Abstract
Bioactive, patterned micro- and nanoscale surfaces that can be spatially engineered for three-dimensional ligand presentation and sustained release of signaling molecules represent a critical advance for the development of next-generation diagnostic and therapeutic devices. Lithography is ideally suited to patterning such surfaces due to its precise, easily scalable, high-throughput nature; however, to date polymers patterned by these techniques have not demonstrated the capacity for sustained release of bioactive agents. We demonstrate here a class of lithographically-defined, electropolymerized polymers with monodisperse micro- and nanopatterned features capable of sustained release of bioactive drugs and proteins. We show that precise control can be achieved over the loading capacity and release rates of encapsulated agents and illustrate this aspect using a fabricated surface releasing a model antigen (ovalbumin) and a cytokine (interleukin-2) for induction of a specific immune response. We further demonstrate the ability of this technique to enable three-dimensional control over cellular encapsulation. The efficacy of the described approach is buttressed by its simplicity, versatility, and reproducibility, rendering it ideally suited for biomaterials engineering.
Keywords: Bionanotechnology, Biomedical Applications, Drug Delivery, Tissue Engineering, Microstructures
1. Introduction
Entrapment and/or release of various bioactive agents—including low molecular weight drugs, proteins, and cells—from patterned micro- and nanomaterials are critical for the creation of sophisticated bioactive surfaces for multiple biomedical applications.[1-6] Spatial control over ligand presentation and soluble factor release are important features for the realization of biomimicry and recapitulation of complex cell-microenvironment interactions.[7-11] Electropolymerization is ideally suited for this purpose because it is a mild encapsulation procedure that enables high retention of the activity of a labile encapsulated species.[2, 12-14] Due to this feature, this technique has been routinely used in biosensor fabrication for the encapsulation of active enzymes or cells within a porous polymer network for small molecule sensing.[12, 15, 16] Removable or adherent films can be created in this manner,[17] and polymers used in these applications, typically polypyrrole or other conductive derivatives, have demonstrated biocompatibility in vitro[14, 15] and in vivo.[17, 18] Lithographic processing techniques have been used by a number of groups to pattern electrodeposited polymers, although neither sustained release of bioactive agents nor release of bioactive compounds that stimulate cellular function from such polymers has been explicitly demonstrated.[8-10, 13, 19] We hypothesized that lithographical processing, in conjunction with polymer electropolymerization in the presence of active agents, could be used to address this shortcoming. Here, we describe this methodology for the creation of precisely patterned polymers loaded with small molecule agents such as drugs, macromolecules such as proteins, and cells. The attractiveness of this approach lies in the spatial control afforded by lithography, which enables the fabrication of monodisperse systems. Thus, when used in concert with electropolymerization, which enables the creation of multilayered structures, control over the spatiotemporal release of molecules can be achieved by a robust fabrication process.
Electropolymerization proceeds by free radical polymerization of dissolved monomers, which is initiated by electric potential-induced oxidation or reduction at the conductive surface.[11, 16] Pyrroles (Fig. 1A) yield conducting polymers[12, 17] while acrylates (Fig. 1B) produce relatively insulating polymers.[20, 21] Molecules containing these electropolymerizable groups, in addition to reactive,[22] degradable,[23, 24] or amphiphilic[25] moieties, can be polymerized to yield polymers with specific properties and activities. Monomers used in this study are shown in Table 1. Deposition time and electric potential can be tuned to afford precise control over polymer thickness, ranging from the nanoscale to micron thick surfaces. By patterning an insulating layer of photoresist on the chip surface, the lateral dimensions of the electropolymerization reaction are confined to the exposed conductive areas.[14] Thus, sequential electropolymerizations of polymers with different release profiles incorporating different molecules or macromolecules can yield surfaces with precise spatiotemporal control over multiple molecules.
Figure 1.
Elecropolymerization schematics. A) Polypyrrole and B) polyacrylic structure. C) Photoresist (red) patterned indium tin oxide (ITO)-coated glass slide. D) Addition of a gasket to the slide in C filled with the electrodeposition solution consisting of monomer, dopant, molecule to be entrapped and buffer. The working electrode (lower left) contacts the ITO outside the gasket and the counter electrode (upper right) is placed in the deposition solution. Not shown in the schematic is a third, silver/silver chloride reference electrode, which is also placed in the solution. E) During electropolymerization the black polypyrrole film is deposited only on the exposed ITO. F) After gasket removal the patterned polymer can be released from the slide with compressed air. Although the photoresist is shown to be stripped in the image, this step is not required for particle removal, enabling templates to be reused.
Table 1.
Monomers used for electropolymerization experiments (next page).
