Abstract
The zebrafish provides a useful experimental system for investigations of aural development. To permit the controlled expression of transgenes in developing hair cells, we isolated the genomic control regions of the parvalbumin 3a (pvalb3a) and parvalbumin 3b (pvalb3b) genes. Deletion analysis and somatic-cell transgenesis restricted the cis-acting control regions for hair cells to as little as 484 base pairs for pvalb3a and 650 base pairs for pvalb3b. Using both meganuclease-mediated and standard methods, we produced transgenic animals that transmit transgenes through their germ lines. These fish express GFP in hair cells in the inner ear and lateral line. Two stable transgenic lines express GFP prior to hair-bundle formation, so the associated promoter constructs are suitable for manipulating gene expression during bundle development. We additionally identified a transgenic line that offers variable labeling of supporting cells.
Keywords: balance, hearing, lateral line, mantle cell, parvalbumin, supporting cell
Introduction
Hair cells are sensory receptors that represent mechanical stimuli as electrical responses that are transmitted to the brain (Hudspeth, 1989). These cells underlie mechanotransduction for the senses of hearing and equilibrium in the ear and for the detection of water movement in the lateral-line system. Extending from the apical surface of each hair cell is an actin-based hair bundle that is essential for mechanosensitivity. The bundle consists of an erect cluster of stereocilia, rigid rods containing parallel actin bundles and crosslinking proteins, that are graded in height. Embedded in a pseudostratified sensory epithelium, hair cells are surrounded and underlain by supporting cells (Jørgensen and Flock, 1973; Flock and Jørgensen, 1974; Jones and Corwin, 1993). In neuromasts of the lateral line, mantle cells form a circumferential ring around the hair and supporting cells. In fish, birds, and amphibians, supporting and mantle cells are required for hair-cell regeneration and thus include progenitors (Corwin et al., 1991; Rubel et al., 1991; Cotanche and Lee, 1994; Tsue et al., 1994; Corwin and Oberholtzer, 1997).
The larval zebrafish is well-suited for studies of the development and operation of hair cells (Nicolson, 2005), which occur in the five sensory epithelia of the otic vesicle and stippled across the animal’s surface in the lateral-line neuromasts. The sensory organs of the larval ear include two maculae that are associated with calcium carbonate-based otoliths. These organs are used for sensing sound and linear accelerations including gravity. The embryonic ear also contains three cristae for the detection of angular accelerations. Among the zebrafish’s favorable attributes is rapid aural development: the first hair cells arise only one day post fertilization (1 dpf; Haddon and Lewis, 1996). Zebrafish embryos are optically transparent and develop externally, features that facilitate the microscopic analysis of aural development. Genetic screens in the zebrafish have been instrumental in identifying mutations that affect hair-cell development and function (Nicolson et al., 1998; Kappler et al., 2004) and that are conserved between humans and fish (Whitfield, 2002).
Investigations of the ear’s development require the establishment of genetic tools for visualizing and manipulating hair cells. In particular, it is desirable to have available promoters for the specific expression of fluorescent markers and transgenes in hair cells. Although two useful promoters have already been isolated, neither is ideal. The brn3c promoter drives expression in receptor cells of the eye as well as the ear (Xiao et al., 2005). The myosin 6b promoter regulates expression in hair cells but its expression in other tissues has not been characterized (Obholzer et al., 2008). The temporal expression patterns for these promoters are unknown. Moreover, their control regions are at least 6.0 kb in length. It would be useful to have smaller promoters for more efficient subcloning.
Parvalbumin 3, or in mammals oncomodulin (Hsiao et al., 2002), is an abundant, proteinaceous Ca2+ buffer that is expressed with high specificity in hair cells (Heller, 2002; Lopez-Schier and Hudspeth, 2005). In hair cells of the bullfrog’s sacculus its concentration is as great as 3 mM. In the chicken, the protein occurs in the cochlea and cerebellum. In the zebrafish there are two, paralogous parvalbumin 3 genes (Hsiao et al., 2002), parvalbumin 3a (pvalb3a) and parvalbumin 3b (pvalb3b).
An understanding of the functions of proteins in hair-cell development and transduction requires genetic manipulation. Because pvalb3a and pvalb3b are strongly and specifically expressed in hair cells, their promoters are promising candidates for the expression of transgenes. We have therefore isolated the regions of genomic DNA associated with the two genes that contain cis-acting elements required for transcription in hair cells.
Materials and Methods
Fish husbandry
Zebrafish of the Tübingen (Tü) strain were used in these experiments. They were maintained and bred at 28°C by standard procedures (Nüsslein-Volhard and Dahm, 2002).
DNA manipulations
DNA manipulations were carried out by standard procedures (Sambrook and Russell, 2001). Restriction endonucleases and other enzymes for DNA manipulations were purchased from New England Biolabs (Beverly, MA).
