Abstract
Poly(γ-glutamic acid) (γ-PGA) nanoparticles (NPs) have previously been reported as an efficient antigen delivery system with adjuvant activity. In this study, the gene expression in murine bone marrow-derived dendritic cells (DCs) treated with γ-PGA NPs was examined by oligonucleotide microarray analysis and compared with that in cells treated with other adjuvants. The gene expression of proinflammatory chemokines, cytokines, and costimulatory molecules was upregulated considerably in DCs treated with γ-PGA NPs. The upregulation pattern was similar to that in DCs treated with lipopolysaccharide (LPS) but not to that in DCs treated with unparticulate γ-PGA. The activation of DCs by γ-PGA NPs was confirmed by real-time reverse transcriptase PCR (RT-PCR) analysis of genes related to Toll-like receptor (TLR) signaling. The effect of γ-PGA NPs on DCs was not annihilated by treatment with polymyxin B, an inhibitor of LPS. Furthermore, the immunization of mice with γ-PGA NPs carrying ovalbumin (OVA) as an antigen significantly induced antigen-specific CD8+ T cells and antigen-specific production of interleukin-2, tumor necrosis factor alpha, and gamma interferon from the cells. Such activities of γ-PGA NPs were more potent than those obtained with immunization with OVA plus aluminum hydroxide or OVA plus complete Freund's adjuvant. These results suggest that γ-PGA NPs induce a CD8+ T-cell response by activating innate immunity in a fashion different from that of LPS. Thus, γ-PGA NPs may be an attractive candidate to be developed further as a vaccine adjuvant.
Adjuvants are essential to enhance antigen-specific immune responses to vaccination. Various substances have been evaluated for immunomodulatory effects in vitro and in vivo (24). However, since most substances have proved to have unacceptable levels of toxicity, aluminum hydroxide (alum) is an adjuvant clinically approved for use in humans. Alum is generally safe, but it induces modest antibody production and does not generate sufficient cellular immunity. Consequently, many efforts have been made to develop novel vaccine adjuvants capable of inducing potent antigen-specific humoral and cellular immune responses (12, 24, 33). MF59 and AS04 have been licensed in Europe. MF59 has been used for the influenza vaccine for a decade, and its safety and potency have also been established (19). AS04 is used for vaccines against human hepatitis B virus (5) and human papillomavirus (21). AS01 and AS02 are currently under development as adjuvants for antimalaria vaccines (17).
Immature dendritic cells (DCs) reside in nonlymphoid tissues, including skin and mucosal membranes, and take up antigens by endocytosis (3). Immature DCs develop into mature DCs accompanying the upregulation of major histocompatibility complex (MHC) and costimulatory molecules, which play an important role in effective induction of antigen-specific immune responses. The maturation of DCs occurs in the presence of various stimuli, such as allergens, cytokines, bacterial products, and viral components (2, 4, 14). Nanoparticles (NPs) are considered to be an efficient antigen carrier and are widely investigated for their biological potential (6, 9). Since DCs are professional cells capable of presenting processed antigens to naïve T cells to generate effector T cells (2), efficient antigen delivery to DCs by NPs is a promising strategy for potent induction of antigen-specific immune responses.
In our previous studies, it was found that poly(γ-glutamic acid) (γ-PGA) NPs acted as a potent vaccine adjuvant as well as an efficient antigen carrier to DCs (32, 33, 35, 37). γ-PGA NPs were predominantly taken up by DCs, and the cells started to produce tumor necrosis factor alpha (TNF-α) and interleukin-12β (IL-12β) abundantly. The expression of CD40, CD80, and CD86 on the surface was also highly upregulated, resulting in strong induction of antigen-specific immune responses (33, 35, 37). γ-PGA is a capsular exopolymer produced by certain strains of bacteria. γ-PGA NPs are created by self-assembly of amphiphilic graft copolymers composed of γ-PGA and l-phenylalanine ethylester (PAE) (1). γ-PGA NPs can carry various proteins and peptides on and inside the particles. γ-PGA NPs are digested by γ-glutamyl transpeptidase, which is widely distributed in the whole body, indicating that γ-PGA NPs are biodegradable (1). Thus, γ-PGA NPs appear to be suitable for medical use as a safe and effective vaccine adjuvant. Although the maturation of DCs induced by γ-PGA NPs has been demonstrated by the expression of surface markers, comprehensive gene expression remains to be determined. In the present study, we analyzed the gene expression of murine bone marrow-derived DCs by oligonucleotide microarray analysis after treatment with either γ-PGA NPs, lipopolysaccharide (LPS), or unparticulate γ-PGA. The effect of γ-PGA NPs on gene expression was also assessed by real-time reverse transcriptase PCR (RT-PCR) and compared to those of LPS, CpG, zymosan, and poly(I:C). Furthermore, to evaluate the adjuvant activity of γ-PGA NPs, antigen-specific T cells and their production of IL-2, TNF-α, and gamma interferon (IFN-γ) were determined for the spleen cells of mice immunized with ovalbumin (OVA)-carrying γ-PGA NPs (OVA-NPs), OVA plus complete Freund's adjuvant (CFA), or OVA plus alum.
MATERIALS AND METHODS
Mice.
Female C57BL/6 mice (H-2Kb; 6 to 8 weeks old) were purchased from Charles River (Yokohama, Japan). Experiments were carried out in accordance with the guidelines for animal experimentation of Kagoshima University.
Reagents.
γ-PGA (average molecular weight = 380,000) was kindly provided by Meiji Seika (Tokyo, Japan). PAE, polymyxin B (PmB), and OVA were purchased from Sigma (St. Louis, MO).
Preparation of γ-PGA NPs.
The synthetic procedures for γ-PGA NPs and OVA-NPs have been described in our previous reports (1, 36). The concentration of bacterial endotoxin in γ-PGA NPs was determined by a Limulus amoebocyte lysate assay (Seikagaku, Tokyo, Japan) and was found to be <10 endotoxin units/ml.
Preparation of DCs.
Murine bone marrow-derived DCs were generated as previously described (26, 27, 36). Briefly, femurs and tibias were removed from mice and purified from the surrounding muscle tissues. Both ends of the bones were cut with scissors, and the marrow was flushed with phosphate-buffered saline (PBS), using a syringe with a 26-gauge needle. The suspension was passed through a 50-μm cell strainer. Bone marrow cells (2.5 × 106 cells/10 ml) were suspended in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin G, 100 μg/ml streptomycin, 2-mercaptoethanol, and 20 ng/ml recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF; PeproTech, London, United Kingdom) and were placed into a petri dish. Fresh culture medium was added to the dish on day 3. On day 7, nonadherent or loosely adherent cells were harvested and used as immature DCs.
Oligonucleotide microarray.
Immature DCs (1 × 106 cells/ml) were incubated with either 300 μg/ml γ-PGA NPs, 1 μg/ml LPS, or 300 μg/ml unparticulate γ-PGA at 37°C. Six, 12, and 24 h after incubation, total RNA was extracted from the cells by use of an extraction kit (RNeasy; Qiagen, Hilden, Germany). The quality of the total RNA was examined by a model 2100 bioanalyzer (Agilent, Santa Clara, CA). Microarray experiments with the samples were carried out according to Agilent's protocol. Briefly, the total RNA (500 ng) was converted to cDNA with Moloney murine leukemia virus RT and the T7 promoter primer. The cDNA was transcribed and amplified with T7 RNA polymerase to produce a cRNA labeled with cyanine 3. The cyanine 3-labeled cRNA was purified with an RNeasy kit and examined for its concentration and labeling quality by use of a spectrophotometer. The cRNA was fragmented and hybridized to an Agilent whole-mouse-genome oligonucleotide microarray (4 × 44,000 slide format). After hybridization, the microarray was washed thoroughly and scanned with a microarray scanner (Agilent). The microarray scan data were processed with Future Extraction software (version 9.5.1; Agilent) according to the instructions in the manual. Cell culture and microarray experiments were repeated three times.
