Abstract
We present an outline of the Critical Assessment of Protein Structure Prediction (CASP) infrastructure implemented at the University of California, Davis, Protein Structure Prediction Center. The infrastructure supports selection and validation of prediction targets, collection of predictions, standard evaluation of submitted predictions, and presentation of results. The Center also supports information exchange relating to CASP experiments and structure prediction in general. Technical aspects of conducting the CASP8 experiment and relevant statistics are also provided.
Keywords: CASP, protein structure prediction
Introduction
We present an update on the operations of the CASP Prediction Center in form of a short note. The standard prediction evaluation methods are discussed in a separate paper in this issue1. The purpose of this article is to guide the readers through some of the technical aspects of conducting the CASP experiments, as well as to familiarize them with the available tools and resources. We also briefly discuss the infrastructure components introduced since the last experiment.
Prediction targets
For CASP8, we have designed and implemented an administrative system for a more effective handling of protein sequences submitted by crystallographers and NMR spectroscopists as potential prediction targets. Many intermediate tasks that formerly were performed manually are now automated. During CASP8 the system proved to be more reliable and operated more smoothly than in previous experiments. The system was integrated into CASP infrastructure, facilitating data flow between sequence screening, target assignment, and target release processes.
During the course of the experiment the Prediction Center received over 200 sequences submitted as potential prediction targets. These sequences were pre-screened and if necessary verified with the providers, leading to 128 sequences being selected as targets. These numbers correspond to more than 20% increase in the number of available prediction targets compared to the previous round CASP (see Fig.1).
Figure 1.
The number of targets released for prediction in eight consecutive CASP experiments. In CASP8, 128 targets were split into 57 human/server and 71 server-only targets.
According to the recommendations of the CASP7 Predictors’ Meeting, the selected target sequences were split into two categories: (1) targets for prediction by all groups (human/server targets), and (2) targets for server prediction (server-only targets). Prediction targets were posted at the CASP8 website (for human-expert groups), as well as automatically forwarded to the participating servers through a distribution system, that for CASP8 allowed simultaneous sending of the same sequence to many servers. The daily release package of targets normally consisted of one human/server target (typically selected from among the more challenging modeling targets), and one or two server-only targets (typically selected from among the easier targets). Targets were released 5 days a week. Following this schedule, the aim of having over 100 targets total, including 50–60 human/server targets over the 11 weeks of target release (May 5 through July 18) was attained. We have set up an automatic system to track the weekly PDB releases for CASP targets, implemented as BLAST searches of CASP sequences versus latest ftp://ftp.wwpdb.org/pub/pdb/derived_data/pdb_seqres.txt file. We also periodically checked the SGI centers’ websites manually to catch any unexpected structure releases prior to the end of the prediction time window. These precautionary measures necessitated canceling of one target (T0472) for the human/server prediction and moving this target to the server-only category. Two other targets (T0484 and T0500) appeared to be mainly disordered and were assessed only in the disorder prediction category. Organizers and assessors additionally canceled 5 targets as unsuitable for structure evaluation2. In all, structural models for 121 targets were assessed.
