Abstract
The analysis of health data and putative covariates, such as environmental, socioeconomic, demographic, behavioral, or occupational factors, is a promising application for geostatistics. Transferring methods originally developed for the analysis of earth properties to health science, however, presents several methodological and technical challenges. These arise because health data are typically aggregated over irregular spatial supports (e.g., counties) and consist of a numerator and a denominator (i.e., rates). This article provides an overview of geostatistical methods tailored specifically to the characteristics of areal health data, with an application to lung cancer mortality rates in 688 U.S. counties of the southeast (1970–1994). Factorial Poisson kriging can filter short-scale variation and noise, which can be large in sparsely populated counties, to reveal similar regional patterns for male and female cancer mortality that correlate well with proximity to shipyards. Rate uncertainty was transferred through local cluster analysis using stochastic simulation, allowing the computation of the likelihood of clusters of low or high cancer mortality. Accounting for population size and rate uncertainty led to the detection of new clusters of high mortality around Oak Ridge National Laboratory for both sexes, in counties with high concentrations of pig farms and paper mill industries for males (occupational exposure) and in the vicinity of Atlanta for females.
Introduction
Cancer is a major public health problem in the United States. In spite of the recent decline in cancer mortality, which is largely attributed to the reduction in smoking and improvements in cancer screening, cancer is still the second most common cause of death. Geographic Information Systems (GISs) are used increasingly for cancer control activities and resource allocation. Cancer atlases are now published by national and state health agencies and have proved useful for quantifying patterns in cancer rates such as incidence and mortality, documenting access to health care, providing tools for risk communication, and assessing disparities in cancer burdens in underserved populations (Devesa et al. 1999; Pickle et al. 1999; Greiling et al. 2005). The major difficulty in the analysis of health outcomes is that the patterns observed reflect the influence of a complex combination of demographic, social, economic, cultural, and environmental factors that are likely to change through time and space, and that interact with the different types and scales of places where people live (Tunstall, Shaw, and Dorling 2004). Thus, there is a need for an integrated approach that allows the mapping of regional trends in cancer rates, and the accurate estimation of rates over the small census areas commonly used in contextual analysis. Another challenge for environmental epidemiology is the analysis and synthesis of spatial data collected at different spatial scales and over different spatial supports. For example, one might want to explore relationships between health outcomes aggregated to the zip code level, census-tract demographic covariates, and air pollution data measured at a few monitoring stations. This article discusses the application of geostatistics to a few key steps in the analysis of cancer rates: estimation of the underlying disease risk, mapping of regional trends and local departures, and detection of areas with significantly higher or lower risk.
Mapping and interpreting cancer mortality or incidence rates face three major hurdles: (1) the presence of unreliable rates that occur for sparsely populated areas and/or rare cancers, (2) the visual bias caused by the aggregation of health data within administrative units of widely different sizes and shapes, and (3) the mismatch of spatial supports for cancer rates and explanatory variables that prevent their direct use in correlation analysis (Goovaerts 2009). Smoothing methods, ranging from simple deterministic techniques to sophisticated full Bayesian models, have been developed to improve the reliability of observed rates by using (or “borrowing”) information from neighboring entities (Waller and Gotway 2004). Bayesian methods are increasingly popular, but they often require large amounts of computer time and careful fine-tuning, making their application and interpretation challenging for nonstatisticians (Woodward 2005). Another shortcoming is that the random effect associated with spatial autocorrelation is typically defined using the conditional autoregressive (CAR) model. This is computationally convenient but not well suited to situations where geographical entities have different sizes and shapes and are not arranged in a regular pattern (Kelsall and Wakefield 2002). Last, according to simulation studies Bayesian disease-mapping models, in particular, the so-called BYM model (based on Besag, York, and Mollie 1991), cause strong smoothing of observed rates, which limits their ability to detect localized increases in risk (Richardson et al. 2004).
Geostatistics provides a model-based approach that is of intermediate difficulty in terms of implementation and computer requirements. Although it was developed the same year as the BYM model, the first initiative to tailor variogram and kriging to the analysis of disease rates (Lajaunie 1991) went largely unnoticed. The method, known as binomial cokriging, was used in only a few studies, such as mapping the risk of childhood cancer in the West Midlands of England (Oliver et al. 1992, 1998; Webster et al. 1994) and lung cancer mortality in Long Island (Goovaerts 2005a). A similar approach, known as Poisson kriging, was developed more recently in the field of marine ecology (Monestiez et al. 2006) and generalized to the analysis of cancer mortality and cholera incidence data (Goovaerts 2005b; Ali et al. 2006). Unlike the CAR model, the geostatistical models (Goovaerts 2006b) can easily incorporate the geometry of administrative units and the spatial repartition of the population at risk, leading to more precise and accurate estimates of the risk than the Bayesian BYM model (Goovaerts and Gebreab 2008). Area-to-point (ATP) Poisson kriging enables isopleth maps of mortality risk to be produced, which attenuates the visual bias associated with the interpretation of choropleth maps. With the latter, the user tends to assign more importance to larger polygons even though they typically correspond to rural areas with smaller populations at risk. Another major advantage of the geostatistical method is that it goes beyond the filtering of noise and can decompose the structure in the variation according to the spatial scales identified. Mapping local and regional spatial components can help identify potential factors responsible for the spatial distribution of mortality rates at different scales (Goovaerts, Jacquez, and Greiling 2005).
A limitation of all least-squares estimators, including Poisson kriging, is the loss of local detail in the spatial variation of the risk in the maps. This smoothing has serious implications for local cluster analysis (LCA): the sizes of clusters of low or high cancer risk tend to be inflated, and most of the spatial outliers are filtered out. Single maps of estimated risk and kriging variance also do not allow the propagation of rate uncertainty through multiple-point statistics, that is, statistics computed from observations made at multiple locations, such as the local Moran's I that measures the correlation between each rate and the rates measured in adjacent geographical units. Goovaerts (2006a) proposed combining Poisson kriging with a geostatistical simulation algorithm to generate multiple realizations of the spatial distribution of risk values. The set of simulated fields enables quantification of how the spatial uncertainty about rates translates into uncertainty about the location of disease clusters (Goovaerts 2006a), the presence of significant boundaries (Goovaerts 2008a), and the relationship between health outcomes and putative risk factors (Goovaerts 2009).
Another value of geostatistical simulation is the generation of more realistic null hypotheses for statistical tests that are performed routinely by health scientists; for example, to detect clusters of adjacent geographical units with high cancer mortality or to identify putative sociodemographic or environmental factors. Most tests are still based on the null hypothesis of spatial independence (SI) of observed rates and, provided the population sizes of areal units are fairly homogeneous, on the assumption of constant or spatially uniform risk. Some spatial pattern is almost always present; therefore, rejecting this hypothesis has little scientific value. The concept of a “neutral model” (Waller and Jacquez 1995; Fortin and Jacquez 2000; Goovaerts and Jacquez 2004) means that more interesting hypotheses can be tested by replacing the null hypothesis of spatial randomness and uniform risk with models that account for spatial patterns and a priori information on the underlying risk. Geostatistical neutral models proved useful for many types of applications, such as (1) the detection of significant clusters or outliers of breast cancer rates above and beyond the risk inferred from environmental covariates on Long Island, NY (Goovaerts 2005a); (2) the identification of significant spatio-temporal changes in cervical cancer mortality rates above and beyond past spatial patterns (Goovaerts and Jacquez 2005); (3) the assessment of significant clustering of residential histories in a case–control study of bladder cancer in Michigan (Jacquez et al. 2006); (4) the detection of significant changes in pancreatic cancer mortality across county boundaries (Goovaerts 2008a); and (5) the study of the impact of demographic and economic factors on cervical cancer mortality in the western United States (Goovaerts 2009).