BAC modification
The modification was conducted according to a published protocol (Gong et al., 2002). A zebrafish genomic library of bacterial artificial chromosome (BAC) clones, which included clones PV_2:189j02, PV_4:16d05, PV_5:39l03, and PV_6:71m16 representing portions of the pvalb3a gene, were ordered from Incyte Genomics (Palo Alto, CA; no longer available). A recombination shuttle vector was created to contain a 500-bp fragment of the pvalb3a gene amplified by the PCR from a region just upstream of the start codon. The primer sequences used for constructing the shuttle vector contained the pvalb3a gene sequence (bold) as well as restriction enzyme recognition sites (italics) and a four-base-pair overhang:
| PV3A BACMF | GAGAGGATCCGGCGCGCCCGGGGAGAACGTGCATACTC |
| PV3A BACMR | CACACCATGGTGATCCTGGACAAGAGGAAGATTCAC |
After the amplified PCR fragment had been cloned with BamHI and NcoI into a modified pBS-based building vector (pBV-EGFP1), the fragments of the pvalb3a and GFP genes were transferred into the pLD53 vector (S96 version). In the shuttle vector with the PCR fragment insertion, the start codon of the GFP gene replaced the normal pvalb3a start codon. A transcription stop site was situated after the GFP coding sequence to avoid transcription of the downstream exons of the pvalb3a gene. The shuttle vector was electroporated into bacterial cells containing the BAC to be modified. For subcloning of the minimal pvalb3a promoter region, we digested the modified BAC clone with the GFP gene insertion with various restriction enzymes and performed southern blotting with isotope-labeled GFP DNA as a probe to determine which restriction fragments included the GFP gene and the upstream region.
The pvalb3b gene was localized to BAC 126 of the CHORI-211 library by use of the zebrafish Ensembl Genome Browser (http://www.ensembl.org/Danio_rerio/index.html). After purification with the ProCipitate purification reagent (Ligochem Inc.), the BAC DNA was used as a template in PCR reactions designed to amplify genomic regions upstream of and including the gene’s start codon. Restriction sites for XhoI and XmaI were added to respectively the 5′ and 3′ ends of the PCR products. The fragments were subcloned into pCR-TOPO vector (Invitrogen Inc.) and subsequently moved into pEGFP-1 (Clontech Inc.) for expression analyses using the XhoI and XmaI restrictions sites.
The following primers were employed:
| Parv3b(5′)2 | ctcgagtgcttttctccttttgtttccaca |
| Parv3b(5′)3 | ctcgagtgcttttctccttttgtttccaca |
| Parv3b(5′)4 | ctcgaggattcagacaacagaggtcaagca |
| Parv3b(5′)5 | ctcgagcacgtcggagatttttgatttctt |
| Parv3b(3′)2 | cccgggggaagtaagtgacatattcaaact |
| Ppv3b-5 | aactcgagcatggagtaaatataatgagctcca |
| Ppv3b-6 | aactcgagattacgtagtggatacctgaacca |
| Ppv3b-7 | aactcgaggaagtgcaggcacatgtgcgc |
| Ppv3b-8 | aactcgagctcattgtccgtcccattagcat |
| Parv3b(3′)1B | aacccgggttcaatggcttcagc |
Generation of transgenic animals
In accord with standard protocols (Nüsslein-Volhard and Dahm, 2002), plasmid DNA was digested with restriction enzymes to liberate the cloned genomic fragment fused to a cDNA encoding GFP. These fragments were purified and injected into zebrafish embryos at the one- or two-cell stage. Animals at 4–5 dpf were anesthetized with 650 μM 3-aminobenzoic acid ethyl ester methanesulfonate and viewed in a fluorescence microscope to screen for GFP expression in hair cells of the acousticolateralis system. Larvae that displayed GFP expression in hair cells were allowed to recover from anesthesia and raised to sexual maturity. Their progeny were then screened for germline transmission.
To increase the efficiency of stable transgenesis, we also employed a second method of producing transgenic animals. After situating 18-bp I-SceI meganuclease restriction sites on both sides of the 7.5-kb Ppv3a-1 promoter, we injected the resultant construct into single-cell embryos in 0.5× I-SceI buffer solution lacking BSA but containing 10% I-SceI enzyme (New England Biolabs) and 0.5% phenol red.