2. Combining electropolymerization with lithography for the fabrication of micro- and nanopatterned surfaces and particlulates
Patterned electropolymerized surfaces can be achieved by the steps depicted in the schematic shown in Fig. 1C-F. Photoresist is first patterned on a conductive substrate, Fig. 1C, and a gasket placed on the slide is used to form a reservoir into which the electrodeposition solution is added, Fig. 1D. The conducting surface serves as the working electrode and the counter-electrode is placed in the solution. Induction of the electric potential results in electropolymerization of dissolved monomers on exposed areas of the chip, Fig. 1E. Thus, molecules are encapsulated during the polymerization process, which occurs in a buffered aqueous solution, resulting in significant retention of bioactivity. Subsequent removal of the electrodes and gasket followed by chip washing, drying, and photoresist stripping yields a patterned polymer. For particulate fabrication, the patterned polymer is simply removed from the chip with compressed air, Fig. 1F. Particles can also be removed in this way with the photoresist still in place, Supp. Fig. 1A-B, enabling mold reuse, a critical feature for electron-beam (E-beam) lithography, since the required E-beam resist cannot be easily removed.[26] A significant advantage of this method is that it facilitates batch processing—here, chips (a single gasket on a chip is used in the schematics for illustration purposes) attached to a working electrode can be repeatedly used and processed with the same deposition solution resulting in negligible loss of bioactive encapsulant, since the solution can be continuously reused. Particle lateral dimensions can be controlled by resist patterning, while height can be adjusted by deposition time. The scanning electron micrographs in Fig. 2 illustrate this point, depicting control over the fabrication of micro- and nanoscale monodisperse particles using this technique. By varying lithographic dimensions and the duration of electropolymerization, different particle geometries and sizes as small as lithographically attainable (~10-20 nm using E-beam lithography), shown to be important for drug delivery applications,[13, 27] can be precisely engineered.
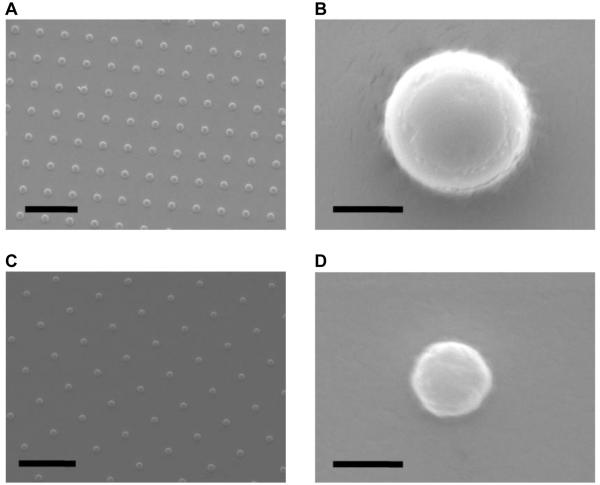
Figure 2.
Images of electropolymerized surfaces. A) Scanning electron micrograph (SEM) of an array of 1 μm-diameter polypyrrole particles (bar = 10 μm). B) SEM of a single 1 μm-diameter polypyrrole particle (bar = 1 μm). C) SEM of an array of 500 nm-diameter polypyrrole particles (bar = 5 μm). D) SEM of a single 500 nm-diameter polypyrrole particle (bar = 1 μm).
3. Control over encapsulation and release of small molecule drugs and analogs from electrodeposited polymers
To assess the capacity of electrodeposited polymers for encapsulation and release of molecules, fluorescent dyes were used as model drugs. A fluorescent micrograph of 1 μm diameter poly-2-methylene-1,3-dioxepane (4, Table 1)[24] particles encapsulating the dye AlexaFluor 568 prior to removal from the chip is shown in Fig. 3A. Monodispersity of the particles produced was assessed by dynamic light scattering (Fig. 3B), and is consistent with those produced by other lithographic methods.[19] Release of sulforhodamine (a hydrophilic dye) from polypyrrole films was strongly dependent on the oxidation potential used for the electropolymerizations (Fig. 3C). By tuning this parameter, both the mass released and the release profile can be adjusted, facilitating flexible controlled release of the molecules. Loadings in the polymers must be estimated because polypyrrole cannot be degraded, preventing precise experimental determinations. For this reason, all dataplots show cumulative mass released. Profilometry was used to determine film thicknesses and loadings are estimated in the text and figure legends based on these values and the concentration of encapsulant present in the elctropolymerization solution. Sulforhodamine loading of the film polymerized at 0.7V was calculated to be 2.24 μg, indicating that incorporated molecules can diffuse out of the polymer and that loading can be controlled by varying the starting drug concentration. The thicknesses of the the three films grown at the three different potentials shown in Fig. 3C are similar (± 0.4 μg), thus loadings should be similar as well. These data suggest, therefore, that the potential has a critical affect on polymerization and enables temporal control over release profiles. Additionally, varying the dopant from adhesion-promoting sodium dodecylbenzenesulfonate (NaDBS) to removable polystyrene sulfonate (PSS)[17] for a set oxidative potential (0.7V) has a minimal effect on loading and release, Supp. Fig. 2B. Taken together, these data show that the variables governing release from electropolymerized films can be easily controlled using this method, suggesting that multiple patterned films or multilayered films can be fabricated and made to release multiple molecules at different rates.
Figure 3.