The problem
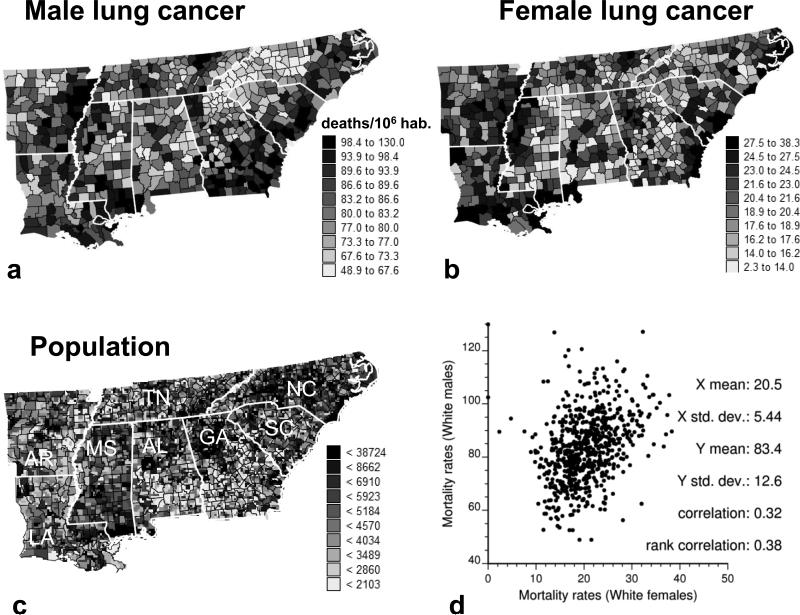
The geostatistical analysis of health data is illustrated with an example for lung cancer, which has been the leading cause of cancer deaths in the United States for several decades. Figure 1a and b show maps of age-adjusted mortality rates for White males and White females recorded over the period 1970–1994 for 688 counties of the southeastern United States. The population-weighted average mortality rate for males is 82.7 deaths per 100,000 person-years, which is almost four times larger than for females (22.4). Although both maps have some similar features, the correlation between mortality rates is rather weak: 0.37 (Fig. 1d). The aims of the analysis are as follows:
create a reliable map of the spatial distribution of cancer mortality that accounts for small population sizes and the counties’ geographies,
separate regional trends in cancer mortality from local patterns and assess similarities between male and female spatial patterns at these two different scales, and
identify groups of adjacent counties (i.e., local clusters) with significantly correlated low or high mortality rates.
Figure 1.
Lung cancer mortality rates recorded. (a) Top left: White males. (b) Top right: White females for the period 1970–1994 in 688 counties. (c) Bottom left: population at risk assigned to 100 km2 cells. (d) Bottom right: scatterplot illustrates the moderate correlation between mortality rates for both sexes. Thick white lines delineate eight state boundaries.
Mapping the risk of cancer
Spatial analysis of cancer risk typically starts with a posting of the data, except when too few deaths are recorded to compute meaningful rates (e.g., rates based on small counts are masked and recorded as “sparse data” in the cancer atlas published by the National Cancer Institute). Creating choropleth maps of mortality appears straightforward because they do not include any missing values. Maps of raw rates can show erratic fluctuation among adjacent geographical units, as illustrated by the county maps of Fig. 1a and b. Although this noise might reflect local factors that change abruptly across county boundaries, it is typically caused by the “small number problem” (Waller and Gotway 2004) whereby rates computed for sparsely populated areas tend to be less reliable. Ignoring the small number problem might lead to spurious conclusions when investigating the existence of local clusters and outliers of high or low cancer mortality. Map interpretation is also biased by the large variation in the spatial support (e.g., county area and shape) of observations across the study area. For example, the areas and population sizes for 688 counties in Fig. 1 vary by one and two orders of magnitude, respectively. Area-to-area (ATA) and ATP Poisson kriging filter the spatially varying noise and account for the heterogeneity in shape, size, and population distribution among counties.
Poisson kriging
For a given number, N, of geographical units vα (e.g., counties), denote the observed mortality rates (areal data) as z(vα) = d(vα)/n(vα), where d(vα) is the number of deaths and n(vα) is the size of the population at risk. The death count d(vα) is interpreted as a realization of a random variable D(vα) that follows a Poisson distribution with one parameter (expected number of counts) that is the product of the population size n(vα) and local risk r(vα). This local risk can be estimated as the following linear combination of the rate z(vα) and the rates observed in (K – 1) neighboring entities vi:
| (1) |
The weights λi assigned to the K rates are computed by solving the following system of linear equations, known as the Poisson kriging system:
| (2) |
where δij = 1 if i = j and 0 otherwise, and m* is the population-weighted mean of the N rates. The term μ(vα) is a Lagrange parameter that results from the minimization of the estimation variance subject to the unbiasedness constraint on the estimator. The “error variance” term, m*/n(vi), leads to smaller weights for less reliable data (i.e., rates measured over smaller populations). This system can be viewed as a particular case of kriging with nonsystematic errors (Chiles and Delfiner 1999, p. 210) where the objective is to filter the noise due to the small number problem from the observed rates. In addition to the population size, the kriging system accounts for the spatial correlation among geographical units through the ATA covariance terms C̄R(vi, vj)= Cov{Z(vi), Z(vj)} and C̄R(vi, vα). Those covariances are numerically approximated by averaging the point-support covariance C(h) computed between any two locations discretizing the areas vi and vj:
| (3) |
where Pi and Pj are the number of points used to discretize the two areas vi and vj, respectively. In this study the discretizing points were the centroids of the grid shown in Fig. 1c. The weights, wss′, are computed as the product of population sizes assigned to each discretizing point us and :
The uncertainty about the cancer mortality risk prevailing within the geographical unit vα can be modeled using the conditional cumulative distribution function (CCDF) of the risk variable R, Prob{R(vα) ≤ r|{z(vi),i = 1, . . . , K}}. Under the assumption of normality of the prediction errors, that CCDF is modeled as a Gaussian distribution with the mean and variance corresponding to the Poisson kriging estimate and variance computed as
| (4) |
where C̄R(vα, vα) is the within-area covariance that is computed according to equation (3) with vi = vj = vα.
ATP Poisson kriging adopts similar expressions for the kriging estimate and variance (equations (1) and (4)), except that the prediction support is now a point us instead of the area vα. An important property of the ATP kriging estimator is its coherence (Kyriakidis 2004): the population-weighted average of the risk values estimated at the Pα points us discretizing a given entity vα gives the ATA risk estimate for this entity
| (5) |
Constraint (5) is satisfied if the same K areal data are used for both the ATA kriging of r̂PK(vα) and the ATP kriging of each of the point values r̂PK(us).