Data analysis.
The expression level of each gene was analyzed by GeneSpring GX software (version 7.3.1; Agilent). Briefly, after importing the processed data into the software, they were normalized based on the default normalizing settings for one-color experiments, according to the GeneSpring 7.3 user's guide (Agilent). The normalized data were filtered on the basis of parameters in certain specific columns of the original data files to remove the control and other inappropriate spots.
Real-time RT-PCR.
DCs (1 × 106 cells/ml) were incubated with either 300 μg/ml γ-PGA NPs, 1 μg/ml LPS, 5 μg/ml zymosan (InvivoGen, San Diego, CA), 1 μg/ml class B CpG (InvivoGen), 1 μg/ml poly(I:C) (InvivoGen), or PBS at 37°C. Six hours after incubation, total RNA was extracted from the cells with an RNeasy kit. The quality of the total RNA was examined by a model 2100 bioanalyzer. Real-time RT-PCR analysis of the samples was carried out using a mouse Toll-like receptor (TLR) signaling pathway PCR array (SuperArray; SuperArray Biosciences, Frederick, MD), which includes primer pairs specific to 84 genes related to TLR-mediated signaling pathways, according to the manufacturer's protocol. Briefly, cDNA was synthesized from the total RNA (1 μg) by use of an RT2 first-strand kit (SuperArray Biosciences) and was subjected to real-time PCR with an RT2 real-time PCR SYBR green/ROX kit (SuperArray Biosciences) on an ABI Prism 7700 sequence detector (Applied Biosystems, Foster City, CA). The PCR consisted of 1 cycle of 95°C for 10 min and 50 cycles of 95°C for 15 s and 60°C for 1 min. The expression levels of target genes were normalized to those in PBS-treated DCs (control).
Effect of PmB on DC activation.
DCs (1 × 106 cells/ml) were incubated with either 100 μg/ml γ-PGA NPs or 1 μg/ml LPS in the absence or presence of 10 μg/ml PmB. Twenty-four hours after incubation, the cells and culture supernatants were collected. The TNF-α level in the culture supernatants was determined by use of a sandwich enzyme-linked immunosorbent assay (ELISA) kit (Invitrogen, Carlsbad, CA). For phenotypic analysis, the cells were washed with PBS and stained with a phycoerythrin (PE)-conjugated anti-CD40 monoclonal antibody (MAb; BD Biosciences, San Jose, CA) or a PE-conjugated anti-CD80 MAb (BD Biosciences) for 30 min at 4°C. After being washed, the cells were analyzed by flow cytometry (FACSCalibur; BD Biosciences).
Analysis of spleen cells.
Mice (3 to 5 mice in each group) were immunized subcutaneously with either PBS (100 μl), OVA alone (10 μg of OVA in 100 μl of PBS), CFA + OVA (10 μg of OVA in 100 μl of CFA), alum + OVA (10 μg of OVA in 100 μl of alum), or OVA-NPs (10 μg of γ-PGA NPs carrying 10 μg of OVA in 100 μl of PBS) on day 0. On day 10, spleen cells were collected by use of lympholyte-M (Cedarlane, Burlington, Ontario, Canada) and were analyzed by pentamer staining, intracellular cytokine staining, and enzyme-linked immunospot (ELISPOT) assay. For pentamer staining, the cells were stained with a fluorescein isothiocyanate (FITC)-conjugated anti-CD8 MAb (Proimmune, Oxford, United Kingdom) and an allophycocyanin-conjugated pentameric H-2Kb complex (BD Biosciences) with the OVA257-264 cytotoxic T-lymphocyte (CTL) epitope. After being washed with PBS containing 0.1% sodium azide and 0.1% bovine serum albumin, the cells were fixed with PBS containing 2.5% formaldehyde (Wako, Osaka, Japan), washed with PBS, and analyzed by flow cytometry. For intracellular cytokine staining, the cells were incubated with BD GolgiPlug (BD Biosciences) and either OVA257-264 peptide (10 μg/ml) or medium alone for 10 h. After being washed, the cells were incubated with a purified anti-mouse CD16/32 MAb (BD Biosciences) followed by staining with a PE-conjugated anti-CD8 MAb (BD Biosciences). The cells were permeabilized with Cytofix/Cytoperm Plus (BD Biosciences) and stained with an FITC-conjugated anti-IFN-γ MAb, an FITC-conjugated anti-IL-2 MAb, or an FITC-conjugated anti-TNF-α MAb (all from BD Biosciences) for 30 min at 4°C. The cells were washed with PBS and subjected to flow cytometric analysis. For ELISPOT assay, the cells were cultured with either medium alone, the OVA257-264 peptide (10 μg/ml), or the control peptide (10 μg/ml) in an ELISPOT plate (BD Biosciences). After incubation for 24 h at 37°C, the plate was washed, and IFN-γ-producing cells were measured by ELISPOT assay according to the manufacturer's protocol.
Microarray data accession numbers.
The microarray data are available at the Gene Expression Omnibus (GEO). The accession numbers are GSE15087 and GSE15089, which contain the data on comparative gene expression modulated by γ-PGA NPs, LPS, and unparticulate γ-PGA and the data on the time course of gene expression modulated by γ-PGA NPs, respectively.
RESULTS
Gene expression in DCs after treatment with γ-PGA NPs.
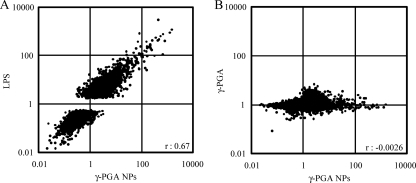
DC maturation involves coordinated regulation of several genes. To identify the genes modulated during the maturation process of DCs, comprehensive gene expression in bone marrow-derived DCs was examined by use of a whole-mouse-genome oligonucleotide microarray after treatment with either γ-PGA NPs, LPS, or unparticulate γ-PGA and was compared to that in DCs treated with PBS (control). Among the genes that could be analyzed (41,267 genes), the expression of 8,006 and 13,282 genes was found to be modulated in DCs, with statistical significance (P < 0.05), after treatment with γ-PGA NPs and LPS, respectively (data not shown). However, the expression of only 20 genes was significantly upregulated or downregulated in DCs after treatment with unparticulate γ-PGA. The correlation coefficient for the relative expression levels of analyzed genes was 0.67 between γ-PGA NP-treated DCs and LPS-treated DCs (Fig. 1A). However, no such positive correlation was observed (r = −0.0026) between γ-PGA NP-treated DCs and unparticulate γ-PGA-treated DCs (Fig. 1B).
FIG. 1.