Particle homogeneity and utility for small molecule release. A) Fluorescent micrograph of 1 μm-diameter poly-MDX particles loaded with AlexaFluor 568 after photoresist removal (bar = 10 μm). B) Normalized particle diameters of 1 μm-diameter-patterned polypyrrole particles. C) Release profiles of sulforhodamine-loaded polypyrrole films electropolymerized at different potentials with NaDBS as the dopant. All films had estimated loadings of 2.2 ± 0.4 μg, thus the released percentages after seven days are 89%, 36%, and 27% for the 0.7V-, 1.1V-, and 1.4V-electropolymerized films, respectively. D) Release profiles of multilayer, 1 μm-diameter polypyrrole particles consisting of an initial AlexaFluor 488-loaded layer electropolymerized at 0.7V and a second layer loaded with AlexaFluor 568, electropolymerized at 1.4V. In both layers PSS was incorporated as the dopant. The theoretical loadings of the 488 and 568 fluorophores were 0.25 μg (~78% released) and 0.28 μg (~32% released), respectively. E) Release profiles of doxorubicin-loaded 1 μm-diameter particles doped with PSS and electrodeposited at 0.7V. The theoretical loading of doxorubicin was 587 ng, thus ~62% was released within seven days. F) Cytotoxicity results for unloaded (MP Alone) and doxorubicin-loaded (MP Dox) 1 μm-diameter particles compared with free doxorubicin (Free Dox). The particles were doped with PSS and electrodeposited at 0.7V and were incubated with A20 cells for 24 hours. The 10 μM MP Dox concentration corresponds to ~2 × 107 particles. Error bars in all plots represent the mean ± 1 standard deviation of three separate batches of polymer.
4. Temporal control over the release of multiple molecules
We exploit the previously described temporal control over release by fabricating particles consisting of two polypyrrole layers, one electropolymerized at 0.7V encapsulating AlexaFluor 488 and the second at 1.1V encapsulating AlexaFluor 568. We used PSS as the dopant in both layers, enabling the fabricated 1 μm diameter particles to be released from the surface. The release profiles of the two fluorophores from these particles are shown in Fig. 3D and are similar to the corresponding datasets in Fig. 3C, demonstrating control over the release of different species at different rates, an important feature for applications involving combination therapies.
To establish the utility of this approach for encapsulation and release of small therapeutic drug molecules, we employed doxorubicin, a broad-spectrum fluorescent chemotherapeutic widely used in clinical settings for a variety of conditions.[29] Sustained release of this small molecule was observed from particles over seven days, Fig. 3E, consistent with release studies of the fluorophores. To assess the effect of the encapsulation procedure on the activity of the drug, we compared the cytoxicity of the released drug and free drug (not encapsulated) on a B cell lymphoma model in vitro. Compared to doxorubicin-loaded particles, blank polypyrrole particles have no cytotoxic effect on the cells over a time period of 24 hours at the doses used in this study, Fig. 3F, indicating polypyrrole biocompatibility.[14, 17, 18] Cytotoxicity of the drug-loaded particles was comparable to free drug, demonstrating that the encapsulation process retained the bioactivity of the free drug. This spatial control over lateral dimensions afforded by electropolymerization, together with its ability to deposit multicomponent stacks, renders the technique ideal for many delivery applications.
5. Creation of complex surfaces regulating cell-cell interactions via ligand patterning and release of signaling molecules: An artificial vaccine node
Spatial regulation over cell adhesion and cell-cell interactions, in concert with the exposure of cells to soluble factors, would provide a fundamentally important technology useful for a wide array of basic science studies and biomedical applications of materials. We next examined the ability of our system to enhance cell-cell interactions by controlling both patterned surface-bound ligand presentation and local factor release. We created a “smart,” adaptable, vaccine-like surface that activates dendritic cells (DCs) to prime an antigen-specific T-cell response. Appropriate antigen-specific T-cell activation requires the interaction of these cells with activated DCs presenting the desired antigen (here ovalbumin, OVA) in the presence of a cytokine molecule (here interleukin-2, IL-2),[30] thus a surface capable of spatial control over delivery of both OVA and IL-2 is required. We modeled our patterned surface using cues from lymph node architecture,[31] which optimizes DC-T-cell interactions. The schematic in Fig. 4A illustrates the organization of our multipolymer surface. The central 100 μm-diameter circular region binds and stimulates T-cells by presenting anti-CD3, an antibody that binds and crosslinks the CD3 antigen receptor present on all T-cells, while simultaneously releasing the costimulant IL-2 in a paracrine fashion.[30] The surrounding region releases OVA and promotes DC binding by presenting anti-CD11c, an antibody that binds the CD11c adhesion ligand present on DCs. Devices consist of arrays of 50 × 50 T-cell binding regions.
Figure 4.
