Abstract
Image-guided near infrared spectroscopy (IG-NIRS) can provide high-resolution vascular, metabolic and molecular characterization of localized tissue volumes in-vivo. The approach for IG-NIRS uses hybrid systems where the spatial anatomical structure of tissue obtained from standard imaging modalities (such as MRI) is combined with tissue information from diffuse optical imaging spectroscopy. There is need to optimize these hybrid systems for large-scale clinical trials anticipated in the near future in order to evaluate the feasibility of this technology across a larger population. However, existing computational methods such as the finite element method mesh arbitrary image volumes, which inhibit automation, especially with large numbers of datasets. Circumventing this issue, a boundary element method (BEM) for IG-NIRS systems in 3–D is presented here using only surface rendering and discretization. The process of surface creation and meshing is faster, more reliable, and is easily generated automatically as compared to full volume meshing. The proposed method has been implemented here for multi-spectral non-invasive characterization of tissue. In phantom experiments, 3–D spectral BEM-based spectroscopy recovered the oxygen dissociation curve with mean error of 6.6% and tracked variation in total hemoglobin linearly.
Keywords: BEM, image-guided NIRS
1. INTRODUCTION
The field of diffuse optical tomography (DOT) has evolved significantly over the past two decades as a method for probing biological tissue in-vivo to gain a better understanding of vascular and metabolic progressions in disease. DOT has undergone several preliminary clinical trials [1–3] by demonstrating the ability to study physiology of tissue through unique mechanisms not available through existing modalities. These include molecular, structural and vascular characterization of tissue using intrinsic contrast processes. Tumor to background contrasts are typically 100% or more [4,5] and use of extrinsic contrasts has further increased the biological and molecular contrasts available in-vivo [6,7]. A major focus of on-going studies has been to increase the spatial resolution and create complementary imaging systems by combining different modalities, such as MRI-DOT [8], X Ray-DOT [9], and Ultrasound-DOT [10]. These hybrid systems have bypassed the resolution limit of DOT (3–5 mm) in favor of the sub-millimeter resolution possible from MRI, X-Ray and Ultrasound by utilizing a-priori structural information to guide the DOT image recovery. This approach merges the strengths of structural information provided by a high resolution modality with the strengths of functional content provided by optical spectroscopy Researchers are now at the threshold of larger-scale multi-institutional clinical trials, especially in radiology (for diagnosis of breast cancer [2]) and oncology (for neo-adjuvant treatment monitoring [11]) across patient populations and geographies. As hybrid DOT systems make their way into the clinic through large-scale trials, algorithms behind spectral image recovery must be optimized for imaging and processing large datasets. This is complicated by the challenge of optical imaging in 3D, which is mandatory for clinicians who are accustomed to viewing volumetric images from the existing standard imaging modalities. It is also well-known that imaging in 3–D is more accurate than 2–D for DOT systems [12]. As such, this study focuses on providing a solution to the proposed problem of image-guided spectroscopy of tissue.
To date, most 3D optical imaging studies have been limited to stand-alone DOT systems. 3–D image reconstruction was employed by Enfield et al [13] in time-resolved optical mammography without priors. In this work, the authors used a hemispherical fluid-filled cup as the breast interface, creating hemispherical meshes for computation on clinical data.
Eppstein et al [14] showed the feasibility of 3–D optical imaging by using simulated data from 3–D slab-shaped volumes which were approximated by a domain decomposition technique that used subdomains. Srinivasan et al [15] used a data-subset algorithm to minimize the computational burden by dynamically choosing optimal data-sets which carried information regarding major features in the domain. Their work presented simulations and experiments also in slab shapes. Dehghani et al[16] used an underdetermined version of the Moore-Penrose generalized inverse to compute 3-D DOT estimates for a clinical subject using a conical shaped mesh. All of these studies have been for stand-alone DOT systems and using regular shapes (slab or cylinder) with numerical volume-based methods such as finite element or finite difference.
