Abstract
The par locus of the Enterococcus faecalis plasmid pAD1 is an RNA-regulated addiction module encoding the peptide toxin Fst. Homology searches revealed that Fst belongs to a family of at least nine related peptides encoded on the chromosomes and plasmids of six different Gram-positive bacterial species. Comparison of an alignment of these peptides with the results of a saturation mutagenesis analysis indicated regions of the peptides important for biological function. Examination of the genetic context of the fst genes revealed that all of these peptides are encoded within par-like loci with conserved features similar to pAD1 par. All four Ent. faecalis family members were demonstrated to produce the expected toxin-encoding and regulatory RNA products. The locus from the Ent. faecalis plasmid pAMS1 was demonstrated to function as an addiction module and Fst was shown to be toxic to Staphylococcus aureus, suggesting that a plasmid-encoded module in that species is performing the same function. Thus, the pAD1-encoded par locus appears to be the prototype of a family of related loci found in several Gram-positive species.
INTRODUCTION
Toxin–antitoxin (TA) systems were first identified on bacterial plasmids, where they function as stability determinants by programming for death any daughter cells that fail to inherit a copy (for recent reviews see Gerdes & Wagner, 2007; Hayes, 2003). These systems, originally called post-segregation killing systems or addiction modules, encode a stable toxin and an unstable antitoxin. As long as the plasmid is properly inherited, antitoxin is continually replenished and the toxin remains inactive. If the plasmid is lost, the antitoxin is degraded and the toxin is free to exert its effect. Toxins have a variety of targets including gyrase (Bahassi et al., 1999; Jiang et al., 2002), ribosome-bound and free mRNA (Condon, 2006; Zhang et al., 2004), and the cell membrane (Gerdes et al., 1986). In most TA systems, both components are proteins, with the antitoxin targeted by a specific cellular protease (Gerdes et al., 2005). In two well-studied cases, the hok/sok system of Escherichia coli plasmid R1 (Gerdes et al., 1990) and the par system of Enterococcus faecalis plasmid pAD1 (Greenfield & Weaver, 2000; Greenfield et al., 2001, 2000), the antitoxin is a small regulatory RNA that represses the translation of the toxin. The RNA-regulated TA systems are sometimes referred to as type I TA loci while the protein-regulated systems are designated type II.
Paradoxically, numerous TA systems have been identified on bacterial chromosomes and have been the subject of numerous recent reviews (Buts et al., 2005; Engelberg-Kulka et al., 2006; Fozo et al., 2008; Gerdes & Wagner, 2007; Gerdes et al., 2005; Van Melderen & Saavedra De Bast, 2009). Various roles have been either demonstrated or proposed for these systems, including stabilization of integrated mobile genetic elements, protection from plasmid-encoded addiction modules, programmed cell death, growth modulation during stress response, persistence, and developmental processes. It seems likely that these ubiquitous modules play a variety of roles in different contexts. As in bacterial plasmids, most of the characterized chromosomal TA systems are type II. However, recent reviews (Fozo et al., 2008; Gerdes & Wagner, 2007) have suggested that type I chromosomal systems may be more common than currently appreciated. Interestingly, most of the toxins encoded in these systems share similar features: they are generally small (less than 60 aa), hydrophobic, and contain a potential transmembrane domain. These features are shared with their plasmid-encoded counterparts, including both the hok/sok Hok toxin and the par Fst toxin. Despite their structural conservation, however, the effects of toxin expression are not always similar. Overexpression of Hok leads to the formation of ‘ghost’ cells resulting from damage to the cell membrane (Gerdes et al., 1986). Overexpression of Fst and the E. coli LdrD toxin leads to chromosomal condensation and, in the case of Fst, subsequent chromosomal segregation and cell division defects (Kawano et al., 2002; Patel & Weaver, 2006; Weaver et al., 2003).
In addition to the par Fst toxin, at least three other Fst-like peptides have been identified. The Orf35 protein from Lactobacillus gasseri phage ϕgaY (Yokoi et al., 2005) has been identified during whole-genome sequencing of strain JCM1131T as an Fst homologue linked to a phage-encoded cell lysis system and its attachment sites. Two peptides from Ent. faecalis strain V583, one on plasmid pTEF2 and one on the chromosome at locus EF0409, have been identified in a genome-wide survey of short coding sequences in streptococci (Ibrahim et al., 2007) using the V583 sequence (Paulsen et al., 2003). Interestingly, V583 also carries a pAD1 homologue, pTEF1, that encodes a locus identical to par. Using these peptides as bait in homology searches, genes encoding five additional Fst homologues on Ent. faecalis plasmids as well as chromosomes and plasmids from Lactobacillus and Staphylococcus species were identified. Furthermore, examination of the DNA sequence surrounding these genes suggests that, as in pAD1 par, they are all regulated by a convergently transcribed, partially complementary RNA. Phylogenetic analysis along with saturation mutagenesis of pAD1 Fst indicated residues important for function. Functional analysis revealed that par-like RNAs are made from all of the Ent. faecalis loci, including the chromosomal locus, and that at least one of the plasmid loci functions as an addiction module. In addition, it was demonstrated that overproduction of Fst is toxic in Staphylococcus aureus, suggesting that the par-like locus identified in this organism performs the same function. Therefore, the pAD1 par locus appears to be the prototype of a family of TA systems with both plasmid and chromosomally encoded members.
METHODS
Bacterial strains, media and culture conditions.
Ent. faecalis strain OG1X (Ike et al., 1983), a streptomycin-resistant, gelatinase-negative derivative of OG1, was used for Fst mutagenic analyses as well as for assessment of RNA production and function of parpAMS1. Sequenced Ent. faecalis strain V583 (Paulsen et al., 2003), provided as a gift (purified DNA from Dr Michael Gilmore, Harvard Medical School; strain from Dr Barbara Murray, University of Texas Health Sciences Center, Houston) was used to assess concurrent production of RNAs from parpTEF1 (identical to parpAD1), parpTEF2 and parEF0409. Assessment of parEF0409 RNA production in the absence of plasmid systems was performed in OG1RF (Oliver et al., 1977), kindly provided as a gift by Dr Murray, which has been recently sequenced (Bourgogne et al., 2008). S. aureus strain UAMS-1 (Gillaspy et al., 1995) was used for assessment of Fst toxicity. E. coli strain DH5α (Invitrogen) was used for production of various constructs. S. aureus strain RN4220 (Kreiswirth et al., 1983) was used as an intermediate (restriction-deficient, modification-proficient) strain in transferring plasmid constructs from DH5α to UAMS-1. S. aureus strains were kindly provided by Dr Mark Smeltzer, University of Arkansas. Ent. faecalis and S. aureus strains were cultured in Todd–Hewitt broth (THB) (Sigma) and E. coli in Luria–Bertani medium at 37 °C. Antibiotics were added at the following concentrations: chloramphenicol, 10–25 μg ml−1; spectinomycin, 100 μg ml−1; ampicillin, 100 μg ml−1; tetracycline, 10 μg ml−1; erythromycin, 100 μg ml−1.
