Introduction
Helicases are enzymes that can separate duplex oligonucleotides in a NTP-dependent fashion and are essential in all aspects of DNA and RNA metabolism. Amino acid sequence analysis identified several conserved sequence motifs in DNA and RNA helicases allowing their classification into 5 major groups (Super families SF1–SF5) [1]. DExD/H helicases share eight conserved sequence motifs, whereas the DEAD box helicase subgroup has an additional ninth conserved sequence motif [2]. These sequence motifs encompass an approximately 300–400 amino acid core region involved in ATP-binding/hydrolysis and RNA binding (Part 2: Figure 1A). Structural analyses of several DEAD-box proteins show this core region forms two RecA-like globular domains [2].
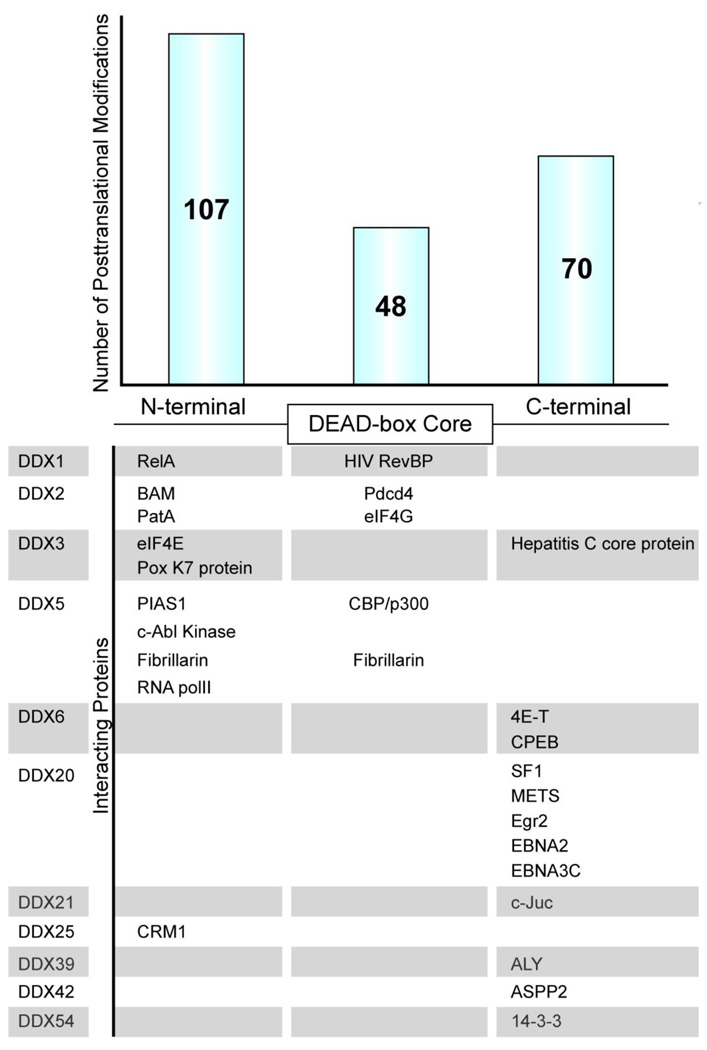
Figure 1. Human DEAD-box protein (DDX) posttranslational modifications and protein-protein interactions.
A schematic representation of the DDX protein domain architecture features the conserved central DEAD-box core region along with the unique flanking N-terminal and C-terminal regions. Above shows the number of posttranslational documented in Table 1 with respect to their location within the DDX protein. Below lists known DDX-binding proteins, as well as where the interaction occurs on their respective DDX open reading frame.