Predictions
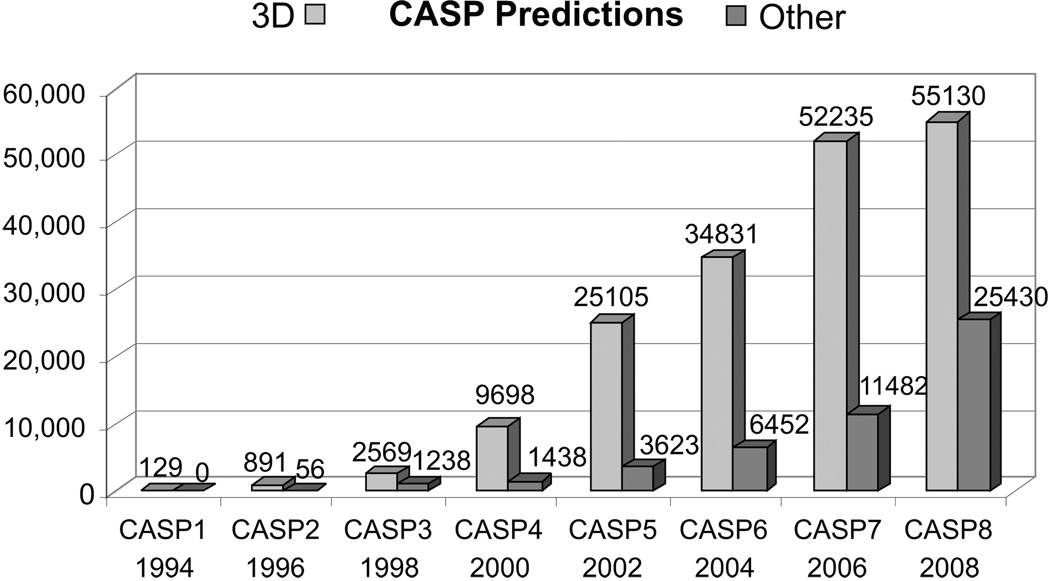
In CASP8, participants submitted over 80,000 predictions in seven prediction categories (http://predictioncenter.org/casp8/index.cgi?page=format), including over 56,000 tertiary structure predictions. This is almost twice as many predictions as submitted in CASP6, only 4 years ago. The last two rounds of CASP were especially challenging, with approximately 20,000 submissions increase per CASP cycle (see Fig. 2). To deal with the growing number of predictions we have improved the existing infrastructure and implemented new strategies for prediction processing, storage, evaluation, and results visualization. Although for the end user the procedures for submitting predictions to CASP remained the same, on the Prediction Center’s side the dataflow was redesigned and the implementation of prediction processing essentially rewritten to accommodate changes in prediction authentication methods associated with the new registration system (described in more detail below). A new, more comprehensive system for checking the status of predictions for both server and human-expert groups was also implemented.
Figure 2.
The number of predictions submitted for evaluation in eight consecutive CASP experiments. Compared to CASP7, CASP8 registered a more than two-fold increase in the number of predictions in the non-tertiary structure categories.
The CASP8 predictions can be downloaded from http://predictioncenter.org/download_area/CASP8/predictions/.
Experimental structures and automatic evaluation of models
Of the 128 targets released for prediction, the coordinates of all but 4 structures were made available to the organizers in time for assessment (i.e. before September 15, 2008). These reference target structures were then evaluated for sequence consistency, structure disorder, and alternative side chain conformations (procedures are described in more detail in an earlier paper3). As CASP assessments are performed for entire structures as well as separate structure domains, domain definitions were arrived at by consensus among the assessors. For some targets, multiple reference structures were identified to accommodate assessors’ requests for additional evaluations based on the alternative domain definitions or alternative reference models selected from the NMR ensembles. All model-model superpositions were performed to enable establishing the originality of submitted predictions (data available through the Results tables – see http://www.predictioncenter.org/casp8/doc/help.html#PC01 for description). The analysis toolkit was also broadened by adding the DALI structure superposition program4 to the tools already used in CASP7 (LGA5, DAL3,6, MAMMOTH7, ACE8). Calculations were conducted on a dedicated cluster of evaluation servers connected to the model database. As several million superpositions had to be performed using five different superposition methods, the effective usage of the available computational resources as well as the reliability of the obtained results became of a paramount concern. To cope with these issues, the process management system, including downloading models to the evaluation servers, distributing tasks between processors, collecting the results, and uploading them back to the web-server database, was expanded to include a number of data verification tools.
Results and Organization of the Website
The CASP evaluation results were made available to the independent assessors on a continuing target by target basis, as soon as the calculations were completed at the Prediction Center. A special password-protected gateway enabled secure access to these data for the assessors. An infrastructure facilitating assessor communications with other assessors and with the organizers was implemented. In particular, we have improved the assessors’ discussion forum and integrated the domain definition and categorization facility with the results database. A week before the CASP8 meeting, the data were released to the public through the Prediction Center website (http://predictioncenter.org/casp8/results.cgi). The software for the presentation of the results uses the system originally developed for CASP6 and later improved for CASP7 (see3 for details). The goal of the changes introduced for CASP8 was making the system even more transparent and intuitive. For example, it is now possible to switch between target-based and group-based views directly from the results pages. Results browsers feature easier navigation throughout, including a one-click selection of the data for the user-specified group/target/model subsets. Also new for CASP8 are pages showing the relative cumulative performance of prediction groups (http://predictioncenter.org/casp8/groups_analysis.cgi), according to the several most popular evaluation measures1.