Gene expression in DCs treated with γ-PGA NPs versus LPS (A) and γ-PGA NPs versus unparticulate γ-PGA (B). Murine bone marrow-derived DCs (1 × 106 cells/ml) were treated with either 300 μg/ml γ-PGA NPs, 1 μg/ml LPS, or PBS for 6 h. Total RNA was extracted from the treated DCs, purified, and subjected to oligonucleotide microarray analysis. The expression level of each gene was normalized to that of the control (PBS treatment). The expression profiles are shown by scatter plots. The genes upregulated >5-fold were plotted.
The expression levels of the genes related to immune responses are summarized in Table 1. Among the genes for cytokines and cytokine receptors, 8 genes were upregulated >100-fold 6 h after treatment with γ-PGA NPs. These gene products included IL-6, CSF2 (GM-CSF), IL-12β, IL-1α, CSF3 (granulocyte colony-stimulating factor [G-CSF]), IL-1 family member 6, IL-23α, and inhibin β. In addition, 14 genes were upregulated >10-fold. A similar result was observed in DCs treated with LPS. The expression of some chemokine and chemokine receptor genes was also highly upregulated by treatment with γ-PGA NPs or LPS (Table 1). An interesting finding is that the genes related to Th1 cytokines, such as IL-12β, IL-23α, IL-12α, and IFN-γ, were equally or even more upregulated in γ-PGA NP-treated DCs than in LPS-treated DCs, whereas those related to Th2 cytokines, such as IL-10, IL-33, and IL-19, were less upregulated in γ-PGA NP-treated DCs than in LPS-treated DCs. Except for a few genes, the expression of most chemokine and chemokine receptor genes was enhanced in both γ-PGA NP-treated DCs and LPS-treated DCs (Table 1). In particular, the expression of the genes related to the neutrophil-attracting chemokines CXCL1, CXCL2, and CXCL5 was remarkably enhanced in γ-PGA NP-treated DCs. It has been demonstrated that CCR7 is upregulated during DC maturation and does play an important role in leading DCs to lymph nodes (16). In fact, the CCR7 gene was upregulated 5.5- and 11.4-fold in γ-PGA NP-treated DCs and LPS-treated DCs, respectively. On the other hand, the CCR2 gene was expressed only in immature DCs (Table 1). The expression of the genes related to costimulatory molecules (CD40, CD80, and CD86) and adhesion molecule (CD54) was also upregulated in γ-PGA NP-treated DCs as well as LPS-treated DCs.
TABLE 1.
Relative gene expression in DCs treated with γ-PGA NPs, LPS, or unparticulate γ-PGAa
| Gene category and GenBank accession no. | Gene name | Relative gene expression (fold change)b |
Ratio for DCs treated with γ-PGA NPsc |
Gene product | |||
|---|---|---|---|---|---|---|---|
| γ-PGA NPs | LPS | γ-PGA | 12 h/6 h | 24 h/6 h | |||
| Cytokine and cytokine receptor genes | |||||||
| NM_031168 | Il6 | 681.8 | 770 | 1 | 0.7 | 0.2 | Interleukin-6 |
| NM_009969 | Csf2 | 660.3 | 112.4 | 0.8 | 0.4 | 0.2 | Colony-stimulating factor 2 |
| NM_008352 | Il12b | 540.7 | 636.8 | 0.9 | 1.5 | 0.7 | Interleukin-12b |
| NM_010554 | Il1a | 418.8 | 429.4 | 0.9 | 0.6 | 0.3 | Interleukin-1 alpha |
| NM_009971 | Csf3 | 315.7 | 99.4 | 0.8 | 1 | 0.2 | Colony-stimulating factor 3 |
| NM_031252 | Il23a | 248.1 | 174.6 | 0.8 | 0.3 | 0.2 | Interleukin-23, alpha subunit p19 |
| NM_019450 | Il1f6 | 215.5 | 292.1 | 1 | 0.4 | 0 | Interleukin-1 family, member 6 |
| NM_008380 | Inhba | 107 | 118.9 | 1 | 0.7 | 0.3 | Inhibin beta A |
| NM_010548 | Il10 | 89.2 | 242.8 | 1.1 | 1.1 | 0.6 | Interleukin-10 |
| NM_133775 | Il33 | 71.8 | 379.1 | 0.8 | 1.3 | 0.7 | Interleukin-33 |
| NM_013693 | Tnf | 63.6 | 48.3 | 1 | 0.5 | 0.1 | Tumor necrosis factor alpha |
| NM_009704 | Areg | 59.8 | 51.3 | 0.7 | 0.2 | 0.1 | Amphiregulin preproprotein |
| NM_008351 | Il12a | 42.7 | 49 | 1 | 1.8 | 0.8 | Interleukin-12a |
| NM_021367 | Tslp | 40.8 | 74.8 | 0.6 | 0.2 | 0.2 | Thymic stromal lymphopoietin |
| NM_008337 | Ifng | 23.2 | 9.3 | 1.4 | 1.3 | 0.3 | Gamma interferon |
| NM_010735 | Lta | 20.9 | 13.5 | 1.2 | 0.9 | 0.3 | Lymphotoxin A |
| NM_001009940 | Il19 | 20 | 75.3 | 1.8 | 0.5 | 0.7 | Interleukin-19 |
| NM_008361 | Il1b | 17.8 | 20.1 | 1 | 0.7 | 0.2 | Interleukin-1 beta |
| NM_153511 | Il1f9 | 15.4 | 13.5 | 1 | 0.6 | 0.2 | Interleukin-1 family, member 9 |
| NM_010171 | F3 | 15.3 | 5.7 | 1 | 0.8 | 0.5 | Coagulation factor III |
| NM_001013365 | Osm | 14.1 | 7.2 | 1.2 | 0.8 | 0.7 | Oncostatin M |
| NM_009404 | Tnfsf9 | 10.4 | 18.2 | 0.7 | 0.6 | 0.3 | Tumor necrosis factor (ligand) superfamily, member 9 |
| NM_016971 | Il22 | 8 | 10.3 | 0.8 | 1.1 | 1 | Interleukin-22 |
| NM_009452 | Tnfsf4 | 5.7 | 9 | 1 | 0.6 | 0.4 | Tumor necrosis factor (ligand) superfamily, member 4 |
| NM_015766 | Ebi3 | 5.5 | 6.5 | 1 | 0.7 | 0.4 | Epstein-Barr virus-induced gene 3 protein precursor |
| NM_011019 | Osmr | 3.4 | 12.4 | 0.5 | 1.1 | 1 | Oncostatin M receptor |