Fluorescent labeling and imaging
Transgenic larvae were fixed overnight at 4°C in 4% paraformaldehyde in phosphate-buffered saline solution (PBS), rinsed in PBS, permeabilized overnight at room temperature in 3% Triton X-100 in PBS, blocked with 5% goat serum diluted with PBS for 2 hr, labeled overnight with primary antisera towards parvalbumin 3 (Heller, 2002) diluted 1:500 in 5% goat serum, washed in 5% goat serum with PBS for 6 hr at room temperature, incubated overnight with anti-rabbit secondary antibody linked to Alexa 543 (Invitrogen) diluted 1:200 in 5% goat serum diluted with PBS and Alexa 633 phalloidin (2.5 units/ml; Invitrogen), and washed with 5% goat serum in PBS. For labeling of hair bundles in transgenic animals, Alexa 543 phalloidin (2.5 units/ml; Invitrogen) was used. Imaging was performed using a DM IRE2 (Leica Micro-systems) or a MRC1024ES (Bio-Rad) laser-scanning confocal inverted microscope with a 20× or 40× objective lens.
Neuromasts were labeled with the styryl fluorophore FM 4–64 as described previously (Nagiel et al, 2008).
Cryostat sectioning
The heads of adult zebrafish about three months old were fixed with 4% paraformaldehyde in PBS for 4 hr at 4°C. After two washes with PBS containing 0.2% Tween20, the heads were equilibrated to 30% sucrose overnight at 4°C. OTC (Tissue Tek Inc.) was used for freezing the heads in dry ice and methanol. Sections were cut at a thickness of 20 μm and placed on coated glass slides. For immunolabeling, sections were permeabilized for 5 min in 0.1% Triton X-100 in PBS with 1% BSA and 1% DMSO, then blocked for 20 min with 2% normal serum in the same solution lacking detergent. After exposure to a monoclonal antibody against chicken neurofilament 200 at a dilution of 1:20, sections were stored at 4°C overnight. After a wash, the secondary antibody at a dilution of 1:100 was applied for 1 hr at room temperature. Finally, sections were washed in 1× PBS containing 0.2% Tween20, placed on coverslips, and examined immediately.
Results
Localizing control regions of parvalbumin promoters
To isolate the promoter responsible for the expression of parvalbumin 3a in hair cells, we first identified BAC clones containing the pvalb3a gene. The coding region was then replaced with a cDNA encoding GFP by homologous recombination in bacteria (Gong et al., 2002). We confirmed by somatic-cell transgenesis that this modified BAC could drive GFP expression in hair cells of the inner ear and lateral line.
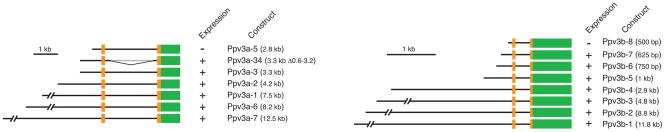
To identify a minimal but sufficient region for expression of GFP in hair cells, we subcloned from the modified BAC fragments that extended 12.5 kb, 8.2 kb, 7.5 kb, 4.2 kb, 3.3 kb, and 2.8 kb upstream of the start codon (Figure 1, left). By microinjection of constructs into single-cell embryos and imaging at least 200 embryos for each construct at 5 dpf, we determined that the constructs Ppv3a-1, Ppv3a-2, Ppv3a-3, Ppv3a-6, and Ppv3a-7 caused consistent GFP expression in hair cells of the inner ear and lateral line, but also in peridermal cells that ordinarily do not express pvalb3a. Comparison of Ppv3a-3 with Ppv3a-34, an internally deleted derivative of Ppv3a-3, and of Ppv3a-5 with Ppv3a-34 suggested that intron 1 is not necessary for transcriptional activation and that the 484-bp region that is present in Ppv3a-34 but absent from Ppv3a-5 is sufficient for transcriptional activation. This region contains a consensus TATA-box sequence, TATAAA, which commences 27 bp upstream of the 5′ end of exon 1 and may be important for driving transcription in hair cells.
Figure 1.
Schematic diagrams of the deletion constructs for the promoter regions of the pvalb3a (left) and pvalb3b (right) genes in the zebrafish. Solid black lines represent genomic fragments. The orange and green boxes indicate respectively the first two exons of the parvalbumin 3 genes and the cDNA encoding green-fluorescent protein. The constructs that showed specific GFP expression in hair cells are marked “+” and those that did not are designated “−.”
Because the expression pattern of pvalb3b as determined by in situ hybridization is more restricted in larval zebrafish than that of pvalb3a (Hsiao et al., 2002), we also isolated the pvalb3b promoter by PCR amplification. According to the zebrafish Ensembl genomic database, the eight regions chosen were predicted to extend 11.8 kb, 8.8 kb, 4.8 kb, 2.9 kb, 1.0 kb, 0.75 kb, 0.625 kb, and 0.5 kb upstream of the endogenous start codon (Figure 1, right). The products were then fused in-frame with the cDNA encoding GFP to allow reporter-gene expression.