Hybrid systems combining MRI,×ray or ultrasound with NIR systems provide superior spatial resolution at millimeter scales. Several algorithms have been developed to incorporate the anatomical tissue structure from an alternate modality into the NIR image recovery. These studies have predominantly been in 2–D exploring Bayesian [17] and regularized methods [18,19] to improve image resolution. In 3-D, Dehghani et al [20] used exact knowledge of boundaries and assumption of homogeneous piece-wise regions in a cylindrical shape in 3–D and showed nearly 100% recovery in experiments. Further work by Yalavarthy et al incorporated structural information in a generalized framework for optical imaging [21,22]. Azar et al [23] have demonstrated co-registration of non-concurrent NIR and MRI images and segmentation of MR images to enable priors to be applied in 3-D and have applied this to phantom models and three clinical cases. None of these have obtained the 3–D imaging volume itself from MRI, instead using assumptions about the shape based on a 2–D slice in the plane of the optical detectors to reconstruct. Hence these do not address the issues of volumetric meshing of complex arbitrary shapes that arise from brain and breast MRI segmentation that is a necessary step to compute on MRI-derived volumes.
In our experience, one of the key challenges faced by hybrid systems towards routine clinical use lies is in image-segmentation and volume rendering of complex arbitrary shapes. Volume meshing is not a simple task. It is a complex procedure and can involve several manual interventions on a case-by-case basis, which severely limit the clinical workflow. It is practically impossible, using current resources, to obtain volume breast meshes with interior boundaries preserved, in a regular manner. With a pressing need to optimize IG-NIRS for large-scale clinical trials, our motivation here is to evaluate the potential of an alternate numerical method that can circumvent the issues of volume meshing and enable processing of large data-sets in the clinic.
Our previous work in combining MRI priors into DOT recovery has indicated that the use of hard priors in image reconstruction provides the most accurate results [24]. Hard priors impose the tissue boundaries obtained from MRI into the image recovery process with the assumption of piece-wise constant tissue types and can be described as Image-guided NIR spectroscopy (IG-NIRS) allowing region-based localized and non-invasive characterization of tissue types. This use of hard priors allows us to investigate alternate numerical methods for IG-NIRS that may be more efficient. One such method is the boundary element method (BEM) [25] which presents an attractive alternate to the existing finite-element method by using only surface description to model light propagation in tissue. It is well known that surfaces can be generated easily and reliably as compared to volume rendering. Most commercial software packages automatically create surfaces with minimal user intervention.
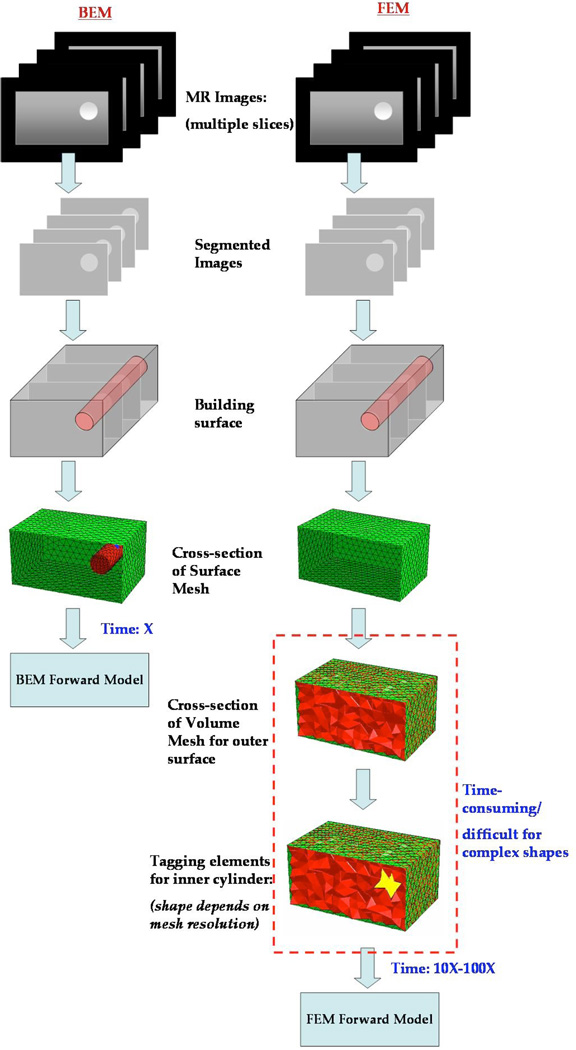
The flowchart in Figure 1 shows an illustration of the basic steps involved in obtaining surface and volume meshes for computation. Both require segmentation of the MR images followed by surface discretization. BEM can be used directly following the surface tessellation whereas FEM requires additional steps to build a volume mesh suitably tagged with boundaries delineating the volume into different tissue types. This has implication in the clinical workflow especially when processing large number of data sets. A drawback in BEM is the use of full matrices as compared to sparse matrices in FEM. However, the faster mesh generation and lower mesh sizes allow for faster computational speed.
