Summary
Huntington's disease (HD) is a genetic neurodegenerative disorder resulting from polyglutamine (polyQ) expansion (> 36Q) within first exon of Huntingtin (Htt) protein. Here we applied X-ray crystallography to determine the secondary structure of the first exon (EX1) of Htt-17Q. The structure of Htt17Q-EX1 consists of an amino-terminal α-helix, a poly17Q region, and a polyproline helix formed by the proline-rich region. The poly17Q region adopts multiple conformations in the structure, including α-helix, random coil and extended loop. The conformation of the poly17Q region is influenced by the conformation of neighbouring protein regions, demonstrating importance of the native protein context. We propose that the conformational flexibility of the polyQ region observed in our structure is a common characteristic of many amyloidogenic proteins. We further propose that the pathogenic polyQ-expansion in the Htt protein increases the length of the random coil, which promotes aggregation and facilitates abnormal interactions with other proteins in cells.
Introduction
Huntington's disease (HD) is an autosomal-dominant neurodegenerative disorder. Neuropathological analysis of HD patients reveals selective and progressive neuronal loss in the striatum (Vonsattel and DiFiglia, 1998), particularly affecting the GABAergic medium spiny striatal neurons (MSN). At the molecular level, the cause of HD is a polyglutamine (polyQ) expansion (≥36Q) near the amino terminus of Huntingtin (Htt), a 350 kDa ubiquitously expressed cytoplasmic protein (The Huntington's Disease Collaborative Research Group, 1993). The cellular mechanisms that link Huntingtin polyQ expansion (Httexp) with the disease are under intense investigation. Most evidence is consistent with the hypothesis that polyQ-expanded Htt acquires a “toxic gain of function” (Tobin and Signer, 2000). A number of toxic functions have been assigned to Httexp, including formation of toxic aggregates, effect on gene transcription, induction of apoptosis, and disruption of key neuronal functions such as proteosomal function, ubiquitination, axonal transport, endocytosis, synaptic transmission and Ca2+ signaling (Bezprozvanny, 2009; Cha, 2007; Li and Li, 2004; Ross, 2002; Rubinsztein, 2002; Tobin and Signer, 2000; Truant et al., 2008). Many of the proposed mechanisms suggest that the mutant Httexp is involved in pathological interactions with other signaling proteins in cells, leading to neuronal dysfunction and death.
Information about the structure of Huntingtin's polyQ region is critical for understanding Httexp toxicity and may aid in the development of potential HD therapies. However, the structure determination of the polyQ region of Htt has proven to be an extremely difficult problem (Temussi et al., 2003). In the aggregated form polyQ sequence most likely adopts β-sheet structure (Bevivino and Loll, 2001; Chen et al., 2001; Perutz, 1996; Perutz et al., 2002; Singer and Dewji, 2006; Takahashi et al., 2008; Tanaka et al., 2001). The biophysical studies of soluble monomeric form of polyQ fragments of various length provided evidence for random coil conformation (Altschuler et al., 1997; Bennett et al., 2002; Chen et al., 2001; Masino et al., 2002), for α-helical conformation (Bhattacharyya et al., 2006; Nagai et al., 2007), β-sheet (Nagai et al., 2007) and extended helix (Chellgren et al., 2006). Random coil, α and β-sheet, α-helix, μ–helix, and π-helix conformations have been predicted using computational methods (Armen et al., 2005; Khare et al., 2005; Lathrop et al., 1998; Tsukamoto, 2006; Wang et al., 2006). Recently the crystal structure of the synthetic peptide GQ10G in the extended conformation was determined in the complex with a monoclonal antibody (Li et al., 2007). Although these studies provided important insights into potential conformations of the polyQ sequence, most of them have been performed in the absence of a native protein context or yielded low resolution data. Here we present the secondary structure of the first exon (EX1) of Htt, containing 17 glutamines (Htt17Q) determined by X-ray crystallography at atomic resolution. We discovered that the poly17Q region adopts multiple conformations in the structure, including α-helix, random coil and extended loop. We concluded that the conformation of the poly17Q region are influenced by the conformation of neighbouring protein regions, demonstrating the importance of the native protein context. We propose that the conformational flexibility of the polyQ region observed in our structure is a common characteristic of many amyloidogenic proteins.