In order to determine whether the cis-acting control regions lay in the subcloned fragments of DNA, we used somatic-cell expression in zebrafish. This involved the injection of constructs into single-cell embryos and fluorescence imaging at 3–4 dpf. We observed expression in hair cells using constructs Ppv3b-1, Ppv3b-2, Ppv3b-3, Ppv3b-4, Ppv3b-5, Ppv3b-6, and Ppv3b-7. However, no labeling of hair cells could be seen with Ppv3b-8. Most animals that expressed GFP in hair cells also displayed ectopic expression in peridermal cells. In larvae that expressed GFP in hair cells, the coexpression rate in at least one skin cell was 85% for Ppv3b-1 (N = 20), 90% for Ppv3b-2 (N = 30), 88% for Ppv3b-3 (N = 50), 92% for Ppv3b-4 (N = 50), 86% for Ppv3b-5 (N = 50), 86% for Ppv3b-6 (N = 50), and 84% for Ppv3b-7 (N = 50). Expression was occasionally seen in other tissues.
Expression in stable transgenic animals
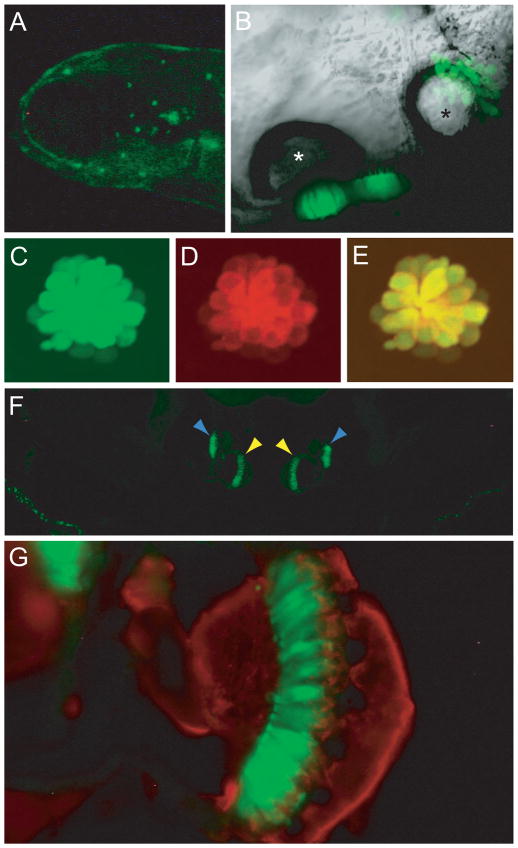
Transgenic larvae that stably express a fluorescent protein in cells of interest are useful in defining the spatial and developmental distribution of these cells and in mutagenesis screening. To obtain stable lines expressing GFP in hair cells, we used the Ppv3a-3 construct that includes a 3.3-kb fragment upstream of the pvalb3a start codon. Injected animals that expressed GFP were raised and their offspring screened; two of the 70 injected fish transmitted the transgene through their germ lines. We analyzed the pattern of GFP expression in these transgenic zebrafish. There was no difference in the expression pattern between these lines. By 5 dpf, GFP-positive hair cells occurred in both maculae of the inner ear as well as in neuromasts (Figure 2A, B). All the lateral-line hair cells that were labeled with FM4-64, a styryl fluorophore that enters hair cells through mechanotransduction channels (Gale et al., 2001; Meyers et al., 2003), also expressed GFP (Figure 2C–E). In addition to hair cells, the olfactory epithelium, anterior pituitary, and mucous cells expressed GFP, a pattern consistent with the results of in situ hybridization with a pvalb3a probe (Hsiao et al., 2002).
Figure 2.
Cellular distribution of GFP in the Ppv3a-3 transgenic line. (A) A fluorescent image of a 5-dpf larva head demonstrates labeling of neuromasts in the anterior lateral line. (B) In a ventral view of an otocyst in a 5-dpf larva, sensory epithelia containing GFP-positive hair cells are located beneath the two otoliths. The anterior otolith is marked with a white asterisk, the posterior otolith with a black one. (C) Transgenically expressed GFP marks hair cells in a 5-dpf larval neuromast. (D) Labeling with FM4-64, a fluorescent dye that passes through functional mechanotransduction channels, reveals the complement of mature hair cells. (E) The merged image of GFP and FM4-64 signals confirms that the transgenic label marks all the functional hair cells. (F) A transverse section of the head of an adult fish portrays the GFP-positive sensory maculae of the medial sacculi (yellow arrowheads) and the lateral lagenae (blue arrowheads). (G) A higher-magnification image of one sacculus shows GFP-positive hair cells (green) and nerve fibers labeled with an antiserum against neurofilaments (red).
To examine the utility of the stable line in investigations of abnormal aural development, we injected into Ppv3a-3 embryos a morpholino against the atrophin2 gene (Asai et al., 2006). Imaging of a live morphant showed that the two sensory maculae as well as the otoliths were fused (data not shown), a phenotype characteristic of animals lacking Atrophin2.
