Abstract
Chlamydia pneumoniae is a frequent pathogen of the respiratory tract, and persistent infections with this obligate intracellular bacterium have been associated with different severe sequelae. Although T-cell activation during acute C. pneumoniae infections has been described, little is known about the frequency or the role of the C. pneumoniae-specific memory T cells that reside in the human body after the resolution of the infection. In the present study, the C. pneumoniae-induced T-cell responses in peripheral blood mononuclear cells of 56 healthy volunteers were analyzed and compared to the donor's serum antibody reactivity toward whole C. pneumoniae as well as recombinant C. pneumoniae antigens. Following short-term stimulation with C. pneumoniae, both gamma interferon (IFN-γ)- and interleukin-2 (IL-2)-producing CD4+ T-cell responses could be detected in 16 of 56 healthy individuals. C. pneumoniae-activated CD4+ T cells expressed CD154, a marker for T-cell receptor-dependent activation, and displayed a phenotype of central memory T cells showing dominant IL-2 production but also IFN-γ production. Interestingly, individuals with both IFN-γ- and IL-2-producing responses showed significantly decreased immunoglobulin G reactivity toward C. pneumoniae RpoA and DnaK, antigens known to be strongly upregulated during chlamydial persistence, compared to IgG reactivity of seropositive individuals with no T-cell response or CD4+ T-cell responses involving the production of a single cytokine (IFN-γ or IL-2). Our results demonstrate that memory CD4+ T cells responding to C. pneumoniae stimulation can be detected in the circulation of healthy donors. Furthermore, among seropositive individuals, the presence or the absence of dual IFN-γ- and IL-2-producing T-cell responses was associated with distinct patterns of antibody responses toward persistence-associated C. pneumoniae antigens.
Besides primary respiratory infection, Chlamydia pneumoniae is thought to establish persistent infections when the bacteria are not eliminated by the host (15, 20). Unresolved C. pneumoniae infections may represent a risk factor for chronic inflammatory diseases (2, 6, 9, 21), but they are difficult to diagnose, since serologic tests are still unable to discriminate between past and persistent infections (1, 6, 15). We recently described novel C. pneumoniae antigens, some of which might prove to be useful serologic markers for the detection of persistent C. pneumoniae infections (8). Unlike for antibody responses, little is known about the frequency and role of C. pneumoniae-specific T cells in humans. Effector CD4+ T cells responding to C. pneumoniae could be isolated from C. pneumoniae-positive atherosclerotic plaque (4, 5), and by using major histocompatibility complex (MHC) class I tetramers, C. pneumoniae-specific CD8+ T cells were detected in the peripheral blood mononuclear cells (PBMCs) of infected patients (10). Studies of mice revealed a protective role for CD4+ and CD8+ T cells during primary C. pneumoniae infection (33, 34, 39) and during reinfection (31, 32, 34), suggesting the development of specific memory T cells. Pathogen-specific memory T cells were found to have a key role in the immune control of persisting viruses, like cytomegalovirus (CMV), Epstein-Barr virus, varicella-zoster virus, and HIV. Notably, the cytokine profile of antiviral T-cell responses was found to reflect the degree of efficiency of control of the viral infection. Whereas CD4+ T-cell responses with predominantly gamma interferon (IFN-γ) production were found during uncontrolled viral infections with high virus titers, the production of both IFN-γ and interleukin-2 (IL-2) by virus-specific CD4+ T cells reflected the efficient immune control of persistent viral infections in association with low or moderate virus titers (16, 17, 30).
In the present study, we investigated C. pneumoniae-induced T-cell activation in the PBMCs of 56 healthy donors and correlated the results with their serum immunoglobulin A (IgA) and IgG reactivities toward whole C. pneumoniae and toward C. pneumoniae antigens known to be upregulated during persistent chlamydial infection (3, 26, 28). For donors with dual IFN-γ- and IL-2-producing CD4+ T-cell responses, we further analyzed the cytokine profile and CD154 expression of activated T cells, as well as their expression of CD45RA and CCR7, to discriminate between effector memory T cells (TEM) and central memory T cells (TCM). Our data demonstrate that after stimulation with C. pneumoniae, activated memory CD4+ T cells producing IFN-γ, IL-2, and CD154 can be detected in the peripheral blood of healthy human donors. Furthermore, in the case of seropositive individuals, the presence of C. pneumoniae-induced CD4+ T-cell responses involving the production of both IFN-γ and IL-2 was found to be associated with decreased serum IgG reactivity toward persistence-associated chlamydial antigens compared to the reactivity in individuals with no response or a response involving the production of only a single cytokine.
MATERIALS AND METHODS
Volunteers and PBMC preparation.
Whole blood and sera were obtained from 57 healthy volunteers (University of Konstanz, Konstanz, Germany). The donors stated that they were healthy, had had no chronic or severe respiratory illnesses during the last 2 years, and had not taken immunosuppressive medication. To determine whether the volunteers had possible acute C. pneumoniae infections, as defined by a 4-fold rise in the specific IgG or IgA titers of consecutive serum samples, serum samples were taken from each donor at the start of the study as well as at 2, 4, and 6 months after the initiation of the study and analyzed for specific antibodies. According to the criteria indicated above, one donor fulfilled the serologic parameters of an acute C. pneumoniae infection (a 4-fold rise in the IgG and IgA titers) and was excluded from the study. Acute infections with unrelated pathogens could further be excluded from the differential blood cell counts by use of a Pentra 60 apparatus (ABX, Montpellier, France). The ages of the 56 donors without evidence of acute infection varied from 24 to 61 years (mean, 40.6 years), and the ratio of men to woman was 30 to 26. PBMCs were isolated with Vacutainer CPT cell preparation tubes (BD Biosciences), according to the manufacturer's instructions. The cells were washed and resuspended in RPMI 1640 supplemented with ultraglutamine, 2.5 IU/ml heparin (Liquemin; Hoffmann-La Roche), and 10% autologous serum at a concentration of 8 × 106 cells/ml. Then, 0.4-ml aliquots of PBMCs were equilibrated in 15-ml polypropylene tubes at 37°C in a humidified 5% CO2 atmosphere for 20 h before they were stimulated.
Stimulation of PBMCs.