| NM_009367 | Tgfb2 | 3.2 | 9.2 | 2.4 | 1.2 | 2.6 | Transforming growth factor beta 2 |
| NM_009425 | Tnfsf10 | 2.9 | 17.7 | 1.2 | 0.3 | 0.1 | Tumor necrosis factor (ligand) superfamily, member 10 |
| NM_008360 | Il18 | 2.3 | 6.5 | 1 | 0.7 | 0.8 | Interleukin-18 |
| NM_008357 | Il15 | 2 | 5.2 | 1 | 0.6 | 0.5 | Interleukin-15 |
| NM_008373 | Il9 | 1.2 | 5.5 | 0.7 | 0.8 | 2.3 | Interleukin-9 |
| NM_011614 | Tnfsf12 | 1 | 0.2 | 0.6 | 1.5 | 2.3 | Tumor necrosis factor (ligand) superfamily, member 12 |
| NM_013584 | Lifr | 0.2 | 0.2 | 1 | 0.3 | 0.1 | Leukemia inhibitory factor receptor |
| Chemokine and chemokine receptor genes | |||||||
| NM_008176 | Cxcl1 | 782.7 | 412.8 | 0.8 | 0.8 | 0.2 | Chemokine (C-X-C motif) ligand 1 |
| NM_009140 | Cxcl2 | 164.2 | 94.9 | 0.9 | 0.6 | 0.2 | Chemokine (C-X-C motif) ligand 2 |
| NM_009141 | Cxcl5 | 156 | 25.9 | 0.8 | 1.7 | 1 | Chemokine (C-X-C motif) ligand 5 |
| NM_011329 | Ccl1 | 27.9 | 19.5 | 1 | 0.2 | 0.2 | Chemokine (C-C motif) ligand 1 |
| NM_009916 | Ccr4 | 22.5 | 36.2 | 0.8 | 1.2 | 0.4 | Chemokine (C-C motif) receptor 4 |
| NM_009137 | Ccl22 | 21.9 | 28.4 | 0.8 | 1.1 | 0.3 | Chemokine (C-C motif) ligand 22 |
| NM_011337 | Ccl3 | 21 | 23.5 | 1 | 0.7 | 0.3 | Chemokine (C-C motif) ligand 3 |
| NM_013653 | Ccl5 | 16.1 | 18.3 | 0.9 | 1.5 | 1.3 | Chemokine (C-C motif) ligand 5 |
| NM_013652 | Ccl4 | 12.6 | 14.4 | 0.8 | 0.3 | 0.2 | Chemokine (C-C motif) ligand 4 |
| NM_017466 | Ccrl2 | 8 | 14.2 | 1.1 | 0.9 | 0.4 | Lipopolysaccharide-inducible C-C chemokine receptor related |
| NM_007719 | Ccr7 | 5.5 | 11.4 | 1 | 1.6 | 1.4 | Chemokine (C-C motif) receptor 7 |
| NM_013654 | Ccl7 | 5 | 9.7 | 1 | 0.5 | 1.1 | Chemokine (C-C motif) ligand 7 |
| NM_008599 | Cxcl9 | 4.1 | 20.6 | 1 | 0.3 | 0 | Chemokine (C-X-C motif) ligand 9 |
| NM_011333 | Ccl2 | 3.6 | 6.4 | 1.1 | 0.6 | 1 | Chemokine (C-C motif) ligand 2 |
| NM_011331 | Ccl12 | 2.7 | 7.8 | 1.1 | 0.4 | 0.3 | Chemokine (C-C motif) ligand 12 |
| NM_019494 | Cxcl11 | 2.6 | 27.4 | 0.9 | 0.3 | 0 | Chemokine (C-X-C motif) ligand 11 |
| NM_021274 | Cxcl10 | 2 | 9.4 | 1 | 0.2 | 0 | Chemokine (C-X-C motif) ligand 10 |
| NM_009909 | Il8rb | 0.2 | 0.1 | 0.8 | 1.1 | 1.7 | Interleukin-8 receptor beta |
| NM_009835 | Ccr6 | 0.1 | 0.1 | 1 | 1.2 | 1.2 | Chemokine (C-C motif) receptor 6 |
| NM_009915 | Ccr2 | 0.04 | 0.05 | 0.9 | 0.4 | 0.3 | Chemokine (C-C motif) receptor 2 |
| Genes for cell surface molecules | |||||||
| AK020012 | Cd150 | 154.5 | 148.2 | 1.9 | 1.2 | 0.4 | |
| U96752 | Q1b | 80.8 | 27.9 | 0.8 | 1 | 0.6 | Major histocompatibility complex Q1b |
| BC052824 | Cd247 | 43.6 | 31.1 | 0.8 | 2 | 0.2 | CD247 protein |
| NM_170701 | Cd40 | 40.4 | 61.1 | 1 | 1 | 0.2 | Tumor necrosis factor receptor superfamily, member 5 isoform 3 |
| NM_011521 | Sdc4 | 37.3 | 52.9 | 1.1 | 0.8 | 0.4 | Syndecan 4 |
| NM_010818 | Cd200 | 25.5 | 43.4 | 0.9 | 2.7 | 3.5 | CD200 antigen |
| NM_007646 | Cd38 | 13.2 | 15.9 | 1 | 1.4 | 1 | CD38 antigen |
| NM_028523 | Dcbld2 | 11.2 | 9.7 | 1.1 | 0.7 | 0.4 | Discoidin, CUB and LCCL domain containing 2 |
| NM_013730 | Slamf1 | 9.6 | 10.6 | 1.2 | 0.4 | 0.4 | Signaling lymphocytic activation molecule family member 1 |
| NM_011610 | Tnfrsf1b | 8.3 | 12.8 | 1 | 1 | 0.5 | Tumor necrosis factor receptor superfamily, member 1b |
| XM_132882 | Cd69 | 7.8 | 19.4 | 1 | 0.3 | 0.1 | CD69 antigen |
| NM_009855 | Cd80 | 7.6 | 13.1 | 1 | 0.7 | 0.5 | CD80 antigen precursor |
| NM_009851 | Cd44 | 7.4 | 12.3 | 2.2 | 0.4 | 0.3 | CD44 antigen isoform a |
| NM_007654 | Cd72 | 7.2 | 16.1 | 0.9 | 1.2 | 0.4 | CD72 antigen |
| BC059059 | MMP20 | 5.8 | 10.3 | 1 | 1.3 | 1.1 | |
| NM_021893 | Cd274 | 5.6 | 7.2 | 0.9 | 1 | 0.7 | CD274 antigen |
| AK019885 | Cd137 | 5.2 | 9.3 | 1 | 1.3 | 0.7 | |
| NM_007640 | Cd1d2 | 4.5 | 10.2 | 0.9 | 1.6 | 0.6 | CD1d2 antigen |
| NM_009856 | Cd83 | 4 | 5.2 | 1 | 0.8 | 0.6 | CD83 antigen |
| NM_010493 | Cd54 | 3.9 | 4.3 | 0.9 | 0.5 | 0.2 | Intercellular adhesion molecule |
| NM_053129 | Pcdhb4 | 2.9 | 5.9 | 1.9 | 1 | 1.1 | Protocadherin beta 4 |
| NM_019388 | Cd86 | 2.8 | 4.7 | 0.8 | 0.4 | 0.2 | CD86 antigen |
| NM_177584 | Btla | 2.8 | 10 | 0.6 | 0.8 | 0.2 | B- and T-lymphocyte-associated isoform 2 |
| AF106008 | Cd159a | 0.4 | 4.9 | 1 | 0.6 | 0.4 | Natural killer cell protein group 2-A1 |
| NM_009690 | Cd5l | 0.4 | 0.1 | 0.9 | 0.3 | 0 | CD5 antigen-like |
| NM_018729 | Cd244 | 0.3 | 0.1 | 0.9 | 0.9 | 0.6 | CD244 natural killer cell receptor 2B4 |
| NM_013487 | Cd3d | 0.3 | 0.2 | 1 | 0.7 | 0.5 | CD3 antigen, delta polypeptide |
| NM_134158 | Cd300d | 0.2 | 0.2 | 0.9 | 1.7 | 2.2 | CD300D antigen |
| BC024571 | Cd49f | 0.2 | 0.2 | 1 | 1.8 | 9.1 | |
| NM_009852 | Cd6 | 0.2 | 0.2 | 1 | 1 | 0.7 | CD6 antigen isoform 2 |
| NM_053094 | Cd163 | 0.2 | 0.2 | 1.1 | 0.5 | 0.5 | CD163 antigen |