We used cryosectioning to examine GFP expression in the ears of adult transgenic zebrafish. The three cristae of the semicircular canals, the utricle, the saccule, and the lagena all contained GFP-positive hair cells (Figure 2F and G). Peridermal cells expressed GFP ectopically but at much lower levels than hair cells (Figure 2A and F).
Coinjection of meganuclease I-SceI improves the efficiency of DNA integration into the fish genome (Grabher et al., 2004; Thermes et al., 2002). From two lines that were derived by this method, we identified a single founder. The GFP expression pattern was consistent with that of the Ppv3a-3 line, but the expression was stronger in both hair cells and skin cells (data not shown).
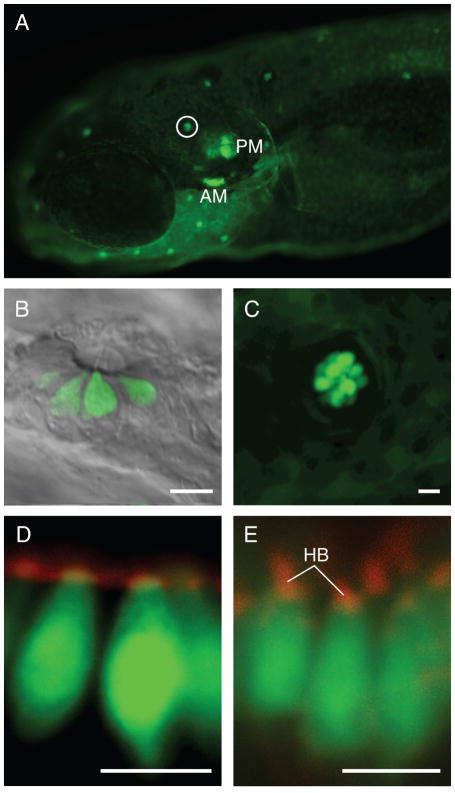
We additionally generated two stable lines, Ppv3b-3 and Ppv3b-4, with sequences extending respectively 4.8 kb and 2.9 kb upstream of the start codon for pvalb3b. To generate these stable lines, we screened the offspring of 87 and 105 somatic transgenics generated from Ppv3b-3 and Ppv3b-4 constructs, respectively. Robust expression of the reporter gene could be observed in the sensory patches of the otic vesicle and neuromasts of Ppv3b-4 transgenic larvae (Figure 3A). Peridermal cells again expressed GFP at a much lower level. We consistently observed expression in hair cells of the anterior maculae, posterior maculae, and neuromasts (Figure 3). This expression persisted in adults (data not shown). The hair-cell expression pattern was the same for the stable line generated using the Ppv3b-3 construct (Supplemental Figure S1) except that there was expression in the chevron-shaped somites (data not shown). This result indicates that the pattern of expression in hair cells is a feature of the cis-acting elements contained within these constructs and not the result of the positions at which the transgenes integrated into the genome. Neither of these lines had apparent GFP expression in pronephric ducts, as had been observed by in situ hybridization studies (Hsiao et al., 2002).
Figure 3.
Labeling pattern in strain generated with the Ppv3b-4 construct. (A) In the Ppv3b-4 line, GFP expression occurs in the anterior macula (AM), posterior macula (PM) and lateral-line neuromasts, of which a representative example is circled. Ectopic peridermal expression is also apparent. (B) At 5 dpf, fluorescence is observed in hair cells of infraorbital neuromast 3 (Raible and Kruse, 2000). (C) In a 5-dpf transgenic larva, the L1 neuromast of the posterior lateral line (Ghysen and Dambly-Chaudiere, 2004) shows much stronger GFP expression in hair cells than in the periderm. (D) The anterior macula of a transgenic fish fixed at 2 dpf and labeled with phalloidin (red) shows GFP expression in the presumptive hair cells prior to hair-bundle formation. The weak phalloidin signal may represent microvilli on the apical surfaces of hair cells and supporting cells. (E) Phalloidin labeling (red) of the anterior macula in a 5-dpf larva reveals the presence of hair bundles (HB) on transgene-labeled hair cells. The scale bars represent 5 μm.
A key feature of a promoter that may be used to label or perturb proteins involved in hair-bundle development is expression prior to bundle assembly. To determine whether Ppv3b-4 larvae display such early expression, we labeled the hair bundles of transgenic animals with fluorophore-coupled phalloidin. At 2 dpf, when the anterior macula had few mature hair cells, we observed vase-shaped presumptive hair cells expressing GFP but lacking stereocilia (Figure 3D). At 5 dpf, instead, most of the hair cells displayed mature hair bundles (Figure 3E). The expression of GFP in presumptive hair cells of the anterior macula was the same for the stable line generated using the Ppv3b-3 construct (Supplemental Figure S1). In addition, we inquired whether GFP expression preceded bundle formation in presumptive hair cells in organs other than the anterior maculae of the Ppv3b-3 stable line. In the crista, GFP expression anticipated bundle formation in hair cells (Supplemental Figure S2). The same held true for expression in the crista of Ppv3b-4 stable transgenic larvae (data not shown). The Ppv3b-3 and Ppv3b-4 promoters should therefore be useful in the expression of genetically encoded fluorescent proteins during bundle genesis. These promoters may also be used to express dominant-negative proteins to determine the role of particular proteins in hair-bundle development.