Equilibrated PBMC aliquots of 0.4 ml (3.2 × 106 cells) were adjusted to 1 ml with RPMI 1640 containing ultraglutamine, 2.5 IE/ml heparin (Hoffmann-La Roche), and 1 μg of the costimulatory anti-CD28 antibody (BD Biosciences). The cells were stimulated with 1 × 109 C. pneumoniae cells, which was found to be the optimal dose for activation, or with staphylococcal enterotoxin B (SEB; 100 ng or 1 μg; Sigma-Aldrich) as a positive control for stimulation. PBMC samples from CMV-seropositive donors were also stimulated with a peptide pool (1 μg per peptide) specific for the CMV pp65 protein (22). Control stimulations with 1 μg and 10 μg LPS from Salmonella enterica serovar Abortus Equi were carried out under identical conditions to analyze the influence of T-cell receptor (TCR)-independent T-cell activation. After application of the stimuli, the PBMCs were incubated at 37°C in a humidified 5% CO2 atmosphere for 6 h; the last 4 h of incubation was done in the presence of 10 μg/ml brefeldin A (Sigma-Aldrich). The PBMCs were then incubated with 2 mM EDTA for 15 min and roughly vortexed to detach adherent cells. The cells were fixed with 4.5 ml fluorescent-activated cell sorter (FACS) lysing solution (BD Biosciences) for 10 min and stored at −70°C until analysis by flow cytometry.
Staining of surface markers and intracellular cytokines.
The fixed PBMCs were thawed, transferred to 5-ml polystyrene tubes, and centrifuged at 1,100 × g for 10 min. The cells were then permeabilized for 10 min at room temperature by using permeabilizing solution 2 (BD Biosciences). For the detection of surface markers, the cells were stained with anti-human CD3-allophycocyanin (APC) and CD4-phycoerythrin (PE) (both from BD Pharmingen) and CD8-peridinin chlorophyll (PerCP) or CD4-PerCP (both from BD Bioscience), CD45RA-fluorescein isothiocyanate (FITC) and CCR7-PE (both from BD Pharmingen), or CD4-PerCP alone. For the staining of intracellular cytokines, FITC-conjugated antibodies against human IL-2 and/or IFN-γ (both from BD Biosciences) and anti-human IFN-γ-PE (BD Pharmingen) were used. Intracellular CD154 was detected by using an anti-human CD154-APC antibody (BD Pharmingen).
Flow cytometric analysis.
Four-color flow cytometry was performed on a FACSCalibur flow cytometer. Data files were analyzed with the CellQuest software package (BD Biosciences). For the determination of IFN-γ-positive (IFN-γ+) or IL-2-positive (IL-2+) T cells, 80,000 to 100,000 CD3+ lymphocytes were analyzed and further characterized according to their expression of CD4 and CD8. The frequencies of IFN-γ+ or IL-2+ T cells were determined by the use of quadrant statistics. To determine the frequencies of specifically activated T cells, the portion of activated T cells of the stimulated samples was subtracted by the respective values of the unstimulated samples (level of noise, always between 0.00% and 0.02%). Frequencies of specifically activated T cells of ≥0.03%, corresponding to 3 per 10,000 cells, were considered positive T-cell responses, and frequencies below this threshold were considered negative responses. To analyze the expression of CD154 among activated CD4+ T cells, 100,000 CD4+ lymphocytes were assessed for the production of IFN-γ and/or IL-2 and further characterized for intracellular CD154. As before, the values of the stimulated samples were corrected by subtraction of the proportion of cytokine-positive (maximum, 0.02%) and CD154-positive cells of the unstimulated samples. To determine the memory phenotype and the cytokine expression profile of CD154+ CD4+ T cells responding to different stimuli, 100,000 to 200,000 CD4+ T cells were analyzed according to their expression of CCR7 and CD45RA as well as of intracellular IL-2 and IFN-γ.
Determination of C. pneumoniae- and CMV-specific antibodies.
The sera of all donors were tested in a blinded fashion for the prevalence of total anti-C. pneumoniae IgG and IgA antibodies by a microimmunofluorescence (MIF) assay and the SeroCP Quant IgA enzyme-linked immunosorbent assay (ELISA) (both from Savyon Diagnostics, St. Ashdod, Israel), respectively. Both assays are based on purified elementary bodies of C. pneumoniae TW-183. The MIF assay was also used to assess the prevalence of IgG antibodies against C. trachomatis and C. psittaci to exclude donors who had previously been infected with these pathogens. The CMV status of some donors was determined in a diagnostic laboratory (Labor Brunner, Konstanz, Germany) by use of the Liaison IgG ELISA (DiaSorin, Saluggia, Italy), according to the manufacturer's instructions.
C. pneumoniae culture, preparation, and quantification.
C. pneumoniae TW-183 was propagated and harvested as described previously (8). In brief, semiconfluent monolayers of HEp-2 cells were infected with C. pneumoniae TW-183 (about 500 genomic equivalents per cell) and cultivated for 72 h. The infected cells were scraped off and disrupted by vortexing with glass beads, and the bacteria were purified by use of a discontinuous gradient containing 25/50% sodium amidotrizoate and meglumine amidotrizoate (Gastrografin; Schering). Purified C. pneumoniae was finally suspended in SPS buffer [10 mM Na(H)PO4, pH 7.4; 250 mM saccharose] at a concentration of ≥1011 per ml, repeatedly passed through a syringe, and stored at −70°C. The amount of C. pneumoniae genomic equivalents was determined by real-time PCR with primers specific for the 16S rRNA (forward primer, ATGTGGCTCTCAACCCCAT; backward primer, GGCGCCTCTCTCCTATAAATAGG) and was calculated according to the amounts on a standard curve of genomic DNA which was prepared with a defined amount of C. pneumoniae elementary bodies, as published previously (27).
Recombinant expression of C. pneumoniae antigens.
The C. pneumoniae antigens RNA polymerase alpha, CpB0704, and polymorphic membrane protein 21 (Pmp21m), which corresponds to the partial amino acid sequence from positions 636 to 1142 of the full-length Pmp21, were cloned and expressed as described previously (8). In brief, the genomic sequences of the antigens were amplified by PCR with Pfu polymerase (Fermentas) and primers flanked with BamHI and SalI restriction sites and ligated into the pQE30 expression vector, which encodes a hexahistidine tag (Qiagen). Protein expression was carried out in Escherichia coli M15 cells (Qiagen) for 6 h at 30°C. The cells were then disrupted in buffer containing 8 M urea, and the proteins were purified with HisTrap HP columns (Amersham Biosciences). The same protocol was used to clone and express the full-length genomic sequence of C. pneumoniae molecular chaperone dnaK after amplification with gene-specific primers (forward primer, CCGGATCCATGAGTGAACACAAAAAATCAAG; backward primer, CCGTCGACTCGTCGTTATCAATAATTTCTAC).
Immunoblot analysis.