Fig. 1.
An illustration of the steps involved in obtaining a test grid for computation with BEM and FEM is shown. Both require segmentation of the MR dicom images followed by surface discretization. BEM can be used directly following the surface tessellation whereas FEM requires additional steps to build a volume mesh suitably tagged with boundaries delineating the volume into different tissue types. The shape of the interior boundaries will then depend on the mesh resolution. Alternately, the volume mesh can be created to preserve the interior boundaries (not shown here), however, this approach becomes difficult when resolving features at different scales (e.g. breast vs. tumor boundary). The volume meshing is a demanding task that takes up to 10–100 times the effort required to create surface meshes.
BEM was applied in simulations using 3–D MRI segmented head models by Zacharopoulos et al [26] to reconstruct for shape and optical properties. In the work here, we have assumed the shape of heterogeneities to be known from MRI and reconstructed directly for chromophore concentrations (oxy and deoxy-hemoglobin and water) and scattering parameters. Results from phantom experiments show accurate monitoring of oxygenation as described by the oxygen dissociation curve, and a linear increase was observed with variation in total hemoglobin.
2. MATERIALS AND METHODS
2.1 Instrumentation
The phantom experiments were carried out in a stand-alone DOT system developed at Dartmouth, described in detail in Mcbride et al [27]. Briefly, the automated NIR tomography device was constructed to perform cross-sectional imaging of the breast. Optical fibers arranged in a circular array (with both radial and vertical degrees of freedom) have the capability to collect data from sets of 16 source and 15 detector locations per source. Intensity modulated light (at 100MHz) at six discrete wavelengths between 660 and 850 nm is detected by photomultiplier tubes. The detected light is electrically mixed with a reference signal to yield a low-frequency (500 Hz) signal from which measurements of amplitude and phase shift were recorded.
2.2 Data Calibration
The measured data from the tissue/phantom was first calibrated to account for small offsets due to source–detector fiber transmission, alignment characteristics, and errors in discretization or model–data mismatch. This was accomplished using a homogeneous fitting algorithm [28] which utilized Newton Raphson method for fitting of two parameters: the slope of the phase (log(θ)) with respect to distance from the source location, and the slope of the logarithm of intensity times distance with respect to distance (log(rI)). This fitting procedure used two solutions: (1) analytical solution for infinite medium, followed by (2) numerical solution to the diffusion equation on the homogeneous domain of relevant size. By using the homogeneous fitting algorithm on both reference phantom measurements and unknown tissue measurements, the calibrated measurements and initial estimates for optical properties were obtained. The initial estimates of the optical properties were converted into concentrations of oxyhemoglobin, deoxyhemoglobin and water and scatter using spectral fitting and used as the starting point in the NIRS reconstruction.
2.3 BEM Spectral Reconstruction for IG-NIRS in 3–D
Reconstruction in DOT comprises of the forward and the inverse problem. The forward problem is defined as follows: Given the tissue parameter distribution [µ], to find the light fluence distribution [Φ] within the tissue domain Ω. The inverse problem is defined as: Given a distribution of sources [q] and a distribution of measurements [M] giving a measure of [Φ] at the boundary dΩ, finding the tissue parameter distribution [μ] within domain Ω. For the forward problem, we used the frequency domain diffusion equation to model light propagation in tissue. In the diffusion approximation, the assumption is that the diffuse intensity encounters many particles and is scattered almost uniformly in all directions, and therefore its angular distribution is almost uniform isotropic[29]. This equation exists under the assumption that scatter dominates over absorption which is true in the case of several tissue types, including the human breast, in the wavelength region of 650–1350 nm [30]. This differential equation is written as[29,31]:
| (1) |
where Φ(r,ω) is the isotropic fluence at modulation frequency ω and position r, κ (r) is the diffusion coefficient, µa (r) is the absorption coefficient, c is the speed of light in the medium and q0(r,ω) is an isotropic source. The diffusion coefficient can be written as
| (2) |
where µs is the reduced scattering coefficient. Under the assumptions that the internal surfaces of the various tissue types in the domain are known a priori, and the optical properties are piece-wise constant in each tissue type, the equation can be recast into a modified Helmholtz equation given by:
| (3) |
where kl is the wave number, constant in sub-domain l [32,33], and for a particular frequency .