Results
Crystallization of Htt-17Q exon 1 fragment
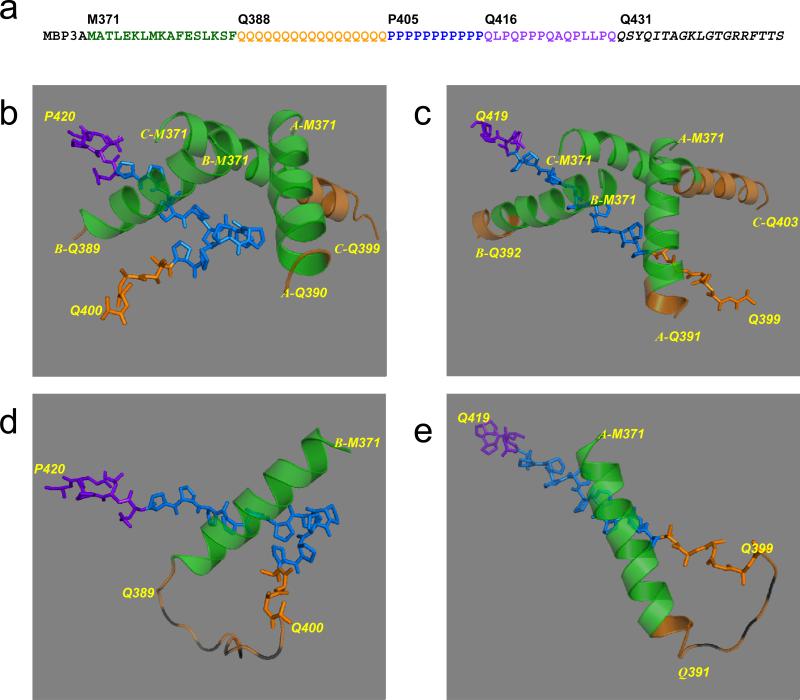
The first exon of Huntingtin protein containing 17 glutamines (Htt17Q-EX1) was expressed in bacteria and crystallized as maltose-binding protein (MBP) fusion protein (see Experimental Procedures for details). The amino-terminal MBP molecule was connected with Htt17Q-EX1 via 3 alanines (3A) linker. The amino-acid (aa) numbering in MBP-Htt17Q-EX1 construct starts with MBP, so that Met1 in the Htt17Q corresponds to Met371 in the MBP-Htt17Q-EX1. The sequence of Htt17Q-EX1 can be subdivided into several distinct regions (Fig 1a). The N-terminal region of Htt17Q-EX1 starting at Met371 (Met1 in Htt sequence) extends for 17aa until Phe387. The next region Gln388 - Gln404 contains a stretch of 17Gln (poly17Q), which is immediately followed by a stretch of 11Pro from Pro405 until Pro415 (poly11P). The proline stretch is followed by 15 aa mixed Pro and Gln region (polyP/Q) that spans from Gln416 until Gln430. The native Htt sequence ends at Gln430 and is followed by 19aa C-terminal tag which was added to facilitate crystallization (Fig. 1a).
Fig. 1. Secondary structure of Htt17Q-EX1.
(a) Amino acid sequence of MBP-Htt17Q-EX1. MBP3A denotes the maltose binding protein followed by a 3Ala linker. M371 to G429 is the sequence of Htt17Q-EX1, which is subdivided into 17 aa N-terminal region (M371 to F387), poly17Q region (Q388 to Q404), poly11P region (P405 to P415), and 15 aa mixed P/Q region (Q416 to Q430). The sequence from Q431 to the C-terminus is the 19 aa tag added to facilitate crystallization. (b) The structure of Htt17Q-EX1 trimer from c95 crystal. The structures of MBP and 3A linker are removed for clarity. The amino-terminal α-helix of Htt17Q-EX1 extends from Met371 to Phe387 (Green). The following poly17Q region (Orange) is α-helical and unstructured (random coil). The poly11P region (Blue) adopts PP-helix (shown as stick model) in the “kinked” conformation. The initial part of polyP/Q region (Purple) is also in PP-helix conformation (shown as stick model). The terminal part of poly17Q region (Orange) is in the extended conformation (shown as stick model) (c) The structure of Htt17Q-EX1 trimer from cHg99 crystal. Same as on panel b but the Poly11P region is in the “straight” conformation. (d) The complete structure of B molecule of Htt17Q-EX1 monomer from c95 crystal. The striped orange loops are for the random coil region between Gln389 and Gln399, which is invisible on the map. (e) The complete structure of A molecule of Htt17Q-EX1 monomer from cHg99 crystal. The striped orange loops are for the random coil region between Gln391 and Gln398, which is invisible on the map.
X-ray diffraction data were collected for 30 crystals of MBP-Htt17Q-EX1. The diffraction for all crystals was anisotropic, extending to 2.8 Å in the high resolution directions, [h, 0, 0] and [0, k, 0] and 4.5 Å in the low resolution direction, [0, 0, l]. All crystals were monoclinic, space group C121, with slightly different unit cell size and β angles in the range from 90° to 99°. The asymmetric unit contained 3 molecules of MBP-Htt17Q-EX1 (designated as molecules A, B, C) in all crystals. The phasing was done by molecular replacement (MR) using MBP structural models (see Experimental Procedures for details). MR gave solutions for 7 crystals with complete maps for MBP and partial maps for Htt17Q-EX1. The β angles and other crystallographic parameters for these 7 crystals are listed in Table 1. Following MR, the models of Htt17Q-EX1 fragments in these 7 crystals were built separately but simultaneously. The final refinement statistics for the 7 crystals used in Htt17Q-EX1 structure determination are summarized in Table 1. The PDB accession numbers for each of the seven data sets are also included in Table 1. The final 7 structures of Htt17Q-EX1 were very similar despite differences in β angles in the corresponding crystals.
Table 1.
Diffraction data collection and refinement statistics for seven crystals used for Htt17Q-EX1 structure determination.