For immunoblotting, recombinant RNA polymerase alpha, molecular chaperone dnaK, polymorphic membrane protein 21 (the middle part), and CpB0704 were separately applied to sodium dodecyl sulfate-polyacrylamide gels by using 12% polyacrylamide gels and 10 μg of protein per 7 cm in width. After electrophoresis, the proteins were transferred to a Bio Trace NT nitrocellulose membrane (Pall) by use of a semidry blotting system (Bio-Rad). The membranes were blocked with TBST (50 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20, pH 7.4) containing 5% nonfat dry milk for 2 h at room temperature (RT). The membranes were then cut into 4-mm strips, and each strip was incubated with a serum sample from each of the 56 donors at a dilution of 1:1,000 in blocking buffer overnight at 4°C. The strips were washed with TBST four times for 15 min each time and incubated with a peroxidase-conjugated rabbit anti-human IgG secondary antibody (diluted 1:2,000; DakoCytomation) for 45 min at RT. After four subsequent washing steps, immunoreactive bands were visualized by enhanced chemiluminescence detection, followed by 5-min exposure to a LAS-3000 imaging system (Fuji). The maximal signal intensities of the immunoreactive bands for each strip were determined by use of the AIDA software package (Raytest/Fuji). An intensity threshold of 1,000 counts above the background level was used as the cutoff.
Statistics.
Statistical analysis was performed with GraphPad Prism (version 4) software. Significant differences in the frequencies of activated T cells and mean antibody titers between different donor groups were tested by analysis of variance (ANOVA) and by the Kruskal-Wallis test followed by Dunn's posttest, respectively.
RESULTS
C. pneumoniae-induced CD4+ T-cell responses can be detected among healthy donors.
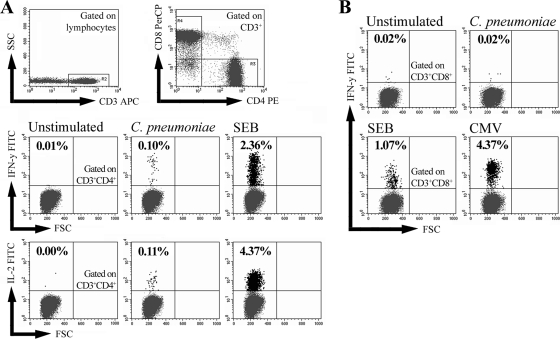
In the present study we investigated whether, in addition to C. pneumoniae-specific antibodies, memory T cells responding to C. pneumoniae could also be detected among healthy individuals. Therefore, the PBMCs of 56 donors without evidence of ongoing infection were stimulated ex vivo with whole C. pneumoniae or control stimuli, and the production of IFN-γ and IL-2 by CD4+ and CD8+ T cells was analyzed by flow cytometry (Fig. 1A and B). Following stimulation with C. pneumoniae, activated CD4+ T cells producing either IFN-γ or IL-2, or both cytokines, at frequencies of ≥3 per 10,000 cells, defined here as a positive response, could be detected in 26 of 56 donors (Fig. 1A). For the remaining 30 donors, no or only marginal activation with IFN-γ+ or IL-2+ CD4+ T-cell frequencies below 3 per 10,000 cells was observed. The 56 donors were classified into three groups, according to their individual C. pneumoniae-induced CD4+ T-cell responses: those with a polyfunctional response, which involved the production of both IFN-γ and IL-2 (n = 16); those with a monofunctional response, which involved the production of either IFN-γ or IL-2 (n = 10); and those with no response (n = 30), respectively (Table 1). Among the three donor groups, no significant difference in the sex ratios, mean ages, and smoking status were observed. The frequencies (means ± standard errors of the means [SEMs]) of C. pneumoniae-activated IFN-γ+ and IL-2+ CD4+ T cells for the polyfunctional responders were 5.4 ± 2.3 and 6.2 ± 3.1 per 10,000 cells, respectively. For the monofunctional responders showing only IFN-γ production (n = 5) or only IL-2 production (n = 5), the frequencies were 3.7 ± 0.5 and 4.5 ± 0.5 per 10,000 cells, respectively. In contrast to CD4+ T-cell activation, C. pneumoniae-induced activation of CD8+ T cells could not be detected among the 56 donors (Fig. 1B). The ability of the donors' CD8+ T cells to respond to exogenous antigens was confirmed by stimulating PBMCs with SEB, as well as with a CMV-specific peptide mix (protein pp65), in the case of CMV-seropositive donors (Fig. 1B).
FIG. 1.
C. pneumoniae-induced T-cell activation in healthy blood donors. The PBMCs of 56 healthy volunteers were stimulated for 6 h with whole C. pneumoniae or control stimuli and assessed for intracellular IFN-γ and IL-2 by flow cytometry. For each donor, 80,000 to 100,000 CD3+ lymphocytes were gated on CD4+ or CD8+ T cells and analyzed for cytokine-positive cells via quadrant analysis. (A) The frequencies of IFN-γ- and IL-2-producing CD3+ CD4+ T cells were determined in the PBMCs of an exemplary donor stimulated with C. pneumoniae or 100 ng SEB or with no stimulation. SSC, side scatter. (B) The frequencies of IFN-γ-producing CD3+ CD8+ T cells in the PBMCs of an exemplary CMV-seropositive donor were determined after stimulation with C. pneumoniae, 100 ng SEB, or CMV-specific peptides.
TABLE 1.
C. pneumoniae-induced CD4+ T-cell activation of 56 healthy donors
| Characteristic | Result for the following C. pneumoniae-specific CD4+ T-cell response: |
||
|---|---|---|---|
| Polyfunctional (production of IFN-γ + IL-2) | Monofunctional (production of IFN-γ or IL-2) | None (no IFN-γ or IL-2 production) | |
| No. of donors (n = 56) | 16 | 10 | 30 |
| Mean no. of IFN-γ+ cells per 10,000 cells ± SD | 5.4 ± 2.3 | 3.7 ± 0.5a | NAb |
| Mean no. of IL-2+ cells per 10,000 cells ± SD | 6.2 ± 3.1 | 4.5 ± 0.5a | NA |
| No. of males (n = 30) | 9 | 7 | 14 |
| No. of smokers (n = 8) | 2 | 2 | 4 |
| Mean age (yr) | 40.9 | 40.3 | 40.4 |
Data were calculated for five donors with either IFN-γ+ and IL-2− or IFN-γ− and IL-2+ cells.
NA, not applicable.
Activation of CD4+ T cells by C. pneumoniae depends on TCR engagement.