Work in a variety of eukaryotes has identified the biological functions of many DEAD-box helicases. The genome of the yeast Saccharomyces cerevisiae encodes 25 DEAD-box proteins. Counterparts for each of these, along with 11 additional DEAD-box genes, are found in the human genome. Although some of the shared DEAD-box genes have similar functions in both humans and yeast, it is clear that several human DEAD-box proteins have acquired additional functions [3, 4]. How these functions are regulated within cellular or developmental contexts is less understood. The N-terminal and C-terminal sequences flanking the DEAD-box core regions are considerably more divergent among DEAD-box proteins and are thought to interact with RNA substrates or cofactors. Such interactions can thereby target and regulate their helicase activity or perform completely independent functions [3]. Although the structures of these divergent flanking sequences are largely unknown, a growing body of evidence suggests they are regulatory hot-spots for posttranslational modifications and protein-protein interactions (Figure 1). Despite DEAD-box helicase conservation throughout the animal kingdom, the most comprehensive data on their posttranslational regulation comes from the human DEAD-box helicase family (DDX proteins). For the purposes of this review, we shall focus primarily on the data concerning human DDX proteins and look at our current understanding on how posttranslational modifications and protein-protein interactions regulate DEAD-box protein functions.
DDX1
DDX1 was originally identified as an overexpressed gene in retinoblastoma and neuroblastoma tumors and cell lines [5]. In its normal context, DDX1 is a nuclear protein expressed in many cell types early in development that later becomes restricted, indicative of a specific role in differentiated cells. While much of DDX1 function remains unknown, a number of studies suggest that interaction with other proteins directs its function as a cofactor in various biological processes. It is also the only human DEAD-box gene with a SPRY-domain, which may facilitate some of its binding properties [5, 6]. The DDX1 interaction with the RelA (p65) subunit of NF-κB recruits it to NF-κB-binding promoter sequences. A mammalian-two hybrid binding assay suggests that amino acids in both the conserved DEAD-box core region and the N-terminal amino acids are important for this interaction [7]. A yeast two-hybrid screen identified DDX1 residues located in the DEAD-box conserved core region (amino acids 189–333) that bound HIV Regulator of virion (Rev) protein, which is required for nuclear export of viral transcripts [8]. Further in vitro binding analysis and in vivo co-immunoprecipitation in HEK293 cell extracts confirmed the interaction between Rev and DDX1 [9]. Overexpression of DDX1 in HIV infected cells corresponded to increased viral production, whereas siRNA knock-down of DDX1 altered Rev localization suggesting that DDX1 is an important cofactor for Rev function [9]. Following ionizing radiation, DDX1 protein colocalizes with ATM (ataxia telangiectasia mutated) to multiple foci within the nucleus. DDX1 is phosphorylated by ATM both in vitro and in vivo and phosphorylation of endogenous DDX1 is enhanced by ionizing radiation, supporting a role for DDX1 in the repair of double-strand DNA breaks within transcriptionally active regions of the genome [10]. Therefore, it is clear that posttranslational regulation plays an important role in DDX1 function.
DDX2 (eIF4A)
DDX2 (eIF4A) is an essential and universally conserved RNA helicase that plays a key role in initiation of translation by relieving secondary mRNA structure and allowing ribosome scanning [11]. The interaction of eIF4A with several factors is essential for its canonical role in the translation initiation complex. The ATPase activity of eIF4A is greatly enhanced when it is part of a multiprotein complex with initiation factors eIF4G, eIF4E, eIF4B and eIF4H compared to the ATPase activity of free eIF4A [3]. The helicase activity of eIF4A is stimulated by the presence of either eIF4B or eIF4H, presumably through a direct interaction. Binding of programmed cell death factor 4 (Pdcd4) to eIF4A inactivates its helicase activity thereby inhibiting cap-dependent translation and competes for eIF4A binding with a C-terminal portion of eIF4G [3]. Mammalian two-hybrid and in vitro binding assays of eIF4A point mutants suggest Pdcd4 and eIF4G bind to partially overlapping sites within both N-terminal and C-terminal globular core domains of eIF4A [12]. Modification of eIF4A with the lipid molecule 15-deoxy-delta 12,14-prostaglandin J2 (15d-PGJ2) blocks the interaction between eIF4A and eIF4G thereby inhibiting translation. This is consistent with the signaling activity of 15d-PGJ2 that directs inhibition of cell proliferation [13]. Recent work in Drosophila demonstrates that eIF4A plays a role in germline stem cell maintenance by directly interacting and antagonizing the function of Bag of marbles (BAM). BAM preferentially interacts with an N-terminal portion of eIF4A in a yeast two-hybrid binding assay and also co-immunoprecipitates with eIF4A in Drosophila S2 cell extracts. The small marine natural product pateamine A (PatA) inhibits eukaryotic translation initiation by directly targeting eIF4AI and II. In vitro binding analyses demonstrate the N-terminal amino acids 1–220 of eIF4A are sufficient for its PatA interaction [14].
