Abstract
Vps8 is a subunit of the CORVET tethering complex, which is involved in early-to-late endosome fusion. Here, we examine the role of Vps8 in membrane fusion at late endosomes in Saccharomyces cerevisiae. We demonstrate that Vps8 associates with membranes and that this association is independent of the class C/HOPS core complex and, contrary to a previous report, also independent of the Rab GTPase Vps21. Our data indicate that Vps8 makes multiple contacts with membranes. One of these membrane binding regions could be mapped to the N-terminal part of the protein. By two-hybrid analysis, we obtained evidence for a physical interaction between Vps8 and the Rab5 homologue Vps21. In addition, the interaction with the HOPS core complex was confirmed by immunoprecipitation experiments. By deletion analysis, the Vps21 and HOPS binding sites were mapped in Vps8. Deletions that abrogated HOPS core complex binding had a strong effect on the turnover of the endocytic cargo protein Ste6 and on vacuolar sorting of carboxypeptidase Y. In contrast, deletions that abolished Vps21 binding showed only a modest effect. This suggests that the Vps21 interaction is not essential for endosomal trafficking but may be important for some other aspect of Vps8 function.
The compartments of the exocytic/endocytic membrane system are dynamic structures that continuously exchange materials by budding and fusion of transport vesicles. Despite this continuous exchange, the compartments maintain their specific identities. A basic machinery consisting of tethering factors, Rab GTPases, SNARE proteins, and Sec1/Munc18 (SM) proteins accomplishes membrane targeting and fusion. For each individual membrane fusion event, a characteristic set of proteins is used.
We are interested in a particular membrane fusion step, the fusion of early endosome-derived vesicles with late endosomes. Screening for vps (vacuolar protein sorting) mutants in Saccharomyces cerevisiae identified factors involved in this fusion step (3). Mutants defective in the early-to-late endosome trafficking step belong to the class D group of vps mutants, whose hallmark is an enlarged vacuole (21). Among the class D functions, representatives of the main groups of targeting and fusion factors can be found. The Q-SNARE protein Pep12, for instance, a member of the syntaxin family, serves as a marker for late endosomal membranes (2). Together with the Q-SNAREs Vti1 and Syn8 or Tlg1, it forms two alternative t-SNARE complexes on late endosomal membranes (17). These t-SNAREs combine with the v-SNARES Snc1/Snc2 or Ykt6 to form functional trans-SNARE complexes. Pep12 functionally interacts with another class D protein, the SM protein Vps45 (4). Another component of the basic fusion machinery at late endosomes is the class D protein Vps21, a member of the Rab GTPase family and the yeast homologue of mammalian Rab5 (8, 12, 30). Rab proteins are key regulators of membrane fusion (9). They are involved in the recruitment of tethering and docking factors, and by their interplay with Rab effectors they contribute to the establishment of specific membrane domains. Another class D protein connected to Rab function is Vps9, a guanidine nucleotide exchange factor (GEF) for Vps21 (11).
Additional class D proteins are involved in vesicle tethering at late endosomes. Basically, there are two kinds of tethers, proteins containing extensive coiled-coil domains and large multisubunit complexes (33). The prototype of the coiled-coil tethers is p115, with its yeast homologue Uso1, involved in tethering of vesicles to Golgi apparatus membranes (25). Another member of this class is EEA1, which is involved in tethering of vesicles to endosomes. The yeast class D protein Vps19/Pep7/Vac1 could be functionally similar to EEA1 (16). Two further class D proteins, Vps3 and Vps8, are part of the multisubunit (class C core vacuole/endosome tethering) CORVET tethering complex (20, 32). This complex shares core components with the HOPS (homotypic fusion and vacuole protein sorting) tethering complex involved in homotypic vacuolar fusion (28). This core complex, the class C Vps complex, consists of Vps11/Pep5, Vps16, Vps18/Pep3, and the SM protein Vps33 (26). Instead of Vps3 and Vps8, HOPS contains two additional subunits, Vps39/Vam6 and Vps41 (35), which appear to be functionally equivalent to Vps3 and Vps8 (20). In addition to bridging donor and acceptor membranes, tethers appear to be involved in coordinating Rab and SNARE functions. This was suggested by the finding that the equivalent CORVET/HOPS subunits Vps3 and Vps39/Vam6 both display GEF activity toward their respective Rab proteins, Vps21 and Ypt7 (20, 35). In addition, whole tethering complexes act as Rab effectors by binding to activated Rab-GTP and interact with the corresponding SNARE complexes (6, 20, 31).