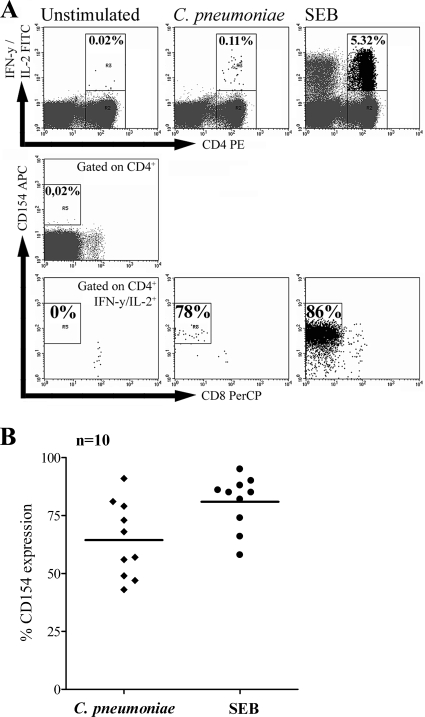
CD154 represents a sensitive marker for CD4+ T cells specifically activated with antigen (11, 14), since its expression strictly depends on engagement of the TCR (24). To exclude TCR-independent activation through chlamydial components, such as C. pneumoniae lipopolysaccharide (LPS), that stimulate innate immune receptors, CD154 expression among C. pneumoniae-activated CD4+ T cells was analyzed. Therefore, the PBMCs of donors showing polyfunctional CD4+ T-cell responses, i.e., the production of IFN-γ and IL-2, were stimulated with C. pneumoniae and assessed for CD154 expression. As shown for an exemplary donor in Fig. 2A, upon stimulation with both C. pneumoniae and SEB, a dominant population of IFN-γ+ and/or IL-2+ CD4+ T cells coexpressed CD154. Compared to the proportion of CD154-expressing IFN-γ+ and/or IL-2+ CD4+ T cells upon C. pneumoniae stimulation (mean, 64%), the proportion was slightly higher after stimulation with SEB (mean, 81%). Our findings indicate that upon stimulation with both C. pneumoniae and SEB, the majority of IFN-γ+ and/or IL-2+ CD4+ T cells is activated in a TCR-dependent manner, leading to CD154 expression. It is important to note that CD4+ T-cell activation was independent of C. pneumoniae infection, as demonstrated by the comparable activation of CD4+ T cells after stimulation with a cell lysate containing corresponding amounts of sonicated C. pneumoniae cells which were unable to infect HEp-2 host cells (data not shown). Furthermore, during the short incubation time of our assay, we observed no TCR-independent T-cell activation when the PBMCs were stimulated with 10 μg LPS from Salmonella serovar Abortus Equi, a potent Toll-like receptor stimulus (always less than 2 IFN-γ+ or IL-2+ cells per 10,000 CD4+ T cells; data not shown).
FIG. 2.
CD154 expression among CD4+ T cells activated by C. pneumoniae. The expression of CD154 among activated CD4+ T cells in the PBMCs of donors with C. pneumoniae-induced CD4+ T-cell responses producing IFN-γ and IL-2 was determined after stimulation with C. pneumoniae. As controls, unstimulated PBMCs or PBMCs stimulated with 1 μg SEB were analyzed. For the analysis, 100,000 CD3+ CD4+ T cells were assessed for intracellular IFN-γ and/or IL-2 and were further assessed for the presence of intracellular CD154. (A) The coexpression of CD154 among IFN-γ+ and/or IL-2+ CD4+ T cells following stimulation with C. pneumoniae or SEB is shown for one exemplary donor. CD4+ CD8+ T cells were not analyzed. (B) Proportions of IFN-γ+ and/or IL-2+ CD4+ T cells from 10 different donors coexpressing CD154 after stimulation with C. pneumoniae or SEB. The horizontal lines indicate the mean proportion of CD154 coexpression.
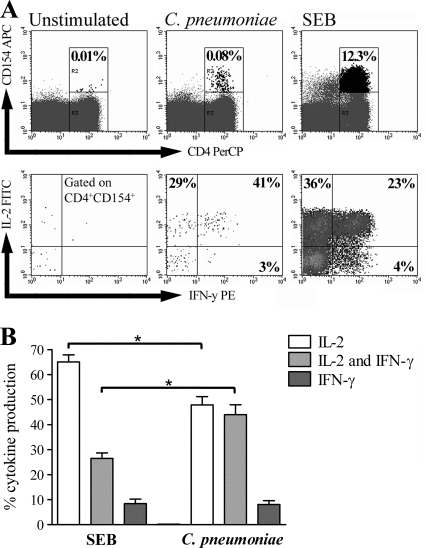
To further characterize the cytokine profile of C. pneumoniae-activated CD154+ CD4+ T cells, we analyzed the distributions of only IFN-γ+ cells, only IL-2+ cells, and both IFN-γ+ and IL-2+ cells. As shown for an exemplary donor in Fig. 3A, C. pneumoniae-activated CD154+ CD4+ T cells were predominantly only IL-2+ cells or both IFN-γ+ and IL-2+ cells. For donors with polyfunctional responses, C. pneumoniae-activated CD154+ CD4+ T cells consisted of 8% only IFN-γ+ cells, 48% only IL-2+ cells, and 44% both IFN-γ+ and IL-2+ cells (Fig. 3B). Compared to the results of stimulation with SEB, stimulation with C. pneumoniae led to a significant shift from the production of only IL-2 to the production of both IFN-γ- and IL-2 by CD154+ CD4+ T cells, arguing for the prominent production of effector cytokines among C. pneumoniae-activated CD4+ T cells.
FIG. 3.
Cytokine profile of C. pneumoniae-activated CD154+ CD4+ T cells. The distributions of only IL-2+, only IFN-γ+, and both IL-2+ and IFN-γ+ CD154+ CD4+ T cells in the PBMCs of donors with C. pneumoniae-induced CD4+ T cell responses producing IFN-γ and IL-2 were determined after stimulation with C. pneumoniae or SEB. For the analysis, 200,000 CD4+ T cells were measured. (A) The frequencies of only IL-2+, only IFN-γ+, and both IL-2+ and IFN-γ+ CD154+ CD4+ T cells after stimulation with C. pneumoniae or SEB are shown for one exemplary donor. The frequencies of the respective subpopulations were determined by quadrant analysis. (B) Mean frequencies (± SEMs) of only IL-2+, only IFN-γ+, and both IL-2+ and IFN-γ+ CD154+ CD4+ T cells of seven different donors. For comparison, the frequencies of the three subpopulations were normalized to 100% in sum for each donor. Asterisks, significant differences (P < 0.01, tested by ANOVA).
Memory phenotype determination of C. pneumoniae-activated CD4+ CD154+ T cells.