DDX3 (PL10)
DDX3 is a nucleocytoplasmic shuttling protein with several reported functions including mRNA translocation, RNA splicing, innate immunity, as well as its cooption in viral pathology [15, 16]. The activity of DDX3 is modulated by several protein-protein interactions and posttranslational modifications. DDX3 amino acids Thr204 and Thr323, in the DEAD-box core region, are phosphorylated in vitro by cyclinB/cdc2 [17]. Both in vitro and in vivo binding analyses suggest the × chromosomal isoform of DDX3 (DDX3X) is a substrate for Tank binding kinase 1 (TBK1). DDX3 is also phosphorylated at 11 sites that are scattered throughout the DEAD-box core region. These TBK1-dependent phosphorylations are required for DDX3X to stimulate the type-I interferon (IFN)-β production through a transcriptional activation mechanism [18]. DDX3 interacts with eIF4E through a conserved eIF4E consensus binding sequence in its N-terminal residues 38–44 and this interaction represses translation [19]. Biochemical and structural analysis show DDX3 also interacts with the vaccinia virus K7 protein through N-terminal amino acids 61–90 [20]. DDX3 C-terminal amino acids 553–622 directly and specifically interact with the hepatitis C core protein. The vaccinia virus K7 protein and hepatitis C virus core protein interactions evade the innate immunity response by preventing DDX3 from inducing IFN-β transcription [21].
DDX4 (Vasa)
DDX4 is the human Vasa ortholog, which is a highly conserved DEAD-box helicase involved in germline formation and fertility. Nine conserved sequence motifs typify all DEAD-box genes [3]. Biochemical analyses show how these motifs, in Vasa and other DEAD-box proteins, confer its ATP-dependant RNA helicase catalytic activity. Structural data also suggests that Vasa unwinds duplex RNA in a non-processive manner [2, 3].
The mouse Vasa homolog is required for spermatogenesis and male fertility. Several studies indicate a functional relationship between Vasa and both the small interfering RNA and micro-RNA processing pathways. One essential component in both of these pathways is the RNase III endonuclease Dicer, which, in mice, colocalizes with Vasa in nuage [22]. Ectopically expressed Vasa and Dicer protein interact in COS cell lysate and this interaction requires the C-terminal portion of Vasa. The C-terminal RNaseIII region of Dicer is sufficient to interact with Vasa and the remaining N-terminal ATPase/helicase-PAZ domain region was unable to bind Vasa [22].
Vasa protein binds to MIWI and MILI, which are mouse PIWI homologs. In vitro binding data suggest these interactions occur through the N-terminal portion of Vasa protein. MILI and Vasa knockout mice have similar phenotypes and defects in spermatogenesis indicative of cooperative molecular functions [23]. In MIWI knockout mice, Vasa protein does not localize to the nuage structures [23]. However, it is still unknown whether MIWI is required for nuage and ultrastructural studies in MIWI knockout mice are needed. Exactly how these specific interactions influence Vasa, MIWI or MILI function is unclear.
Recent work has identified Maelstrom as a nuage component that interacts with both mouse Vasa and MIWI, is required for spermatogenesis and also is involved in silencing transposable elements [24]. In Drosophila, Maelstrom protein localizes to nuage in a Vasa-dependent manner. In maelstrom mutant oocytes, a higher molecular weight Vasa protein species is evident indicating that Maelstrom is required for proper Vasa modification or processing.
Mouse Vasa also interacts with RanBPM The N-terminal portion of Vasa is sufficient for this interaction and both proteins colocalize to nuage in maturing spermatocytes [25]. RanBPM is believed to be involved in recruiting Ran-GTP to microtubule assembly sites. These results suggest a functional relationship between Vasa and microtubule nucleation during meiosis. Much like another Vasa-interacting protein, Gustavus, RanBPM has contains a SPRY domain. It is unclear, however, whether the Vasa-RanBPM interaction occurs through its SPRY domain.