The fundamental Green’s function solution is readily available for this equation and can be used to derive the BEM formulation[25]. The details of this formulation can be found elsewhere [34]. The forward solver was implemented in Matlab using Fortran code for building the internal matrices and parallelized using Matlab Distributive computing toolbox. The computational time for a two region forward problem with a mesh size of 3015 nodes using 4 processors was 51 seconds.
The inverse problem was solved using a gradient-based iterative procedure based on Newton’s method which minimizes the least-squares functional:
| (4) |
where M is the total number of measurements at each wavelength, and are the measured and calculated fluence respectively, at the boundary for each measurement point j. The update in the optical properties ∂µ is given by,
| (5) |
where ∂Φ refers to the change in boundary data, ℑ is the Jacobian, the matrix containing the sensitivity of the boundary data to a change in optical property µa and diffusion coefficient κ given by and α is the regularization parameter chosen empirically based on phantom experiments. The Jacobian was calculated using a perturbation approach to approximate the required derivatives by perturbing µa and κ in each region in turn and calculating the resulting change in the boundary measurements.
By coupling the measurements obtained at all the wavelengths together and applying spectral relationships defining absorption characteristics of oxyhemoglobin, deoxyhemoglobin and water as well as scattering relationships defined empirically, into the reconstruction, it is possible to obtain the NIRS parameters directly [35]. This use of spectral constraints is known to improve accuracy of recovered spectroscopic parameters and reduce cross-talk [35–37]. We implemented this into the BEM-based reconstruction so that the update in the chromophore concentrations and scatter could be recovered directly. The structure of the Jacobian is changed due to the addition of spectral features and is detailed in Srinivasan et al [35]. The spectral BEM recovery, implemented for the first time in DOT, was used to obtain IG-NIRS estimates from phantoms and human subjects.
2.4 Phantom experiments
The phantom experiments presented in this paper were carried out using liquid homogeneous phantoms (for obtaining oxygen dissociation curve) and gelatin heterogeneous phantoms (for characterizing changes in total hemoglobin). For the first experiment, a solution with 1% Intralipid and 1% whole blood was placed in a plastic cup of 70 mm diameter. The pO2 was measured independently using a chemical microelectrode, after calibration of the electrode overnight in saline solution. The 1% whole blood was found to have 18µM hemoglobin, and the oxygenation of the solution was reduced by varying the pO2 values from 150mm to 0mm Hg by addition of yeast. Using the spectral BEM toolbox, we reconstructed values of oxygen saturation, total hemoglobin, water concentration and scatter.
For the second experiment, a phantom using gelatin with whole blood added for absorption and Titanium dioxide for scatter, was made. This phantom of diameter 82 mm had a 2.5cm hole drilled 1cm from the boundary which was filled with a saline solution containing blood with 0.75% Intralipid for scattering. The inclusion’s blood concentration was varied systematically from 20 to 44µM and measurements were taken. The dataset was used to reconstruct spectral BEM estimates of the inclusion and background.