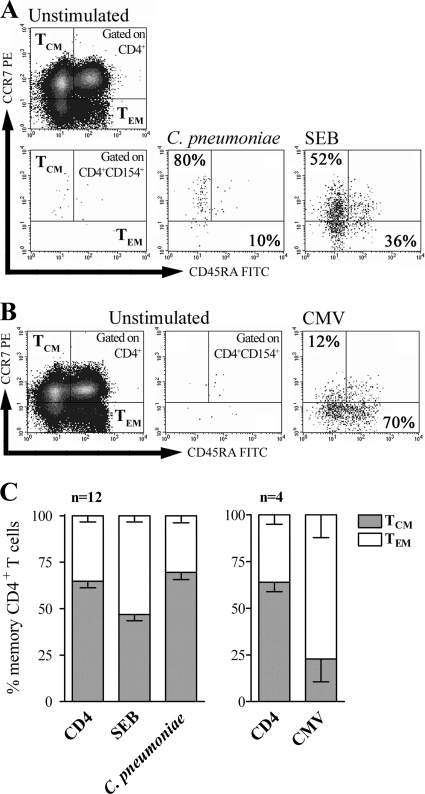
The high proportion of CD154+ CD4+ T cells producing only IL-2 observed in C. pneumoniae-stimulated PBMCs indicates the presence of pathogen-specific memory CD4+ T cells. To determine the phenotype of these memory CD4+ T cells, we investigated the cells for the expression of CCR7 and CD45RA to distinguish between CCR7-negative (CCR7−) and CD45RA-positive (CD45RA+)/CD45RA-negative (CD45RA−) TEM and CCR7+/CD45RA− TCM (17, 18, 35). Besides the analysis of C. pneumoniae- and SEB-activated CD154+ CD4+ T cells, we also evaluated total CD4+ T cells from unstimulated PBMC samples for CCR7 and CD45RA expression (Fig. 4A). Compared to total CD4+ T cells, which mainly consisted of TCM (mean TCM, 65%; mean, TEM, 35%), increased frequencies of TEM were observed among SEB-activated CD154+ CD4+ T cells (mean TCM, 47%; mean TEM, 53%) (Fig. 4C). In contrast to that finding, C. pneumoniae-activated CD154+ CD4+ T cells were found to be predominantly TCM (mean TCM, 70%; mean TEM, 30%). To confirm these findings, we further characterized the distribution of TEM and TCM among CMV-specific CD154+ CD4+ T cells of CMV-seropositive donors after stimulation with CMV-specific peptides (Fig. 4B). In line with previously published results (17, 37), the vast majority of CMV-specific CD154+ CD4+ T cells were found to be TEM (mean TCM, 23%; mean TEM, 77%) (Fig. 4C). We typically observed about 10 to 20% activated CD154+ CD4+ T cells which were positive for the expression of both CCR7 and CD45RA, thus representing naïve CD4+ T cells.
FIG. 4.
Memory phenotype analysis of C. pneumoniae-activated CD154+ CD4+ T cells. The levels of expression of CCR7 and CD45RA among total CD4+ T cells and CD154+ CD4+ T cells producing IFN-γ and IL-2 in the PBMCs of donors with C. pneumoniae-induced CD4+ T-cell responses were analyzed after stimulation with C. pneumoniae, SEB, or CMV-specific peptides. For the analysis, 100,000 to 150,000 CD4+ T cells were measured. The quadrants for analysis of CCR7+ and CD45RA− TCM and CCR7− and CD45RA+/− TEM were determined for each donor according to the expression of these markers among total CD4+ T cells. (A and B) Proportions of TCM and TEM among CD154+ CD4+ T cells determined after stimulation of PBMCs with C. pneumoniae or SEB (A) or CMV-specific peptides (B) of one exemplary donor and another exemplary CMV-seropositive donor, respectively. (C) Mean frequencies (± SEMs) of TCM and TEM among total CD4+ T cells and CD154+ CD4+ T cells for 12 donors and 4 CMV-seropositive donors stimulated with C. pneumoniae, SEB, or CMV-specific peptides. For comparison, the sum of the frequencies of TCM and TEM was normalized to 100% for each donor.
Association between C. pneumoniae-induced CD4+ T-cell response and serum antibody reactivity toward whole C. pneumoniae or recombinant antigens.
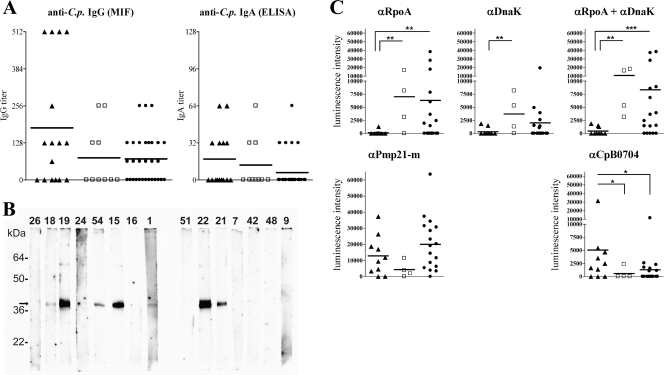
The presence of C. pneumoniae-induced memory T-cell responses in the peripheral blood of apparently healthy donors suggests a history of infection. To address this, we analyzed the IgG and IgA antibody reactivities toward whole C. pneumoniae in the sera of all seropositive donors using MIF and ELISA, respectively (Fig. 5A). Of the 56 donors, 31 (55%) had detectable anti-C. pneumoniae IgG antibodies in their serum (titer ≥ 64). In addition, the sera of 14 (25%) donors tested positive for IgA antibodies (titer ≥ 32), all of whom were found to be IgG positive. Interestingly, there was no significant difference between the number of seropositive samples for donors with undetectable CD4+ T-cell responses (IgG, 57%; IgA, 17%), donors with only IFN-γ- or IL-2-producing responses (IgG, 40%; IgA, 30%), and donors with both IFN-γ- and IL-2-producing responses (IgG, 63%; IgA, 38%). For the last group, however, the mean anti-C. pneumoniae antibody titer was the highest. The absence of C. pneumoniae-induced CD4+ T-cell activation in 17 of 31 seropositive donors was unexpected, and we investigated whether this finding was related to individual differences in the serological response patterns of these donors. To compare the serological responses between donors with or without detectable CD4+ T-cell activation, we analyzed the antibody reactivity toward the recombinant C. pneumoniae antigens RpoA, DnaK, and Pmp21m, which are upregulated during chlamydial persistence in vitro (3, 26, 28). In addition, we analyzed the antibody reactivity toward control C. pneumoniae antigen CpB0704, which lacks sequence homology to proteins of other Chlamydia species and which represents a major target of antibodies contributing to reactivity in the MIF assay (8). Using quantitative strip immunoblot analysis (Fig. 5B), we found significantly increased IgG reactivity toward RpoA and DnaK, as well as higher mean reactivity toward Pmp21m, among donors with undetectable T-cell responses or only single cytokine-producing CD4+ T-cell responses compared to that among donors showing both IFN-γ- and IL-2-producing responses. In contrast, for subjects with undetectable or single-cytokine-producing responses, the IgG reactivity toward the CpB0704 control antigen was significantly lower than that for donors with dual IFN-γ- and IL-2-producing CD4+ T-cell responses (Fig. 5C), indicating distinct patterns of anti- C. pneumoniae antibody responses among these donor groups.