DDX5 (p68), DDX17 (p72)
The human DDX5 (p68) and DDX17 (p72) are very similar RNA helicases required for splicing that also play roles in transcriptional regulation [26]. p68 is important for normal cell growth, differentiation and proliferation (Stevenson et al., 1998; Heinlein, 1998). Various types of posttranslational regulation, including covalent modifications and protein-protein interactions, appear to coordinate and control these activities. Colorectal tumors are associated with increased expression and polyubiquitylation of p68. However, it is unknown which portions of the p68 protein are ubiquitylated. Addition of the small ubiquitin-like modifier (SUMO) to proteins is also known to alter their localization, binding capabilities and function [27]. PIAS1-mediated SUMO modification of p68 on single site Lys53 modulates its transcriptional activity and promotes its interaction with HDAC1 [28]. Platelet-derived growth factor stimulates c-Abl kinase phosphorylation of p68 on Tyr593 in the nucleus. This phosphorylation is detected in seven different cancer cell lines. Phosphorylated p68 interacts with nuclear β-catenin which then promotes cell proliferation by activating transcription of cyclin D1 and c-Myc genes and also promotes an epithelial-mesenchymal transition [29, 30]. Several studies suggest that the localization and function of p68 are regulated by interactions with various proteins. For instance, fibrillarin and p68 interact in HeLa cell extracts and colocalize in nascent nucleoli during late telophase. This interaction requires p68 amino acids 67–483, suggesting that both N-terminal, C-terminal and conserved core regions are involved. During interphase, p68 can localize to the nuclear matrix by binding A-kinase-anchoring protein (AKAP95). The transcriptional coactivator CREB-binding protein (CBP/p300) and RNA polymerase II bind p68 in vivo, suggesting p68 mediates this multiprotein complex. In vitro binding analyses suggests the CBP/p300 interaction occurs through p68 amino acids 176–388 in the DEAD-box core region and RNA polymerase II interacts with amino acids 1–80 on the p68 N-terminus. p68 and p72 preferentially form hetero-dimers [31]. This interaction regulates their activities as transcriptional coactivators.
DDX6 (RCK/p54)
The DDX6-like p54 genes are implicated in several biological processes including translational regulation and function in various cytoplasmic bodies such as C. elegans P-bodies, Drosophila sponge bodies and Xenopus Balbiani bodies [32]. Human RCK/p54 interacts with AGO1 and AGO2 in vitro and in vivo and facilitates P-body formation [33]. Much of the regulatory data comes from its Xenopus homolog Xp54. Potential nuclear export sequence and protein kinase CK2 sites were identified in N-terminal region and a C-terminal 44 amino acid segment respectively in Xenopus p54. Xenopus p54 interacts with cytoplasmic polyadenylation element-binding protein (CPEB) [34, 35]. A C-terminal portion of p54 interacts with 4E-T and embryonic poly(A)-binding protein (ePAB) and is important for its P-body localization [36]. It is still unclear, however, if similar posttranslational regulatory events occur in humans.
DDX20 (DP103)
Functional analyses of DP103 knock out mice show that it is required for embryonic development as well as ovarian development and function [37]. The DP103 C-terminus interacts with several proteins that target its function. DP103 represses transcription by interacting with SUMOylated steroidogenic factor 1 (SF1), but it is unknown whether DP103 itself is SUMOylated [38]. This interaction was mapped to amino acids 721 to 825 within the nonconserved C-terminal region of DP103. This same region interacts with the Ets repressor METS to form a transcription corepressor complex. A yeast two-hybrid screen for early growth response 2 (Egr2) interacting protein also identified an interacting clone containing the C-terminal amino acids 612–825 [38]. The C-terminal region of DP103 containing amino acids 666–824 interacts with Epstein Barr virus nuclear antigens EBNA2 and EBNA3C. Together, these data suggest that the transcriptional corepressor activity of DP103 is important for embryonic and ovarian development.
DDX21 (RHII/Gu)
DDX21 functions in ribosomal RNA processing and is normally localized in the nucleolus. DDX21 interacts with c-Jun through its C-terminal 749 to 801 amino acids and functions as a transcriptional coactivator. Recent work shows that c-Jun also regulates DDX21 function. Depletion of c-Jun inhibits 18S and 28S rRNA accumulation and results in a mislocalization of DDX21 out of the nucleolus and into the nucleoplasm [39].