Development and demonstration of an antigen-specific T cell stimulation platform. A) Schematic representation (not to scale) of the completed multicomponent device. The central polymer (black) releases IL-2 and presents anti-CD3 on its surface and the surrounding polymer (gray) releases OVA and presents anti-CD11c. Devices consist of 50 × 50 arrays of 100 μm-diameter regions of the central polymer surrounded by the second polymer. B) Release characteristics of BSA-FITC-loaded polypyrrole (Polypyr) and polypyrrole/poly-1 (Polypyr/Poly-1) films electropolymerized at 0.7V and 1.4V, respectively, with NaDBS as the dopant. BSA-FITC was measured by fluorescence. C) Release characteristics of IL-2-loaded polypyrrole/poly-1 films electropolymerized at 1.4V with NaDBS as the dopant. IL-2 was measured by ELISA (black) and bioactive IL-2 (red) was determined using a splenocyte stimulation assay. D) Fluorescent optical micrograph (FITC and TRITC filters) overlay showing segregation of IgG-AlexaFluor 568 bound to pyrrole/1/2 (1 : 1 : 0.1 ratio) in the central regions and IgG-AlexaFluor 488 bound to 4/acrylic acid (1 : 0.1 ratio) in the periphery. The scale bar represents 100 μm. E) Fluorescent optical micrograph (FITC and TRITC filters) and brightfield overlay of cellular localization of B3Z (stained red) and DC2.4 (green) cells after 72 h incubation with chip described in the text. The array points are polypyrrole/poly-1/poly-2 and the scale bar represents 100 μm. F) IFN-γ release from stimulated B3Z cells measured by ELISA. The Blank Chip consisted of polymers presenting no surface molecules and with no encapsulants that were incubated with cells in the prescence of IL-2 previously treated with OVA. The Chip + IL-2 + OVA was a fully-formed device encapsulating IL-2 and OVA and presenting both anti-CD3 and anti-CD11c. Cells incubated with these devices were not pre-treated with OVA and no exogenous IL-2 was added. Cells Alone were incubated similarly to the other groups but without the presence of a chip, IL-2, or OVA. Cellular stimulation from the loaded and blank chips was statistically significant at the 99.9% confidence level (CL). Error bars represent the mean ± 1 standard deviation.
Fabrication of this complex multicomponent surface requires the sequential electrodeposition of two patterned polymers, each capable of releasing an encapsulated protein (OVA or IL-2) and presenting a bound antibody (anti-CD3 or anti-CD11c). Critically, polyacrylates were the first electrodeposited polymer to ensure these regions were sufficiently insulating such that electrodeposition of the second polymer, performed after photoresist removal, occurred only in surrounding areas and not over the first polymer.[20, 21, 24] To demonstrate the applicability of electropolymerized films in releasing proteins, we used a fluorescently tagged albumin (BSA-FITC) as a model protein and monitored its release from the polymer over three days. We observed that polypyrrole is capable of BSA-FITC release over this time period, Fig. 4B. Previous studies by Mousty et al. showed that the ampiphilic, bifunctional crosslinker 1,1′-(3,3′-(2,2′-oxybis(ethane-2,1-diyl)bis(oxy))bis(propane-3,1-diyl)) bis(1H-pyrrole) (1, Table 1) significantly increased the encapsulation of glucose oxidase in electropolymerized films,[25] thus use of this molecule would be expected to improve loading. As seen in Fig. 4B, the 1 : 1 polypyrrole/poly-1 significantly increased BSA-FITC loading and also improved the release characteristics of the film. The calculated BSA-FITC loading in polypyrrole/poly-1 film is 4.3 μg, suggesting that ~29% of encapsulated proteins are released. This multicomponent polymer is also capable of releasing bioactive IL-2 in quantities required for T cell stimulation[30, 32] over three days, Fig. 4C. The ~27% of encapsulated IL-2 released within three days is consistent with the amount of BSA-FITC released, indicating the robustness of the technique in encapsulation of protein molecules. Poly-4 films are also capable of releasing encapsulated BSA-FITC and IL-2 over seven days, Supp. Fig. 4C-D. By including monomers with reactive sidegroups [such as N-(3-aminopropyl) pyrrole, 2, and acrylic acid, Table 1] during electropolymerization and antibody-monomer conjugates, we created films capable of protein release and ligand presentation (see Supp. Info.).