Computer analysis.
Fst homologues were retrieved from the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) by standard blast searching using FstpAD1, FstϕgaY (ϕgaY Orf 35), FstpTEF2 and FstEF0409 as query. Alignments were constructed using the CLC Combined Workbench sequence analysis software at default settings. DNA sequences were then obtained from either the linked NCBI sequences or the Comprehensive Microbial Resource (cmr.jcvi.org) and 400 bp were retrieved from either side of the fst gene. Alignments were performed as above and sequence elements identified by eye.
fst mutagenesis and toxicity assessment.
Most fst mutants were constructed by the three-step site-specific PCR mutagenesis procedure previously described (Shokeen et al., 2008), using end, mutagenic and overlapping primers shown in Supplementary Table S1. PCR was performed with PCR Supermix Hi Fidelity (Invitrogen) according to the manufacturer's instructions. PCR products were cut with SalI and BamHI restriction enzymes (New England Biolabs), sites for which were introduced on the end primers (EPRNASR34, 5′-ATGTCGGGATCCCAGCTAGATAAATTGTTGAAGG-3′; 3′RNASal1, 5′-ATGTGCGTCGACAAAAGCAATCCTACGGCGA-3′; restriction sites in bold type), and ligated into the E. coli–Ent. faecalis shuttle vector pAM401 (Wirth & Clewell, 1987) using T4 DNA ligase (New England Biolabs). Subcloning Efficiency DH5α chemically competent cells (Invitrogen) were transformed with the ligation mixtures according to the manufacturer's instructions with selection for vector-encoded chloramphenicol resistance and screened for loss of the insert-disrupted tetracycline resistance. Plasmid DNA was purified by either the Bio-Rad Miniprep kit or the Qiagen-tip 100 gravity-flow column according to the manufacturers' instructions, and the presence of an appropriately sized band was confirmed by agarose gel electrophoresis. Inserts were then sequenced at either Lone Star Laboratories (Houston, TX, USA) or the University of South Dakota (USD) sequencing core facility using the pAM401 sequencing primer (Table 1) to ensure that only the intended mutation was present. DNA was then introduced into OG1X cells by electroporation (Greenfield & Weaver, 2000). RNA was purified from cells successfully transformed with mutated constructs and subjected to Northern blotting as previously described (Weaver et al., 2004) with the oligonucleotide probes shown in Table 1 to confirm that the construct was indeed producing the toxin-encoding RNA I transcript. Constructs consistently able to transform OG1X and producing RNA I at levels observed with the non-toxic E19stop mutant (Weaver et al., 2004) were considered to be non-toxic. Mutated constructs that could not transform OG1X were introduced into OG1X cells containing pDAK611 (Weaver et al., 1996), a pDL278 construct containing the RNA II gene. Constructs consistently unable to transform OG1X but able to transform OG1X(pDAK611) at levels similar to pDAK704, a pAM401 construct containing wild-type RNA I, were considered to retain toxicity. Mutant K2F was constructed by error-prone PCR as previously described (Shokeen et al., 2008), and its toxicity assessed as above.
Table 1.
Oligonucleotide probes and primers
| Primer | Sequence (5′–3′)* | Restriction site |
|---|---|---|
| Oligonucleotide probes for Northern blots | ||
| 5S rRNA probe | AACAGGTGTATCCTTCTCGCTAT | |
| pAD1 RNAI (SP) | ATAACCAACGACATTAAATCTTTCAC | |
| pAD1 RNAII (SP) | TGTGTTATCTGTACGATTTAATGTCG | |
| pTEF2 RNAI probe | CTAACCAATGGTCAACTAGTAAGATTA | |
| pTEF2 RNAII probe | GTGCTGTTCATATCGAACTTATGTCTT | |
| EF0409 RNAI probe | CAGGACAATCCCGACAAAAATCGGCACAAGAAT | |
| EF0409 RNAII probe | GCATCCATGCACGACTTTGTCACAAGTTACTTT | |
| pAMS1 RNAI probe | AGACCCACAAGTAGCGGTGCGATAATC | |
| pAMS1 RNA II probe | AGTATAACATATTTTTAAAGGAGGTCAACC | |
| PCR primers for cloning and sequencing | ||
| 5′pAMS1 | ATGTGCGTCGACATAAAGTACTGAAGGTTTTAAAAGTCCA | SalI |
| 3′pAMS1 | ATGTGCGGATCCAAGGGAAGGGCCTATCTGCTT | BamHI |
| 5′SalI-pCN51 | GCCGATCGGTCGACGCGGCAGCTCGCCTCGATTGGA | SalI |
| 3′EcoRI-pCN51 | CGCGATGCGAATTCCACAAAAAGCAATCCTACGGCGAA | EcoRI |
| pAM401 sequencing primer | GAGATTACGCGCAGACC | |
*Restriction sites of PCR primers are indicated in bold type.
Isolation and functional analysis of parpAMS1 and detection of other par RNAs.