Deconvolution of the variogram of the risk
Both ATA and ATP kriging require knowledge of the point-support covariance of the risk C(h), or equivalently the variogram γ(h). This function cannot be estimated directly from the observed rates, because only areal data are available. Thus, only the areal variogram of the risk can be estimated
| (6) |
where N(h) is the number of pairs of areas (vα, vβ) whose population-weighted centroids are separated by the vector h. The different spatial increments [z(vα) – z(vβ)]2 are weighted by a function of their respective population sizes, n(vα)n(vβ)/[n(vα)+n(vβ)], which is a term that is inversely proportional to their standard deviations (Monestiez et al. 2006). Thus, more importance is given to the more reliable data pairs (i.e., smaller standard deviations).
Derivation of a point-support variogram γ(h) from the variogram γR(h) fitted to areal data is known as deconvolution. The reverse operation, that is, the derivation of the areal variogram from a point-support variogram, is called regularization. Although deconvolution and regularization are common operations in geostatistics, the methods available were developed for regular areas or blocks, as in mining applications (e.g., Journel and Huijbregts 1978). In this article, the iterative procedure introduced for rate data measured over irregular geographical units is adopted whereby the point-support model sought, once regularized, is the closest to the model fitted to the areal data. See Goovaerts (2006b, 2008b) for more detail and the results of simulation studies.
Mapping regional background and local patterns
For large areas, such as the region formed by the group of 688 counties analyzed in this article, the spatial distribution of cancer mortality rates is likely to be influenced by a series of factors related, for example, to demography, economy, diet, smoking behavior, and environment. If the scales at which these different factors operate are very different from one another, then they should be apparent in the parameters of the model fitted to the variogram of the risk. For lung cancer, both the areal and the point-support models fitted for each sex is the sum of two models (a nested model): γ(h) = γlocal(h)+γregional(h). The range of the small-scale component of the point-support model is 46 km for males and 42 km for females, which is one order of magnitude smaller than for the long-range component of 428 km for males and 920 km for females. Based on the nested point-support variogram model, the estimate of risk (equation [1]) can be decomposed into the sum of a local and a regional component
| (7) |
The spatial components are still estimated as linear combinations of rates recorded in neighboring counties (equation [1]), but the weights are computed by solving the following two systems of equations
| (8) |
| (9) |
This decomposition is simply a generalization of factorial kriging analysis (Wackernagel 1998) to Poisson kriging. The unbiasedness constraints result in zero mean local components, whereas the regional component incorporates the local trend.
Detection of local clusters of high and low mortality
A major goal of spatial analysis in public health is to detect local clusters (regions where adjacent areas have similar values) of high or low cancer mortality. Similarity between the rate measured within area vα and those recorded in J(vα) adjacent areas vβ (e.g., units sharing a common border or vertex with the kernel vα) is often quantified by the local Moran's I statistic (Anselin 1995) defined as
| (10) |
where m and s are the mean and standard deviation of the set of N rates. This local indicator of spatial association (LISA) is simply the product of the kernel rate and the average of neighboring rates; it can detect both positive and negative autocorrelations. It exceeds zero if the kernel and neighborhood averaged rates jointly exceed the global mean m (High–High, HH cluster) or are jointly below m (Low–Low, LL cluster). Despite its widespread use, this statistic suffers from several limitations, such as the arbitrary use of the global mean to detect local clusters of low or high values, the lack of power compared to other clustering tests (Song and Kulldorff 2003), and the use of predefined neighborhoods like first- or second-order adjacencies, which makes it less sensitive to the detection of clusters of different shapes or that occur at different spatial scales (Greiling et al. 2005).
Propagating the uncertainty through the LCA
Equation (10) is limited because it ignores the uncertainty attached to the mortality rates. Several modifications of the local Moran's I hypothesis test have been proposed to take into account the small number problem (e.g., see Oden 1995; Waldhör 1996; Assunçao and Reis 1999). More recently, Anselin, Syabri, and Kho (2006) suggested transforming or standardizing the rates before applying the test, thereby removing much of the noise caused by the small population. This approach, however, tends to inflate artificially the size of the clusters because the smoothing imparts autocorrelation to the risk estimates through the averaging of mortality rates recorded in neighboring geographical units.
Goovaerts (2006a) proposed a p-field simulation-based approach to account for rate uncertainty in LCA. First, the uncertainty attached to the spatial distribution of mortality risk values is modeled through the generation of a set of L equally probable simulated maps, {r(l)(vα), α = 1, . . . , N; l = 1, . . . , L}, each consistent with the information available, such as a histogram or a spatial correlation function. Second, the uncertainty is propagated through the LISA statistic by replacing the rates z(vα) in equation (10) by the simulated risk values, leading to a set of L simulated LISA values {I(l)(vα), l = 1, . . . L}. In other words, the correlation of each county with adjacent counties is tested L times, enabling the determination of the probability of a county belonging to an LL or HH cluster. Unlike estimation, simulation reproduces the spatial variability of the data; therefore, it avoids the aforementioned smoothing effect and inflation of cluster size.
The p-field simulation approach proceeds in two steps: (1) the uncertainty about the cancer mortality risk prevailing within each of the N geographical units vα is modeled using the CCDF of the risk variable R(vα), and (2) risk values r(l)(vα) are simulated through the sampling of the set of N CCDFs by a set of N spatially correlated probability values {p(l)(vα), α = 1, . . . , N}, known as a probability field or p-field. Because the CCDFs are Gaussian—recall equation (4)—each risk value r(l)(vα) is simply computed as r(l)(vα) = r̂(vα) + σ(vα)y(l)(vα), where r̂(vα) and σ(vα) are the kriging estimate and kriging standard deviation computed according to equations (1) and (4), and y(l)(vα) is the quantile of the standard normal distribution corresponding to the cumulative probability p(l)(vα). The L sets of normal scores, {y(l)(vα), α = 1, . . . N}, are generated by nonconditional sequential Gaussian simulation with the variogram of the risk, γR(h), rescaled to a unit sill (see Goovaerts 2006a for a detailed description of the p-field simulation algorithm).
Identifying significant clusters
To test whether any test statistic, I(l)(vα), is significantly greater than 0 (i.e., presence of spatial autocorrelation), one needs to know its probability distribution under the null hypothesis of SI. The common way to generate such reference distributions is to shuffle the set of simulated rates randomly and then to use the shuffled values to compute the neighborhood average in statistic (10) while the kernel rate remains the same. In other words, the LISA statistic is computed for randomly distributed rates in adjacent areas. The main drawback of this randomization procedure is that both the underlying mortality risk and the population size are assumed uniform across the study area. To account for the population size, the random shuffling is replaced by the random sampling of a Poisson distribution Po(n(vj) × m), where n(vj) is the size of the population at risk and m is the population-weighted average of rates. This operation is repeated K times (K = 999 in this article) to compute the P-value of the test. Because the statistical test is repeated for each county, there is an increased likelihood of false positives (i.e., risk of rejecting the null hypothesis when it is true). In this article, the multiple testing correction was done using the false discovery rate (FDR) approach, which aims to control the expected proportion of true null hypotheses that will be rejected (Castro and Singer 2006); in other words, the objective is to limit the risk of false positives.