| Crystal |
c90 |
ca92 |
cb92 |
c94 |
c95 |
c99 |
cHg99 |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Wavelength (λ) | 1.55 | 0.98 | 0.98 | 1.55 | 0.98 | 0.98 | 0.98 |
| Space group | C121 | C121 | C121 | C121 | C121 | C121 | C121 |
| Unit Cell parameter ( Å, ° ) | |||||||
| a | 162 | 163 | 163 | 160 | 163 | 163 | 101 |
| b | 101 | 101 | 101 | 100 | 100 | 101 | 162 |
| c | 142 | 138 | 137 | 140 | 137 | 134 | 139 |
| β | 90 | 92 | 92 | 94 | 95 | 99 | 99 |
| Resolution limit ( Å ) | 3.8-41 / 3.8-4.1 | 4.0-37 / 4.0-4.6 | 3.5-37 / 3.5-3.7 | 3.7-40 / 3.7-3.8 | 3.5-37 / 3.5-3.6 | 43-3.5/3.5-3.6 | 3.5-40/3.5-3.7 |
| Rsym(%) | 12.8 / 24.1 | 6.3 / 12.8 | 21.2 / 30.1 | 12.4 / 47.1 | 9.9 / 33.2 | 8.6 / 35.0 | 16.0 / 24.8 |
| I/σI (overall /outer shell) | 13.2 / 3.1 | 13.2 / 5.9 | 8.4 / 2.3 | 14.3 / 3.1 | 9.7 / 2.9 | 10.0 / 1.3 | 15.0 / 4.8 |
| Redundancy (overall/outer shell) | 2.0 / 1.9 | 1.8 / 1.8 | 3.2 / 2.8 | 2.4 / 1.7 | 2.7 / 1.9 | 2.8 / 1.5 | 3.7 / 3.2 |
| Overall completeness ( % ) | 64 | 92 | 69 | 83 | 82 | 87 | 92 |
| Refinement | |||||||
| Overall mean B-factor ( Å2 ) | 104 | 81 | 56 | 70 | 56 | 82 | 48 |
| No of data | 16075 | 21188 | 18671 | 19565 | 20472 | 20629 | 24515 |
| RWork / RFree | 0.251/0.303 | 0.245/ 0.292 | 0.247 / 0.295 | 0.272/0.298 | 0.257 / 0.267 | 0.263 / 0.280 | 0.243 / 0.280 |
| RMS_D ( Å ) | 0.009 | 0.006 | 0.009 | 0.007 | 0.007 | 0.007 | 0.006 |
| RMS_A ( ° ) |
1.180 |
0.937 |
1.149 |
1.171 |
1.335 |
0.979 |
0.961 |
| PDB ID | 3IO4 | 3IO6 | 3IOT | 3IOU | 3IOR | 3IOV | 3IOW |
The crystals are sorted by β angle of unit cell (C121). The values of β angles are assigned to each crystal name in superscript. For the same β angles crystals are noted by letters of the alphabet in subscript. The mercury socked crystal is indicated by Hg in subscript. PDB accession numbers for the data collected with each crystal are shown in the last raw.
Secondary structure of the Htt17Q-EX1 fragment
MBP-Htt17Q-EX1 forms a trimer, composed of molecules A, B and C and packed sandwich-like in the crystal (Supplementary Figure 1). The trimer is likely to be an artifact of crystallization since MBP-Htt17Q-EX1 is a monomer in solution according to dynamic light scattering and size exclusion chromatography analysis (data not shown). Two representative models of Htt17Q-EX1 trimer are shown on Figs 1b (crystal c95) and 1c (crystal cHg99). The models consist of the N-terminal region of Htt17Q-EX1 starting with Met371 (Green), the poly17Q region (Orange), and stretches of poly11P (Blue) and a mixed polyP/Q region (Purple). The N-terminal region of Htt17Q-EX1 (Green) is resolved as α-helix for all 3 molecules (A, B, C) in the asymmetric unit. The structures of N-terminal α-helical trimers are identical in all seven crystals and can be superimposed on each other. The initial portions of poly17Q region (Orange) on Figs 1b and 1c can be identified as α-helix or loop for all 3 molecules. The rest of these poly17Q regions is not visible on the map and not shown on Figs 1b and 1c. Only a single poly11P region (Blue) and initial fragment of polyP/Q region (Purple) is resolved on the map in PP helix form and the same region is invisible for other 2 molecules in the asymmetric unit. The structure of poly11P in crystals c94, c95, and c99 is kinked in the middle (Fig 1b, Table 2). In crystals c90, cb92, and cHg99 the poly11P is straight (Fig 1c, Table 2). In crystal ca92 the poly11P region adopts a mixture of “kinked” and “straight” conformations and can not be clearly resolved on the map (Table 2). The last 4 glutamine residues from the poly17Q region immediately preceding poly11P reappear on the map in the extended conformation (Orange).
Table 2.
The last visible residue in poly17Q α-helix and conformation of poly11P in different MBP-Htt17Q-EX1 crystals.
| Crystal |
c90 |
ca92 |
cb92 |
c94 |
c95 |
c99 |
cHg99 |
|---|---|---|---|---|---|---|---|
|
β angle
|
90 |
92 |
92 |
94 |
95 |
99 |
99 |
| α-helix A | 395 | 388 | 388 | 388 | 388 | 390 | 389 |
| B | 397 | 401 | 386 | 390 | 388 | 396 | 392 |
|
C
|
395 |
396 |
391 |
400 |
396 |
393 |
402 |
| PP-helix | Straight | * | Straight | Kinked | Kinked | Kinked | Straight |
Not resolved on the map
Examples of A and B molecules of Htt17Q-EX1 are shown on Figs 1d and 1e. The B molecule from crystal c95 is shown on Fig 1d. The structure of this molecule includes N-terminal α-helix (Green), polyQ stretch (Orange), “kinked” poly11P region (Blue), and initial part of polyP/Q region (Purple). The unstructured random coil portion of polyQ region (Gln389-Gln399) is shown on Fig 1d as a striped orange loop. The remaining carboxy-terminal polyQ region (Gln400-Gln404) is resolved in the extended loop conformation (Orange stick model). The A molecule from cHg99 is shown on Fig 1e. This structure includes N-terminal α-helix (Green), polyQ stretch (Orange), “straight” poly11P region (Blue), and initial portion of the polyP/Q region (Purple). The beginning of the polyQ region (Gln388-Gln390) is α-helical and the carboxy-terminal portion of polyQ region (Gln399-Gln404) is in the extended loop conformation (Orange stick model). The random coil portion of polyQ region in this molecule (Gln391-Gln398) is shown by the striped orange loop.