Next we verified that our platform could be used to activate T-cells (B3Z) via DCs (DC2.4) presenting model antigens. OVA was encapsulated in the polyacrylate layer (1 : 0.1 ratio of 4 : acrylic acid) and anti-CD11c was bound to the surface. The central polymer (1 : 1 : 0.1 ratio of pyrrole : 1 : 2-anti-CD3) was loaded with IL-2 (see Supp. Info.). Despite the spatial segregation of bound antibodies (Fig. 4D), fluorescence imaging after three days of incubation revealed that both cell populations (B3Z and DC2.4) were predominantly present on the polypyrrole islands, Fig. 4E. Both cell populations migrated to the central regions when the polymers were reversed, Supp. Fig. 6A, suggesting that IL-2 stimulation of T-cells may have enhanced production of chemotactic factors by T-cells,[33] propelling the DCs away from the anti-CD11c and towards the IL-2 rich islands. More importantly, the platform was highly effective at stimulating B3Z cells (which are specific to the DC presenting the OVA antigen) as indicated by interferon-γ (IFN-γ) secretion[32] well above non-stimulated controls after three days, Fig. 4F. The device additionally outperformed a control device consisting only of electrodeposited polymers (no proteins) to which DCs previously treated with OVA were added. This finding indicates that the local delivery of OVA to DCs via anti-CD11c sequestering to the antigen presenting substrate led to antigen-specific immune responses. These results are consistent with previous findings from our laboratory, which show that T-cell stimulation is substantially enhanced by paracrine IL-2 delivery from anti-CD3-functionalized microparticles[30] and from surfaces capable of presenting high densities of T-cell stimulating ligands in clusters.[34] By creating a surface capable of both high density ligand presentation and soluble factor paracrine delivery, we can effectively stimulate lymphocytes against a variety of antigens, positioning this technology as a significant advance for the creation of substrates for enhancing ex vivo expansion of lymphoctyes for adoptive immunotherapy applications or vaccine platforms.[35]
6. Cell encapsulation in electropolymerized materials
Administering cells as therapeutics is a very promising new mode of therapy with a wide variety of clinical applications (e.g., cancer, regenerative medicine). Thus, materials capable of encapsulating cells and providing a controlled microenvironment are crucial for the effectiveness of cell therapies.[7, 36, 37] To demonstrate the applicability of the technique for cellular encapsulation important for tissue engineering applications, we utilized alginate-pyrole conjugates (3, Table 1). Alginate-pyrrole conjugates have previously been demonstrated to be electropolymerizable,[22] and we chose this modified polymer for cellular immobilization studies, given the demonstrated utility of alginate in tissue engineering applications.[5, 38] Using BSA-FITC as a model protein, we first demonstrated that poly-3 hydrogels, which are >1 mm in thickness, are capable of sustained BSA-FITC release and release profiles can be readily controlled by post-electropolymerization crosslinking with divalent calcium or zinc,[39] Fig. 5A. Next, we demonstrate the ability of poly-3 gels to immobilize NIH 3T3 cells and human primary osteoblasts. After cellular entrapment, the hydrogels were further crosslinked with calcium chloride at 37°C. Live/dead staining confirmed that cell viability is not compromised by electropolymerization with PSS as the dopant, Fig. 5B-C. A viability assay demonstrated that cells of both types could be immobilized under such conditions with a negligible (< 5%) loss in viability, Fig. 5D. This demonstrated ability to entrap live cells, in concert with the three-dimensional control afforded by electropolymerization, opens the potential for future studies to demonstarate the creation of complex scaffold geometries.
Figure 5.
Scaffold synthesis and osteoblast immobilization. A) Release profile of BSA-FITC-loaded poly-3 hydrogels. After elecctropolymerization labeled groups were further crosslinked with 0.2 M solutions of either Ca2+ or Zn2+. All hydrogels were fully dissolved within five days, thus released masses correspond directly to encapsulated masses. B-C) Fluorescent micrographs (FITC filter) showing live osteoblasts immobilized in poly-3 hydrogels by electropolymerization. After cellular immobilization the gels were further crosslinked with 0.2 M Ca2+ at 37°C and the scaffolds were subsequently incubated in media for a day at 37°C. Live/dead cell staining was performed on B, primary or C, immortalized (3T3) osteoblast samples prepared similarly to those in B. The insets are fluorescent micrographs (TRITC filter) showing dead cells. The insets were taken with the same exposure time. The scale bar represents 100 μm and the inset scale bar represents 100 μm. D) Cytotoxicity results for 3T3 and primary osteoblasts immobilized in hydrogels fabricated as described in A. The results were normalized to cells plated in two-dimensions and treated similarly. Error bars in all plots represent the mean ± 1 standard deviation. All experiments were repeated three times with similar results.
7. Conclusions
Control over patterns of ligand presentation and release of soluble factors from biocompatible material surfaces is a long sought-after capability for multiple biomedical applications. In this article, we demonstrate the utility of electropolymerization in the flexible fabrication of biomaterials for delivery of small molecules and macromolecules and cellular therapeutic applications. The power of this approach is its simplicity and versatility in addition to its amenability to scaling and manufacturability. Thus, the scaling of fabricated particles and multi-component surfaces to any dimension is trivial and arrays of engineered micro- and nanostructures for multiple applications can be easily created.
Traditional methods for encapsulation of labile products, such as proteins and small molecule drugs, often require conditions that result in deactivation or denaturing of the encapsulated agent. Additionally, particulate size dispersity, encapsulation efficiency, and release rates are difficult to control using conventional techniques. Given these limitations, widespread application of particulate preparations and delivery of therapeutic products such as cells and proteins have met with many challenges. Clearly, a facile technique that can be easily implemented based on scalable conventional lithographic approaches and utilizing mild encapsulation conditions is highly desirable. The mild conditions and batch nature of processing using the technique presented in this work results in negligible loss of encapsulant during the encapsulation procedure and indeed makes feasible the entrapment of labile molecules in polymer particulates. This technology additionally enables the immobilization of living cells in electropolymerized hydrogels, rendering the three-dimensional control over cell patterning crucial for many cellular therapy applications possible. Furthermore, the spatiotemporal control over encapsulation and release of multiple agents ranging from small molecules to cells demonstrated here is ideally suited for combination therapies, in which multiple agents, such as small molecules drugs and proteins, are administered to achieve significant therapeutic outcomes.
8. Experimental
Materials
Pyrrole, acrylic acid, doxorubicin, sulforhodamine, BSA-FITC and all other chemicals, unless otherwise noted, were purchased from Sigma. AlexaFluor dyes were purchased from Invitrogen. Alginate was purchased from EMC BioPolymer.