Case study
Mapping lung cancer mortality
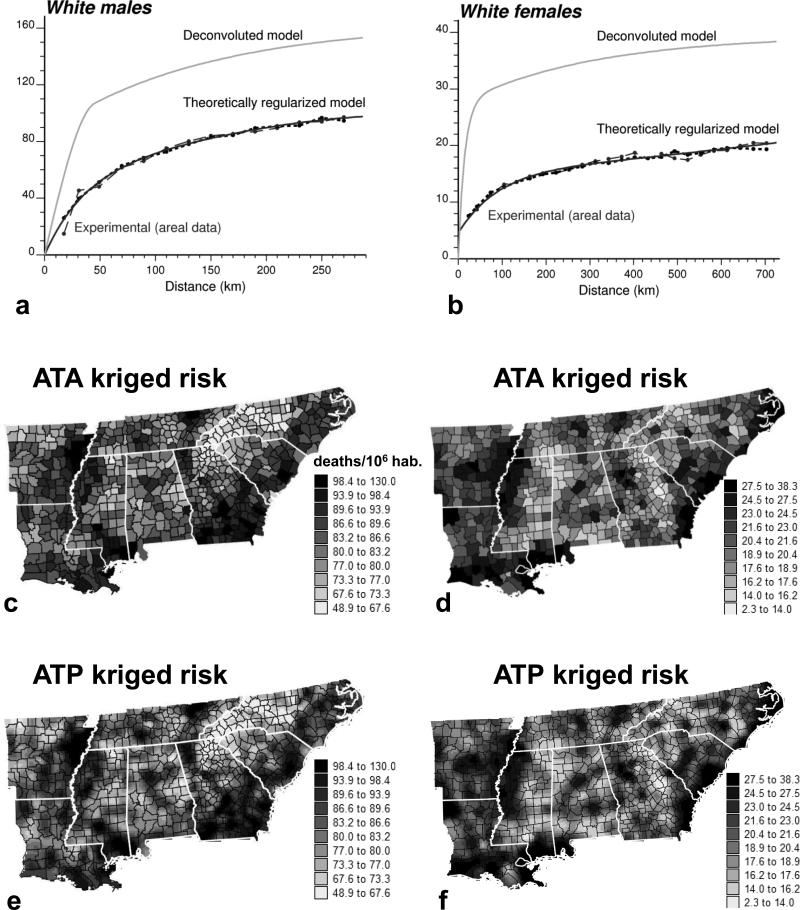
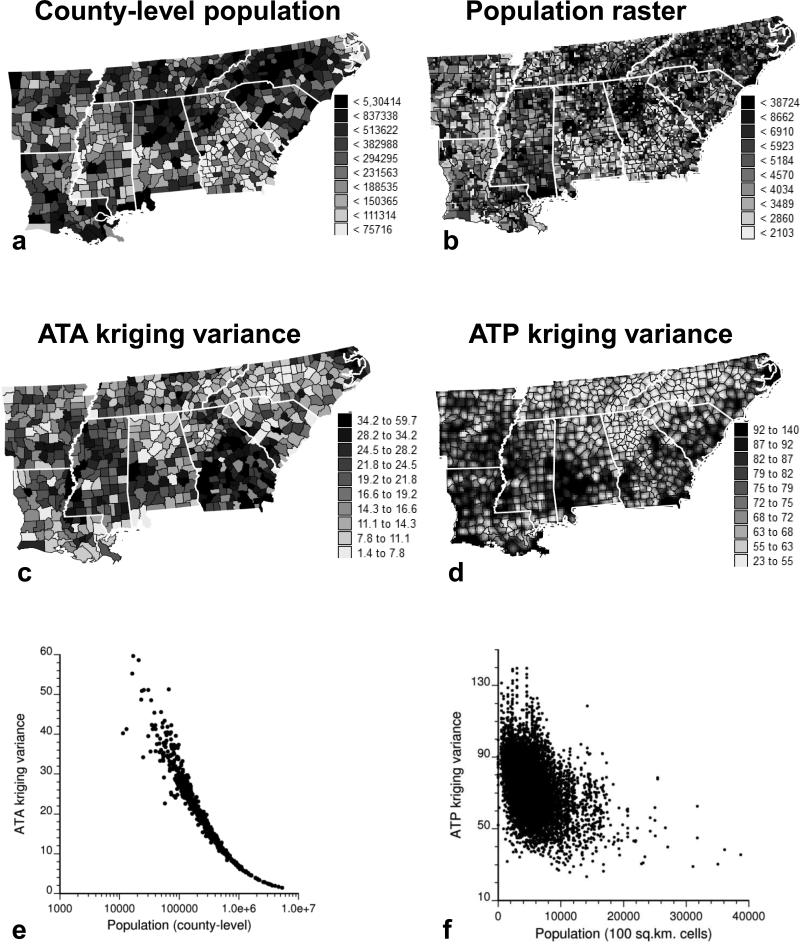
Figure 2a and b show the areal and point-support models inferred from 688 rate data for males and females, respectively. As expected, the point-support model (light gray curve) has a larger sill because the point process is always more variable than the aggregated counterpart. Its regularization (short dashed line) gives a variogram model that is close to the one fitted to experimental values, which validates the consistency of the deconvolution. This model was used to estimate mortality risk at the county level (ATA kriging) and to map the spatial distribution of risk within counties (ATP kriging) using the 32 nearest data in each case. All maps of kriged estimates are smoother than those of the raw rates because the noise related to small population sizes is filtered. High mortality can be observed along the Mississippi valley or Delta region (AR, MS), the southern Atlantic region (GA, SC), and along the Gulf Coast (Fig. 2c and d). Smoking patterns largely account for the regional variation in lung cancer mortality. For example, smoking habits, including the greater use of hand-rolled cigarettes, contributed to the high rates in southern Louisiana (LA), especially in the Cajun population (Devesa et al. 1999). In the 1970s and early 1980s, studies in coastal Georgia (GA), northeast Florida, and southern Louisiana revealed an excess risk of lung cancer associated with work in shipyards primarily during World War II (Blot et al. 1978; Jemal, Grauman, and Devesa 2000). The rates for North Carolina (NC) show a clear east-west trend, with lower mortality in the more rural western counties that are part of the Smoky Mountains compared to the coastal region. As expected from theory (equation [5]), aggregating the ATP kriged estimates within each county using the population density map (Fig. 1c) gives the ATA kriging map. The maps of kriging variance in Fig. 3 essentially reflect the lower confidence that can be placed in the risk estimated for sparsely populated counties and over smaller spatial supports, such as the 100 km2 raster cells used for ATP kriging.
Figure 2.