3. RESULTS
3.1 Oxygen dissociation curve
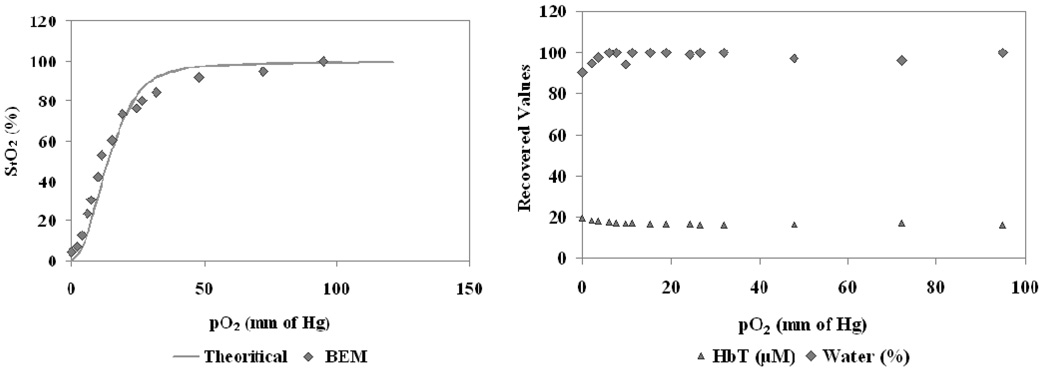
A surface mesh was created for the tissue cup phantom comprising of 2595 nodes from 5186 triangular elements using Netgen [38]. The BEM model was used to obtain homogeneous estimates of NIR optical properties using the homogeneous fitting procedure described in Section 2.2 using Newton-Raphson method. This minimization procedure successfully updated the optical properties which were then used with spectral fitting to obtain homogeneous values for total hemoglobin, oxygen saturation, water content and scatter parameters. The results are shown in Figure 2 for the NIRS estimates. The recovered oxygen saturation followed the expected values from literature with a mean error of 6.6%. The total hemoglobin and water stayed constant with the variation in pO2 as expected with some crosstalk between them at low pO2 values (below 8mm Hg). The average for recovered total hemoglobin was 17µM with a standard deviation of 0.9µM compared to expected value of 18µM measured for the whole blood using a clinical co-oximeter. The water recovered average was 98% with standard deviation of 3% comparable to expected 100% for liquid solution. The reduced scattering coefficient at 785nm stayed constant with average of 1.29 mm−1 and standard deviation of 0.03 mm-1.
Fig. 2.
3-D BEM estimates are shown from homogeneous fitting algorithm for a phantom with varying oxygen partial pressure (pO2) of the solution, induced by addition of yeast. The oxygen saturation is shown in (a) along with the theoretical estimate from the Hill curve. The total hemoglobin and % water are shown in (b), with the theoretically estimated values of 18 µM and 100% respectively, and should not vary with changes in oxygenation. The reduced scattering coefficient at 785 nm also stayed constant as expected (not shown here).
3.2 Variation in total hemoglobin
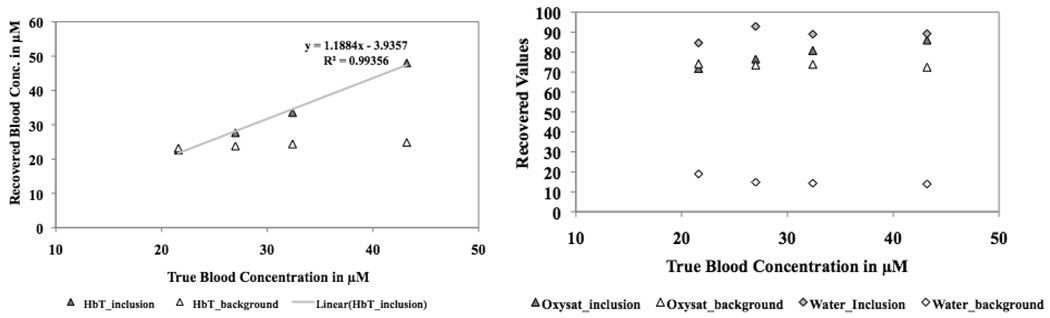
A surface mesh comprising of two surfaces (outer and inclusion) for the gelatin phantom was created using Netgen with mesh size of 2154 nodes and 4300 triangles. The datasets obtained with varying hemoglobin were calibrated using the homogeneous fitting algorithm and used to reconstruct for the NIRS parameters using the 3–D BEM spectral reconstruction for the two region problem. The recovered values in the inclusion and the background are shown in Figure 3(a). As expected, the total hemoglobin in the inclusion increased linearly with change in concentration while the background stayed fixed. The other NIRS parameters (oxygen saturation, water and scattering) stayed constant with this variation in hemoglobin as expected, as shown in Figure 3(b).
Fig. 3.