Expression in supporting cells
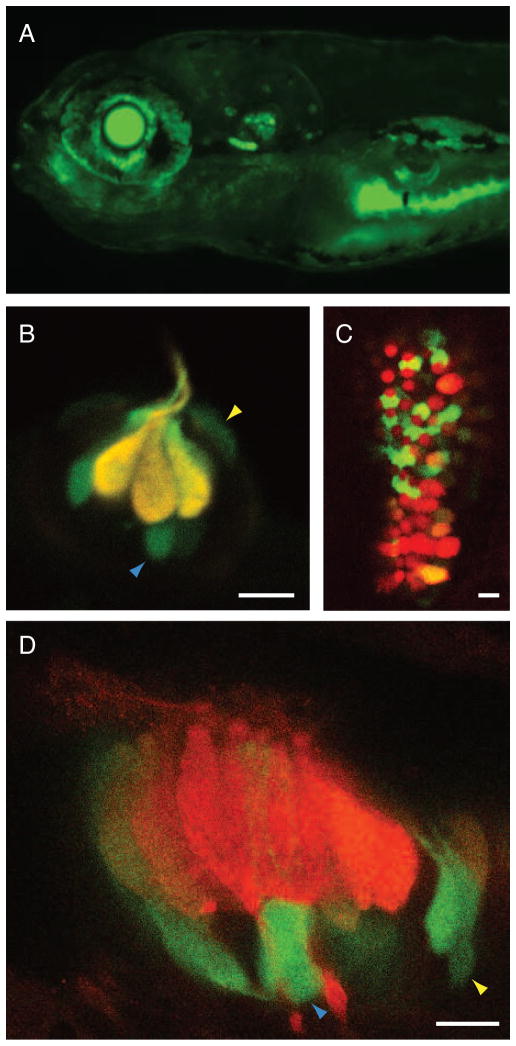
In the course of examining transgenic larvae, we encountered one line, Ppv3b-4-12825, in which GFP was expressed in the eye and gut as well as in maculae, cristae, neuromasts, and periderm (Figure 4A). Close inspection of the hair cell-containing organs disclosed robust but variegated labeling of supporting and mantle cells as well as hair cells (Figure 4). In neuromasts, the fluorescence intensity in hair cells exceeded that in the other cells (Figure 4B). By contrast, the GFP fluorescence in the anterior and posterior maculae appeared primarily in supporting cells (Figure 4C). The labeling pattern remained similar in larval crista at 30 dpf (Figure 4D).
Figure 4.
Transgene expression in larvae of strain Ppv3b-4-12825. (A) The transgenic line Ppv3b-4-12825 displays a pattern of expression similar to that of the Ppv3b-4 line in the ear and lateral line; however, labeling also extends to the eye and gut. (B) In supraorbital neuromast 3 of a 7-dpf embryo, the overlap of GFP expression (green) and immunolabeling for parvalbumin 3 (red) causes hair cells to appear yellow. Some supporting cells (blue arrowheads) and mantle cells (yellow arrowheads) also express GFP. (C) Similar labeling of the posterior macula at 7 dpf reveals GFP expression (green) primarily in the supporting cells separating hair cells labeled for parvalbumin 3 (red). (D) In the crista of a horizontal semicircular canal at 30 dpf, labeling like that above demonstrates robust expression of GFP (green) in supporting cells and of parvalbumin 3 in hair cells (red). Blue and yellow arrowheads designate supporting and mantle cells, respectively. The scale bars represent 5 μm.
Discussion
Cell-specific promoters provide a means of producing transgenic animals that express reporter genes in cells of interest and of analyzing gene function through the expression of dominant-negative proteins. Using both somatic and stable transgenesis, we have shown that genomic fragments upstream of the coding regions of the zebrafish pvalb3a and pvalb3b genes direct expression in hair cells. These promoters offer two advantages over others now available for this purpose. First, the pvalb3b promoter is active prior to hair-bundle formation, making it useful for manipulating proteins involved in bundle development. And second, because the reduced control regions are small, they are suitable for subcloning.
Using these constructs, we have generated stable transgenic lines that display specific expression of a reporter construct in all hair cells of a larva. Use of the Ppv3a-3 transgenic line permits observation without staining of the normal or abnormal development of the sensory maculae in the inner ear. This is a substantial advantage in understanding the mechanisms of a gene’s function in development.