How exactly the tethers coordinate Rab and SNARE functions during membrane fusion is at present unclear. Here, we examine the function of the CORVET subunit Vps8 (5, 13) in membrane fusion at late endosomes in yeast. We demonstrate that Vps8 directly associates with membranes. Contrary to a previous report (13), we show that this membrane association is not dependent on Vps21. We further investigate the functional relationship between Vps8 and Vps21. We found that Vps21 physically interacts with Vps8 but that this interaction does not appear to be absolutely required for endosomal trafficking. Finally, we speculate that Vps8 could be part of a higher-order structure.
MATERIALS AND METHODS
Yeast strains and plasmids.
The yeast strains used are listed in Table 1. Deletion strains were derived from the wild-type strain JD52. They were constructed by one-step gene replacement with PCR-generated cassettes (18). The deletions were verified by PCR. Site-directed mutagenesis of VPS21 was performed by QuikChange PCR mutagenesis (Stratagene, La Jolla, CA). Yeast cells were grown in yeast extract-peptone-dextrose medium (1% yeast extract, 2% Bacto peptone, 2% glucose) or in synthetic dextrose-Casamino Acids medium (0.67% yeast nitrogen base without amino acids, 1% casein hydrolysate, 2% glucose).
Table 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| GPY398.1 | MATa ura3-52 his4 and/or his6 leu2-3,112 trp1 chc1 Ts | Greg Payne, Los Angeles, CA |
| JD52 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 | Jürgen Dohmen, Köln, Germany |
| NYY0-1 | MATa ura3 his3 leu2 trp1 lys2 ade2::[ARF1 ADE2] arf1::HIS3 arf2::HIS | 36 |
| NYY13-1 | MATa ura3 his3 leu2 trp1 lys2 ade2::[arf1-13 ADE2] arf1::HIS3 arf2::HIS3 | 36 |
| NYY16-1 | MATa ura3 his3 leu2 trp1 lys2 ade2::[arf1-16 ADE2] arf1::HIS3 arf2::HIS3 | 36 |
| NYY18-1 | MATa ura3 his3 leu2 trp1 lys2 ade2::[arf1-18 ADE2] arf1::HIS3 arf2::HIS3 | 36 |
| PJ69-4A | MATa ura3-52 his3-Δ200 leu2-3, 112 trp1-901 gal4Δ gal80Δ LYS2::GAL1-HIS GAL2-ADE2 met2::GAL7-lacZ | 14 |
| RKY1577 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 sec18-1 | This study |
| RKY1603 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ lys2-801 Δvps18::TRP1 | This study |
| RKY1843 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δvps8::HIS3 | This study |
| RKY1920 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δvps21::LEU2 | This study |
| RKY1921 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δpep12::kanMX6 | This study |
| RKY1926 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δvps8::HIS3 Δvps21::LEU2 | This study |
| RKY2038 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δypt53::kanMX6 | This study |
| RKY2040 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 vps45tsf-3 | This study |
| RKY2046 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δvps8::HIS3 VPS11-3HA::kanMX6 | This study |
| RKY2053 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δvps34::HIS3 | This study |
| RKY2069 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δypt52::HIS3 Δypt53::kanMX6 | This study |
| RKY2071 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δypt52::kanMX6 | This study |
| RKY2124 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δvps21::LEU2 Δypt52::HIS3 Δypt53::kanMX6 | This study |
| RKY2427 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δvps11::HIS3 Δvps21::LEU2 | This study |
| Y00000/BY4741 | MATa ura3 his3-Δ1 leu2-Δ met15Δ | EUROSCARF |
| Y00817 | MATa ura3 his3-Δ1 leu2-Δ met15Δ vps11Δ::kanMX4 | EUROSCARF |
| Y02783 | MATa ura3 his3-Δ1 leu2-Δ met15Δ vps16Δ::kanMX4 | EUROSCARF |
| Y03625 | MATa ura3 his3-Δ1 leu2-Δ met15Δ vps19Δ::kanMX4 | EUROSCARF |
| Y04105 | MATa ura3 his3-Δ1 leu2-Δ met15Δ vps18Δ::kanMX4 | EUROSCARF |
| Y04329 | MATa ura3 his3-Δ1 leu2-Δ met15Δ vps33Δ::kanMX4 | EUROSCARF |
| Y05305 | MATa ura3 his3-Δ1 leu2-Δ met15Δ vps3Δ::kanMX4 | EUROSCARF |
| YES95 | MATa ura3-52 his3-Δ200 leu2-Δ1 trp1-Δ1 lys2-801a ade2-101 pik1-63::TRP1 | 10 |
| YES102 | MATa ura3-52 his3-Δ200 leu2-Δ1 trp1-Δ1 lys2-801a ade2-101 pik1-83::TRP1 | 10 |
Flotation gradients.