| NM_133654 | Cd34 | 0.1 | 0.2 | 1 | 0.5 | 0.3 | CD34 antigen |
| NM_133238 | Cd209a | 0.1 | 0.1 | 0.8 | 1 | 1.4 | CD209a antigen |
| NM_021476 | Cysltr1 | 0.1 | 0.2 | 0.9 | 0.9 | 1.4 | Cysteinyl leukotriene receptor 1 |
| NM_027987 | Cd300lg | 0.1 | 0.1 | 0.9 | 1.1 | 1 | CMRF-35-like molecule-9 |
| Signal transduction genes | |||||||
| NM_007707 | Socs3 | 65.8 | 75.5 | 0.9 | 0.9 | 0.5 | Suppressor of cytokine signaling 3 |
| NM_011488 | Stat5a | 8.5 | 9.1 | 1 | 0.7 | 0.2 | Signal transducer and activator of transcription 5A |
| NM_009421 | Traf1 | 8.3 | 13.1 | 0.9 | 1.1 | 0.6 | TNF receptor-associated factor 1 |
| NM_009896 | Socs1 | 7.5 | 17.2 | 1 | 1.3 | 0.7 | Suppressor of cytokine signaling 1 |
| NM_011489 | Stat5b | 6.5 | 8.1 | 1 | 0.8 | 0.6 | Signal transducer and activator of transcription 5B |
| NM_028679 | Irak3 | 5.8 | 6.3 | 1 | 1.1 | 0.8 | Interleukin-1 receptor-associated kinase 3 |
| NM_019408 | Nfkb2 | 5.2 | 6.7 | 0.9 | 0.9 | 0.5 | Nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| NM_008689 | Nfkb1 | 4.2 | 5.4 | 0.9 | 0.7 | 0.4 | Nuclear factor kappa B, subunit 1 |
| NM_033601 | Bcl3 | 3.9 | 6.9 | 1 | 0.8 | 0.4 | B-cell leukemia/lymphoma 3 |
| NM_213659 | Stat3 | 2.4 | 5.2 | 0.9 | 0.9 | 0.7 | Signal transducer and activator of transcription 3 isoform 1 |
| NM_011487 | Stat4 | 2 | 8.1 | 0.9 | 2.3 | 4.4 | Signal transducer and activator of transcription 4 |
| NM_009283 | Stat1 | 1.3 | 5.8 | 1 | 1.2 | 0.9 | Signal transducer and activator of transcription 1 |
Selected genes whose expression levels were modulated are listed and sorted by their expression level in γ-PGA NP-treated DCs.
All data represent means for three separate microarray experiments.
Expression levels in DCs 12 and 24 h after treatment with γ-PGA NPs were compared with those at 6 h.
γ-PGA NPs modulate the expression of TLR signaling-related genes.
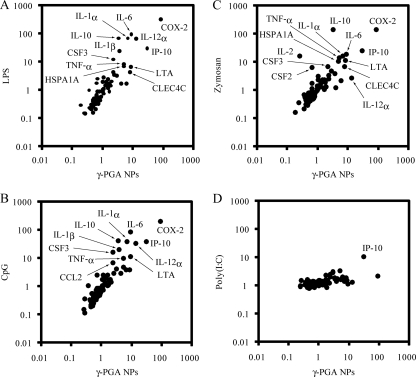
Since the microarray analysis revealed that like LPS, γ-PGA NPs could modulate gene expression in DCs, the effect of γ-PGA NPs on the expression of TLR signaling-related genes was examined by quantitative real-time RT-PCR and compared to that of LPS, CpG, zymosan, and poly(I:C). Consistent with the results of microarray analysis, γ-PGA NPs and LPS upregulated the gene expression of IL-1α, IL-1β, IL-6, IL-10, IL-12α, lymphotoxin A (LTA), TNF-α, and CXCL10 (Fig. 2A). The correlation coefficient between γ-PGA NP-treated DCs and LPS-treated DCs was high (r = 0.920). A high correlation coefficient (r = 0.918) for gene expression was also obtained between γ-PGA NP-treated DCs and CpG-treated DCs (Fig. 2B). In addition, significant increases in cyclooxygenase-2 (COX-2) gene expression were observed in DCs treated with γ-PGA NPs, LPS, CpG, and zymosan. The correlation coefficient for gene expression between γ-PGA NP-treated DCs and zymosan-treated DCs was 0.708 (Fig. 2C), while there was no correlation (r = 0.394) between γ-PGA NP-treated DCs and poly(I:C)-treated DCs (Fig. 2D).
FIG. 2.
Gene expression related to TLR signaling in DCs treated with γ-PGA NPs versus LPS (A), γ-PGA NPs versus CpG (B), γ-PGA NPs versus zymosan (C), and γ-PGA NPs versus poly(I:C) (D). DCs (1 × 106 cells/ml) were treated with either 300 μg/ml γ-PGA NPs, 1 μg/ml LPS, 1 μg/ml CpG, 5 μg/ml zymosan, 1 μg/ml poly(I:C), or PBS for 6 h. Total RNA was extracted from the treated DCs, purified, and subjected to real-time RT-PCR analysis. The expression level of each gene was normalized to that of the control (PBS treatment). The genes upregulated >2-fold are indicated by their product names.
γ-PGA NP-induced DC maturation is not due to LPS contamination.
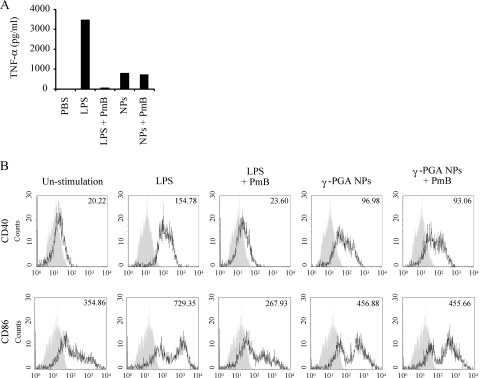
Although γ-PGA is a bacterial product, γ-PGA NPs contain LPS at a concentration far below the threshold that activates cytokine production and costimulatory molecule expression in DCs (33, 35). To confirm that LPS contamination is indeed not responsible for the activation of DCs by γ-PGA NPs, the cells were treated with γ-PGA NPs or LPS after preincubation with PmB and analyzed for protein expression by ELISA and flow cytometry. TNF-α production from DCs was enhanced by treatment with both LPS and γ-PGA NPs (Fig. 3A). However, such enhancement was significantly reduced by preincubation of LPS with PmB but not by preincubation of γ-PGA NPs with PmB. A similar result was obtained for the surface expression of costimulatory molecules, where CD40 and CD86 expression was strongly upregulated by treatment with both LPS and γ-PGA NPs (Fig. 3B). Again, the upregulation was annihilated by preincubation of LPS with PmB but not by preincubation of γ-PGA NPs with PmB. These results indicate that the induction of DC maturation by γ-PGA NPs is due not to LPS contamination but to their own effect on DCs.
FIG. 3.