Conformation of the polyglutamine region
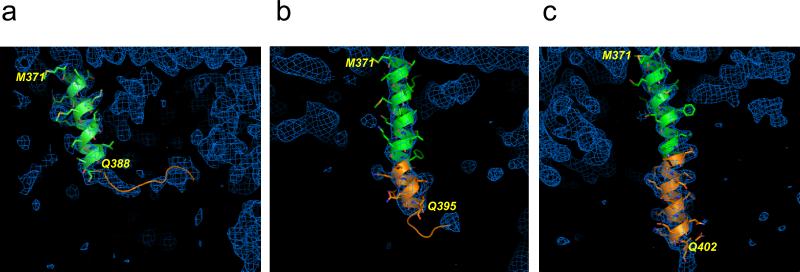
For all 3 molecules (A, B, and C) in all seven crystals the N-terminal portion of Htt17Q-EX1 (Green) is α-helical (Figs 1b and 1c). In all molecules N-terminal α-helix continue until Phe387 but eventually lose helical conformation within the polyQ region (Orange). The remainder of polyQ region following the α-helixes form “random coil” conformation. The polyQ region is connected to the poly11P region in “kinked” (e.g. Fig 1d) or “straight” (e.g. Fig 1e) PP-helical conformations. The last 4 Gln residues at the carboxy-terminal end of poly17Q region (Gln400-Gln404) are in the extended conformation (Figs 1d and 1e). The transition from α-helix to a random coil within poly17Q region occurs at variable positions for the six molecules shown in Fig. 1b and 1c. The examples of three α-helixes with various lengths chosen from three different crystals and corresponding electron density maps are presented on Fig 2. The shortest helix is in the A molecule in crystal c95. The α-helix in this crystal continues only until Gln388, the first residue of the poly17Q region, and then becomes a loop (Fig 2a). The diminishing electron density map of the polyQ loop of the short α-helix on Fig. 2a approaches hydrophilic residues Asp41, Lys42, Glu45 and Gln49 in MBP-B or MBP-A molecules. The medium length α-helix is C molecule of crystal c90, which extends until Gln395 and then becomes a loop (Fig 2b). The longest α-helix is observed in the C molecule of crystal cHg99; it extends until Gln402 and then becomes random coil which is invisible on the map (Fig 2c). Table 2 indicates the last residues of the poly17Q α-helixes for molecules A, B and C in each of the solved 7 crystals with different β angles.
Fig. 2. Structure of amino-terminal and poly17Q regions of Htt17Q-EX1.
The structures of amino-terminal region (Green) and poly17Q region (Orange) of Htt17Q-EX1 are shown for three different molecules with variable length of poly17Q α-helix. Also shown are corresponding regions of electron density maps contoured at 1.0σ (Blue). (a) The short poly17Q helix makes transition to loop at Gln388 (molecule A, crystal c95). (b) The medium poly17Q helix makes transition to loop at Gln395 (molecule C, crystal c90). (c) The long poly17Q helix extends for the length of polyQ region until Gln402 (molecule C, crystal cHg99).
Conformation of the polyproline region
The poly11P region is resolved as PP-helix in the crystals. Only single poly11P helix is resolved in the trimer, which adopts “kinked” or “straight” conformation in different crystals (Table 2) The beginning of poly11P in the “kinked” conformation is positioned at the equal distance between A and B N-terminal α-helices (Fig 1b) and the beginning of poly11P in the “straight” conformation is positioned between A and C N-terminal α-helices (Fig 1c). The poly11P region is connected with the N-terminal α-helices via the polyQ random coil as described above. Following poly11P region, a few residues from the polyQ/P mixed region (shown in purple on Figs 1b and 1c) maintain an extended conformation until the structure is no longer visible. The parts of poly11P helix are stabilized by interactions with hydrophobic surface of the α-helical trimers in the crystals (Figs 1b and 1c).
Discussion
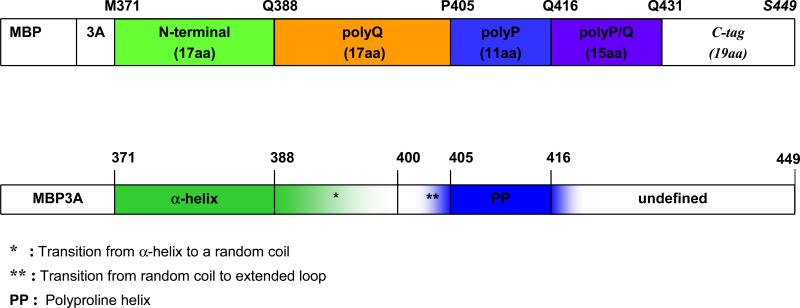
The two parallel bar diagrams on Fig. 3 show the protein sequence of MBP-Htt17Q-EX1 and the corresponding secondary structure of each sequence region as determined from our crystallographic data. The N-terminal region of Htt (M371 to F387) is α-helical in all crystals (bold Green). The N-terminal α-helix ends at variable residues within poly17Q region between Q388 and Q400 (shaded Green) and the rest of poly17Q region continues as a random coil (not colored). The polyQ residues immediately preceding poly11P region (Q400 to Q404) adopt extended loop conformation (shaded Blue). The poly11P region exists as PP helix (bold Blue) in all crystals, either in the “kinked” or “straight” conformation. The secondary structure beyond poly11P region continues as PP helix for several residues into mixed P/Q region (shaded Blue) until structure can no longer be resolved.
Fig. 3. Schematic diagram of the secondary structure elements of Htt17Q-EX1.