FIG. 5.
Analysis of serum antibody reactivity toward whole C. pneumoniae and different antigens which either are or are not associated with persistent infection. The sera of donors with IFN-γ+ and IL-2+ C. pneumoniae-induced CD4+ T-cell responses (▴), either an IFN-γ+ or an IL-2+ response (□), or no response (•) were analyzed for anti-C. pneumoniae (anti-C.p.) antibodies. *, **, and ***, significant differences representing P values of 0.05, 0.01, and 0.001, respectively (tested by the Kruskal-Wallis test). (A) IgG and IgA antibody titers specific for whole C. pneumoniae elementary bodies were determined in the sera of 56 donors by MIF and ELISA, respectively. The horizontal lines represent the mean titers of each donor group. (B) Strip immunoblots with sera from 15 different exemplary donors obtained by use of the control C. pneumoniae antigen CpB0704. Each number above the lanes represents an individual serum sample, and the arrow indicates the location of CpB0704. (C) Quantitative comparison of chemiluminescence signals of the strip immunoblots for all 31 seropositive donors. The IgG reactivities toward C. pneumoniae antigens RpoA, DnaK, and Pmp21m as well as CpB0704 were determined.
DISCUSSION
Antigen-specific T cells play an important role in the immune control of persisting pathogens, including viruses and bacteria. For C. pneumoniae, a pathogen that can establish persistent infections (15, 20), CD4+ T-cell responses have not yet been studied. In our study, C. pneumoniae-induced CD4+ T-cell responses involving the production of IFN-γ and/or IL-2 could be detected in the PBMCs of 26 of 56 healthy human individuals without any evidence of acute respiratory infection. Following restimulation with C. pneumoniae, the majority of cytokine-producing CD4+ T cells expressed CD154, a marker for CD4+ T cells specifically activated with antigen (11, 14), indicating a TCR-restricted activation of cells by C. pneumoniae. Among C. pneumoniae-activated CD154+ CD4+ T cells, we found two major populations of cells expressing either only IL-2 or both IFN-γ and IL-2, but we also found a small population of cells expressing only IFN-γ. The production of IL-2 among the vast majority of C. pneumoniae-activated IFN-γ+ CD4+ T cells suggests the presence of self-renewing populations of effector cells. Compared to the C. pneumoniae-induced cytokine responses, the cytokine profile of SEB-activated CD154+ CD4+ T cells showed a significant shift from both IFN-γ+ and IL-2+ cells toward only IL-2+ cells, corresponding to reports demonstrating that SEB-activated CD154+ CD4+ T cells produce IL-2 rather than IFN-γ (11, 14). Corresponding to their dominant IL-2 expression, the majority of C. pneumoniae-activated CD154+ CD4+ T cells displayed the CCR7+ CD45RA− phenotype of TCM. In contrast, the vast majority of CMV-specific CD154+ CD4+ T cells were CCR7−/CD45RA+/−, characteristic for TEM, which is in line with previous findings (17, 37). Interestingly, while they had a predominant CCR7+ TCM phenotype, more than 50% of the C. pneumoniae-activated CD154+ CD4+ T cells produced IFN-γ, indicating the presence of CD4+ TCM cells producing the effector cytokine IFN-γ. Even though, according to proposed models, CCR7+ TCM typically produce IL-2 and not IFN-γ (23, 35), our data are consistent with those presented in other reports showing that human antigen-specific TCM are able to produce effector cytokines such as IFN-γ (12, 13, 36, 38). It is also possible that C. pneumoniae-activated TCM represent a transitional population of cells in the process of differentiation from TCM to TEM, which was described to be the predominant population of memory CD4+ T cells during persistent viral infections (17). In our experiments, C. pneumoniae-induced CD8+ T-cell responses could not be detected. This was also observed in previous studies, which demonstrated the inability of recombinant C. pneumoniae proteins or whole bacteria to activate CD8+ T cells (29, 39). We cannot exclude the possibility that stimulation of PBMCs with whole C. pneumoniae was more effective at activating CD4+ T cells than CD8+ T cells, since stimulation of CD8+ T cells by exogenous antigens requires MHC class I cross-presentation of antigens by dendritic cells or macrophages (7, 19), which are rare in PBMCs. Interestingly, studies of mice demonstrated an impaired development of Chlamydia-specific memory CD8+ T cells after primary and secondary C. trachomatis infections (25), a process that might also account for the lack of anti-C. pneumoniae CD8+ T-cell responses observed in our study.
In our study, the proportions of C. pneumoniae IgG- and IgA-seropositive individuals were not significantly different between donors with and without T-cell responses, although responders had higher mean antibody titers. Interestingly, compared to seropositive individuals with CD4+ T-cell responses (IFN-γ- and IL-2-production), seropositive donors lacking these responses showed different patterns of antibody reactivity toward recombinant C. pneumoniae antigens (8) associated with persistent chlamydial infection. We found increased IgG reactivity toward RpoA, DnaK, and Pmp21m, known to be upregulated during persistent chlamydial infection (3, 26, 28), in the sera of subjects with no or only single cytokine-producing CD4+ T-cell responses compared to the IgG reactivity in the sera of donors showing dual IFN-γ- and IL-2-producing responses. There could be different explanations for the finding that a portion of seropositive donors lacked CD4+ T-cell responses and displayed an altered antibody recognition pattern compared to that of seropositive donors with detectable CD4+ T-cell responses (IFN-γ and IL-2 production). These findings could be related to several factors. (i) They could be related to the presence of cross-reactive antibodies that bind to C. pneumoniae in the MIF assay but that were gained during an infection(s) with an unrelated pathogen(s). The presence of cross-reactive antibodies would also explain why seropositive individuals lacking CD4+ T-cell responses showed only low levels of reactivity toward C. pneumoniae CpB0704, the more specific antigen, but high levels of reactivity toward RpoA and DnaK, both of which represent rather conserved bacterial proteins. (ii) The findings could be related to individual differences in the repertoire of C. pneumoniae antigens targeted by the host immune response or expressed by C. pneumoniae during infection (e.g., infections with different C. pneumoniae strains). In both cases, the B- and T-cell responses of the donors might have targeted a set of C. pneumoniae antigens that allow antibody binding and, thus, positive MIF assay results but only the inefficient restimulation of T cells in our assay (e.g., due to the rare expression in cultivated C. pneumoniae). (iii) Finally, the findings could be related to a direct influence of T cells on the course of infection. The impaired development of T-cell responses during C. pneumoniae infection might favor the establishment of persistent infections, in which the production of antibodies with a preferential specificity for persistence-associated C. pneumoniae antigens, such as RpoA and DnaK, might even take place in the absence of strong T-cell help due to the prolonged antigen exposure. Regarding the last possibility, our data demonstrate that the production of both IFN-γ and IL-2 among C. pneumoniae-activated CD4+ T cells was associated with low levels of reactivity to persistence-associated C. pneumoniae antigens. Recent studies analyzing IFN-γ and IL-2 production among antigen-specific CD4+ T cells during persistent viral infections observed distinct functional T-cell heterogeneity, which could be attributed to different levels of virus load (16, 30). Interestingly, the authors described that only polyfunctional CD4+ T-cell responses, characterized by the production of both IFN-γ and IL-2, reflected the efficient immune control of viral infection. It could be speculated whether, in our study, the presence of polyfunctional C. pneumoniae-induced T-cell responses also represents a signature of an effective immune response that is able to prevent the development or improve the control of persistent C. pneumoniae infections. Unfortunately, we could not test this hypothesis due to the lack of reliable tools for diagnosing persisting C. pneumoniae.