3–D BEM-based spectral recovery was used to track changes in total hemoglobin in a phantom experiment. The recovered values are shown in (a) for the inclusion and the background. The hemoglobin is expected to vary linearly in the inclusion and stay constant in the background. The other parameters did not vary as shown in (b) for oxygen saturation and water. Not shown here: the reduced scattering coefficient at 785 nm also stayed constant.
4. DISCUSSION
A key advantage to using BEM is the ease in meshing since only surface discretization is necessary. Volume meshing is a demanding task that takes up to 10–100 times the computational effort required to create surface meshes. This has implication in the clinical work-flow especially when processing large number of data sets. As IG-NIRS systems move into clinical trials, optimization for large scale trials become especially important and the use of BEM facilitates this need. The use of spectral constraints ensures accuracy in the recovered chromophores and minimal cross talk between the parameters.
We have validated this approach in phantom experiments by showing linear tracking of changes in total hemoglobin and recovering the hill-curve response of oxygen saturation with changes in partial pressure of oxygen. The next stage of demonstration of this approach will likely involve processing a complete cohort of data to analyze the biological parameter, rather than the computational parameters, as was the focus here.
ACNOWLEDGEMENTS
This work was supported by NIH grants PO1CA80139 and R01EB007966-01A1.
REFERENCES
- 1.Chance B, Nioka S, Zhang J, Conant EF, Hwang E, Briest S, Orel SG, Schnall MD, Czerniecki BJ. Breast cancer detection based on incremental biochemical and physiological properties of breast cancers: a six-year, two-site study. Acad Radiol. 2005;12(8):925–933. doi: 10.1016/j.acra.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 2.Poplack SP, Tosteson TD, Wells WA, Pogue BW, Meaney PM, Hartov A, Kogel CA, Soho SK, Gibson JJ, Paulsen KD. Electromagnetic breast imaging: results of a pilot study in women with abnormal mammograms. Radiology. 2007;243(2):350–359. doi: 10.1148/radiol.2432060286. [DOI] [PubMed] [Google Scholar]
- 3.Cerussi A, Shah N, Hsiang D, Durkin A, Butler J, Tromberg B. In vivo absorption, scattering, and physiologic properties of 58 malignant breast tumors determined by broadband diffuse optical spectroscopy. Journal of Biomed Opt. 2006;11(4):044005–04400516. doi: 10.1117/1.2337546. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan S, Pogue BW, Brooksby B, Jiang S, Dehghani H, Kogel C, Wells WA, Poplack SP, Paulsen KD. Near-infrared characterization of breast tumors in-vivo using spectrally-constrained reconstruction. Technology in Cancer Research and Treatment. 2005;4(5):513–526. doi: 10.1177/153303460500400505. [DOI] [PubMed] [Google Scholar]
- 5.Tromberg BJ, Shah N, Lanning R, Cerussi A, Espinoza J, Pham T, Svaasand L, Butler J. Neoplasia. 1–2. Vol. 2. New York: 2000. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy; pp. 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452:580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alacam B, Yazici B, Intes X, Nioka S, Chance B. Pharmacokinetic-rate images of indocyanine green for breast tumors using near-infrared optical methods. Physics in Medicine and Biology. 2008;53:837–859. doi: 10.1088/0031-9155/53/4/002. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter C, Pogue BW, Jiang S, Dehghani H, Wang X, Paulsen KD, Wells WA, Forero J, Kogel C, Weaver J, Poplack SP, Kaufman PA. Image-guided optical spectroscopy provides molecular-specific information in vivo: MRI-guided spectroscopy of breast cancer hemoglobin, water & scatterer Size. Optics Letters. 2007;32(8):933–935. doi: 10.1364/ol.32.000933. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Q, Brukilacchio TJ, Li A, Stott JJ, Chaves T, Hillman E, Wu T, Chorlton M, Rafferty E, Moore RH, Kopans DB, Boas DA. Coregistered tomographic x-ray and optical breast imaging: initial results. J Biomed Opt. 2005;10(2):024033–0240339. doi: 10.1117/1.1899183. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Q, Cronin EB, Currier AA, Vine HS, Huang M, Chen N, Xu C. Benign versus malignant breast masses: optical differentiation with US-guided optical imaging reconstruction. Radiology. 2005;237(1):57–66. doi: 10.1148/radiol.2371041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerussi A, Hsiang D, Shah N, Mehta R, Durkin A, Butler J, Tromberg BJ. Predicting response to breast cancer neoadjuvant chemotherapy using diffuse optical spectroscopy. PNAS. 2007;104(10):4014–4019. doi: 10.1073/pnas.0611058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schweiger M, Arridge SR. Comparison of two- and three-dimensional reconstruction methods in optical tomography. Applied Optics. 1998;37(31):7419–7428. doi: 10.1364/ao.37.007419. [DOI] [PubMed] [Google Scholar]
- 13.Enfield LC, Gibson AP, Everdill NL, Delpy DT, Schweiger M, Arridge SR, Richardson C, Keshtgar M, Douek M, Hebden JC. Three-dimensional time-resolved optical mammography of the uncompressed breast. Applied Optics. 2007;46(17):3628–3638. doi: 10.1364/ao.46.003628. [DOI] [PubMed] [Google Scholar]
- 14.Eppstein MJ, Dougherty DE, Hawrysz DJ, Sevick EM. Three-dimensional bayesian optical image reconstruction with domain decomposition. IEEE Trans Med Imaging. 2001;20(3):147–162. doi: 10.1109/42.918467. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan S, Pogue BW, Dehghani H, Leblond F, Intes X. A Data Subset Algorithm for Computationally Efficient Reconstruction of 3-D Spectral Imaging in Diffuse Optical Tomography. Optics Express. 2006;14(12):5394–5410. doi: 10.1364/oe.14.005394. [DOI] [PubMed] [Google Scholar]
- 16.Dehghani H, Pogue BW, Poplack SP, Paulsen KD. Multiwavelength three-dimensional near-infrared tomography of the breast: initial simulation, phantom, and clinical results. Applied Optics. 2003;42(1):135–145. doi: 10.1364/ao.42.000135. [DOI] [PubMed] [Google Scholar]
- 17.Guven M, Yazici B, Intes X, Chance B. Diffuse optical tomography with apriori anatomical information. Physics in Medicine and Biology. 2005;50:2837–2858. doi: 10.1088/0031-9155/50/12/008. [DOI] [PubMed] [Google Scholar]
- 18.Brooksby B, Jiang S, Dehghani H, Pogue BW, Paulsen KD, Weaver J, Kogel C, Poplack SP. Combining near infrared tomography and magnetic resonance imaging to study in vivo breast tissue: implementation of a Laplacian-type regularization to incorporate MR structure. Journal of Biomedical Optics. 2005;10(5) doi: 10.1117/1.2098627. 051504-1:10. [DOI] [PubMed] [Google Scholar]
- 19.Li A, Miller EC, Kilmer ME, Brukilacchio TJ, Chaves T, Stott J, Zhang Q, Wu T, Chorlton M, Moore RH, Kopans DB, Boas DA. Tomographic optical breast imaging guided by three-dimensional mammography. Applied Optics. 2003;42(25):5181–5190. doi: 10.1364/ao.42.005181. [DOI] [PubMed] [Google Scholar]
- 20.Dehghani H, Pogue BW, Shudong J, Brooksby B, Paulsen KD. Three-dimensional optical-tomography: resolution in small-object imaging. Applied Optics. 2003;42(16):3117–3128. doi: 10.1364/ao.42.003117. [DOI] [PubMed] [Google Scholar]
- 21.Yalavarthy PK, Lynch DR, Pogue BW, Dehghani H, Paulsen KD. Implementation of a computationally efficient least-squares algorithm for highly under-determined three-dimensional diffuse optical tomography problems. Medical Physics. 2008;35(5):1682–1697. doi: 10.1118/1.2889778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yalavarthy PK, Pogue BW, Dehghani H, Paulsen KD. Weight-matrix structured regularization provides optimal generalized least-squares estimate in diffuse optical tomography. Med Phys. 2007;34(6):2085–2098. doi: 10.1118/1.2733803. [DOI] [PubMed] [Google Scholar]