The parpAMS1 sequence was amplified using the primers shown in Table 1 under the following PCR conditions: 2 min at 94 °C, followed by 35 cycles of 45 s at 94 °C, 45 s at 42 °C, and 1 min at 72 °C with a final extension of 10 min at 72 °C. The PCR product was then cut with SalI and BamHI, sites for which were inserted into the primers, and ligated into the cognate sites of E. coli–Ent. faecalis shuttle vectors pAM401 and pWM401 using T4 DNA ligase. DH5α cells were transformed with the ligation mixture with selection for chloramphenicol resistance and screening for tetracycline sensitivity. DNA was purified from transformants, examined for the appropriately sized fragment and sequenced as described above. OG1X was transformed with constructs containing the appropriate sequence by electroporation and transformants were selected for chloramphenicol resistance. RNA was purified from transformants and subjected to Northern blotting using the specific RNA IpAMS1 and RNA IIpAMS1 probes shown in Table 1. For stability tests, cultures were grown overnight in THB supplemented with 25 μg chloramphenicol ml−1 (THB/Cm). Cultures were diluted 1×10−4 in antibiotic-free medium and allowed to grow for 8 h. Cultures were then diluted 1×10−6 in antibiotic-free medium and allowed to grow overnight. The overnight culture was then diluted 1×10−4 in antibiotic-free medium and allowed to grow for another 8 h. At each dilution, cultures were diluted and plated on THB/Cm and THB plates to determine the percentage of plasmid-containing cells remaining. For long-term growth curve analysis, cultures grown overnight in THB/Cm were diluted 1×10−6 in antibiotic-free medium and OD600 was determined on a Milton Roy spectronic 21D spectrophotometer.
For detection of par RNAs from other putative par homologues, RNA was purified from the appropriate strains, subjected to Northern blotting as above and probed with the specific oligonucleotides listed in Table 1.
Assessment of Fst toxicity in S. aureus.
The RNA I gene was amplified by PCR using the primers listed in Table 1 under the same cycling conditions as above. The PCR product was then cut with SalI and EcoRI, sites for which were inserted into the primers and ligated into the cognate sites of pCN51, an E. coli–S. aureus shuttle vector with a cadmium-inducible promoter (Charpentier et al., 2004). DH5α was transformed with the ligation mixture and transformants selected for vector-encoded erythromycin resistance. Colonies were then screened for insert, and DNA was purified and introduced into S. aureus UAMS-1 after passage through RN4220 by electroporation (Schenk & Laddaga, 1992). DNA was purified (Dyer & Iandolo, 1983) from selected transformants and sequenced to ensure that no unintended mutations were present and that the fst gene was properly fused to the inducible promoter. To determine fst toxicity, cells were cultured overnight in THB supplemented with erythromycin and then diluted 1 : 100 in antibiotic-free THB. After 30 min growth, induced cultures were exposed to cadmium chloride added at 10 μM. Growth was followed spectrophotometrically as above. For electron microscopy, cultures were grown as described above and harvested 1 h after addition of cadmium chloride. Cells were washed twice with PBS and fixed with 3 % glutaraldehyde (in PBS). Samples were further fixed in 1 % osmium tetroxide in 0.1 M phosphate buffer (pH 7.4) for 1 h, then dehydrated in a graduated ethanol series and embedded in Epon 812 (Electron Microscopic Sciences). Thin sections (80 nm) were stained with uranyl acetate and lead citrate and observed under a transmission electron microscope (Hitachi H7500-I) at the University of Nebraska, Lincoln, Microscopy Core Facility.
RESULTS
Phylogenetic and mutagenic analysis of Fst
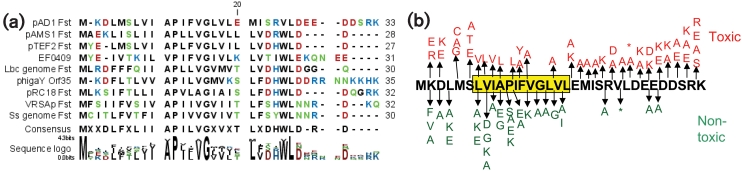
Using the pAD1 par-encoded Fst toxin and three other Fst-like proteins identified in earlier studies (Ibrahim et al., 2007; Yokoi et al., 2005) as blast queries, a total of nine small peptides related to Fst were identified in the NCBI database. In addition to pAD1 Fst, the family includes peptides encoded on four plasmids, pTEF2 (Ibrahim et al., 2007; Paulsen et al., 2003) and pAMS1 (Flannagan et al., 2008) from Ent. faecalis, pRC18 (Cuozzo et al., 2000) from Lactobacillus curvatus, and VRSAp (Kuroda et al., 2001) from S. aureus strain Mu50; one phage, ϕgaY (Yokoi et al., 2005) from L. gasseri; and one each on the chromosomes of Ent. faecalis strain V583 (EF0409) (Paulsen et al., 2003), Lactobacillus casei strain ATCC334 (LSEI 2682) (Makarova et al., 2006) and Staphylococcus saprophyticus strain ATCC15305 (SSP0870) (Kuroda et al., 2005). An alignment of all nine of these peptides is shown in Fig. 1(a).
Fig. 1.
The Fst family of peptides and identification of important residues. (a) Alignment of nine identified Fst-like peptides. Included are, from top to bottom, four Ent. faecalis sequences from the plasmids pAD1 (accession no. AAF03910), pAMS1 (ABW08104) and pTEF2 [peptide identified in Ibrahim et al. (2007), no accession no.], and from the EF0409 locus of the sequenced V583 strain (NP_814198); three Lactobacillus sequences from the genome of L. casei (YP_807843), the L. gasseri phage ϕgaY (BAD44723) and the L. curvatus plasmid pRC18 (NP_542225); and two Staphylococcus sequences from S. aureus plasmid VRSAp (NP_115298) and the S. saprophyticus genome (YP_300960). Residues are coloured according to polarity with non-polar residues coloured black, polar residues green, basic residues blue and acidic residues red. Note that while the annotated EF0409 sequence contains a 20 aa N-terminal extension, the DNA sequence upstream of the annotated initiation codon contains no recognizable ribosome-binding site, while the alternate initiation codon used in this alignment contains a strong ribosome-binding site. This observation and the identification of other features that fit the pAD1 par pattern described in the text and shown in Fig. 2(b) indicate that the methionine residue shown here is the true peptide start position. (b) Results of saturation mutagenesis of the pAD1 Fst toxic peptide. All mutations disrupting the toxicity of the peptide are shown below the sequence in green, while those not affecting toxicity are shown above the sequence in red. Mutations introducing termination codons are indicated by asterisks. The conserved hydrophobic sequence derived from the alignment is highlighted in yellow.