In conclusion, this study demonstrates the presence of circulating memory CD4+ T cells in the PBMCs of healthy human blood donors that, upon stimulation with C. pneumoniae, produce IFN-γ, IL-2, and CD154. Furthermore, the presence or absence of dual IFN-γ- and IL-2-producing CD4+ T-cell responses was found to be associated with distinct differences in the patterns of IgG reactivity toward C. pneumoniae antigens that are upregulated during chlamydial persistence.
Acknowledgments
This study was supported by the Eliteprogramm für Postdoktorandinnen und Postdoktoranden der Landesstiftung Baden-Württemberg GmbH, Zukunftskolleg, and the Young Scholar Fund of the University of Konstanz, as well as, in part, by the Schwerpunktprogramm Autoimmunität of the University of Luebeck.
We thank the volunteers who participated in the study, Pauline Maria van Helden for critical reading of the manuscript, and Thomas Hartung as well as Thomas Meergans for helpful discussions.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Apfalter, P. 2006. Chlamydia pneumoniae, stroke, and serological associations: anything learned from the atherosclerosis-cardiovascular literature or do we have to start over again? Stroke 37:756-758. [DOI] [PubMed] [Google Scholar]
- 2.Belland, R. J., S. P. Ouellette, J. Gieffers, and G. I. Byrne. 2004. Chlamydia pneumoniae and atherosclerosis. Cell. Microbiol. 6:117-127. [DOI] [PubMed] [Google Scholar]
- 3.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 100:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benagiano, M., A. Azzurri, A. Ciervo, A. Amedei, C. Tamburini, M. Ferrari, J. L. Telford, C. T. Baldari, S. Romagnani, A. Cassone, M. M. D'Elios, and G. Del Prete. 2003. T helper type 1 lymphocytes drive inflammation in human atherosclerotic lesions. Proc. Natl. Acad. Sci. U. S. A. 100:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benagiano, M., M. M. D'Elios, A. Amedei, A. Azzurri, R. van der Zee, A. Ciervo, G. Rombola, S. Romagnani, A. Cassone, and G. Del Prete. 2005. Human 60-kDa heat shock protein is a target autoantigen of T cells derived from atherosclerotic plaques. J. Immunol. 174:6509-6517. [DOI] [PubMed] [Google Scholar]
- 6.Boman, J., and M. R. Hammerschlag. 2002. Chlamydia pneumoniae and atherosclerosis: critical assessment of diagnostic methods and relevance to treatment studies. Clin. Microbiol. Rev. 15:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brode, S., and P. A. Macary. 2004. Cross-presentation: dendritic cells and macrophages bite off more than they can chew! Immunology 112:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunk, S., I. Susnea, J. Rupp, J. T. Summersgill, M. Maass, W. Stegmann, A. Schrattenholz, A. Wendel, M. Przybylski, and C. Hermann. 2008. Immunoproteomic identification and serological responses to novel Chlamydia pneumoniae antigens that are associated with persistent C. pneumoniae infections. J. Immunol. 180:5490-5498. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, L. A., and C. C. Kuo. 2004. Chlamydia pneumoniae—an infectious risk factor for atherosclerosis? Nat. Rev. Microbiol. 2:23-32. [DOI] [PubMed] [Google Scholar]
- 10.Carralot, J. P., C. Dumrese, R. Wessel, R. Riessen, I. Autenrieth, S. Walter, O. Schoor, S. Stevanovic, H. G. Rammensee, and S. Pascolo. 2005. CD8+ T cells specific for a potential HLA-A*0201 epitope from Chlamydophila pneumoniae are present in the PBMCs from infected patients. Int. Immunol. 17:591-597. [DOI] [PubMed] [Google Scholar]
- 11.Chattopadhyay, P. K., J. Yu, and M. Roederer. 2005. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat. Med. 11:1113-1117. [DOI] [PubMed] [Google Scholar]
- 12.Debes, G. F., U. E. Hopken, and A. Hamann. 2002. In vivo differentiated cytokine-producing CD4(+) T cells express functional CCR7. J. Immunol. 168:5441-5447. [DOI] [PubMed] [Google Scholar]
- 13.Fontenot, A. P., B. E. Palmer, A. K. Sullivan, F. G. Joslin, C. C. Wilson, L. A. Maier, L. S. Newman, and B. L. Kotzin. 2005. Frequency of beryllium-specific, central memory CD4+ T cells in blood determines proliferative response. J. Clin. Invest. 115:2886-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frentsch, M., O. Arbach, D. Kirchhoff, B. Moewes, M. Worm, M. Rothe, A. Scheffold, and A. Thiel. 2005. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat. Med. 11:1118-1124. [DOI] [PubMed] [Google Scholar]
- 15.Hammerschlag, M. R. 2002. The intracellular life of chlamydiae. Semin. Pediatr. Infect. Dis. 13:239-248. [DOI] [PubMed] [Google Scholar]
- 16.Harari, A., V. Dutoit, C. Cellerai, P. A. Bart, R. A. Du Pasquier, and G. Pantaleo. 2006. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol. Rev. 211:236-254. [DOI] [PubMed] [Google Scholar]
- 17.Harari, A., F. Vallelian, P. R. Meylan, and G. Pantaleo. 2005. Functional heterogeneity of memory CD4 T cell responses in different conditions of antigen exposure and persistence. J. Immunol. 174:1037-1045. [DOI] [PubMed] [Google Scholar]
- 18.Harari, A., F. Vallelian, and G. Pantaleo. 2004. Phenotypic heterogeneity of antigen-specific CD4 T cells under different conditions of antigen persistence and antigen load. Eur. J. Immunol. 34:3525-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heath, W. R., G. T. Belz, G. M. Behrens, C. M. Smith, S. P. Forehan, I. A. Parish, G. M. Davey, N. S. Wilson, F. R. Carbone, and J. A. Villadangos. 2004. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol. Rev. 199:9-26. [DOI] [PubMed] [Google Scholar]