CASP8 results can be viewed in one of the three modes: Target based view, Group based view and Table Browser view.
The Target based view (http://predictioncenter.org/casp8/results.cgi) is the default and provides access to the results on target-by-target basis. Miniature plots allow an at-a-glance comparison between all evaluated targets/domains. Results for each target are collected in “information cells” consisting of six clickable pictograms (see “TARGET PERSPECTIVE” in the center of Fig. 3). The upper left-hand-side pictogram displays the target/domain number, difficulty category (TBM for template-based modeling or FM for free modeling), and the range of target residues used in evaluation. The available data analysis tools are described following Fig. 3, top-to-bottom.
Figure 3.
An overall view of the CASP8 standard evaluation results available at the Prediction Center. For the analysis performed by the CASP8 independent assessors the reader is referred to other papers in this issue. RASMOL and SPICE can be referenced in9 and10, respectively.
1. 3D Model Tables present results for all predictions accepted on a given target. The results are available in the form of dynamic tables/plots (designed for interactive on-site analysis) or plain text computer-readable files that can be generated by using the Text File link. Tables may be sorted by each column and may be expanded or contracted with the Full/Brief option available for every evaluation measure. In addition to the scores presented in CASP7, we have calculated new scores suggested by the assessors and predictors. Among these are the results of DALI structural superpositions and the Z-scores indicating the relative quality of a prediction with respect to the average prediction submitted on a given target. The Z-scores were calculated for several evaluation measures (GDT_TS, GDT_HA, AL0) and subsets of data (different model difficulty categories, different target types).
2. The visualization tool, based on the SPICE software [10], allows viewing several structures in the same frame of reference. It works under all major operational systems and is particularly useful when CASP targets, corresponding predictions, and the closest templates are analyzed simultaneously. A 3D protein structure display, a structure /model selection list, and a sequence display are all interconnected to facilitate analysis.
3. The value-added plots help identify model features that are not available from a single template. A sequence-independent protocol is used to superimpose best templates onto the target structure. Template/model selection, model strip charts, and template strip charts allow relating quality of every submitted model to the structural information available from the 20 best structural templates.
4. The alignment summary strip charts indicate percentages of correctly aligned residues in the model relative to target. The bars under the position-specific alignment tab show the distribution of the correctly and incorrectly aligned residues along the target protein sequence. 3D interactive renderings of the superimposed model and target in the best rigid body sequence independent superposition are also available.
5. GDT plots provide model quality estimates by finding the largest subsets of residues fitted to the target in a series of rigid-body sequence dependent superpositions. The calculation is performed for a series of cutoffs from 0.5 to 10.0 Å. Results are plotted as a line for each model separately and selected models may be identified in the context of all submissions on a given target.
6. The 3D interactive representations of target structures (lower left-hand side corner of the “information cell”) may be viewed with Rasmol [9]. Selected target domains or their subsets, as defined by the assessors, are shown in green.
The Groups view allows assessing performance of a particular prediction group. It is possible to retrieve dynamically generated tables and graphical results over all targets (or their subset) predicted by that group. Results are shown in the context of all other submissions.
The Table Browser view adds additional flexibility in generating custom comparisons of numerical results, where prediction groups, targets, and measures may be independently selected.
Separately, refinement results for the twelve CASP8 refinement category targets are also available. For each target, strip charts show improvements over the starting model. Finally, the model quality assessment provides results on both the overall and residue-by-residue comparisons of the model quality predictions with the actual modeling performance recorded in CASP.
Notes on CASP8 participation
CASP8 registration was open from late March until the end of August, 2008. In all, 234 predictor groups representing 25 countries registered and submitted predictions. The number of participating server groups grew relative to the previous CASPs (121 in CASP8 vs. 93 in CASP7 and 63 in CASP6) and for the first time exceeded the number of participating human-expert groups (112). The increase in the number of servers came mainly from prediction categories other than tertiary structure prediction, with the largest contribution from the assessments of model quality. The number of servers predicting tertiary structure remained approximately constant (72 in CASP8 vs. 68 in CASP7).