Chemical syntheses
The Supplementary Information section contains a detailed description of chemical syntheses performed in this study.
Lithographic processing
All lithographic processing was performed in the Yale Center for Microelectronic Materials and Structures and the Harvard Center for Nanoscale Systems cleanrooms. All chemicals used for lithographic processing were cleanroom grade. Platinum (99.99%, Kurt J. Lesker Co.) and gold (99.999%, Kurt J. Lesker Co.) depositions were performed using a Sharon Systems Electron-beam evaporator on 2- and 4-inch silicon wafers (Silicon Quest International), which were used for studies requiring adherent films. ITO-coated glass slides (Sigma) were cut into 1 inch × 1 inch squares for processing and were used for all in vitro studies. Clariant AZ5214-E and AZ5218-E photoresists (MicroChem) and an EV Group 620 maskaligner were used for pattern definitions. We found that S1808 and S1813 (MicroChem) did not effectively withstand electropolymerization. Photomasks were purchased from Photo Sciences or Benchmark Technologies. PMMA 950 A4 (MicroChem) was used for E-beam processing using a JEOL 6400 SEM converted to perform direct write.
Electrodepositions
A Gamry Femtostat was used for all electrodepositions, which were performed in a 5 mL vial into which the counter (Pt, Earnest Fullham Inc.) and reference electrodes (Ag/AgCl) were inserted. The reference electrode was fabricated by cathodic deposition of Cl- (from a saturated NaCl solution) on Ag wire (Earnest Fullham Inc.). The working electrode was clipped to an exposed, conductive region of the chip and was inserted into a 2 mL plastic vial. The use of an electrodeposition vial enabled batch processing, minimizing loss of encapsulants. Depositions were performed by constant-voltage amperometry to enable mass calculations. Depositions of pyrrole and pyrrole-terminated monomers were performed at 0.7-1.4 V and depositions of acrylate-terminated monomers were performed at −1.7 V. Electrodepositions were performed serially, thus each datapoint represents a separate “batch.” Details of the layered particle and multicompoent chip fabrication are included in the Supplementary Information section. IL-2 was a generous gift from the S Wrzesinski and R Flavell laboratories.
Release studies
Release studies were performed in the dark at 37°C with constant agitation. Phosphate buffered saline (PBS, 1X; Sigma) was used for all studies except those performed on poly-3, for which PBS with Ca2+ and Mg2+ (Sigma) was used. At each time point the complete volume was removed and replaced with fresh buffer. Samples with particles were centrifuged prior to buffer removal. Fluorescence measurements were taken with a Beckman Coulter platereader and IL-2 and IFN-γ ELISAs (BD Biosciences) were performed according to the manufacturer’s instructions.
Cytotoxicity assays
Cell titer blue (Invitrogen) was used as a cell viability marker according to the manufacturer’s instructions. CHO cells were incubated at 37°C in DMEM (Gibco) with 10% fetal calf serum and 2% penicillin/streptomycin with or without serial dilutions of particles for 24 h after which the cell titer blue reagent was added (20 μL/100 μL volume). The cells were incubated for 4 h at 37°C after which they were pelleted and the fluorescence of the supernatant was measured. A serial dilution of cells was used as the control to quantify numbers of cells. All samples were run in triplicate.
Bioactivity assay
Cumulative releases of 24, 48, and 72 hours of IL-2-loaded 1 : 1 polypyrrole/poly-1 films were performed in complete media [RPMI media (Gibco) with 10 % (v/v) fetal bovine serum (Atlanta Biological) and 2% (v/v) penicillin/streptomycin (Sigma) supplemented with L-glutamine (Sigma), non-essential amino acids (Gibco), Hepes buffer (Sigma), gentamicin (Sigma), and β-mercaptoethanol (Sigma)]. Splenocytes were isolated from a C57BL/6 mouse and 1 × 106 cells were added in 500 μL T cell media to each well of a 24 well plate previously coated with 10 μg/mL anti-CD3 (BD Biosciences, coated in PBS overnight at 4°C). The media from release studies was filtered through a 0.22-μm syringe filter (Whatman) and 500 μL was added to the wells. All wells additionally contained 5 μg/mL soluble anti-CD28 (BD Biosciences). Soluble IL-2 was added in varying concentrations to control wells to create a standard. Cells were incubated at 37°C and cellular stimulation was assessed after 72 hours using an IFN-γ ELISA (BD Biosciences). IFN-γ production was correlated with IL-2 concentration to determine bioactivity.
T-cell activation studies
5 × 105 DC2.4 and 5 × 105 B3Z cells (T hybridoma cells specific for the OVA peptide OVA257-264) were cultured on UV-sterilized devices in each well of a 12-well plate in RPMI media with 10% fetal calf serum and 2% penicillin/streptomycin. Where applicable, exogenous IL-2 (50 ng/well; BD Biosciences) and OVA (2 mg/well; Sigma) were added to the media. At 72 hours, supernatant was analyzed for IFN-γ by ELISA. Each sample was performed in triplicate. For imaging studies, DC2.4 cells were loaded with carboxyfluorescein diacetate, succinimidyl ester (CFDA-SE; Molecular Probes) according to the Cell Tracer Kit specifications prior to use. Post-incubation, devices were washed in 1X PBS, incubated for 1 hour on ice in 2 μg/ml CD8-PE (BD Biosciences), washed thrice in 1X PBS for 5 minutes, and fixed for 20 minutes in 4% paraformaldehyde. Devices were mounted on cover slips with hard-set Vectashield for visualization by fluorescence microscopy.