- 20.Hogan, R. J., S. A. Mathews, S. Mukhopadhyay, J. T. Summersgill, and P. Timms. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 72:1843-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ieven, M. M., and V. Y. Hoymans. 2005. Involvement of Chlamydia pneumoniae in atherosclerosis: more evidence for lack of evidence. J. Clin. Microbiol. 43:19-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kern, F., T. Bunde, N. Faulhaber, F. Kiecker, E. Khatamzas, I. M. Rudawski, A. Pruss, J. W. Gratama, R. Volkmer-Engert, R. Ewert, P. Reinke, H. D. Volk, and L. J. Picker. 2002. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals. J. Infect. Dis. 185:1709-1716. [DOI] [PubMed] [Google Scholar]
- 23.Lanzavecchia, A., and F. Sallusto. 2005. Understanding the generation and function of memory T cell subsets. Curr. Opin. Immunol. 17:326-332. [DOI] [PubMed] [Google Scholar]
- 24.Lindgren, H., K. Axcrona, and T. Leanderson. 2001. Regulation of transcriptional activity of the murine CD40 ligand promoter in response to signals through TCR and the costimulatory molecules CD28 and CD2. J. Immunol. 166:4578-4585. [DOI] [PubMed] [Google Scholar]
- 25.Loomis, W. P., and M. N. Starnbach. 2006. Chlamydia trachomatis infection alters the development of memory CD8+ T cells. J. Immunol. 177:4021-4027. [DOI] [PubMed] [Google Scholar]
- 26.Molestina, R. E., J. B. Klein, R. D. Miller, W. H. Pierce, J. A. Ramirez, and J. T. Summersgill. 2002. Proteomic analysis of differentially expressed Chlamydia pneumoniae genes during persistent infection of HEp-2 cells. Infect. Immun. 70:2976-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller, M., S. Postius, J. G. Thimm, K. Gueinzius, I. Muehldorfer, and C. Hermann. 2004. Toll-like receptors 2 and 4 do not contribute to clearance of Chlamydophila pneumoniae in mice, but are necessary for the release of monokines. Immunobiology 209:599-608. [DOI] [PubMed] [Google Scholar]
- 28.Mukhopadhyay, S., R. D. Miller, E. D. Sullivan, C. Theodoropoulos, S. A. Mathews, P. Timms, and J. T. Summersgill. 2006. Protein expression profiles of Chlamydia pneumoniae in models of persistence versus those of heat shock stress response. Infect. Immun. 74:3853-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mygind, T., B. Vandahl, A. S. Pedersen, G. Christiansen, P. Hollsberg, and S. Birkelund. 2004. Identification of an in vivo CD4+ T cell-mediated response to polymorphic membrane proteins of Chlamydia pneumoniae during experimental infection. FEMS Immunol. Med. Microbiol. 40:129-137. [DOI] [PubMed] [Google Scholar]
- 30.Pantaleo, G., and A. Harari. 2006. Functional signatures in antiviral T-cell immunity for monitoring virus-associated diseases. Nat. Rev. Immunol. 6:417-423. [DOI] [PubMed] [Google Scholar]
- 31.Penttila, J. M., M. Anttila, M. Puolakkainen, A. Laurila, K. Varkila, M. Sarvas, P. H. Makela, and N. Rautonen. 1998. Local immune responses to Chlamydia pneumoniae in the lungs of BALB/c mice during primary infection and reinfection. Infect. Immun. 66:5113-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penttila, J. M., M. Anttila, K. Varkila, M. Puolakkainen, M. Sarvas, P. H. Makela, and N. Rautonen. 1999. Depletion of CD8+ cells abolishes memory in acquired immunity against Chlamydia pneumoniae in BALB/c mice. Immunology 97:490-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothfuchs, A. G., M. R. Kreuger, H. Wigzell, and M. E. Rottenberg. 2004. Macrophages, CD4+ or CD8+ cells are each sufficient for protection against Chlamydia pneumoniae infection through their ability to secrete IFN-gamma. J. Immunol. 172:2407-2415. [DOI] [PubMed] [Google Scholar]
- 34.Rottenberg, M. E., A. C. Gigliotti Rothfuchs, D. Gigliotti, C. Svanholm, L. Bandholtz, and H. Wigzell. 1999. Role of innate and adaptive immunity in the outcome of primary infection with Chlamydia pneumoniae, as analyzed in genetically modified mice. J. Immunol. 162:2829-2836. [PubMed] [Google Scholar]
- 35.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708-712. [DOI] [PubMed] [Google Scholar]
- 36.Stubbe, M., N. Vanderheyde, M. Goldman, and A. Marchant. 2006. Antigen-specific central memory CD4+ T lymphocytes produce multiple cytokines and proliferate in vivo in humans. J. Immunol. 177:8185-8190. [DOI] [PubMed] [Google Scholar]
- 37.Stubbe, M., N. Vanderheyde, H. Pircher, M. Goldman, and A. Marchant. 2008. Characterization of a subset of antigen-specific human central memory CD4+ T lymphocytes producing effector cytokines. Eur. J. Immunol. 38:273-282. [DOI] [PubMed] [Google Scholar]
- 38.Unsoeld, H., S. Krautwald, D. Voehringer, U. Kunzendorf, and H. Pircher. 2002. Cutting edge: CCR7+ and CCR7− memory T cells do not differ in immediate effector cell function. J. Immunol. 169:638-641. [DOI] [PubMed] [Google Scholar]
- 39.Wizel, B., B. C. Starcher, B. Samten, Z. Chroneos, P. F. Barnes, J. Dzuris, Y. Higashimoto, E. Appella, and A. Sette. 2002. Multiple Chlamydia pneumoniae antigens prime CD8+ Tc1 responses that inhibit intracellular growth of this vacuolar pathogen. J. Immunol. 169:2524-2535. [DOI] [PubMed] [Google Scholar]