(a and b) Top: experimental semivariogram of the risk estimated from county-level rates and the results of its deconvolution (top curve). The regularization of the point-support model yields a curve (short dashed line) that is very close to the experimental one. The point-support model is then used to estimate lung cancer mortality risk (deaths/100,000 inhabitants) at the county level (ATA kriging) or at the nodes of a 10 km spacing grid (ATP kriging). (c and d) Middle: respectively for White males. (e and f) Bottom: respectively for White females. Thick white lines delineate state boundaries (see Fig. 1 for state names).
Figure 3.
(a and b) Top: population at risk estimated at the county level or at the nodes of a 10 km spacing grid for White males. Thick white lines delineate state boundaries. (c and d) 7Middle: maps of the prediction variance reflect the larger reliability of risk estimates for heavily populated counties or raster cells. (e and f) Bottom: scatterplots illustrating the impact of the population size on the reliability of risk estimates.
Mapping regional background and local patterns
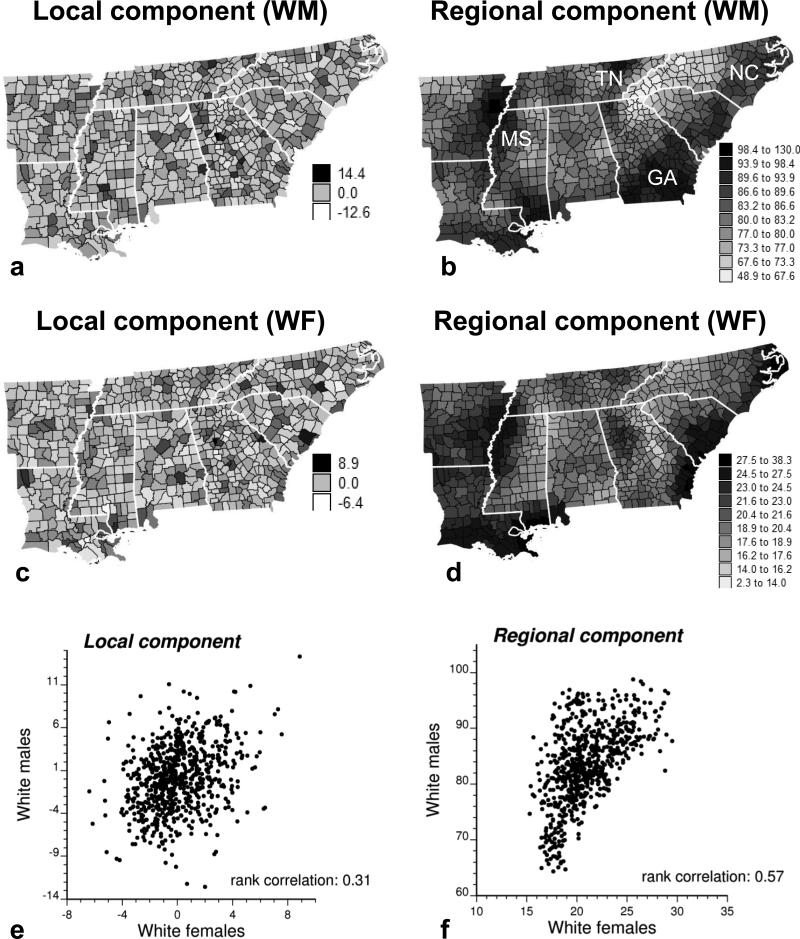
Figure 4 shows the maps of factorially kriged estimates of the local and regional components of lung cancer mortality risk for both sexes. Decomposition into local and regional components facilitates the identification of local hot spots and cold spots using the map of local components (Fig. 4a–c), whereas larger-scale trends are enhanced on the map of regional components (Fig. 4b–d). Although such decomposition is based on the somewhat subjective fitting of a nested variogram model, the presence of these two scales of variation in the variogram supports the application of this technique here. The regional maps for males and females share a similar background of high mortality rates along the Mississippi valley (MS) and the coastal areas of the Atlantic and the Gulf, whereas lower mortality is observed inland (Fig. 4b–d). The highest cancer mortality for White females tends, however, to be confined to coastal counties, whereas for White males high rates extend to low-income rural southern Georgia. This latter observation agrees with recent findings that mortality rates for White males in Georgia are higher in rural than in urban counties, a trend that is opposite to that observed for White females (Singh et al. 2005). Another potential cause of higher mortality from lung cancer is the paper mill industry, which is widespread in the heavily forested parts of southern Georgia and has a mainly male workforce (Harrington et al. 1978). The lowest cancer mortality rates for White males aggregate in western North Carolina (NC) and across the border into Tennessee (TN); this region corresponds to the Smoky Mountains. Similarities between male and female mortality risks are less obvious in the maps of the local spatial components; this visual impression is confirmed by the smaller correlation coefficients between these spatial components (Fig. 4e).
Figure 4.
Maps of the local and regional components of lung cancer mortality risk. (a and b) Top: for White males (WM), (c and d) Middle: for White females (WF). (e and f) Bottom: scatterplots portraying the relationships. The strongest correlation is observed among the regional components, which display the large-scale trend in risk for both sexes; the local component, which reflects local departures from this regional background, appears more sex specific.
Stochastic simulation of lung cancer mortality
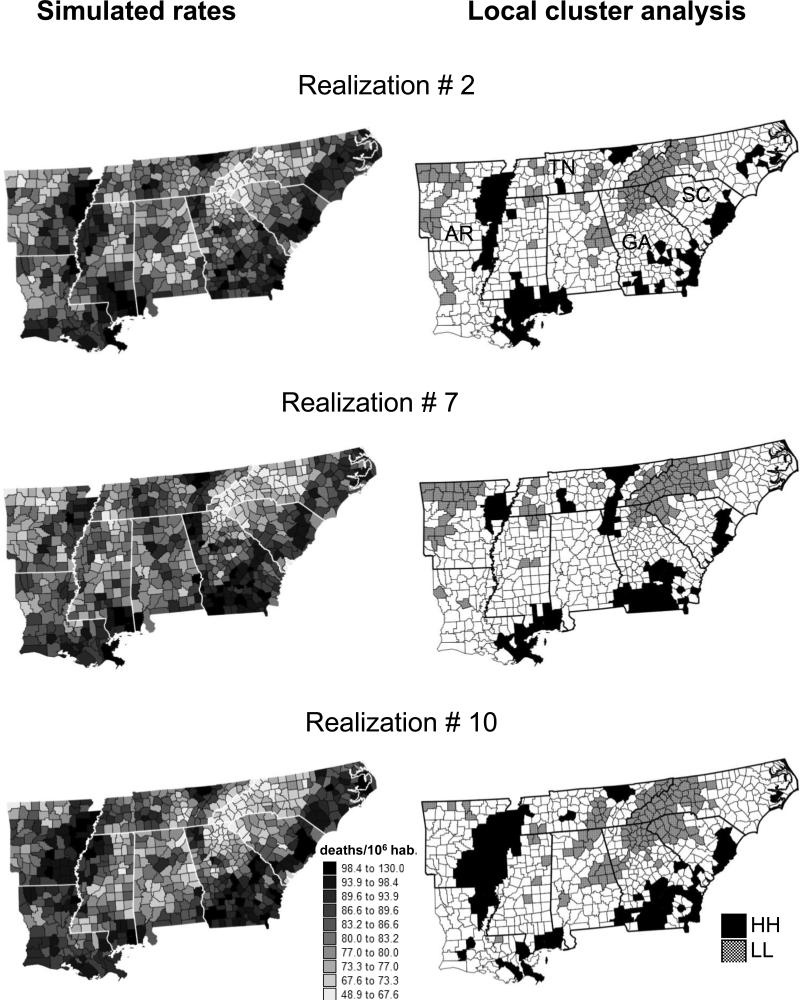
Figure 5a, c, and e shows three realizations of the spatial distribution of mortality risk values generated for males using p-field simulation. Differences among realizations depict the uncertainty attached to the risk map. On the one hand, features that appear consistently on simulated maps, such as low mortality in the Smoky Mountains or high mortality in the South Carolina (SC) coastal region and north-central Tennessee (TN, Oak Ridge National Laboratory area), are considered the most likely to be true. On the other hand, larger differences are observed in the less densely populated southern Georgia (Fig. 1c), where some of the high mortality rates might be less reliable than in other states. This again reflects the small numbers problem.