Effect of PmB on cytokine production and costimulatory molecule expression induced by γ-PGA NPs and LPS. DCs were incubated with either γ-PGA NPs or LPS in the absence or presence of 10 μg/ml PmB. (A) Culture supernatants were collected and their TNF-α level determined by ELISA. (B) The cells were stained with a PE-conjugated anti-CD40 MAb or a PE-conjugated anti-CD86 MAb and analyzed by flow cytometry. The number in each histogram indicates the mean fluorescence intensity.
γ-PGA NPs are more effective than conventional adjuvants.
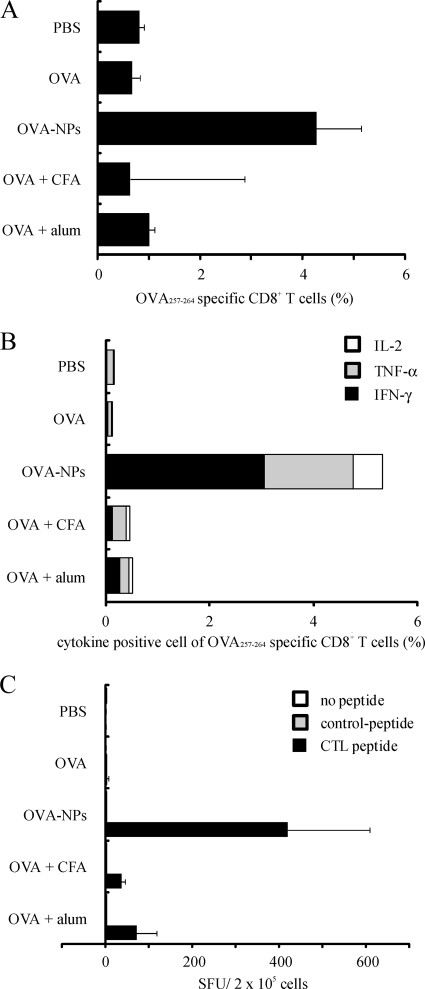
To determine the comparative effects of γ-PGA NPs and conventional adjuvants in inducing antigen-specific cellular immune responses, mice were injected subcutaneously with either PBS, OVA, OVA-NPs, OVA + CFA, or OVA + alum. When the antigen-specific T-cell response was examined by flow cytometry, a marked increase of OVA antigen-specific CD8+ T cells was observed for the cells obtained from the OVA-NP-immunized mice, whereas little increase of the antigen-specific CD8+ T cells was identified for the cells from mice immunized with OVA alone, OVA + CFA, or OVA + alum (Fig. 4A). Since the cytokine production from immune cells is an important marker of their activation, the intracellular cytokine assay is able to detect the state of activation in individual cells. When the production of intracellular cytokines in the antigen-specific CD8+ T cells was examined, marked increases of IFN-γ, TNF-α, and IL-2 production were observed for the cells obtained from the OVA-NP-immunized mice. Again, little increases of such cytokine production were observed for the cells from mice immunized with OVA alone, OVA + CFA, or OVA + alum (Fig. 4B). Furthermore, the most induction of IFN-γ-producing cells was achieved by immunization with OVA-NPs, as determined by ELISPOT assay (Fig. 4C). These results suggest that γ-PGA NPs are a more efficient adjuvant than CFA and alum in inducing cellular immune responses.
FIG. 4.
Antigen-specific T-cell responses induced by OVA-NPs, CFA, and alum. Mice were immunized subcutaneously once with either PBS, OVA, OVA-NPs, OVA + CFA, or OVA + alum. T cells were obtained from the spleen. (A) The antigen (OVA)-specific CD8+ T cells were analyzed by pentamer staining. (B) IL-2, TNF-α, and IFN-γ production of the antigen-specific CD8+ T cells was measured by intracellular cytokine staining. (C) Spleen cells from the immunized mice were restimulated with the control or antigen peptide. The number of IFN-γ-producing cells was determined by ELISPOT assay. SFU, spot-forming units. All results represent the means for three separate experiments.
DISCUSSION
The induction of innate immunity is required for effective adaptive immune responses against various antigens, and activated DCs play a critical role in the initial activation of innate immunity (23). Therefore, the activation of DCs is an important factor for the development of novel adjuvants. In our previous studies, it was demonstrated that γ-PGA NPs induced phenotypic and functional changes in DCs in vitro and in vivo (32, 33, 35). However, there are no previous reports describing genomic analysis of gene expression in DCs treated with γ-PGA NPs. In this study, murine DCs were treated with either γ-PGA NPs, LPS, or unparticulate γ-PGA. Since the effect of γ-PGA NPs on gene expression was quite similar to that of LPS (Fig. 1A), we had to exclude the possibility that the effect of γ-PGA NPs was due to LPS contamination. The results for preincubation of γ-PGA NPs with the LPS antagonist PmB clearly show that the immunomodulatory activity of γ-PGA NPs is their own property (Fig. 3). Furthermore, unparticulate γ-PGA could scarcely modulate gene expression in DCs (Fig. 1B), suggesting that the particulate form of γ-PGA is required to exert biological activity.
In immune responses, DCs represent a major source of IL-12, which is an important cytokine linking the innate and adoptive immune systems and driving Th1 polarization (31). In addition to IL-12, other members of the IL-12 family, such as IL-23 and IL-27, also modulate the Th1 response. IL-23, which is composed of the β subunit of IL-12 and the IL-23-specific α subunit, has been shown to preferentially act on Th1 effector/memory CD4+ T cells and to induce their proliferation and IFN-γ production (20). IL-27 is composed of the IL-27-specific α subunit and the Epstein-Barr virus-induced gene 3 (Ebi-3) product (β subunit) (25). It contributes to Th1 development by promoting the expression of T-bet and IL-12Rβ2 genes (13). In our previous studies, it was found that γ-PGA NPs induced IL-12β production from DCs (33, 35, 37). In this study, the treatment of DCs with γ-PGA NPs increased the transcript levels of the IL-12 cytokine family, especially IL-12β, IL-23α, and IL-12α. Furthermore, the treatment of DCs with OVA-NPs induced antigen-specific IFN-γ producing CD8+ T cells in vivo (Fig. 4C). Although their perforin and granzyme levels were not evaluated, it has been demonstrated that OVA-NPs can induce the antigen-specific cytotoxicity determined by a 51Cr release assay (33). These results suggest that γ-PGA NPs are capable of acting as an adjuvant that potently induces antigen-specific cellular immune responses.
The capacity of DC migration from peripheral organs to the T-cell areas of lymph nodes is a key step in successful induction of protective immune responses (3). CCR7 controls DC migration into afferent lymphatic vessels. In addition, DC migration is also controlled by CD38, since DCs lacking the CD38 gene had a significantly decreased ability to respond to CCR7 ligands (22). The microarray analysis revealed that γ-PGA NPs upregulated the expression of CCR7 and CD38 in DCs. Furthermore, the expression of CCR7 was upregulated in the lymph node DCs of mice subcutaneously immunized with γ-PGA NPs (data not shown). Thus, γ-PGA NPs may be able to induce the migration of DCs to spleen and lymph nodes by upregulating CCR7 and CD38 expression.