DDX23 (Prp28)
DDX23 is a spliceosome component required for splicing of nuclear pre-mRNA. The human DDX23 contains an N-terminal RS domain and can be phosphorylated in vitro by the Clk/Sty and the U1 snRNP-associated kinase, which both are known to phosphorylate RS domains. Consistent with this, DDX23 phosphorylation is required for its spliceosomal B complex association [40].
DDX25 (Gonadotropin-regulated testicular helicase)
DDX25 (GRTH) is a gonadotropin-dependent testis-specific RNA helicase in Leydig and germinal cells essential for spermatogenesis. A leucine-rich sequence in the N-terminal amino acids 61–74 functions as a nuclear export sequence and phosphorylated GRTH displays a cytoplasmic localization, whereas unphosphorylated GRTH interacts with CRM1 [41].
DDX39
DDX39 is a growth-associated RNA helicase involved in pre-mRNA processing and export. DDX39 is also upregulated in lung squamous cell carcinoma [42]. DDX39 interacts with cytokine induced protein 29 (CIP29), is polyubiquitylated when expressed in 293 cells and is degraded by the ubiquitin proteasome pathway. The DDX39 ubiquitylation occurs somewhere on its N-terminal portion [43]. Aly is a splicing factor that links pre-mRNA splicing to nuclear export and interacts with DDX39 but not the C-terminal truncated splice variant DDX39-S, implying that Aly interacts with the C-terminal portion of DDX39 [43]. These data suggest that the posttranslational regulation controling proper levels of DDX39 protein are important for normal cell physiology.
DDX42
DDX42 is a component of 17S U2 snRNP splicing complex and also interacts with the Japanese Encephalitis Virus (JEV) NS4A protein. A functional analysis suggests that the interaction between JEV NS4A and DDX42 impedes an interferon -α/β-mediated innate immune response to JEV infection [44]. The C-terminal 685–938 amino acids interacted with apoptosis-stimulating protein of p53 2 (ASPP2) in a yeast two-hybrid screen. A Ddx42 protein consisting of amino acids 1–737 failed to coprecipitate with ASPP2, confirming that the C-terminal portion was required for its ASPP2 binding ability. DDX42 interaction with ASPP2 interferes with the apoptosis-stimulating properties of ASPP2 [45].
DDX54 (DP97)
DP97 is thought to function in transcriptional regulation, but its other roles are unclear. DP97 interacts with a 14-3-3 protein, in a ligand-dependent manner with estrogen receptors and with other nuclear receptors to represses their transcriptional activity [46]. This interaction was mapped to amino acids 657–865 in the C-terminal region. The transcriptional repression by DP97 maps to amino acids 413–865 in the C-terminal region.
Other DDX proteins
Functional and regulatory data are presently lacking for 23 of the 36 DEAD-box genes in the human genome. However, several recent proteomic analyses have shed light on in vivo posttranslational modification dynamics, phosphorylation in particular, during the cell cycle and in response to a number of different stimuli [47 – 57]. PhosphoSite ® is a curated, sequence-oriented protein database dedicated to in vivo phosphorylation sites that culls all the proteomic data into a searchable platform (http://www.phosphosite.org/). Using this database to search for posttranslational modifications detected on all human DDX proteins, we catalogued their location within either the N-terminal region, DEAD-box helicase conserved core region and C-terminal region (Table 1). While modifications were detected in all regions of the DDX proteins, the substantial majority occurred on their N-terminal and C-terminal regions (Figure 1).
Table 1.