Figure 5.
Three realizations of the spatial distribution of lung cancer mortality for White males, and the significant clusters of low (LL) and high (HH) risk detected using local Moran's I. Thick white or black lines delineate state boundaries.
LCA of lung cancer mortality
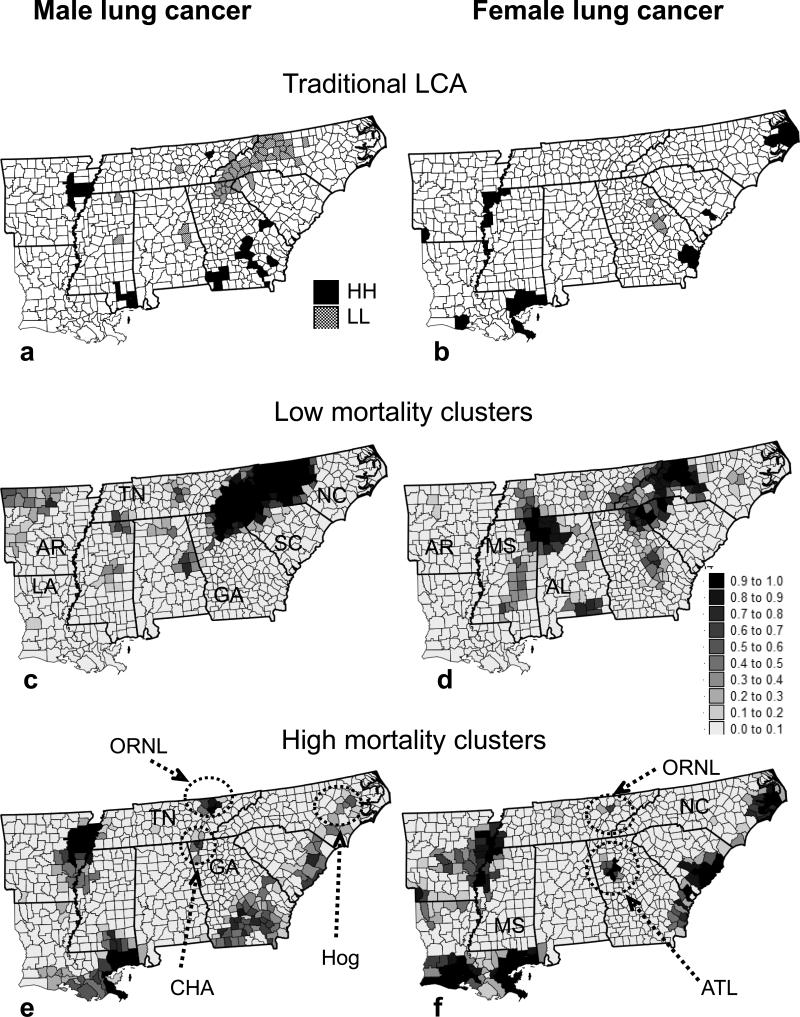
Aggregates of counties with lower or higher mortality risks are easily detected in LCA; the results are shown in Fig. 6a and b. This analysis was conducted using a traditional random shuffle of rates (significance level α = 0.05), followed by the FDR correction of the P-values. The largest cluster corresponds to lower mortality (LL) recorded for males in the Smoky Mountains. Significant clusters of high risks (HH) occur along the Mississippi valley (MS) and the coastal areas of the Atlantic and the Gulf, as well as in southern Georgia for males.
Figure 6.
(a and b) Top: clusters of low (LL) or high (HH) cancer mortality identified using a traditional local cluster analysis (LCA) that ignores population size and rate uncertainty. Likelihood that a county belongs to a cluster computed from the LCA of 100 simulated risk maps for both males and females. (c and d) Middle: low cancer mortality. (e and f) High cancer mortality.
A shortcoming of the LCA in Fig. 6a and b is that it disregards the county population size, hence the uneven reliability of the observed rates. The cluster maps in Fig. 5b, d, and f illustrate how the uncertainty in the risk values identified by the simulated maps translates into uncertainty in the results of the LCA. From one realization to another, the shape and position of local clusters of high mortality can change substantially. For example, the extent of the LL cluster in eastern Arkansas (Mississippi River) or the compactness of the HH cluster in southern Georgia varies greatly among the three realizations. Conversely, the HH cluster in north-central Tennessee and the LL cluster in the Smoky Mountains are very stable across realizations.
One hundred realizations of the spatial distribution of male and female lung cancer mortality risk were generated by p-field simulation, followed by a LCA using the aforementioned random sampling of a Poisson distribution to account for population size. To summarize the information contained in the set of 100 cluster maps, the proportion of realizations with a significant LL or HH cluster was computed for each county and mapped in Fig. 6e and f. Accounting for population size in the randomization reduces the range of risk values that can be simulated in any county, in particular, where the population is large and the uncertainty about mortality risk is thus small. This results in a narrower range for the simulated values of the LISA statistic, leading to smaller P-values (i.e., increased significance of the tests) for heavily populated counties along the coast. Both sexes have large clusters of high mortality around New Orleans (LA), the Mississippi valley (MS), and the southern Atlantic (GA, SC) coastal region. Another smaller HH cluster occurs in north central Tennessee (Oak Ridge National Laboratory area). Several sex-specific clusters might reflect the impact of occupational exposure on male lung cancer mortality, such as in southern Georgia, which is a leading pulp and paper production area in the United States (Prunty 1956) or in Sampson County (NC), which is adjacent to the two largest pig-producing counties in the United States. Male cancer mortality also shows a small HH cluster on the border of Tennessee and Georgia in Chattanooga (CHA), an area given the unwelcome title of having the dirtiest air in the United States by the federal government in 1969. There is a cluster of low mortality for males around Benton County in the northwest corner of Arkansas, which has the second largest population and the lowest poverty rate of any county in the state (Fig. 6c). For females, an HH cluster is centered on Atlanta. For both sexes the largest cluster of low mortality corresponds to the Smoky Mountains in western North Carolina. It is noteworthy that this very large cluster went completely undetected in the LCA of female rates (Fig. 6b). Another large LL cluster for females spans across three states: Tennessee (TN), Alabama (AL), and Missouri (MS).
