Y. H. (2008) PLoS ONE 3, e2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y. H., Yu H. F., Peterlin B. M. (2003) Nat. Cell Biol. 5, 611–618 [DOI] [PubMed] [Google Scholar]
- 22.Dang Y., Siew L. M., Zheng Y. H. (2008) J. Biol. Chem. 283, 13124–13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy B. B., Hu J., Guo X., Russell R. S., Guo F., Kleiman L., Liang C. (2006) J. Biol. Chem. 281, 12625–12635 [DOI] [PubMed] [Google Scholar]
- 24.Fang J., Kubota S., Yang B., Zhou N., Zhang H., Godbout R., Pomerantz R. J. (2004) Virology 330, 471–480 [DOI] [PubMed] [Google Scholar]
- 25.Yedavalli V. S., Neuveut C., Chi Y. H., Kleiman L., Jeang K. T. (2004) Cell 119, 381–392 [DOI] [PubMed] [Google Scholar]
- 26.Robb G. B., Rana T. M. (2007) Mol. Cell 26, 523–537 [DOI] [PubMed] [Google Scholar]
- 27.Cavrois M., De Noronha C., Greene W. C. (2002) Nat. Biotechnol. 20, 1151–1154 [DOI] [PubMed] [Google Scholar]
- 28.Zeng Y., Wagner E. J., Cullen B. R. (2002) Mol. Cell 9, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 29.Chen G., Guo X., Lv F., Xu Y., Gao G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4352–4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harbers K., Schnieke A., Stuhlmann H., Jaenisch R. (1982) Nucleic Acids Res. 10, 2521–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Nature 418, 646–650 [DOI] [PubMed] [Google Scholar]
- 32.Wiegand H. L., Doehle B. P., Bogerd H. P., Cullen B. R. (2004) EMBO J. 23, 2451–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng Y. H., Irwin D., Kurosu T., Tokunaga K., Sata T., Peterlin B. M. (2004) J. Virol. 78, 6073–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neil S. J., Zang T., Bieniasz P. D. (2008) Nature 451, 425–430 [DOI] [PubMed] [Google Scholar]
- 35.Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. (2003) Science 302, 1056–1060 [DOI] [PubMed] [Google Scholar]
- 36.Margottin F., Bour S. P., Durand H., Selig L., Benichou S., Richard V., Thomas D., Strebel K., Benarous R. (1998) Mol. Cell 1, 565–574 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.