The first bar represents the sequence of MBP-Htt17Q-EX1 construct which consists of MBP protein (not shown to scale), 3 Alanines linker (3A), 17 aa long N-terminal region of Htt (green), polyQ region with 17 glutamine residues (poly17Q, orange), polyP region with 11 proline residues (poly11P, blue)), 15 aa long mixed polyglutamine and polyproline region (polyP/Q, purple), and C-terminal tag of 19 aa (not coloured). The second bar is a summary of structural information obtained from analysis of Htt17Q-EX1 structures resolved in the seven crystals. The main secondary structure elements of Htt17Q-EX1 are shown: α-helix (bold green), transition from α-helix to random coil region (shaded green), random coil (uncoloured), transition to extended loop region (shaded blue), PP helix (bold blue), transition from PP helix to unstructured region (shaded blue).
It is informative to compare our results with previous experimental and modelling studies of Huntingtin and other polyQ-containing proteins. The molecular dynamics simulations of Htt amino-terminal headpiece predicted α-helical structure (Kelley et al., 2009) and the α-helical structure of Htt amino-terminal peptide was observed by CD spectra analysis (Atwal et al., 2007). However, predominantly random coil conformation for the same region was reported by another group based on CD and NMR spectra analysis (Thakur et al., 2009). The Htt N-terminal headpiece modulates subcellular localization and aggregation of Httexp (Atwal et al., 2007; Rockabrand et al., 2007; Thakur et al., 2009), and definitive information about its secondary structure is useful for understanding these effects. The N-terminal region of Htt is α-helical in all our crystals (Fig 3), consistent with the molecular dynamics simulations and biophysical measurements (Atwal et al., 2007; Kelley et al., 2009). The agreement with these studies indicate that ability to form α-helix is an intrinsic property of Htt amino-terminal region and not a result of crystallization of Htt17Q-EX1 as amino-terminal MBP-fusion protein.
A range of structures was reported for polyQ region in the previous experimental and modelling studies. Some studies indicated that monomeric polyQ peptide is unstructured and exists as a random coil in solution (Altschuler et al., 1997; Bennett et al., 2002; Chen et al., 2001; Masino et al., 2002). Using CD spectra analysis other groups concluded that polyQ stretch is at least partially α-helical (Bhattacharyya et al., 2006; Nagai et al., 2007), an observation supported by modelling results (Lathrop et al., 1998; Wang et al., 2006). We have found evidence for both random coil and α-helical conformations of polyQ in our structures (Fig 2), in agreement with both sets of observations. We further discovered that transition between α-helical and random coil conformations of polyQ region occurs at random positions (Fig 2, Table 2), indicating a very shallow equilibrium between these two polyQ conformations as suggested by molecular dynamics stimulations and biophysical measurements (Bhattacharyya et al., 2006; Wang et al., 2006). Additional biophysical and modelling studies provided evidence that short polyQ stretches may also exist as the extended conformation (Chellgren et al., 2006; Wang et al., 2006). The ability of polyQ fragments to adopt an extended conformation was further supported by crystal structure of GQ10G peptide bound to a monoclonal antibody (Li et al., 2007). In our structures the last 4 glutamine residues also adopt the extended conformation (Fig 3). The analysis of these structures led us to conclude that the extended conformation at the carboxy-terminal end of polyQ stretch is imposed by PP helix of the neighbouring poly11P region. Similar conclusions were reached in biophysical studies of polyQ-polyP peptides in solution (Darnell et al., 2007). An ability of poly11P region to stabilize the structure of a polyQ stretch in the extended conformation may account for a known role played by poly11P region in reducing aggregation and toxicity of Httexp (Bhattacharyya et al., 2006; Darnell et al., 2007; Dehay and Bertolotti, 2006; Duennwald et al., 2006)
The polyQ in the monomeric state is likely to exist in an equilibrium between α-helical, random coil and extended conformations, which is consistent with the range of structures observed in Htt17Q-EX1 crystals (Fig 3) and with results obtained in biophysical studies performed with the soluble proteins (Altschuler et al., 1997; Bennett et al., 2002; Bhattacharyya et al., 2006; Chellgren et al., 2006; Chen et al., 2001; Darnell et al., 2007; Masino et al., 2002; Nagai et al., 2007). Similar conclusion has been reached in the recent conformational epitope mapping study performed with the mutant Htt-EX1 fragment in solution (Legleiter et al., 2009). The shallow equilibrium between α-helical, loop and extended conformations could be easily affected by the length of polyQ sequence, neighbouring protein context, interactions with other proteins, changes in temperature, or ionic composition. This “conformational flexibility” within Htt-polyQ region observed in our structure might be a common characteristic of many amyloidogenic proteins such as other polyQ-disease proteins and proteins involved in AD, PD and Prion Disease (PrP) (Blondelle et al., 1997; Fraser et al., 1991; Murphy, 2002; Nguyen et al., 1995; Temussi et al., 2003). Similar to Htt-polyQ, the sequences of these proteins also have been predicted to have an ambiguous secondary structure and can easily adopt multiple conformations, some of which may be toxic (Williams and Paulson, 2008).