Chemokines affect the polarization and recruitment of T cells. T cells sense the chemotactic gradient of chemokines by their receptors involving CCR5 and CCR2 and elicit the formation of an immunological synapse (16). CCR5 and CXCR3 are markers for Th1 cells, and CCR2 is specifically expressed on Th2 cells (16). The gene expression of CCR5 ligands (CCL3, CCL4, and CCL5) and CXCR3 ligand (IP-10) in DCs was enhanced equally by γ-PGA NP and LPS treatments (Table 1). Although γ-PGA NP-treated DCs also showed enhanced gene expression of CCR2 ligands (CCL2, CCL7, and CCL12), the expression levels of these genes were lower (<50%) than those in LPS-treated DCs. Thus, in comparison with LPS, γ-PGA NPs may produce the chemotactic gradient of chemokine ligands recruiting Th1 cells rather than Th2 cells, supporting the capability of γ-PGA NPs to induce Th1 immune responses.
The gene expression pattern in γ-PGA NP-treated DCs was quite similar to that in DCs treated with LPS and CpG (Fig. 2A and B), whose receptors are TLR4 and TLR9, respectively. DCs were also stimulated by the TLR2 ligand zymosan, and the gene expression pattern induced by γ-PGA NPs was also similar to that induced by zymosan (Fig. 2C). On the other hand, the gene expression pattern in DCs treated with the TLR3 ligand poly(I:C) differed from that in γ-PGA NP-treated DCs (Fig. 2D). This difference may be due to the facts that TLR4, TLR9, and TLR2 activate the signaling pathway through MyD88 and that the signaling pathway associated with TLR3 is independent of MyD88 (15, 30, 34). TNF-α production from γ-PGA NP-treated DCs was suppressed by a MyD88 inhibitor peptide (33), indicating that γ-PGA NPs activate DCs through the MyD88 signaling pathway. Further studies are in progress to determine the TLR(s) involved in the interaction with γ-PGA NPs.
Suppressor of cytokine signaling (SOCS) family proteins have been identified as inducible feedback inhibitors for the activation of cytokine receptors through JAK/STAT signal transduction (10, 18, 28). SOCS3 and SOCS1 expression is induced in DCs after treatment with CpG (8) and LPS (29). DCs lacking SOCS1 generate aberrant activation of adaptive immunity, resulting in the induction of autoimmunity (11). Moreover, SOCS3 suppresses the activities of G-CSF and GM-CSF (7). Therefore, the expression of SOCS proteins is important for the regulation of innate immune responses caused by TLR signaling. The gene expression of SOCS3 and SOCS1 was activated by γ-PGA NPs as well as by LPS (Table 1), suggesting a feedback mechanism in response to the interaction of γ-PGA NPs with the TLR(s) of DCs.
In conclusion, considering the unique properties of γ-PGA NPs, such as their biodegradable nature, efficient antigen-carrying capacity, and strong induction of DC activation and cellular immune responses to carried antigens, γ-PGA NPs have great potential not only as an antigen delivery system to DCs but also as an effective adjuvant.
Acknowledgments
This work was supported by Core Research for Evolutional Science and Technology, Japan Science and Technology Agency, Japan.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Akagi, T., T. Kaneko, T. Kida, and M. Akashi. 2005. Preparation and characterization of biodegradable nanoparticles based on poly(gamma-glutamic acid) with l-phenylalanine as a protein carrier. J. Control. Release 108:226-236. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau, J., F. Briere, C. Caux, J. Davoust, S. Lebecque, Y. J. Liu, B. Pulendran, and K. Palucka. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767-811. [DOI] [PubMed] [Google Scholar]
- 3.Banchereau, J., and R. M. Steinman. 1998. Dendritic cells and the control of immunity. Nature 392:245-252. [DOI] [PubMed] [Google Scholar]
- 4.Caux, C., C. Massacrier, B. Vanbervliet, B. Dubois, C. Van Kooten, I. Durand, and J. Banchereau. 1994. Activation of human dendritic cells through CD40 cross-linking. J. Exp. Med. 180:1263-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiew Tong, N. K., J. Beran, S. A. Kee, J. L. Miguel, C. Sanchez, J. M. Bayas, A. Vilella, J. R. de Juanes, P. Arrazola, F. Calbo-Torrecillas, E. L. de Novales, V. Hamtiaux, M. Lievens, and M. Stoffel. 2005. Immunogenicity and safety of an adjuvanted hepatitis B vaccine in pre-hemodialysis and hemodialysis patients. Kidney Int. 68:2298-2303. [DOI] [PubMed] [Google Scholar]
- 6.Coester, C., P. Nayyar, and J. Samuel. 2006. In vitro uptake of gelatin nanoparticles by murine dendritic cells and their intracellular localisation. Eur. J. Pharm. Biopharm. 62:306-314. [DOI] [PubMed] [Google Scholar]
- 7.Croker, B. A., D. Metcalf, L. Robb, W. Wei, S. Mifsud, L. DiRago, L. A. Cluse, K. D. Sutherland, L. Hartley, E. Williams, J. G. Zhang, D. J. Hilton, N. A. Nicola, W. S. Alexander, and A. W. Roberts. 2004. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity 20:153-165. [DOI] [PubMed] [Google Scholar]
- 8.Dalpke, A. H., S. Opper, S. Zimmermann, and K. Heeg. 2001. Suppressors of cytokine signaling (SOCS)-1 and SOCS-3 are induced by CpG-DNA and modulate cytokine responses in APCs. J. Immunol. 166:7082-7089. [DOI] [PubMed] [Google Scholar]
- 9.Dinauer, N., S. Balthasar, C. Weber, J. Kreuter, K. Langer, and H. von Briesen. 2005. Selective targeting of antibody-conjugated nanoparticles to leukemic cells and primary T-lymphocytes. Biomaterials 26:5898-5906. [DOI] [PubMed] [Google Scholar]
- 10.Endo, T. A., M. Masuhara, M. Yokouchi, R. Suzuki, H. Sakamoto, K. Mitsui, A. Matsumoto, S. Tanimura, M. Ohtsubo, H. Misawa, T. Miyazaki, N. Leonor, T. Taniguchi, T. Fujita, Y. Kanakura, S. Komiya, and A. Yoshimura. 1997. A new protein containing an SH2 domain that inhibits JAK kinases. Nature 387:921-924. [DOI] [PubMed] [Google Scholar]
- 11.Hanada, T., H. Yoshida, S. Kato, K. Tanaka, K. Masutani, J. Tsukada, Y. Nomura, H. Mimata, M. Kubo, and A. Yoshimura. 2003. Suppressor of cytokine signaling-1 is essential for suppressing dendritic cell activation and systemic autoimmunity. Immunity 19:437-450. [DOI] [PubMed] [Google Scholar]
- 12.Hauguel, T. M., and C. J. Hackett. 2008. Rationally-designed vaccine adjuvants: separating efficacy from toxicity. Front. Biosci. 13:2806-2813. [DOI] [PubMed] [Google Scholar]