To identify regions of Fst important for its toxin function, alanine scanning mutagenesis was performed on the peptide. In addition, at some residues of particular interest, conservative and/or non-conservative amino acid changes were introduced. Mutations were introduced in the complete RNA I gene, cloned into the shuttle vector pAM401 and tested for their ability to transform Ent. faecalis strain OG1X in the presence and absence of RNA II, as described in Methods. Fig. 1(b) shows the results of this saturation mutagenesis analysis. Examination of the alignments and mutagenic data indicated regions of the peptide that were important for its function. Thus, as in the broader family of RNA-regulated peptide toxins (Fozo et al., 2008), all of the Fst-like toxins contained a conserved hydrophobic domain that could span the cytoplasmic membrane (yellow highlighted region of peptide in Fig. 1b). This region of Fst was highly sensitive to mutation. The proline and glycine at Fst positions 11 and 15, respectively (positions 12 and 16 in the alignment because of the single base gap introduced at the N terminus of the Fst sequence), were present in all peptides and were sensitive to mutation in Fst. Interestingly, an APUU(A/V)GUU motif, where U equals an isoleucine, leucine, valine or phenylalanine, is also present in the hydrophobic region of the Ldr family of RNA-regulated toxic peptides in E. coli (Kawano et al., 2002); like Fst (Patel & Weaver, 2006; Weaver et al., 2003), LdrD overexpression leads to chromosomal condensation. Most of the Fst-like peptides contained two charged/polar residues at the extreme N terminus, with the two staphylococcal representatives having only one. In pAD1 Fst, these residues could be altered to the opposite charge without affecting toxicity, but could not be substituted with uncharged amino acids. Each peptide also had a polar residue at position 7 or 8, but mutation of this residue in Fst had no effect. The C-terminal end of all of the peptides was highly charged but also highly variable and therefore of uncertain importance. The toxicity of Fst prematurely terminated after L25 supported the conclusion that the charged tail was not essential for function, although the R23A mutation suggested that at least one charged residue is essential. Why alanine mutations at E28 and D29 eliminated toxicity was not clear. It should be noted that, since toxicity was measured as the inability to transform in the absence of RNA II, it was not possible to measure the degree of toxicity. Therefore, some ‘toxic’ mutants may have reduced toxicity relative to wild-type Fst. It was not possible to measure toxicity in an inducible system since the only inducible expression vector available in enterococci utilizes a nisin-inducible promoter (Bryan et al., 2000), and nisin and Fst act synergistically to kill Ent. faecalis cells (Weaver et al., 2003).
Fst-like peptides are encoded within par-like loci
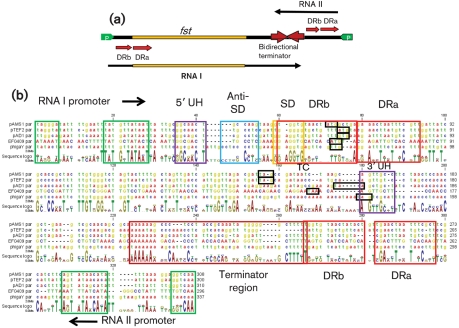
Examination of the DNA sequence surrounding all nine Fst-like genes revealed conservation of the key structural features previously defined in pAD1 par. The basic features of pAD1 par are depicted in Fig. 2(a). An alignment of the four enterococcal sequences and the ϕgaY locus depicting these features is shown in Fig. 2(b). Also indicated are two previously described structural features of RNA I, the upstream helix (UH) which stabilizes the transcript, and the 5′ stem–loop (SL) which suppresses translation of fst (Shokeen et al., 2008, 2009). The salient features defining the par-like loci are: (1) converging promoters (green arrows and boxes) transcribing the toxin mRNA and the smaller regulatory RNA (Weaver et al., 1996); (2) direct repeats (red arrows and boxes) that provide the interacting sequences for the RNAs and, in RNA I, overlap the ribosome-binding site [Shine–Dalgarno (SD) sequence] and initiation codon (yellow and black boxes, respectively) (Greenfield & Weaver, 2000; Greenfield et al., 2001, 2000); (3) an SL (brown arrows and boxes) with runs of As on one side and Ts on the other indicative of a bidirectional ρ-independent terminator (Greenfield et al., 2001); (4) complementary sequences that would form the RNase protective UH (purple boxes) (Shokeen et al., 2009); and (5) a pyrimidine-rich sequence (blue box) that, with the SD, forms the translational inhibitory SL of par (Shokeen et al., 2008). A U-turn motif (Franch et al., 1999) present in the terminator of pAD1 par RNA I has been previously shown to be essential for RNA II-mediated translational inhibition of RNA I and has been demonstrated to be the site of initial interaction between the two RNAs (Greenfield et al., 2001). Similar U-turn motifs were present in either the RNA I- or the RNA II-like component of some of the par homologues but not in others. For example, at consensus position 241 ϕgaY had a 5′-UUUG-3′ sequence within the terminator SL of RNA I at the same position as the pAD1 RNA I 5′-UUCG-3′ previously shown to function as a U-turn (YUNR) motif. pAMS1 par contained a 5′-CAAA-3′ sequence at position 245 that would be transcribed into a U-turn motif on RNA II. However, no U-turn motif was apparent in either RNA I or RNA II from pTEF2 or EF0409, suggesting that different modes of interaction function in different systems.
Fig. 2.
Conserved structural features of Fst-encoding par-like loci. (a) Organization of the pAD1 par locus. Converging promoters (green arrows) transcribe the toxin-encoding RNA I and the antitoxin RNA II (arrows below and above the main line, respectively) toward a bidirectional intrinsic transcriptional terminator (brown converging arrows). The RNAs are transcribed across direct repeats (red arrows labelled DRa and DRb) at which interaction occurs, suppressing translation of the Fst coding sequence (yellow box on line). (b) DNA sequences are, from top to bottom, Ent. faecalis plasmids pAMS1, pTEF2 and pAD1, Ent. faecalis chromosomal locus EF0409, and L. gasseri phage ϕgaY. Nucleotides for the chromosomal sequence are in upper-case type to allow easier orientation. Features are colour coordinated with (a) and described more fully in the text. The regions encoding the DRa and DRb repeats are boxed in red. The individual repeats are located in different places within these regions in the different elements. The sequence and location of the repeats are given in Table 2. The ribosome-binding site (SD) is boxed in yellow. The initiation and termination codons are boxed in black. The Anti-SD and SD sequences form the SL translational inhibitor structure and the 5′ and 3′ UH sequences form the nuclease-protective UH structure. The translational inhibitory structure of ϕgaY has been noted elsewhere (Yokoi et al., 2005).