Conclusions
The major difficulty in the analysis of health outcomes is that the patterns observed reflect the influence of a complex combination of demographic, social, economic, cultural, and environmental factors that change through time and space, and interact with the different types and scales of places where people live. It is essential, therefore, to incorporate the scale and spatial support of the data in the processing of health data and to account for the impact of population sizes on the reliability of the estimates of rates. Geostatistics provides a methodology to model the spatial correlation among rates measured over irregular geographic supports and to compute noise-free estimates of risk over the same units or at much finer scales. It also enables the propagation of uncertainty in rates through the delineation of areas with significantly higher or lower mortality or incidence rates, as well as the analysis of relationships between health outcomes and putative risk factors.
The geostatistical analysis of lung cancer data here indicated the existence of two scales of spatial variation in cancer mortality across 688 counties in southeastern United States. The filtering of short-scale variation and noise, which can be very large in sparsely populated counties, emphasized the similarity in the regional patterns for male and female cancer mortality that correlates well with proximity to shipyards. Accounting for population size and rate uncertainty in LCA revealed new clusters of high and low mortality that were overlooked in a traditional approach. In particular, significantly higher mortality was found around Oak Ridge National Laboratory for both sexes, in counties with high concentrations of pig farms and paper mills for males (occupational exposure), and in the vicinity of Atlanta for females.
Although lung cancer mortality is on average four times greater for males than females, similar environmental and socioeconomic factors are likely to be operating, especially at the regional scale, as shown by a correlation of 0.5 between male and female regional components. Discrepancies between maps of risk can be interpreted as signs of the local impact of sex-specific factors, such as occupational exposure to coal tar fumes in coke production and coal gas production, which converts solid coal into a gas that can be used for power generation, or to the use of asbestos in marine construction and repair, steel and iron mills, power-generating stations, pulp and paper mills, and oil refineries. The spatial distribution of the disparities between the sexes is being explored in relation to potential covariates in research currently underway. Methodological research is also investigating the modeling of nonstationary spatial trends using the hierarchical model recently developed by Bellier, Monestiez, and Guinet (2009) to account for complex spatial structures in the geostatistical analysis of count data.
Acknowledgements
This research was funded by grant R44-CA132347-01 from the National Cancer Institute. The views stated in this publication are those of the author and do not necessarily represent the official views of the NCI. The author thanks the two anonymous referees and the editors for their comments, which greatly improved the presentation of this article.
References
- Ali M, Goovaerts P, Nazia N, Haq MZ, Yunus M, Emch M. Application of Poisson Kriging to the Mapping of Cholera and Dysentery Incidence in an Endemic Area of Bangladesh. International Journal of Health Geographics. 2006;5(45) doi: 10.1186/1476-072X-5-45. Available at http://www.ij-healthgeographics.com/content/5/1/45. [DOI] [PMC free article] [PubMed]
- Anselin L. Local Indicators of Spatial Association—LISA. Geographical Analysis. 1995;27:93–115. [Google Scholar]
- Anselin L, Syabri I, Kho Y. GeoDa: An Introduction to Spatial Data Analysis. Geographical Analysis. 2006;38:5–22. [Google Scholar]
- Assunçao RM, Reis EA. A New Proposal to Adjust Moran's I for Population Density. Statistics in Medicine. 1999;18:2147–62. doi: 10.1002/(sici)1097-0258(19990830)18:16<2147::aid-sim179>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Bellier E, Monestiez P, Guinet C. Geostatistical Modelling of Wildlife Populations: A Non-Stationary Hierarchical Model for Count Data. In: Atkinson P, et al., editors. GeoEN VII—Geostatistics for Environmental Applications. Springer-Verlag; Berlin, Germany: 2009. in press. [Google Scholar]
- Besag J, York J, Mollie A. Bayesian Image Restoration with Two Applications in Spatial Statistics. Annals of the Institute of Statistical Mathematics. 1991;43:1–59. [Google Scholar]
- Blot WJ, Harrington JM, Toledo A, Hoover R, Heath CW, Fraumeni JF., Jr. Lung Cancer after Employment in Shipyards During World War II. The New England Journal of Medicine. 1978;299:620–4. doi: 10.1056/NEJM197809212991202. [DOI] [PubMed] [Google Scholar]
- Castro MC, Singer BH. Controlling the False Discovery Rate: A New Application to Account for Multiple and Dependent Tests in Local Statistics of Spatial Association. Geographical Analysis. 2006;38:180–208. [Google Scholar]
- Chiles JP, Delfiner P. Geostatistics: Modeling Spatial Uncertainty. Wiley; New York: 1999. [Google Scholar]
- Devesa SS, Grauman DJ, Blot WJ, Fraumeni JF., Jr. Cancer Surveillance Series: Changing Geographic Patterns of Lung Cancer Mortality in the United States, 1950 through 1994. Journal of the National Cancer Institute. 1999;91(12):1040–50. doi: 10.1093/jnci/91.12.1040. [DOI] [PubMed] [Google Scholar]
- Fortin MJ, Jacquez G. Randomization Tests and Spatially Autocorrelated Data. Bulletin of the Ecological Society of America. 2000;81:201–5. [Google Scholar]
- Goovaerts P. Detection of Spatial Clusters and Outliers in Cancer Rates Using Geostatistical Filters and Spatial Neutral Models. In: Renard P, Demougeot-Renard H, Froidevaux R, editors. GeoENV V—Geostatistics for Environmental Applications, 149–60. Springer-Verlag; Berlin, Germany: 2005a. [Google Scholar]
- Goovaerts P. Geostatistical Analysis of Disease Data: Estimation of Cancer Mortality Risk from Empirical Frequencies Using Poisson Kriging. International Journal of Health Geographics. 2005b;4(31) doi: 10.1186/1476-072X-4-31. Available at http://www.ij-healthgeographics.com/content/4/1/31. [DOI] [PMC free article] [PubMed]
- Goovaerts P. Geostatistical Analysis of disease data: visualization and propagation of spatial uncertainty in cancer mortality risk using Poisson kriging and p-field simulation. International Journal of Health Geographics. 2006a;5(7) doi: 10.1186/1476-072X-5-7. Available at http://www.ij-healthgeographics.com/content/5/1/7. [DOI] [PMC free article] [PubMed]