Posttranslational DDX modifications
| Protein | N-terminal | DEAD-box helicase core | C-terminal |
|---|---|---|---|
| DDX1 (DBP-RB) | - | T80, T83 | Y628 |
| DDX2 (eIF4A1) | - | Y126, T158, Y197 | - |
| DDX3 (PL10) | Y52, S60, S61, Y68, S63, S75, S77, S81 S91, Y103, S124, S130, Y162, K117 and K129 |
T203, Y242, Y282, Y300, T322, Y342, Y461 and Y465 |
S590, S593, S611 and S613 |
| DDX4 (Vasa) | S200 and S202 | - | - |
| DDX5 (p68) | K32 and K33 | Y202, Y244, Y297 | T438, S480, T507, Y518, S520, Y593 and Y595, R502 |
| DDX6 (RCK/p54) | T36 and K73 | Y312 and Y313 | Y473 |
| DDX10 | T4, S7 | T255 | S539, T569, T577 and S831 |
| DDX17 (p72) | Y77 | Y281, Y323 | T525, T574, Y584, S603, S605, S675, S676, S680, R688 |
| DDX18 (MrDb) | S74, T130, S140 | T225, S227, Y440 and Y442 | - |
| DDX19B (DBP5) | S86, Y89 and S90 | - | - |
| DDX20 (DP103) | S47 and S48 | S187 | S500, S532, T552, S649, S652, S654, S656, S672, S677, S678, T688, S703, T705, S714, Y756 and Y787 |
| DDX21 (RHII/Gu) | S13, S65, S71 S89, S121, S164, S168, S171, S173 |
T296, T315 | S567, S776 |
| DDX23 (Prp28) | S14, S23, T25, S67, S107, S109 | K642 | - |
| DDX24 | S82, S92, S93, S94, S166, S192 | S287, S295, T302 | - |
| DDX26 (DBI-1) | - | Y351 | S804 |
| DDX27 | S54, S56, S79, S166, S171, T173, Y175, S176 and S177 |
- | S766 |
| DDX28 | S99 | - | - |
| DDX29 | S71, S192, S200 | R647, Y811 | - |
| DDX39 (URH49) | Y13, S37, Y38, S40 | T171, K333 | - |
| DDX41 (Abstrakt) | S21, S23, Y33 | - | - |
| DDX42 (SF3b) | T10, K25, S96, T98, S104, S109, S11, Y160, Y183, S185, T193, |
- | S744, S751, S754, T757, S758 |
| DDX43 (HAGE) | - | Y449, S459, S460 | T592 |
| DDX46 (Prp5) | S40, S79, Y294, S295, S296, K271 |
- | K776, K779, S804 |
| DDX47 | - | - | S424, K448, K451 |
| DDX48 (elF4A-III) | S12 | T163, Y202 | - |
| DDX50 (Gu-beta) | S82 | - | S579, T582, S602 |
| DDX51 | S83, S103 | Y304 | - |
| DDX52 (ROK1-like) | Y35, S39, S99 | - | - |
| DDX54 (DP97) | S34, S39, S41, S71, T74, S75 | - | S644, S782, S788 |
| DDX55 | - | S234, Y305 | K517, K519, K523, S544, S594 |
| DDX56 | - | S126 | - |
| DDX59 | S64, S76 S160 | S246 | - |
- Y, S and T indicate phosphorylation on tyrosine, serine and threonine residues respectfully
- K indicates acetylation on lysine residues
- R indicates methylation on argentine residues
The human DEAD-box RNA helicases represent a large family of enzymes important for most, if not all, aspects of RNA function and regulation. Despite a similar and well conserved catalytic core region, DDX proteins have remarkably different and specialized cellular, tissue and developmental functions. We posit that the divergent flanking N-terminal and C-terminal sequences serve as regions essential for proper DDX target recognition, localization and stability. The human DDX N-terminal and C-terminal sequences vary immensely in their sequence lengths and composition and generally contain no predicted structural motifs. The only notable exceptions are the C-terminal GUCT domains in DDX21 and DDX50, as well as the C-terminal CCHC Zn-knuckle motif in DDX41. The GUCT and CCHC Zn-knuckle domains are predicted to facilitate interactions with RNA. Since all DEAD-box proteins are already thought to bind and unwind RNA, these offer little functional insight. The data reviewed here suggests that, while all parts of a DDX protein are subject to posttranslational regulation, the N-terminal and C-terminal portions are substantially preferred sites for such regulation through both posttranslational modifications and protein-protein interactions. Targeted analyses of these divergent regions using yeast two-hybrid, immunoprecipitation or affinity chromatography approaches will hasten our understanding of how these proteins function and how such functions are regulated.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 3.Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdelhaleem M. RNA helicases: regulators of differentiation. Clin Biochem. 2005;38:499–503. doi: 10.1016/j.clinbiochem.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Godbout R, Li L, Liu RZ, Roy K. Role of DEAD box 1 in retinoblastoma and neuroblastoma. Future Oncol. 2007;3:575–587. doi: 10.2217/14796694.3.5.575. [DOI] [PubMed] [Google Scholar]