Cellular immobilization studies
NIH 3T3 and primary osteoblast cells were suspended at a concentration of 2 × 107 cells/mL in phenol red-free Dulbecco’s Modified Eagle Media (DMEM; Gibco). Cells were maintained at 37°C and mixed with a solution of compound 1 and PSS (Sigma) in phenol red-free DMEM (Gibco) directly prior to the onset of electropolymerization, which occurred at room temperature. Chips used for hydrogel electrodeposition were UV-sterilized directly prior to use. The seeded hydrogels, which remained on the chips, were immediately further crosslinked with 0.2 M CaCl2 (Sigma) in phenol red-free DMEM for 5 min at 37°C. The hydrogels were then incubated in fresh media for 24 h at 37°C, at which time either a cell viability assay or live/dead staining (Invitrogen) was performed. Cell titer blue was used to assess viability as described above. Live/dead staining was performed according to the manufacturer’s instructions. Briefly, cells were first stained with a 0.5% solution of calcein-AM and 0.5% dimethylsulfoxide (DMSO; sigma) in PBS. The stain was warmed to 37°C and incubated with the sample for 20 min at room temperature, after which the sample was washed thrice with PBS. The cells were then stained with a 0.5% propidium iodide solution in PBS for 10 min at room temperature and again washed three times with PBS. After staining, cells were fixed in zinc-formalin at 4°C for 20 min. After three washes of 5 min each in PBS, the samples were mounted on cover slips with hard-set Vectashield for fluorescence microscopy imaging.
Supplementary Material
Acknowledgments
[**] This work was performed in part in the Harvard Center for Nanoscale Systems and the Yale Center for Microelectronic Materials and Structures. The authors would like to thank Alec Flyer, Nathan Huebsch, Rebecca Robinson, Stephen Wrezesinski, Andrea Brock, Rosalba Camicia, and Erin Steenblock for experimental assistance; Stephen Wrezesinski and Richard Flavell for the generous gift of recombinant human IL-2; Jason Criscione and James Bertram for many helpful discussions; W Mark Saltzman for departmental support; and David Stern for critical reading of the manuscript. The work was supported in part by NIH through grants R01-HL085416 (M.A.R and T.M.F), R01-EB008260, and R37 DE013033 (D.J.M.) and the Director’s New Innovator Award Program (DP22OD002913; D.A.S.); a Public Health Grant (HL-55397; T.M.); a NSF CAREER grant (T.M.F); and Yale University (D.A.S.). Supporting Information is available online from Wiley InterScience.
This paper is dedicated in loving memory to Alan R Stern
References
- [1].Lahann J. Chem Eng Comm. 2006;193:1457. [Google Scholar]
- [2].Langer R, Tirrell DA. Nature. 2004;428 doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- [3].Mossman KD, Campi G, Groves JT, Dustin ML. Science. 2005;310:1191. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- [4].Patolsky F, Timko BP, Yu G, Fang Y, Greytak AB, Zheng G, Lieber CM. Science. 2007;313:1100. doi: 10.1126/science.1128640. [DOI] [PubMed] [Google Scholar]
- [5].Cao L, Mooney DJ. Adv Drug Del Rev. 2007;59:1340. doi: 10.1016/j.addr.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mrksich M. Curr Opin Chem Biol. 2002;6:794. doi: 10.1016/s1367-5931(02)00362-9. [DOI] [PubMed] [Google Scholar]
- [7].Panda P, Ali S, Lo E, Chung BG, AHatton TA, Khademhosseini A, Doyle PS. Lab Chip. 2008;8:1056. doi: 10.1039/b804234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Glangchai LC, Caldorera-Moore M, Shi L, Roy K. J Control Rel. 2008;125:263. doi: 10.1016/j.jconrel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- [9].Guan J, Ferrell N, Lee LJ, Hansford DJ. Biomaterials. 2006;27:4034. doi: 10.1016/j.biomaterials.2006.03.011. [DOI] [PubMed] [Google Scholar]
- [10].Rolland JP, Maynor BW, Euliss LE, Exner AE, Denison GM, DeSimone JM. J Am Chem Soc. 2005;127:10096. doi: 10.1021/ja051977c. [DOI] [PubMed] [Google Scholar]
- [11].Stern E, Jay S, Bertram J, Boese B, Kretzschmar I, Turner-Evans D, Dietz C, LaVan DA, Malinski T, Fahmy TM, Reed MA. Anal Chem. 2006;78:6340. doi: 10.1021/ac060410r. [DOI] [PubMed] [Google Scholar]