- Goovaerts P. Geostatistical Analysis of Disease Data: Accounting for Spatial Support and Population Density in the Isopleth Mapping of Cancer Mortality Risk Using Area-to-Point Poisson Kriging. International Journal of Health Geographics. 2006b;5(52) doi: 10.1186/1476-072X-5-52. Available at http://www.ij-healthgeographics.com/content/5/1/52. [DOI] [PMC free article] [PubMed]
- Goovaerts P. Accounting for Rate Instability and Spatial Patterns in the Boundary Analysis of Cancer Mortality Maps. Environmental and Ecological Statistics. 2008a;15(4) doi: 10.1007/s10651-007-0064-6. doi 10.1007/s10651-007-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goovaerts P. Kriging and Semivariogram Deconvolution in Presence of Irregular Geographical Units. Mathematical Geosciences. 2008b;40:101–28. [PMC free article] [PubMed] [Google Scholar]
- Goovaerts P. Medical Geography: A Promising Field of Application for Geostatistics. Mathematical Geosciences. 2009;41:243–64. doi: 10.1007/s11004-008-9211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goovaerts P, Gebreab S. How does Poisson Kriging Compare to the Popular BYM Model for Mapping Disease Risks? International Journal of Health Geographics. 2008;7(6) doi: 10.1186/1476-072X-7-6. Available at http://www.ij-healthgeographics.com/content/7/1/6. [DOI] [PMC free article] [PubMed]
- Goovaerts P, Jacquez GM. Accounting for Regional Background and Population Size in the Detection of Spatial Clusters and Outliers Using Geostatistical Filtering and Spatial Neutral Models: The Case of Lung Cancer in Long Island, New York. International Journal of Health Geographics. 2004;3(14) doi: 10.1186/1476-072X-3-14. Available at http://www.ij-healthgeographics.com/content/3/1/14. [DOI] [PMC free article] [PubMed]
- Goovaerts P, Jacquez GM. Detection of Temporal Changes in the Spatial Distribution of Cancer Rates Using LISA Statistics and Geostatistically Simulated Spatial Neutral Models. Journal of Geographical Systems. 2005;7:137–59. doi: 10.1007/s10109-005-0154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goovaerts P, Jacquez GM, Greiling D. Exploring Scale-Dependent Correlations Between Cancer Mortality Rates Using Factorial Kriging and Population-Weighted Semivariograms: A Simulation Study. Geographical Analysis. 2005;37:152–82. doi: 10.1111/j.1538-4632.2005.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiling DA, Jacquez GM, Kaufmann AM, Rommel RG. Space Time Visualization and Analysis in the Cancer Atlas Viewer. Journal of Geographical Systems. 2005;7:67–84. doi: 10.1007/s10109-005-0150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JM, Blot WJ, Hoover RN, Housworth WJ, Heath CW, Jr., Fraumeni JF., Jr. Lung Cancer in Coastal Georgia: A Death Certificate Analysis of Occupation: Brief Communication. Journal of the National Cancer Institute. 1978;60(2):295–8. doi: 10.1093/jnci/60.2.295. [DOI] [PubMed] [Google Scholar]
- Jacquez GM, Meliker JR, AvRuskin G, Goovaerts P, Kaufmann A, Wilson ML, Nriagu J. Case-Control Geographic Clustering for Residential Histories Accounting for Risk Factors and Covariates. International Journal of Health Geographics. 2006;5(32) doi: 10.1186/1476-072X-5-32. Available at http://www.ij-healthgeographics.com/content/5/1/32. [DOI] [PMC free article] [PubMed]
- Jemal A, Grauman D, Devesa S. Recent Geographic Patterns of Lung Cancer and Mesothelioma Mortality Rates in 49 Shipyard Counties in the United States, 1970–94. American Journal of Industrial Medicine. 2000;37:512–21. doi: 10.1002/(sici)1097-0274(200005)37:5<512::aid-ajim7>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Journel AG, Huijbregts CJ. Mining Geostatistics. Academic Press; New York: 1978. [Google Scholar]
- Kelsall J, Wakefield J. Modeling Spatial Variation in Disease Risk: A Geostatistical Approach. Journal of the American Statistical Association. 2002;97(459):692–701. [Google Scholar]
- Kyriakidis P. A Geostatistical Framework for Area-to-Point Spatial Interpolation. Geographical Analysis. 2004;36(2):259–89. [Google Scholar]
- Lajaunie C. Local Risk Estimation for a Rare Noncontagious Disease Based on Observed Frequencies. 1991. Note N-36/91/G. Centre de Géostatistique, Ecole des Mines de Paris.
- Monestiez P, Dubroca L, Bonnin E, Durbec JP, Guinet C. Geostatistical Modelling of Spatial Distribution of Balenoptera physalus in the Northwestern Mediterranean Sea from Sparse Count Data and Heterogeneous Observation Efforts. Ecological Modelling. 2006;193:615–28. [Google Scholar]
- Oden N. Adjusting Moran's I for Population Density. Statistics in Medicine. 1995;14:17–26. doi: 10.1002/sim.4780140104. [DOI] [PubMed] [Google Scholar]
- Oliver MA, Webster R, Lajaunie C, Muir KR, Parkes SE, Cameron AH, Stevens MCG, Mann JR. Binomial Cokriging for Estimating and Mapping the Risk of Childhood Cancer. IMA Journal of Mathematics Applied in Medicine and Biology. 1998;15:279–97. [PubMed] [Google Scholar]
- Prunty M., Jr. Recent Expansions in the Southern Pulp-Paper Industries. Economic Geography. 1956;32(1):51–7. [Google Scholar]
- Richardson S, Thomson A, Best N, Elliot P. Interpreting Posterior Relative Risk Estimates in Disease-Mapping Studies. Environmental Health Perspectives. 2004;112:1016–25. doi: 10.1289/ehp.6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Bayakly AR, McNamara C, Redding K. Lung Cancer in Georgia, 1999-2002. 2005 Georgia Department of Human Resources, Division of Public Health, Chronic Disease, Injury, and Environmental Epidemiology Section. Publication number DPH06/006W. Available at http://health.state.ga.us/pdfs/chronic/cancer/lungCancer99_02.pdf.
- Song C, Kulldorff M. Power Evaluation of Disease Clustering Tests. International Journal of Health Geographics. 2003;2(9) doi: 10.1186/1476-072X-2-9. Available at http://www.ij-healthgeographics.com/content/2/1/9. [DOI] [PMC free article] [PubMed]
- Tunstall HVZ, Shaw M, Dorling D. Places and Health. Journal of Epidemiology and Community Health. 2004;58:6–10. doi: 10.1136/jech.58.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackernagel H. Multivariate Geostatistics. Springer-Verlag; Berlin: 1998. [Google Scholar]
- Waldhör T. The Spatial Autocorrelation Coefficient Moran's I Under Heteroscedasticity. Statistics in Medicine. 1996;15:887–92. doi: 10.1002/(sici)1097-0258(19960415)15:7/9<887::aid-sim257>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Waller LA, Gotway CA. Applied Spatial Statistics for Public Health Data. Wiley; New Jersey: 2004. [Google Scholar]
- Waller LA, Jacquez GM. Disease Models Implicit in Statistical Tests of Disease Clustering. Epidemiology. 1995;6(6):584–90. doi: 10.1097/00001648-199511000-00004. [DOI] [PubMed] [Google Scholar]
- Webster R, Oliver MA, Muir KR, Mann JR. Kriging the Local Risk of a Rare Disease from a Register of Diagnoses. Geographical Analysis. 1994;26:168–85. [Google Scholar]
- Woodward P. BugsXLA: Bayes for the Common Man. Journal of Statistical Software. 2005;14(5) Available at http://www.jstatsoft.org/v14/i05.