- 6.Finn RD, Mistry J, Schuster-Bockler B, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishaq M, Ma L, Wu X, et al. The DEAD-box RNA helicase DDX1 interacts with RelA and enhances nuclear factor kappaB-mediated transcription. J Cell Biochem. 2009;106:296–305. doi: 10.1002/jcb.22004. [DOI] [PubMed] [Google Scholar]
- 8.Yu Z, Sanchez-Velar N, Catrina IE, et al. The cellular HIV-1 Rev cofactor hRIP is required for viral replication. Proc Natl Acad Sci U S A. 2005;102:4027–4032. doi: 10.1073/pnas.0408889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang J, Kubota S, Yang B, Zhou N, Zhang H, Godbout R, Pomerantz RJ. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology. 2004;330:471–480. doi: 10.1016/j.virol.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Monckton EA, Godbout R. A role for DEAD box 1 at DNA double-strand breaks. Mol Cell Biol. 2008;28:6413–6425. doi: 10.1128/MCB.01053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myasnikov AG, Simonetti A, Marzi S, Klaholz BP. Structure-function insights into prokaryotic and eukaryotic translation initiation. Curr Opin Struct Biol. 2009;19:300–309. doi: 10.1016/j.sbi.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Zakowicz H, Yang HS, Stark C, et al. Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI. Rna. 2005;11:261–274. doi: 10.1261/rna.7191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim WJ, Kim JH, Jang SK. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. Embo J. 2007;26:5020–5032. doi: 10.1038/sj.emboj.7601920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low WK, Dang Y, Schneider-Poetsch T, et al. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell. 2005;20:709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 16.Kwong AD, Rao BG, Jeang KT. Viral and cellular RNA helicases as antiviral targets. Nat Rev Drug Discov. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekiguchi T, Kurihara Y, Fukumura J. Phosphorylation of threonine 204 of DEAD-box RNA helicase DDX3 by cyclin B/cdc2 in vitro. Biochem Biophys Res Commun. 2007;356:668–673. doi: 10.1016/j.bbrc.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 18.Soulat D, Burckstummer T, Westermayer S. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. Embo J. 2008;27:2135–2146. doi: 10.1038/emboj.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih JW, Tsai TY, Chao CH, Wu Lee YH. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene. 2008;27:700–714. doi: 10.1038/sj.onc.1210687. [DOI] [PubMed] [Google Scholar]
- 20.Kalverda AP, Thompson GS, Vogel A, et al. Poxvirus K7 protein adopts a Bcl-2 fold: biochemical mapping of its interactions with human DEAD box RNA helicase DDX3. J Mol Biol. 2009;385:843–853. doi: 10.1016/j.jmb.2008.09.048. [DOI] [PubMed] [Google Scholar]
- 21.Schroder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. Embo J. 2008;27:2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 24.Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ. Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum Mol Genet. 15:2324–2334. doi: 10.1093/hmg/ddl158. [DOI] [PubMed] [Google Scholar]
- 25.Shibata N, Tsunekawa N, Okamoto-Ito S, Akasu R, Tokumasu A, Noce T. Mouse RanBPM is a partner gene to a germline specific RNA helicase, mouse vasa homolog protein. Mol Reprod Dev. 2004;67:1–7. doi: 10.1002/mrd.20009. [DOI] [PubMed] [Google Scholar]
- 26.Fuller-Pace FV, Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans. 2008;36:609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- 27.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 28.Jacobs AM, Nicol SM, Hislop RG, et al. SUMO modification of the DEAD box protein p68 modulates its transcriptional activity and promotes its interaction with HDAC1. Oncogene. 2007;26:5866–5876. doi: 10.1038/sj.onc.1210387. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Lin C, Liu ZR. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell. 2006;127:139–155. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, Lin C, Zhao S, Wang H, Liu ZR. Phosphorylation of p68 RNA helicase plays a role in platelet-derived growth factor-induced cell proliferation by up-regulating cyclin D1 and c-Myc expression. J Biol Chem. 2007;282:16811–16819. doi: 10.1074/jbc.M610488200. [DOI] [PubMed] [Google Scholar]