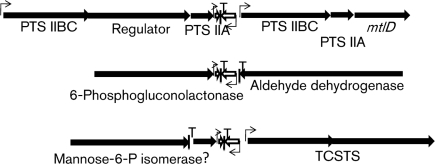
In a broader context, the plasmid-encoded par homologues were generally linked to replicon-associated genes. In pTEF2, which contains genes typical of a pheromone-responsive plasmid, the par homologue was in a context similar to that of pAD1. In pRC18 and VRSAp, the par homologues were linked to both putative replication initiator proteins and transposase genes, suggesting independent mobility. In pAMS1, the par homologue was associated with a cluster of genes involved in bacteriocin production (Flannagan et al., 2008). The complete sequence of the plasmid was not available so its potential linkage to the replicon was not discernible. The ϕgaY par homologue was located on the opposite side of the integration site from the intgY gene required for prophage integration (Yokoi et al., 2005). It was also linked to, but not transcribed with, an operon encoding a holin and endolysin involved in host cell lysis. The chromosomal par homologues were not linked to genes associated with mobile genetic elements but were instead associated with genes involved in carbohydrate metabolism: mannitol uptake and metabolism in Ent. faecalis, mannose utilization in L. casei, and 6-phosphogluconolactonase in S. saprophyticus (Fig. 3). This context suggests that the chromosomal par homologues are not associated with integrated mobile genetic elements but play some role in basic carbohydrate metabolism.
Fig. 3.
Genomic context of the chromosomally encoded par homologues. The genomic contexts of parEF0409 (top), parSSP0870 (middle) and parLSEI2682 (bottom) are shown. The converging white arrows indicate the par locus with the shorter arrow representing RNA II and the longer RNA I. The extent and direction of transcription of neighbouring genes are represented by black arrows. Predicted transcriptional terminators and promoters are indicated by vertical lines with a T and bent arrows, respectively. Genes without apparent promoters may be transcribed in operons with upstream genes. Gene names are based on the genome annotations (see Fig. 1 legend for accession nos). PTS, phosphotransferase components with closest homology to mannitol transport systems; Regulator, putative transcriptional regulator; mtlD, mannitol-1-phosphate 5-dehydrogenase; TCSTS, two-component signal transduction system; ?, hypothetical protein.
Due to the conservation of all of the basic components of the pAD1 par locus in all related loci, throughout the rest of the paper relevant elements will be referred to according to their pAD1 par counterpart with a subscript referring to the locus. Thus, parEF0409 produces RNA IEF0409, RNA IIEF0409 and FstEF0409.
Using oligonucleotide probes specific for the RNA I and RNA II components of parpTEF2 and parEF0409 (Table 1), RNAs of a similar size to RNA IpAD1 and RNA IIpAD1 were detected in strain V583. RNA IEF0409 and RNA IIEF0409 were also detected in the recently sequenced plasmid-free OG1RF strain (Bourgogne et al., 2008), but no bands hybridizing with probes for RNA I and RNA II for parpAD1, parpTEF2 or parpAMS1 were detected, confirming the specificity of the probes (data not shown).
Plasmid-encoded par-like loci likely function as addiction modules
To further assess their function, PCR amplification and cloning of parpTEF2, parEF0409 and parpAMS1 was attempted into shuttle vectors pAM401 and pWM401. Despite numerous attempts, only cloning of parpAMS1 was successful. Previous work with parpAD1 has demonstrated that Fst is toxic in E. coli if translation is increased by mutating the translational inhibitory SL structure (Shokeen et al., 2008). It is possible that the toxins of parpTEF2 and parEF0409 are expressed at high enough levels in E. coli to be toxic in the absence of further modification.
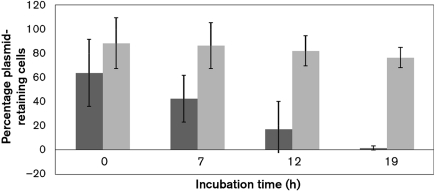
Both pAM401 : : parpAMS1 and pWM401 : : parpAMS1 were introduced into Ent. faecalis strain OG1X and demonstrated to produce RNA IpAMS1 and RNA IIpAMS1 (data not shown). Plasmid stability assessment revealed that, in pAM401, parpAMS1 actually accelerated plasmid loss, and remaining plasmid-containing cells grew very slowly on plates (data not shown). The possibility was considered that pAM401 was too unstable for parpAMS1 to effectively stabilize, leading to a growth disadvantage for plasmid-containing cells and rapid overgrowth by plasmid-free segregants. Since pWM401 is more stable than pAM401 (Weaver et al., 1996), the effect of parpAMS1 was tested on this plasmid. As shown in Fig. 4, pWM401 stability was significantly increased by the presence of parpAMS1. Furthermore, in OG1X(pWM401 : : parpAMS1) cultures maintained in exponential phase for longer than 10 h, generation time increased approximately twofold after 6 h of growth, about the time when plasmid-free cells would be expected to start to accumulate. OG1X(pWM401) cells showed no such change in growth rate.
Fig. 4.
Stabilization function of parpAMS1. Plasmid retention (as measured by retention of the plasmid-encoded chloramphenicol resistance) was determined in OG1X(pWM401) (dark-grey boxes) and OG1X(pWM401 : : parpAMS1) (light-grey boxes). Stability was measured per unit time rather than per generation because growth rate declines over time in OG1X(pWM401 : : parpAMS1), presumably due to effects of the translated toxin. Results shown are the mean of three independent experiments; error bars, sd.