General Registration at Prediction Center
In time for CASP8 we have implemented a new registration system ensuring a simpler and more secure access to the user personal data and to some limited-access sections of the Prediction Center website. With the new system in place, the registration has to be done only once and the submitted information will remain in place for subsequent CASP experiments, between-CASP initiatives, and for accessing local services. A tool for editing registration data and changing/reminding passwords was also implemented. In previous CASPs we have communicated with predictors mainly through email. With the implementation of the new registration system, it is possible to subscribe to the Prediction Center newsletter, as well as to access the updated information through the website’s message board. Automatic, synchronized email notifications are also sent out to people who have subscribed to this service.
CASP8 registration
Having completed the general registration with the Prediction Center, predictors could then proceed with the simplified registration for CASP8. Registrations were possible in several participation roles, including (1) a human-expert group; (2) a server group; (3) an extended timeframe group (for participants relying on computationally intensive prediction methods); (4) a member of a prediction group; or (5) an observer. The group leaders (persons who registered a group) were assigned a registration code allowing submission of predictions according to the group category. The group leaders were solely responsible for submissions from their group, but they could also delegate submission privileges to any registered member of the group. The human-expert groups were allowed to use any combination of knowledge and computational techniques, and had approximately three weeks to submit predictions for a given target. Server groups had to respond to the Prediction Center queries automatically and return models within 72 hours, without any human intervention. Extended timeframe groups (the groups not assessed in the main CASP analysis) could register in addition to the regular groups and had approximately 6 weeks per target.
ACKNOWLEDGMENTS
We acknowledge the crystallographers and NMR spectroscopists taking part in CASP8, especially the researchers from the five structural genomic centers - NESG, MCSG, SGC, JCSG and NYSGXRC – who provided 118 out of the 128 CASP8 prediction targets (see http://predictioncenter.org/casp8/numbers.cgi for the detailed list of contributors). Special thanks are extended to the staff of Protein Data Bank for setting up the system for temporarily putting on hold the release of CASP8 prediction targets. We are indebted to Adam Zemla (LGA) and Andreas Prlic (SPICE) for upgrading their programs for CASP8 and making them available for our extensive use. We are also grateful to the late Angel Ortiz for our use of MAMMOTH. This work was supported by NIH/NLM grant LM07085.
REFERENCES
- 1.Cozzetto DKA, Fidelis K, Moult J, Rost B, Tramontano A. Evaluation of template-based models in CASP8 with standard measures. PROTEINS: Struct Funct Genet. 2009 doi: 10.1002/prot.22561. (this issue):???-??? [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tress M. Target Domain Definition and Classification in CASP8. PROTEINS: Struct Funct Genet. 2009 doi: 10.1002/prot.22497. (this issue):???-??? [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kryshtafovych A, Prlic A, Dmytriv Z, Daniluk P, Milostan M, Eyrich V, Hubbard T, Fidelis K. New tools and expanded data analysis capabilities at the Protein Structure Prediction Center. Proteins. 2007;69 Suppl 8:19–26. doi: 10.1002/prot.21653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24(23):2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31(13):3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kryshtafovych A, Milostan M, Szajkowski L, Daniluk P, Fidelis K. CASP6 data processing and automatic evaluation at the protein structure prediction center. Proteins. 2005;61 Suppl 7:19–23. doi: 10.1002/prot.20718. [DOI] [PubMed] [Google Scholar]
- 7.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11(11):2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zemla A, Venclovas, Moult J, Fidelis K. Processing and evaluation of predictions in CASP4. Proteins. 2001 Suppl 5:13–21. doi: 10.1002/prot.10052. [DOI] [PubMed] [Google Scholar]
- 9.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20(9):374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 10.Prlic A, Down TA, Hubbard TJ. Adding some SPICE to DAS. Bioinformatics. 2005;21 Suppl 2:ii40–ii41. doi: 10.1093/bioinformatics/bti1106. [DOI] [PMC free article] [PubMed] [Google Scholar]