- 31.Wilson BJ, Bates GJ, Nicol SM, Gregory DJ, Perkins ND, Fuller-Pace FV. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol. 2004;5:11. doi: 10.1186/1471-2199-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minshall N, Reiter MH, Weil D, Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J Biol Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- 36.Minshall N, Kress M, Weil D, Standart N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol Biol Cell. 2009;20:2464–2472. doi: 10.1091/mbc.E09-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mouillet JF, Yan X, Ou Q, et al. DEAD-box protein-103 (DP103, Ddx20) is essential for early embryonic development and modulates ovarian morphology and function. Endocrinology. 2008;149:2168–2175. doi: 10.1210/en.2007-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillian AL, Svaren J. The Ddx20/DP103 dead box protein represses transcriptional activation by Egr2/Krox-20. J Biol Chem. 2004;279:9056–9063. doi: 10.1074/jbc.M309308200. [DOI] [PubMed] [Google Scholar]
- 39.Holmstrom TH, Mialon A, Kallio M, et al. c-Jun supports ribosomal RNA processing and nucleolar localization of RNA helicase DDX21. J Biol Chem. 2008;283:7046–7053. doi: 10.1074/jbc.M709613200. [DOI] [PubMed] [Google Scholar]
- 40.Mathew R, Hartmuth K, Mohlmann S, et al. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat Struct Mol Biol. 2008;15:435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 41.Sheng Y, Tsai-Morris CH, Gutti R, Maeda Y, Dufau ML. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is a transport protein involved in gene-specific mRNA export and protein translation during spermatogenesis. J Biol Chem. 2006;281:35048–35056. doi: 10.1074/jbc.M605086200. [DOI] [PubMed] [Google Scholar]
- 42.Sugiura T, Nagano Y, Noguchi Y. DDX39, upregulated in lung squamous cell cancer, displays RNA helicase activities and promotes cancer cell growth. Cancer Biol Ther. 2007;6:957–964. doi: 10.4161/cbt.6.6.4192. [DOI] [PubMed] [Google Scholar]
- 43.Sugiura T, Sakurai K, Nagano Y. Intracellular characterization of DDX39, a novel growth-associated RNA helicase. Exp Cell Res. 2007;313:782–790. doi: 10.1016/j.yexcr.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 44.Lin CW, Cheng CW, Yang TC, et al. Interferon antagonist function of Japanese encephalitis virus NS4A and its interaction with DEAD-box RNA helicase DDX42. Virus Res. 2008;137:49–55. doi: 10.1016/j.virusres.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Uhlmann-Schiffler H, Kiermayer S, Stahl H. The DEAD box protein Ddx42p modulates the function of ASPP2, a stimulator of apoptosis. Oncogene. 2009;28:2065–2073. doi: 10.1038/onc.2009.75. [DOI] [PubMed] [Google Scholar]
- 46.Satoh J, Nanri Y, Yamamura T. Rapid identification of 14-3-3-binding proteins by protein microarray analysis. J Neurosci Methods. 2006;152:278–288. doi: 10.1016/j.jneumeth.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 47.Beausoleil SA, Villen J, Gerber SA, et al. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 48.Olsen JV, Blagoev B, Gnad F, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 49.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 51.Stokes MP, Rush J, Macneill J, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 53.Imami K, Sugiyama N, Kyono Y, et al. Automated phosphoproteome analysis for cultured cancer cells by two-dimensional nanoLC-MS using a calcined titania/C18 biphasic column. Anal Sci. 2008;24:161–166. doi: 10.2116/analsci.24.161. [DOI] [PubMed] [Google Scholar]
- 54.Dephoure N, Zhou C, Villen J, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo A, Villen J, Kornhauser J, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen RQ, Yang QK, Lu BW, et al. CDC25B mediates rapamycin-induced oncogenic responses in cancer cells. Cancer Res. 2009;69:2663–2668. doi: 10.1158/0008-5472.CAN-08-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brill LM, Xiong W, Lee KB, et al. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]