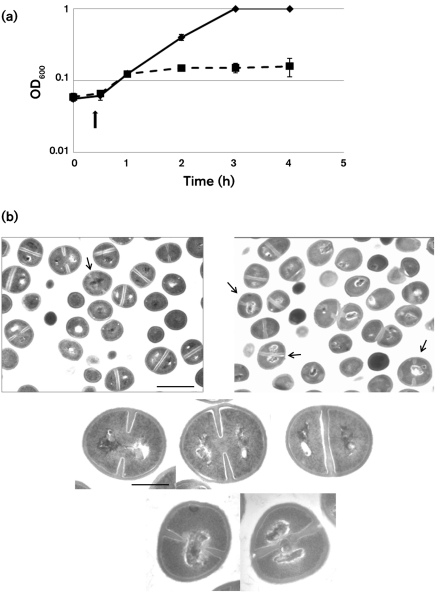
In order to assess the possibility that par-like loci function as addiction modules in species other than Ent. faecalis, the toxicity of FstpAD1 was assessed in S. aureus. The RNA IpAD1 gene was cloned downstream of the cadmium-inducible promoter in the plasmid vector pCN51 and introduced into the S. aureus strain UAMS-1. Induction of the promoter resulted in cessation of growth of the host strain within 30 min (Fig. 5a) and showed an approximately threefold drop in viability after 1 h (data not shown), indicating that Fst was indeed toxic to S. aureus. Induction of the empty vector had no effect on cell growth. Transmission electron micrographs of induced cells revealed that cells induced for pAD1Fst expression showed a variety of chromosome segregation and cell division defects (Fig. 5b). Common defects included compacted nucleoids trapped at the cell centre in deeply invaginated cells, bilobed nucleoids guillotined by completed septa, nucleoids displaced to the edges of cells, and formation of aberrant septa. These results are similar to those previously observed in B. subtilis and Ent. faecalis cells (Patel & Weaver, 2006; Weaver et al., 1996), except that in the latter host, compacted nucleoids were frequently shunted to one or other of the daughter cells. This difference may be due to differences in cell division and chromosome partition functions between Ent. faecalis and S. aureus.
Fig. 5.
Effects of overexpression of Fst on S. aureus. (a) RNA Ipar was introduced into S. aureus UAMS-1 cells under control of the cadmium-inducible promoter of pCN51. Cadmium was added at 10 μM 30 min after culture dilution into antibiotic-free medium. Squares with broken line, pCN51 : : RNA Ipar; diamonds with unbroken line, pCN51. (b) Effects of the Fst toxin on cell morphology. The top row of panels shows a large field of induced pCN51 control (left) and pCN51 : : RNA I (right) cells to provide perspective on the proportion of abnormal cells. The arrow indicates a single cell with recognizable division septa and a centrally located nucleoid in the control panel. Note, however, that this cell is very early in the division process with only very small septal invaginations. In contrast, arrows in the right panel indicate two cells with centrally located nucleoids late in division and a third in which the nucleoid has been guillotined by the septum. Fst-exposed cells also occasionally displayed nucleoids pressed against the side of the cell, similar to those at the top of the image, probably due to completion of division in guillotined cells. Cells showing abnormal division septa, like the centrally located dividing cell, were more rarely seen. The middle row of panels shows the typical progression of nucleoid segregation seen in control cells, with the nucleoids showing some connection early in division (left), completely separating before the completion of division (middle), and well-separated by the time the septum is complete (right). The bottom row of panels shows close-ups of the two most common defects seen in Fst-exposed cells: centrally located nucleoids late in division (left) and guillotined nucleoids at the completion of division (right). Bars: top panels, 1 μm; middle and bottom panels, 0.3 μm.
DISCUSSION
Identification of homologues of the Ent. faecalis plasmid pAD1 par addiction module indicates that par is the prototype of a family of type I toxin–antitoxin systems that are present on plasmids and chromosomes of Enterococcus, Lactobacillus and Staphylococcus species. Given the small size of the Fst homologues (27–33 aa) and the par loci (<350 nt) and the divergence of their sequences, it seems likely that other such loci remain to be identified. All three enterococcal plasmids are pheromone-responsive plasmids and, therefore, likely relatively closely related. The S. aureus VRSAp plasmid is not pheromone-responsive but does encode a replication initiator protein related to that encoded by the pheromone-responsive plasmids (Weaver et al., 2009). The replication initiator protein for L. curvatus plasmid pRC18 has not been identified, but the par homologue is associated with putative transposases and therefore may be mobile. As reported here, the pAMS1 par homologue has effects on plasmid stability consistent with a function as an addiction module. In addition, overexpression of FstpAD1 is toxic in S. aureus, suggesting that parVRSAp performs the same function. Thus, it is likely that all of the plasmid-encoded par homologues function as addiction modules. However, given differences in the presence of U-turn motifs and predicted structures of the RNAs, the mechanisms of RNA interaction and regulation of toxin production may vary in the different plasmid systems.
The function of the chromosomal par homologues is currently unclear. The fact that they are associated with genes involved in carbohydrate metabolism in three different species is intriguing. They appear not to be part of integrated mobile genetic elements but rather closely linked to genes involved in basic metabolism. The possibility that they play a physiological role in nutritional shifts is currently under investigation. Interestingly, chromosomal homologues of the other well-studied type I addiction module, hok/sok, have been identified in E. coli and related enteric bacteria. Unlike the chromosomal par homologues identified here, these loci are frequently inactivated by insertion sequences, point mutations or other rearrangements. Their functions are also unknown (Gerdes & Wagner, 2007).
Overexpression of Fst results in chromosomal compaction and defects in segregation and cell division that, after prolonged exposure, compromise the cell membrane (Patel & Weaver, 2006; Weaver et al., 2003). How Fst causes these effects is not clear. Fozo et al. (2008) have postulated the existence of a family of RNA-regulated toxins that are membrane-localized. The results of the phylogenetic and mutagenic analysis of Fst are consistent with this hypothesis, as the hydrophobic domain is conserved in all Fst homologues and is highly sensitive to amino acid changes. The existence of a conserved APUU(A/V)GUU motif within this region in both Fst and Ldr toxic peptides, which both cause chromosome compaction, suggests the existence of a subgroup of the RNA-regulated toxins. While some peptide toxins, such as Hok, may form membrane pores, it is possible that other toxins have more subtle effects manifested by interaction with specific membrane proteins. Peptides that regulate degradation of membrane proteins, stabilize P-type ATPase transporters and modulate the activity of two-component systems have been described (Alix & Blanc-Potard, 2009). Toxic peptides could affect the function of membrane proteins in such a way as to be detrimental to the cell. Given the association of parEF0409 with genes involved in mannitol uptake, the possibility that FstEF0409 modulates the activity of these membrane-localized transporters is intriguing.