- 23.Azar FS, Lee K, Khamene A, Choe R, Corlu A, Konecky SD, Sauer F, Yodh AG. Standardized platform for co-registration of nonconcurrent diffuse optical and magnetic resonance breast images obtained in different geometries. Journal of Biomedical Optics. 2007;12(5):051902. doi: 10.1117/1.2798630. [DOI] [PubMed] [Google Scholar]
- 24.Brooksby B, Dehghani H, Pogue BW, Paulsen KD. Near infrared (NIR) tomography breast image reconstruction with apriori structural information from MRI: algorithm development for reconstructing heterogeneities. IEEe Journal of Selected Topics in Quantum Electronics on Lasers in Medicine and Biology. 2003;9:199–209. [Google Scholar]
- 25.Brebbia CA, Dominguez J. Boundary elements: an introductory course. Boston: Computational Mechanics Publications; 1992. [Google Scholar]
- 26.Zacharopoulos AD, Arridge SR, Dorn O, Kolehmainen V, Sikora J. Three-dimensional reconstruction of shape and piecewise constant region values for optical tomography using spherical harmonic parametrization and a boundary element method. Inverse Problems. 2006;22:1509–1532. [Google Scholar]
- 27.Mcbride TO, Pogue BW, Jiang S, Osterberg UL, Paulsen KD. A parallel-detection frequency-domain near-infrared tomography system for hemoglobin imaging of the breast in vivo. Review of Scientific Instruments. 2001;72(3):1817–1824. [Google Scholar]
- 28.McBride TO. Dartmouth College Thesis. 2001. Spectroscopic reconstructed near infrared tomographic imaging for breast cancer diagnosis, in Thayer School of Engineering; pp. 1–200. [Google Scholar]
- 29.Ishimaru A. Wave propagation and scattering in random media. New York: Academic Press, Inc.; 1978. [Google Scholar]
- 30.Patterson MS, Wilson BC, Wyman DR. The propagation of optical radiation in tissue II. Optical properties of tissues and resulting fluence distributions. Lasers Med. Sci. 1990;6:379–390. [Google Scholar]
- 31.Farrell TJ, Patterson MS, Wilson BC. A diffusion theory model of spatially resolved, steady-state diffuse reflectance for the noninvasive determination of tissue optical properties. Med. Phys. 1992;19(4):879–888. doi: 10.1118/1.596777. [DOI] [PubMed] [Google Scholar]
- 32.Fedele F, Eppstein MJ, Laible JP, Godavarty A, Sevick-Muraca EM. Fluorescence photon migration by the boundary element method. Journal of computational physics. 2005;210:109–132. [Google Scholar]
- 33.Sikora J, Zacharopoulos A, Douiri A, Schweiger M, Horesh L, Arridge SR, Ripoll J. Diffuse photon propagation in multilayered geometries. Phys Med Biol. 2006;51:497–516. doi: 10.1088/0031-9155/51/3/003. [DOI] [PubMed] [Google Scholar]
- 34.Srinivasan S, Pogue BW, Carpenter C, Yalavarthy PK, Paulsen KD. A boundary element approach for image-guided near-infrared absorption and scatter estimation. Medical Physics. 2007;34(11):4545–4557. doi: 10.1118/1.2795832. [DOI] [PubMed] [Google Scholar]
- 35.Srinivasan S, Pogue BW, Jiang S, Dehghani H, Paulsen KD. Spectrally constrained chromophore and scattering NIR tomography provides quantitative and robust reconstruction. Applied Optics. 2005;44(10):1858–1869. doi: 10.1364/ao.44.001858. [DOI] [PubMed] [Google Scholar]
- 36.Corlu A, Choe R, Durduran T, Lee K, Schweiger M, Arridge SR, Hillman EM, Yodh AG. Diffuse optical tomography with spectral constraints and wavelength optimization. Appl Opt. 2005;44(11):2082–2093. doi: 10.1364/ao.44.002082. [DOI] [PubMed] [Google Scholar]
- 37.Li A, Zhang Q, Culver JP, Miller EL, Boas DA. Reconstructing chromosphere concentration images directly by continuous-wave diffuse optical tomography. Opt Lett. 2004;29(3):256–258. doi: 10.1364/ol.29.000256. [DOI] [PubMed] [Google Scholar]
- 38.Schöberl J. http://www.hpfem.jku.at/netgen/