We have additionally identified a transgenic line, Ppv3b-4-12825, in which GFP is expressed in the supporting cells of the ear and lateral line. Because not all supporting cells are labeled, there may be a random element in the expression. An alternative explanation is that two or more classes of supporting cells exist, each expressing a distinct repertoire of genes. This transgenic line may prove useful in determining which supporting cells of the internal ear and neuromasts are the hair-cell progenitors responsible for regeneration. Like the lines that express GFP in hair cells, this line can serve as a background strain for genetic screens and for the separation of specific cell types by fluorescence-activated cell sorting in preparation for mRNA isolation.
Supplementary Material
GFP expression pattern in a strain generated with the Ppv3b-3 construct. (A) Phalloidin labeling (red) of the anterior macula in a 5-dpf larva shows the presence of a hair bundle on a transgene-labeled hair cell. (B) In a transgenic fish fixed at 2 dpf and labeled with phalloidin (red), GFP expression reveals a presumptive hair cell of the anterior macula prior to hair-bundle formation. The scale bars represent 2 μm.
The pattern of GFP expression in a line generated with the Ppv3b-3 construct. (A) Phalloidin labeling (red) of the posterior crista in a 5-dpf larva demonstrates a hair bundle on a transgene-labeled hair cell. (B) In the posterior crista of a transgenic fish fixed at 5 dpf and labeled with phalloidin (red), GFP expression reveals a presumptive hair cell prior to hair-bundle formation. The scale bars represent 2 μm.
Acknowledgments
The authors thank Dr. Shiaoching Gong for the S96 version of the pBV-EGFP1 construct, Dr. William Yang for the pBV-EGFP1 construct, and the members of their research groups for comments on the manuscript. We also thank Mr. A. Afolalu, Mrs. C. Fernando, and Ms. M. West for fish husbandry and Mrs. C. Fernando for stereoscopic imaging. This work was supported by National Institutes of Health grants DC00241 (A.J.H.) and DC006539 (B.M.M), the University Hospitals Case Medical Center Center for Clinical Research and Technology (B.M.M.), the Case Research Institute Vision Fund (B.M.M.), the National Organization for Hearing Research (B.M.M.), The Rockefeller University’s Science Outreach Program (J.M.M.), and a Basil O’Connor Starter Scholar Research Award from the March of Dimes (B.M.M.). A.J.H. is an Investigator of Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asai Y, Chan DK, Starr CJ, Kappler JA, Kollmar R, Hudspeth AJ. Mutation of the atrophin2 gene in the zebrafish disrupts signaling by fibroblast growth factor during development of the inner ear. Proc Natl Acad Sci U S A. 2006;103:9069–74. doi: 10.1073/pnas.0603453103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin JT, Jones JE, Katayama A, Kelley MW, Warchol ME. Hair cell regeneration: the identities of progenitor cells, potential triggers and instructive cues. Ciba Found Symp. 1991;160:103–20. doi: 10.1002/9780470514122.ch6. discussion 120–30. [DOI] [PubMed] [Google Scholar]
- Corwin JT, Oberholtzer JC. Fish n’ chicks: model recipes for hair-cell regeneration? Neuron. 1997;19:951–4. doi: 10.1016/s0896-6273(00)80386-4. [DOI] [PubMed] [Google Scholar]
- Cotanche DA, Lee KH. Regeneration of hair cells in the vestibulocochlear system of birds and mammals. Curr Opin Neurobiol. 1994;4:509–14. doi: 10.1016/0959-4388(94)90051-5. [DOI] [PubMed] [Google Scholar]
- Flock A, Jorgensen JM. The ultrastructure of lateral line sense organs in the juvenile salamander Ambystoma mexicanum. Cell Tissue Res. 1974;152:283–92. doi: 10.1007/BF00223950. [DOI] [PubMed] [Google Scholar]
- Gale JE, Marcotti W, Kennedy HJ, Kros CJ, Richardson GP. FM1–43 dye behaves as a permeant blocker of the hair-cell mechanotransducer channel. J Neurosci. 2001;21:7013–25. doi: 10.1523/JNEUROSCI.21-18-07013.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudiere C. Development of the zebrafish lateral line. Curr Opin Neurobiol. 2004;14:67–73. doi: 10.1016/j.conb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 2002;12:1992–8. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabher C, Joly JS, Wittbrodt J. Highly efficient zebrafish transgenesis mediated by the meganuclease I-SceI. Methods Cell Biol. 2004;77:381–401. doi: 10.1016/s0091-679x(04)77021-1. [DOI] [PubMed] [Google Scholar]
- Haddon C, Lewis J. Early ear development in the embryo of the zebrafish, Danio rerio. J Comp Neurol. 1996;365:113–28. doi: 10.1002/(SICI)1096-9861(19960129)365:1<113::AID-CNE9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Heller S. Molecular screens for inner ear genes. J Neurobiol. 2002;53:265–75. doi: 10.1002/neu.10122. [DOI] [PubMed] [Google Scholar]