The polyQ expansions of more than 36Q are toxic to cells and cause HD and other neurodegenerative disorders (Gusella and MacDonald, 2000). One explanation for the toxicity of polyQ expansion is aggregation. The proteins containing expanded polyQ repeats are prone to aggregation and some studies suggested that polyQ aggregates are toxic to neurons (Ross, 2002; Truant et al., 2008). It is generally accepted that in the aggregated state polyQ-expanded proteins adopt β-sheet structure (Bevivino and Loll, 2001; Chen et al., 2001; Perutz, 1996; Perutz et al., 2002; Singer and Dewji, 2006; Tanaka et al., 2001). Some modelling and biophysical studies suggested that monomeric polyQ can also adopt β-sheet conformation in solution (Nagai et al., 2007; Wang et al., 2006). We have not observed an evidence of β-strand conformation in the Htt17Q-EX1 crystals of wild type Htt (Fig 3). It is however possible that in the mutant Httexp long polyQ random coil may adopt different conformation, such as for example β-sheet (Nagai et al., 2007), whose nucleation can occur from the random coil state of expanded polyQ region (Uversky and Fink, 2004; Vitalis et al., 2008) or from some other conformation that needs to be determined experimentally. However it has also been reported that pathogenic and non-pathogenic monomeric polyQ tracts have similar structural properties in solution (Klein et al., 2007), arguing against major conformational changes in long polyQ tracks. To address these questions the crystal structure of Htt-EX1 containing pathological polyQ expansion needs to be determined.
Another explanation for the pathogenic effects of polyQ expansion is abnormal protein interactions. For example, toxicity of Httexp may result from pathological interactions between Httexp and signalling proteins in cells (Bezprozvanny, 2009; Cha, 2007; Li and Li, 2004; Ross, 2002; Rubinsztein, 2002; Tobin and Signer, 2000; Truant et al., 2008). From the structure we found that the α-helical form of polyQ is not involved in any strong molecular interactions. The random coil form of polyQ is localized to the solvent area or near the polar residues of MBP, suggesting weak interactions between polyQ random coil and the polar residues of MBP. From these results we conclude that the α-helical and random coil forms of Htt-polyQ region are relatively “inert”. There are two potential mechanisms which may account for interactions between expanded polyQ region and other proteins. First possibility is that mutant polyQ region may adopt a novel conformation, which makes it more “sticky”. As discussed above, this “toxic” conformation can be a β-sheet or some other conformation. Second possibility is that many weak protein interactions mediated by expanded polyQ in the random coil conformation are sufficient to result in pathological effects. In both cases the interactions between expanded polyQ region (in novel “toxic” conformation or in random coil conformations) would be stronger with certain amino acid sequences in target proteins, resulting in some degree of specificity for these interactions.
In conclusion, here we have applied X-ray crystallography to determine the secondary structure of the first exon of Huntingtin protein containing 17Q sequence. The structure of Htt17Q-EX1 consists of an amino-terminal α-helix, a poly17Q region, and a polyproline (PP) helix formed by the poly11P and mixed P/Q regions. The poly11P region can adopt “kinked” or “straight” conformations. The poly17Q region displays “conformational flexibility” by adopting α-helical, random coil and extended loop conformations in our crystals. The conformations of poly17Q region appear to be influenced by the conformation of neighbouring protein regions, demonstrating importance of the native protein context. We propose that the conformational flexibility of the polyQ region observed in our structure is a common characteristic of many amyloidogenic proteins. We further propose that the pathogenic polyQ-expansion in the Httexp protein increases the length of the random coil, which promotes Httexp aggregation and abnormal interactions with other proteins in cells. Similar ideas are likely to be relevant for understanding structure and toxicity of other polyQ-containing proteins and amyloidogenic proteins in general.
Experimental Procedures
Cloning, protein expression, purification, crystallization and X-ray diffraction data collection
The first exon (EX1) fragment of human huntingtin containing 17 CAG repeats (Met1-Gln59) was amplified by PCR and cloned into NotI and PstI sites of the pMAL vector with modified 3Ala-linker (Center et al., 1998). The amino-acid (aa) numbering in the resulting MBP-Htt17Q-EX1 fusion protein is continuous from the MBP protein, so that Met1 in Htt17Q sequence correspond to Met371 and Gln60 in Htt17Q sequence correspond to Gln430. Following Gln430 in the MBP-Htt17Q-EX1 sequence the 19aa carboxy-terminal (C-terminal) tag (QSYQITAGKLGTGRRFTTS) was added to facilitate crystallization. MBP-Htt17Q-EX1 protein was expressed in BL21 bacteria and purified on Maltose-binding affinity column, followed by molecular exclusion chromatography (Superdex™200 from Pharmacia Biotech) using FPLC. The purified MBP-Htt17Q-EX1 was crystallized in hanging drop without removing MBP in order to improve protein solubility. Single crystals 300/300/20 μm in size grew in the mother drop containing 12% polyethylene glycol, 200 mM Zn Acetate, 200 mM Sodium Acetate, 100 mM Sodium Cacodylate pH = 6.5 to 7.4 at 4 to 16°C in 2 to 6 weeks. Synthetic 10Gaunadines RNA was used as an additive in the molar ratio 1:1 to the protein during crystallization. The crystals were frozen in mother liquid in liquid nitrogen and the X-ray diffraction data were collected at the APS 19ID beam line. Some crystals were soaked in 1 mM CH3HgCl for 1 week prior to freezing. The diffraction data collected from 30 crystals were indexed and processed by HKL2000 (Minor et al., 2000).