- [12].Cosnier S. Biosens Bioelectron. 1999;14:443. doi: 10.1016/s0956-5663(99)00024-x. [DOI] [PubMed] [Google Scholar]
- [13].Euliss LE, DuPont JA, Gratton SEA, DeSimone JM. Chem Soc Rev. 2006;35:1095. doi: 10.1039/b600913c. [DOI] [PubMed] [Google Scholar]
- [14].Gomez N, Lee JY, Nickels JD, Schmidt CE. Adv Funct Mater. 2007;17:1645. doi: 10.1002/adfm.200600669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Deshpande MV, Hall EAH. Biosens Bioelectron. 1990;5:431. [Google Scholar]
- [16].Cosnier S. Anal Bioanal Chem. 2003;377:507. doi: 10.1007/s00216-003-2131-7. [DOI] [PubMed] [Google Scholar]
- [17].George PM, Lyckman AW, LaVan DA, Hegde A, Leung Y, Avasare R, Testa C, Alexander PM, Langer R, Sur M. Biomaterials. 2005;26:3511. doi: 10.1016/j.biomaterials.2004.09.037. [DOI] [PubMed] [Google Scholar]
- [18].Jiang X, Marois Y, Traore A, Tessier D, Dao LH, Guidoin R, Zhang Z. Tissue Eng. 2002;8:635. doi: 10.1089/107632702760240553. [DOI] [PubMed] [Google Scholar]
- [19].Gratton SEA, Pohlhaus PD, Lee J, Guo J, Cho MJ, DeSimone JM. J Control Rel. 2007;121:10. doi: 10.1016/j.jconrel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Katz E, de Lacey AL, Fierro JLG, Palacios JM, Fernandez VM. J Electroanal Chem. 1993;358:247. [Google Scholar]
- [21].Wang X, Haasch RT, Bohn PW. Langmuir. 2005;21:8452. doi: 10.1021/la0580053. [DOI] [PubMed] [Google Scholar]
- [22].Abu-Rabeah K, Polyak B, Ionescu RE, Cosneir S, Marks RS. Biomacromolecules. 2005;6:3313. doi: 10.1021/bm050339j. [DOI] [PubMed] [Google Scholar]
- [23].Zelikin AN, Lynn DM, Farhadi J, Martin I, Shastri V, Langer R. Angew Chem Int Ed. 2002;41:141. doi: 10.1002/1521-3773(20020104)41:1<141::aid-anie141>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- [24].Bailey WJ, Ni Z, Wu S-R. J Polym Sci: Polym Chem Ed. 1982;20:3021. [Google Scholar]
- [25].Mousty C, Galland B, Cosnier S. Electroanal. 2001;13:186. [Google Scholar]
- [26].Madou MJ. Fundamentals of microfabrication: The science of miniaturization. CRC Press; New York: 2002. [Google Scholar]
- [27].Muro S, Garnacho C, Champion J, Leferovich J, Gajewski C, Schuchman E, Mitragotri S, Muzykantov V. Mol Ther. 2008;2008:1450. doi: 10.1038/mt.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Holdcroft S, Funt BL. J Electroanal Chem. 1988;240:89. [Google Scholar]
- [29].Furgeson DY, Dreher MR, Chilkoti A. J Control Rel. 2006;110:362. doi: 10.1016/j.jconrel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- [30].Steenblock ER, Fahmy TM. Mol Ther. 2008;16:765. doi: 10.1038/mt.2008.11. [DOI] [PubMed] [Google Scholar]
- [31].Miller MJ, Wei SH, Parker I, Cahalan MD. Science. 2002;296:1869. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- [32].Kruisbeek AM. Curr Protocols Immunol. 2006 Chap 3. [Google Scholar]
- [33].Loetscher P, Seitz M, Baggiolini M, Moser B. J Exp Med. 1996;184:569. doi: 10.1084/jem.184.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fadel TR, Steenblock ER, Stern E, Li N, Wang X, Haller GL, Pfefferle LD, Fahmy TM. Nano Lett. 2008;8:2070. doi: 10.1021/nl080332i. [DOI] [PubMed] [Google Scholar]
- [35].Dudley ME, Rosenberg SA. Semin Oncol. 2007;34:524. doi: 10.1053/j.seminoncol.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Child JA, Morgan GJ, Davies FE, Owen RG, Bell SE, Hawkins K, Brown J, Drayson MT, Selby PJ. New Engl J Med. 2003;348:1875. doi: 10.1056/NEJMoa022340. [DOI] [PubMed] [Google Scholar]
- [37].Couzin J, Vogel G. Science. 2004;304:192. doi: 10.1126/science.304.5668.192. [DOI] [PubMed] [Google Scholar]
- [38].Silva EA, Kim E-S, Kong HJ, Mooney DJ. Proc Natl Acad Sci USA. 2008;105:14347. doi: 10.1073/pnas.0803873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jay SM, Shepherd BR, Bertram JP, Pober J, Saltzman S,WM. FASEB J. 2008;22:2949. doi: 10.1096/fj.08-108803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bott AW, Heineman WR. Current Separations. 2004;20:121. [Google Scholar]
- [41].Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Anal Chem. 1998;70:4974. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- [42].Hermanson GT. Bioconjugate Techniques. Academic Press; New York: 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.