Moritz & Hergenrother (2007) examined the prevalence of various TA systems in vancomycin-resistant enterococci and concluded that while some systems, such as homologues of ε/ζ, axe/txe, MazEF and RelBE, are widespread, par-like systems are virtually absent. However, the PCR-based approach used required a significant amount of conservation at the borders of the element to detect related systems. The par-like loci identified in this study show little nucleotide sequence conservation and would not have been identified by the PCR-based approach used in the Moritz and Hergenrother study. In the current study, sequence homology of the toxic peptides themselves was used to identify par-like loci. This approach is only as good as the ORF-detection systems used to analyse the various genomes, and since small ORFs are frequently missed in these analyses, more par-like systems may await discovery on both chromosomes and plasmids. Clearly, par-like loci are not restricted to enterococcal plasmids and may play a broader role in plasmid stability and metabolic regulation in Gram-positive bacteria.
Table 2.
Location of DRa and DRb repeats
| Locus | DRb sequence* | RNA I† | RNA II | DRa sequence | RNA I | RNA II |
|---|---|---|---|---|---|---|
| parpAMS1 | 5′-tggtgtaactat-3′ | 60–72 | 266–277 | 5′-aactaatttcg-3′ | 81–91 | 285–295 |
| parpTEF2 | 5′-gtgctgtt-3′ | 65–72 | 270–277 | 5′-cgaacttatgtctt-3′ | 79–92 | 283–297 |
| parpAD1 | 5′-tgtgttat-3′ | 64–71 | 266–273 | 5′-gatttaatgtcgtt-3′ | 80–93 | 284–297 |
| parEF0409 | 5′-gtgcatcacatg-3′ | 63–74 | 268–278 | 5′-ttgtcacaa-3′ | 84–92 | 288–296 |
| parϕgaY | 5′-gagttcgttgaa-3′ | 65–78 | 267–280 | 5′-ttctaactt-3′ | 84–92 | 287–296 |
*Repeat sequence may vary by a single nucleotide substitution or insertion between the RNA I and RNA II sequences.
†Location given refers to the nucleotide sequence shown in Fig. 2(b).
Acknowledgments
This work was supported by Public Health Service grant GM55544, the Division of Basic Biomedical Sciences of the Sanford School of Medicine (K. E. W.) and Public Health Service Grant AI038901 (K. W. B.).
Abbreviations
SD, Shine–Dalgarno
SL, stem–loop
TA, toxin–antitoxin
UH, upstream helix
Footnotes
A supplementary table, listing oligonucleotide primers used for mutant construction, is available with the online version of this paper.
References
- Alix, E. & Blanc-Potard, A.-B. (2009). Hydrophobic peptides: novel regulators within bacterial membrane. Mol Microbiol 72, 5–11. [DOI] [PubMed] [Google Scholar]
- Bahassi, E. M., O'Dea, M. H., Allali, N., Messens, J., Gellert, M. & Couturier, M. (1999). Interactions of CcdB with DNA gyrase. Inactivation of GyrA, poisoning of the gyrase–DNA complex, and the antidote action of CcdA. J Biol Chem 274, 10936–10944. [DOI] [PubMed] [Google Scholar]
- Bourgogne, A., Garsin, D., Qin, X., Singh, K., Sillanpaa, J., Yerrapragada, S., Ding, Y., Dugan-Rocha, S., Buhay, C. & other authors (2008). Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol 9, R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, E. M., Bae, T., Kleerebezem, M. & Dunny, G. M. (2000). Improved vectors for nisin-controlled expression in Gram-positive bacteria. Plasmid 44, 183–190. [DOI] [PubMed] [Google Scholar]
- Buts, L., Lah, J., Dao-Thi, M.-H., Wyns, L. & Loris, R. (2005). Toxin–antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci 30, 672–679. [DOI] [PubMed] [Google Scholar]
- Charpentier, E., Anton, A. I., Barry, P., Alfonso, B., Fang, Y. & Novick, R. P. (2004). Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl Environ Microbiol 70, 6076–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon, C. (2006). Shutdown decay of mRNA. Mol Microbiol 61, 573–583. [DOI] [PubMed] [Google Scholar]
- Cuozzo, S. A., Sesma, F., Palacios, J. M., de Ruíz Holgado, A. P. & Raya, R. R. (2000). Identification and nucleotide sequence of genes involved in the synthesis of lactocin 705, a two-peptide bacteriocin from Lactobacillus casei CRL 705. FEMS Microbiol Lett 185, 157–161. [DOI] [PubMed] [Google Scholar]
- Dyer, D. W. & Iandolo, J. J. (1983). Rapid isolation of DNA from Staphylococcus aureus. Appl Environ Microbiol 46, 283–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka, H., Amitai, S., Kolodkin-Gal, I. & Hazan, R. (2006). Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet 2, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan, S. E., Clewell, D. B. & Sedgley, C. M. (2008). A “retrocidal” plasmid in Enterococcus faecalis: passage and protection. Plasmid 59, 217–230. [DOI] [PubMed] [Google Scholar]
- Fozo, E. M., Hemm, M. R. & Storz, G. (2008). Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev 72, 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch, T., Petersen, M., Wagner, E. G. H., Jacobsen, J. P. & Gerdes, K. (1999). Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J Mol Biol 294, 1115–1125. [DOI] [PubMed] [Google Scholar]
- Gerdes, K. & Wagner, E. G. H. (2007). RNA antitoxins. Curr Opin Microbiol 10, 117–124. [DOI] [PubMed] [Google Scholar]
- Gerdes, K., Bech, F. W., Jorgensen, S. T., Lobner-Olesen, A., Rasmussen, P. B., Atlung, T., Boe, L., Karlstrom, O., Molin, S. & von Meyenburg, K. (1986). Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J 5, 2023–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, K., Thisted, T. & Martinussen, J. (1990). Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol Microbiol 4, 1807–1818. [DOI] [PubMed] [Google Scholar]
- Gerdes, K., Christensen, S. K. & Lobner-Olesen, A. (2005). Prokaryotic toxin–antitoxin stress response loci. Nat Rev Microbiol 3, 371–382. [DOI] [PubMed] [Google Scholar]
- Gillaspy, A. F., Hickmon, S. G., Skinner, R. A., Thomas, J. R., Nelson, C. L. & Smeltzer, M. S. (1995). Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun 63, 3373–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield, T. J. & Weaver, K. E. (2000). Antisense RNA regulation of the pAD1 par post-segregational killing system requires interaction at the 5′ and 3′ ends of the RNAs. Mol Microbiol 37, 661–670. [DOI] [PubMed] [Google Scholar]