- Hsiao CD, Tsai WY, Tsai HJ. Isolation and expression of two zebrafish homologues of parvalbumin genes related to chicken CPV3 and mammalian oncomodulin. Mech Dev. 2002;119(Suppl 1):S161–6. doi: 10.1016/s0925-4773(03)00110-2. [DOI] [PubMed] [Google Scholar]
- Hudspeth AJ. How the ear’s works work. Nature. 1989;341:397–404. doi: 10.1038/341397a0. [DOI] [PubMed] [Google Scholar]
- Jones JE, Corwin JT. Replacement of lateral line sensory organs during tail regeneration in salamanders: identification of progenitor cells and analysis of leukocyte activity. J Neurosci. 1993;13:1022–34. doi: 10.1523/JNEUROSCI.13-03-01022.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen JM, Flock A. The ultrastructure of lateral line sense organs in the adult salamander Ambystoma mexicanum. J Neurocytol. 1973;2:133–42. doi: 10.1007/BF01474715. [DOI] [PubMed] [Google Scholar]
- Kappler JA, Starr CJ, Chan DK, Kollmar R, Hudspeth AJ. A nonsense mutation in the gene encoding a zebrafish myosin VI isoform causes defects in hair-cell mechanotransduction. Proc Natl Acad Sci U S A. 2004;101:13056–61. doi: 10.1073/pnas.0405224101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Schier H, Hudspeth AJ. Supernumerary neuromasts in the posterior lateral line of zebrafish lacking peripheral glia. Proc Natl Acad Sci U S A. 2005;102:1496–501. doi: 10.1073/pnas.0409361102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers JR, MacDonald RB, Duggan A, Lenzi D, Standaert DG, Corwin JT, Corey DP. Lighting up the senses: FM1–43 loading of sensory cells through nonselective ion channels. J Neurosci. 2003;23:4054–65. doi: 10.1523/JNEUROSCI.23-10-04054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson T, Rusch A, Friedrich RW, Granato M, Ruppersberg JP, Nusslein-Volhard C. Genetic analysis of vertebrate sensory hair cell mechanosensation: the zebrafish circler mutants. Neuron. 1998;20:271–83. doi: 10.1016/s0896-6273(00)80455-9. [DOI] [PubMed] [Google Scholar]
- Nicolson T. The genetics of hearing and balance in zebrafish. Annu Rev Genet. 2005;39:9–22. doi: 10.1146/annurev.genet.39.073003.105049. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Dahm R. Zebrafish : a practical approach. Oxford University Press; Oxford: 2002. [Google Scholar]
- Obholzer N, Wolfson S, Trapani JG, Mo W, Nechiporuk A, Busch-Nentwich E, Seiler C, Sidi S, Sollner C, Duncan RN, Boehland A, Nicolson T. Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J Neurosci. 2008;28:2110–8. doi: 10.1523/JNEUROSCI.5230-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible DW, Kruse GJ. Organization of the lateral line system in embryonic zebrafish. J Comp Neurol. 2000;421:189–98. [PubMed] [Google Scholar]
- Rubel EW, Oesterle EC, Weisleder P. Hair cell regeneration in the avian inner ear. Ciba Found Symp. 1991;160:77–96. doi: 10.1002/9780470514122.ch5. discussion 96–102. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning : a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2001. [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118:91–8. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- Tsue TT, Oesterle EC, Rubel EW. Hair cell regeneration in the inner ear. Otolaryngol Head Neck Surg. 1994;111:281–301. doi: 10.1177/01945998941113P118. [DOI] [PubMed] [Google Scholar]
- Whitfield TT. Zebrafish as a model for hearing and deafness. J Neurobiol. 2002;53:157–71. doi: 10.1002/neu.10123. [DOI] [PubMed] [Google Scholar]
- Xiao T, Roeser T, Staub W, Baier H. A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development. 2005;132:2955–67. doi: 10.1242/dev.01861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GFP expression pattern in a strain generated with the Ppv3b-3 construct. (A) Phalloidin labeling (red) of the anterior macula in a 5-dpf larva shows the presence of a hair bundle on a transgene-labeled hair cell. (B) In a transgenic fish fixed at 2 dpf and labeled with phalloidin (red), GFP expression reveals a presumptive hair cell of the anterior macula prior to hair-bundle formation. The scale bars represent 2 μm.
The pattern of GFP expression in a line generated with the Ppv3b-3 construct. (A) Phalloidin labeling (red) of the posterior crista in a 5-dpf larva demonstrates a hair bundle on a transgene-labeled hair cell. (B) In the posterior crista of a transgenic fish fixed at 5 dpf and labeled with phalloidin (red), GFP expression reveals a presumptive hair cell prior to hair-bundle formation. The scale bars represent 2 μm.