- 13.Hunter, C. A. 2005. New IL-12-family members: IL-23 and IL-27, cytokines with divergent functions. Nat. Rev. Immunol. 5:521-531. [DOI] [PubMed] [Google Scholar]
- 14.Jakob, T., P. S. Walker, A. M. Krieg, M. C. Udey, and J. C. Vogel. 1998. Activation of cutaneous dendritic cells by CpG-containing oligodeoxynucleotides: a role for dendritic cells in the augmentation of Th1 responses by immunostimulatory DNA. J. Immunol. 161:3042-3049. [PubMed] [Google Scholar]
- 15.Kenny, E. F., and L. A. J. O'Neill. 2008. Signalling adaptors used by Toll-like receptors: an update. Cytokine 43:342-349. [DOI] [PubMed] [Google Scholar]
- 16.Luther, S. A., and J. G. Cyster. 2001. Chemokines as regulators of T cell differentiation. Nat. Immunol. 2:102-107. [DOI] [PubMed] [Google Scholar]
- 17.Mettens, P., P. M. Dubois, M. A. Demoitie, B. Bayat, M. N. Donner, P. Bourguignon, V. A. Stewart, D. G. Heppner, Jr., N. Garcon, and J. Cohen. 2008. Improved T cell responses to Plasmodium falciparum circumsporozoite protein in mice and monkeys induced by a novel formulation of RTS,S vaccine antigen. Vaccine 26:1072-1082. [DOI] [PubMed] [Google Scholar]
- 18.Naka, T., M. Narazaki, M. Hirata, T. Matsumoto, S. Minamoto, A. Aono, N. Nishimoto, T. Kajita, T. Taga, K. Yoshizaki, S. Akira, and T. Kishimoto. 1997. Structure and function of a new STAT-induced STAT inhibitor. Nature 387:924-929. [DOI] [PubMed] [Google Scholar]
- 19.O'Hagan, D. T., A. Wack, and A. Podda. 2007. MF59 is a safe and potent vaccine adjuvant for flu vaccines in humans: what did we learn during its development? Clin. Pharmacol. Ther. 82:740-744. [DOI] [PubMed] [Google Scholar]
- 20.Oppmann, B., R. Lesley, B. Blom, J. C. Timans, Y. Xu, B. Hunte, F. Vega, N. Yu, J. Wang, K. Singh, F. Zonin, E. Vaisberg, T. Churakova, M. Liu, D. Gorman, J. Wagner, S. Zurawski, Y. Liu, J. S. Abrams, K. W. Moore, D. Rennick, R. de Waal-Malefyt, C. Hannum, J. F. Bazan, and R. A. Kastelein. 2000. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 13:715-725. [DOI] [PubMed] [Google Scholar]
- 21.Paavonen, J., P. Naud, J. Salmerón, C. M. Wheeler, S. N. Chow, D. Apter, H. Kitchener, X. Castellsague, J. C. Teixeira, S. R. Skinner, J. Hedrick, U. Jaisamrarn, G. Limson, S. Garland, A. Szarewski, B. Romanowski, F. Y. Aoki, T. F. Schwarz, W. A. J. Poppe, F. X. Bosch, D. Jenkins, K. Hardt, T. Zahaf, D. Descamps, F. Struyf, M. Lehtinen, and G. Dubin. 2009. Efficacy of human papillomavirus (HPV)-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by oncogenic HPV types (PATRICIA): final analysis of a double-blind, randomised study in young women. Lancet 374:301-314. [DOI] [PubMed] [Google Scholar]
- 22.Partida-Sanchez, S., S. Goodrich, K. Kusser, N. Oppenheimer, T. D. Randall, and F. E. Lund. 2004. Regulation of dendritic cell trafficking by the ADP-ribosyl cyclase CD38: impact on the development of humoral immunity. Immunity 20:279-291. [DOI] [PubMed] [Google Scholar]
- 23.Pashine, A., N. M. Valiante, and J. B. Ulmer. 2005. Targeting the innate immune response with improved vaccine adjuvants. Nat. Med. 11:S63-S68. [DOI] [PubMed] [Google Scholar]
- 24.Petrovsky, N., and J. C. Aguilar. 2004. Vaccine adjuvants: current state and future trends. Immunol. Cell Biol. 82:488-496. [DOI] [PubMed] [Google Scholar]
- 25.Pflanz, S., J. C. Timans, J. Cheung, R. Rosales, H. Kanzler, J. Gilbert, L. Hibbert, T. Churakova, M. Travis, E. Vaisberg, W. M. Blumenschein, J. D. Mattson, J. L. Wagner, W. To, S. Zurawski, T. K. McClanahan, D. M. Gorman, J. F. Bazan, R. de Waal Malefyt, D. Rennick, and R. A. Kastelein. 2002. IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4(+) T cells. Immunity 16:779-790. [DOI] [PubMed] [Google Scholar]
- 26.Sato, K., N. Yamashita, M. Baba, and T. Matsuyama. 2003. Modified myeloid dendritic cells act as regulatory dendritic cells to induce anergic and regulatory T cells. Blood 101:3581-3589. [DOI] [PubMed] [Google Scholar]
- 27.Sato, K., N. Yamashita, M. Baba, and T. Matsuyama. 2003. Regulatory dendritic cells protect mice from murine acute graft-versus-host disease and leukemia relapse. Immunity 18:367-379. [DOI] [PubMed] [Google Scholar]
- 28.Starr, R., T. A. Willson, E. M. Viney, L. J. Murray, J. R. Rayner, B. J. Jenkins, T. J. Gonda, W. S. Alexander, D. Metcalf, N. A. Nicola, and D. J. Hilton. 1997. A family of cytokine-inducible inhibitors of signalling. Nature 387:917-921. [DOI] [PubMed] [Google Scholar]
- 29.Stoiber, D., P. Kovarik, S. Cohney, J. A. Johnston, P. Steinlein, and T. Decker. 1999. Lipopolysaccharide induces in macrophages the synthesis of the suppressor of cytokine signaling 3 and suppresses signal transduction in response to the activating factor IFN-gamma. J. Immunol. 163:2640-2647. [PubMed] [Google Scholar]
- 30.Takeda, K., and S. Akira. 2004. TLR signaling pathways. Semin. Immunol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 31.Trinchieri, G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3:133-146. [DOI] [PubMed] [Google Scholar]
- 32.Uto, T., X. Wang, T. Akagi, R. Zenkyu, M. Akashi, and M. Baba. 2009. Improvement of adaptive immunity by antigen-carrying biodegradable nanoparticles. Biochem. Biophys. Res. Commun. 379:600-604. [DOI] [PubMed] [Google Scholar]
- 33.Uto, T., X. Wang, K. Sato, M. Haraguchi, T. Akagi, M. Akashi, and M. Baba. 2007. Targeting of antigen to dendritic cells with poly(gamma-glutamic acid) nanoparticles induces antigen-specific humoral and cellular immunity. J. Immunol. 178:2979-2986. [DOI] [PubMed] [Google Scholar]
- 34.Wagner, H. 2004. The immunobiology of the TLR9 subfamily. Trends Immunol. 25:381-386. [DOI] [PubMed] [Google Scholar]
- 35.Wang, X., T. Uto, T. Akagi, M. Akashi, and M. Baba. 2007. Induction of potent CD8+ T-cell responses by novel biodegradable nanoparticles carrying human immunodeficiency virus type 1 gp120. J. Virol. 81:10009-10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, X., T. Uto, T. Akagi, M. Akashi, and M. Baba. 2008. Poly(gamma-glutamic acid) nanoparticles as an efficient antigen delivery and adjuvant system: potential for an AIDS vaccine. J. Med. Virol. 80:11-19. [DOI] [PubMed] [Google Scholar]
- 37.Wang, X., T. Uto, K. Sato, K. Ide, T. Akagi, M. Okamoto, T. Kaneko, M. Akashi, and M. Baba. 2005. Potent activation of antigen-specific T cells by antigen-loaded nanospheres. Immunol. Lett. 98:123-130. [DOI] [PubMed] [Google Scholar]