- Greenfield, T. J., Ehli, E., Kirshenmann, T., Franch, T., Gerdes, K. & Weaver, K. E. (2000). The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol Microbiol 37, 652–660. [DOI] [PubMed] [Google Scholar]
- Greenfield, T. J., Franch, T., Gerdes, K. & Weaver, K. E. (2001). Antisense RNA regulation of the par post-segregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target, RNAI. Mol Microbiol 42, 527–537. [DOI] [PubMed] [Google Scholar]
- Hayes, F. (2003). Toxins–antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301, 1496–1499. [DOI] [PubMed] [Google Scholar]
- Ibrahim, M., Nicolas, P., Bessieres, P., Bolotin, A., Monnet, V. & Gardan, R. (2007). A genome-wide survey of short coding sequences in streptococci. Microbiology 153, 3631–3644. [DOI] [PubMed] [Google Scholar]
- Ike, Y., Craig, R. A., White, B. A., Yagi, Y. & Clewell, D. B. (1983). Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc Natl Acad Sci U S A 80, 5369–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., Pogliano, J., Helinski, D. R. & Konieczny, I. (2002). ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol 44, 971–979. [DOI] [PubMed] [Google Scholar]
- Kawano, M., Oshima, T., Kasai, H. & Mori, H. (2002). Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol Microbiol 45, 333–349. [DOI] [PubMed] [Google Scholar]
- Kreiswirth, B. N., Lofdahl, S., Betley, M. J., O'Reilly, M., Schlievert, P. M., Bergdoll, M. S. & Novick, R. P. (1983). The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712. [DOI] [PubMed] [Google Scholar]
- Kuroda, M., Ohta, T., Uchiyama, I., Baba, T., Yuzawa, H., Kobayashi, I., Cui, L., Oguchi, A., Aoki, K. & other authors (2001). Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357, 1225–1240. [DOI] [PubMed] [Google Scholar]
- Kuroda, M., Yamashita, A., Hirakawa, H., Kumano, M., Morikawa, K., Higashide, M., Maruyama, A., Inose, Y., Matoba, K. & other authors (2005). Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc Natl Acad Sci U S A 102, 13272–13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., Pavlov, A., Pavlova, N., Karamychev, V. & other authors (2006). Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 103, 15611–15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, E. M. & Hergenrother, P. J. (2007). Toxin–antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc Natl Acad Sci U S A 104, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, D. R., Brown, B. L. & Clewell, D. B. (1977). Analysis of plasmid deoxyribonucleic acid in a cariogenic strain of Streptococcus faecalis: an approach to identifying genetic determinants on cryptic plasmids. J Bacteriol 130, 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, S. & Weaver, K. E. (2006). Addiction toxin Fst has unique effects on chromosome segregation and cell division in Enterococcus faecalis and Bacillus subtilis. J Bacteriol 188, 5374–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen, I. T., Banerjei, L., Myers, G. S. A., Nelson, K. E., Seshadri, R., Read, T. D., Fouts, D. E., Eisen, J. A., Gill, S. R. & other authors (2003). Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299, 2071–2074. [DOI] [PubMed] [Google Scholar]
- Schenk, S. & Laddaga, R. A. (1992). Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett 73, 133–138. [DOI] [PubMed] [Google Scholar]
- Shokeen, S., Patel, S., Greenfield, T. J., Brinkman, C. & Weaver, K. E. (2008). Translational regulation by an intramolecular stem–loop is required for intermolecular RNA regulation of the par addiction module. J Bacteriol 190, 6076–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokeen, S., Greenfield, T. J., Ehli, E. A., Rasmussen, J., Perrault, B. E. & Weaver, K. E. (2009). An intramolecular upstream helix ensures the stability of a toxin-encoding RNA in Enterococcus faecalis. J Bacteriol 191, 1528–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen, L. & Saavedra De Bast, M. (2009). Bacterial toxin–antitoxin systems: more than selfish entities? PLoS Genet 5, e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, K. E., Jensen, K. D., Colwell, A. & Sriram, S. I. (1996). Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol Microbiol 20, 53–63. [DOI] [PubMed] [Google Scholar]
- Weaver, K. E., Weaver, D. M., Wells, C. L., Waters, C. M., Gardner, M. E. & Ehli, E. A. (2003). Enterococcus faecalis plasmid pAD1-encoded Fst toxin affects membrane permeability and alters cellular responses to lantibiotics. J Bacteriol 185, 2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, K. E., Ehli, E. A., Nelson, J. S. & Patel, S. (2004). Antisense RNA regulation by stable complex formation in the Enterococcus faecalis plasmid pAD1 par addiction system. J Bacteriol 186, 6400–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, K. E., Kwong, S. M., Firth, N. & Francia, M. V. (2009). The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids. Plasmid 61, 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth, R. F. & Clewell, D. B. (1987). Highly efficient cloning system for Streptococcus faecalis protoplast transformation, shuttle vectors, and applications. In Streptococcal Genetics, pp. 25–27. Edited by R. Curtis III. Washington, DC: American Society for Microbiology.
- Yokoi, K.-J., Shinohara, M., Kawahigashi, N., Nakagawa, K., Kawasaki, K.-I., Nakamura, S., Taketo, A. & Kodaira, K.-I. (2005). Molecular properties of the two-component cell lysis system encoded by prophage ϕgaY of Lactobacillus gasseri JCM 1131T: cloning, sequencing, and expression in Escherichia coli. Int J Food Microbiol 99, 297–308. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Zhang, Y., Zhu, L., Suzuki, M. & Inouye, M. (2004). Interference of mRNA function by sequence-specific endoribonuclease PemK. J Biol Chem 279, 20678–20684. [DOI] [PubMed] [Google Scholar]