Phasing by molecular replacement
All crystals of MBP-Htt17Q-EX1 had the monoclinic symmetry, space group C121, with three molecules of MBP-Htt17Q-EX1 in the asymmetric unit (ASU). For each crystal, phasing was done by molecular replacement (MR) using the Phaser program. 40 MBP pdb files were downloaded from the protein databank and different permutations of 3 MBP models were used as possible models for three proteins in the ASU. The solutions for seven data sets produced by MR were suitable to build a model of Htt17Q-EX1. The pdb files used in MR for these seven crystals are shown in Supplementary Table 1. The MBP structures described in 1JW4, 1JW5, 10MP, 1LLS represent open forms without maltose and differ primarily in the side chain conformation. The crystals were nearly isomorphous but insufficient for averaging them. Thus, the structure determination was performed separately for each crystal. The electron density maps generated by MR consisted of rich and poor electron density layers, with the gradual transition from rich to poor electron density (Supplementary Fig 1). The rich electron density layer corresponded to the layer of MBP molecules and the poor density layer corresponded to the layer of Htt17Q-EX1 in the crystal. The model for Htt17Q-EX1 was built using Coot. Table 1 summaries the properties of the seven crystals and corresponding diffraction data sets used in solving Htt17Q-EX1 structure.
Model building and refinement
When the seven maps generated by MR were compared, it became apparent that the Htt17Q-EX1 fragments adopt somewhat different configuration in different crystals. To account for this fact, the models of Htt17Q-EX1 fragments in the seven crystals were built separately but simultaneously. Furthermore, to account for the weak diffraction from Htt17Q-EX1 fragment, the occupancy of a corresponding model was chosen in the range between 0.80 - 0.30 to yield the best RWork and RFree factors during refinement using refmac5. In the process of model building and refinement the RWork and RFree were improved and the electron density map of Htt17Q-EX1 became more clear. Table 1 summarizes the final refinement statistics for the seven crystals used for Htt17Q-EX1 structure determination.
Supplementary Material
Acknowledgments
We thank Professor Johann Deisenhofer (JD) for generous support of this project and comments on the manuscript, Dominika Borek for help with data collection and processing, the personnel of ID-19 beamline at the Advanced Photon Source (APS) of Argonne National Laboratory, Dr Bostjan Kobe for the gift of the MBP-3A vector, all members of JD laboratory for help and useful advice, Kimberly Aikman for administrative assistance and Youxing Jiang for comments on the paper. This work was supported by the Howard Hughes Medical Institute (JD), Robert A. Welch Foundation (IB and JD), and NIH grants NS056224 (IB) and GM053163 (ZO).
References
- Altschuler EL, Hud NV, Mazrimas JA, Rupp B. Random coil conformation for extended polyglutamine stretches in aqueous soluble monomeric peptides. J Pept Res. 1997;50:73–75. doi: 10.1111/j.1399-3011.1997.tb00622.x. [DOI] [PubMed] [Google Scholar]
- Armen RS, Bernard BM, Day R, Alonso DO, Daggett V. Characterization of a possible amyloidogenic precursor in glutamine-repeat neurodegenerative diseases. Proc Natl Acad Sci U S A. 2005;102:13433–13438. doi: 10.1073/pnas.0502068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Huey-Tubman KE, Herr AB, West AP, Jr., Ross SA, Bjorkman PJ. Inaugural Article: A linear lattice model for polyglutamine in CAG-expansion diseases. Proc Natl Acad Sci U S A. 2002;99:11634–11639. doi: 10.1073/pnas.182393899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevivino AE, Loll PJ. An expanded glutamine repeat destabilizes native ataxin-3 structure and mediates formation of parallel beta -fibrils. Proc Natl Acad Sci U S A. 2001;98:11955–11960. doi: 10.1073/pnas.211305198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny I. Calcium signaling and neurodegenerative diseases. Trends Mol Med. 2009;15:89–100. doi: 10.1016/j.molmed.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A, Thakur AK, Chellgren VM, Thiagarajan G, Williams AD, Chellgren BW, Creamer TP, Wetzel R. Oligoproline effects on polyglutamine conformation and aggregation. J Mol Biol. 2006;355:524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- Blondelle SE, Forood B, Houghten RA, Perez-Paya E. Polyalanine-based peptides as models for self-associated beta-pleated-sheet complexes. Biochemistry. 1997;36:8393–8400. doi: 10.1021/bi963015b. [DOI] [PubMed] [Google Scholar]
- Center RJ, Kobe B, Wilson KA, Teh T, Howlett GJ, Kemp BE, Poumbourios P. Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein. Protein Sci. 1998;7:1612–1619. doi: 10.1002/pro.5560070715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JH. Transcriptional signatures in Huntington's disease. Prog Neurobiol. 2007;83:228–248. doi: 10.1016/j.pneurobio.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellgren BW, Miller AF, Creamer TP. Evidence for polyproline II helical structure in short polyglutamine tracts. J Mol Biol. 2006;361:362–371. doi: 10.1016/j.jmb.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Chen S, Berthelier V, Yang W, Wetzel R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J Mol Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- Darnell G, Orgel JP, Pahl R, Meredith SC. Flanking polyproline sequences inhibit beta-sheet structure in polyglutamine segments by inducing PPII-like helix structure. J Mol Biol. 2007;374:688–704. doi: 10.1016/j.jmb.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Dehay B, Bertolotti A. Critical role of the proline-rich region in Huntingtin for aggregation and cytotoxicity in yeast. J Biol Chem. 2006;281:35608–35615. doi: 10.1074/jbc.M605558200. [DOI] [PubMed] [Google Scholar]
- Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci U S A. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PE, Nguyen JT, Surewicz WK, Kirschner DA. pH-dependent structural transitions of Alzheimer amyloid peptides. Biophys J. 1991;60:1190–1201. doi: 10.1016/S0006-3495(91)82154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nat Rev Neurosci. 2000;1:109–115. doi: 10.1038/35039051. [DOI] [PubMed] [Google Scholar]
- Kelley NW, Huang X, Tam S, Spiess C, Frydman J, Pande VS. The predicted structure of the headpiece of the Huntingtin protein and its implications on Huntingtin aggregation. J Mol Biol. 2009;388:919–927. doi: 10.1016/j.jmb.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare SD, Ding F, Gwanmesia KN, Dokholyan NV. Molecular origin of polyglutamine aggregation in neurodegenerative diseases. PLoS Comput Biol. 2005;1:230–235. doi: 10.1371/journal.pcbi.0010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein FA, Pastore A, Masino L, Zeder-Lutz G, Nierengarten H, Oulad-Abdelghani M, Altschuh D, Mandel JL, Trottier Y. Pathogenic and non-pathogenic polyglutamine tracts have similar structural properties: towards a length-dependent toxicity gradient. J Mol Biol. 2007;371:235–244. doi: 10.1016/j.jmb.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Lathrop RH, Casale M, Tobias DJ, Marsh JL, Thompson LM. Modeling protein homopolymeric repeats: possible polyglutamine structural motifs for Huntington's disease. Proc Int Conf Intell Syst Mol Biol. 1998;6:105–114. [PubMed] [Google Scholar]
- Legleiter J, Lotz GP, Miller J, Ko J, Ng C, Williams GL, Finkbeiner S, Patterson PH, Muchowski PJ. Monoclonal antibodies recognize distinct conformational epitopes formed by polyglutamine in a mutant huntingtin fragment. J Biol Chem. 2009;284:21647–21658. doi: 10.1074/jbc.M109.016923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Huey-Tubman KE, Gao T, Li X, West AP, Jr., Bennett MJ, Bjorkman PJ. The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model. Nat Struct Mol Biol. 2007;14:381–387. doi: 10.1038/nsmb1234. [DOI] [PubMed] [Google Scholar]
- Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Masino L, Kelly G, Leonard K, Trottier Y, Pastore A. Solution structure of polyglutamine tracts in GST-polyglutamine fusion proteins. FEBS Lett. 2002;513:267–272. doi: 10.1016/s0014-5793(02)02335-9. [DOI] [PubMed] [Google Scholar]
- Minor W, Tomchick D, Otwinowski Z. Strategies for macromolecular synchrotron crystallography. Structure. 2000;8:R105–110. doi: 10.1016/s0969-2126(00)00139-8. [DOI] [PubMed] [Google Scholar]
- Murphy RM. Peptide aggregation in neurodegenerative disease. Annu Rev Biomed Eng. 2002;4:155–174. doi: 10.1146/annurev.bioeng.4.092801.094202. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Inui T, Popiel HA, Fujikake N, Hasegawa K, Urade Y, Goto Y, Naiki H, Toda T. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- Nguyen J, Baldwin MA, Cohen FE, Prusiner SB. Prion protein peptides induce alpha-helix to beta-sheet conformational transitions. Biochemistry. 1995;34:4186–4192. doi: 10.1021/bi00013a006. [DOI] [PubMed] [Google Scholar]
- Perutz MF. Glutamine repeats and inherited neurodegenerative diseases: molecular aspects. Curr Opin Struct Biol. 1996;6:848–858. doi: 10.1016/s0959-440x(96)80016-9. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Finch JT, Berriman J, Lesk A. Amyloid fibers are water-filled nanotubes. Proc Natl Acad Sci U S A. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, Sullivan PG, Steffan JS, Sensi SL, Thompson LM. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. Lessons from animal models of Huntington's disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- Singer SJ, Dewji NN. Evidence that Perutz's double-beta-stranded subunit structure for beta-amyloids also applies to their channel-forming structures in membranes. Proc Natl Acad Sci U S A. 2006;103:1546–1550. doi: 10.1073/pnas.0509892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Kikuchi S, Katada S, Nagai Y, Nishizawa M, Onodera O. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet. 2008;17:345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Morishima I, Akagi T, Hashikawa T, Nukina N. Intra- and intermolecular beta-pleated sheet formation in glutamine-repeat inserted myoglobin as a model for polyglutamine diseases. J Biol Chem. 2001;276:45470–45475. doi: 10.1074/jbc.M107502200. [DOI] [PubMed] [Google Scholar]
- Temussi PA, Masino L, Pastore A. From Alzheimer to Huntington: why is a structural understanding so difficult? EMBO J. 2003;22:355–361. doi: 10.1093/emboj/cdg044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, Byeon IJ, Anjum DH, Kodali R, Creamer TP, Conway JF, et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Tobin AJ, Signer ER. Huntington's disease: the challenge for cell biologists. Trends Cell Biol. 2000;10:531–536. doi: 10.1016/s0962-8924(00)01853-5. [DOI] [PubMed] [Google Scholar]
- Truant R, Atwal RS, Desmond C, Munsie L, Tran T. Huntington's disease: revisiting the aggregation hypothesis in polyglutamine neurodegenerative diseases. FEBS J. 2008;275:4252–4262. doi: 10.1111/j.1742-4658.2008.06561.x. [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Shimizu H, Ishida T, Akiyama Y, Nukina N. Aggregation mechanism of polyglutamine disease revealed using quantum chemical calculations, fragment molecular orbital calculations, molecular dynamics simulations, and binding free energy calculations. Journal of molecular Structure. 2006;778:85–95. [Google Scholar]
- Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim Biophys Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Vitalis A, Wang X, Pappu RV. Atomistic simulations of the effects of polyglutamine chain length and solvent quality on conformational equilibria and spontaneous homodimerization. J Mol Biol. 2008;384:279–297. doi: 10.1016/j.jmb.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Wang X, Vitalis A, Wyczalkowski MA, Pappu RV. Characterizing the conformational ensemble of monomeric polyglutamine. Proteins. 2006;63:297–311. doi: 10.1002/prot.20761. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.