Flotation gradients were essentially performed as described in reference 1. Briefly, cells were grown overnight to logarithmic phase. Ten optical density at 600 nm (OD600) units were harvested by centrifugation, washed with water, and lysed by agitation with glass beads in TNE buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA plus protease inhibitors). After removal of cell debris and intact cells by centrifugation at 500 × g for 5 min, 125 μl of the extract was mixed with 250 μl of 60% Optiprep solution (Axis-Shield, Oslo, Norway). The resulting 40% Optiprep fraction was transferred to the bottom of a centrifugation tube and overlaid with 600 μl of 30% Optiprep in TNE buffer and 100 μl of TNE. Then the gradients were centrifuged at 77,000 rpm and 4°C for 2 h in a TLA 100.2 rotor in a Beckman tabletop ultracentrifuge. Six equal fractions were collected and assayed for the presence of proteins by Western blotting.
Other methods.
Pulse-chase, carboxypeptidase Y (CPY) sorting, and immunoprecipitation experiments were performed as described previously (34).
RESULTS
Membrane association of Vps8.
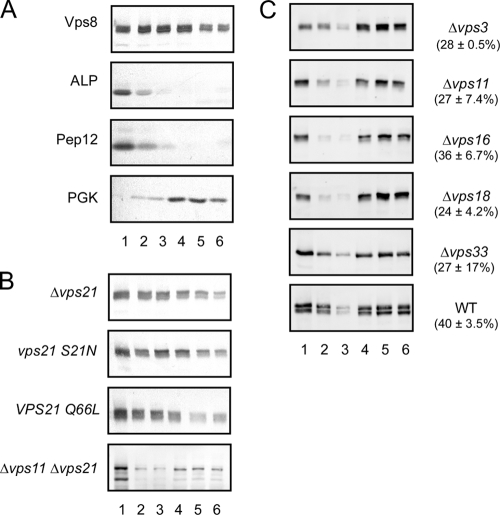
The hydrophilic protein Vps8 can be sedimented by centrifugation. From this, it has been concluded that Vps8 is membrane associated (5, 13). But, sedimentation is not necessarily proof of membrane association, since the protein could also be sedimented as part of a larger protein structure. To distinguish between these possibilities, Vps8 membrane association was examined in flotation experiments (Fig. 1). For detection, Vps8 was 13myc tagged at its C terminus, resulting in a fully functional protein (data not shown). Cell extracts were mixed with a high-density Optiprep solution and were overlaid with Optiprep solutions of lower densities in a centrifuge tube. Due to their low densities, membranes float to the top of the gradient during centrifugation, while proteins not associated with membranes remain in the lower part of the gradient. About half of Vps8 was recovered in the top three fractions of the gradient, along with the membrane markers ALP and Pep12 (Fig. 1A). The soluble protein phosphoglycerate kinase (PGK) was mostly found in the lower three fractions of the gradient. This indicated that indeed a substantial fraction of Vps8 is membrane associated.
Fig. 1.
Fractionation of Vps8 by flotation. (A) Cell extracts of RKY1843 (Δvps8) transformed with pRK827 (VPS8-13myc) were fractionated on Optiprep flotation gradients. Six fractions (lanes 1 to 6) were collected from the gradients (lanes 1 to 3, membrane fraction; lanes 4 to 6, soluble proteins) and analyzed for the presence of marker proteins by Western blotting with specific antibodies as indicated. (B) Additional flotation experiments were performed with (from top to bottom) strain RKY1926 (Δvps8 Δvps21) transformed with pRK827 (VPS8-13myc) and YCplac22 (vector), pRK933 (VPS21 S21N), and pRK935 (VPS21 Q66L) and with strain RKY2427 (Δvps11 Δvps21) transformed with pRK827. (C) Flotations with HOPS/CORVET mutants (in the BY4741 background [see Table 1]). The average percentages of Vps8 in the membrane fraction from three independent experiments are indicated (with standard deviations). WT, wild-type strain BY4741.
Effect of Δvps21 on Vps8 membrane association.
Since Vps8 does not contain any hydrophobic membrane-spanning segments, it appears to be a peripheral membrane protein. We were interested to know how Vps8 associates with membranes and to identify potential factors involved in its membrane association. Previously, it was suggested that membrane association is dependent on the small GTPase Vps21, the yeast Rab5 homologue (13). We therefore tested by flotation whether Vps8 membrane association was abrogated in a Δvps21 strain (Fig. 1B). Controls were performed as described for Fig. 1A (data not shown). In conflict with these earlier results, Vps8 was still found at the membrane in the Δvps21 strain. We further tested whether the nucleotide state of the Vps21 GTPase had an influence on Vps8 membrane association. To this end, the dominant negative Vps21 S21N mutant, locked in the GDP-bound form, and the permanently active Vps21 Q66L mutant, locked in the GTP-bound form (11), were expressed in a Δvps21 strain. But, similar to the Δvps21 strain, Vps8 was still found at the membrane. We also considered the possibility that the Vps21 homologues Ypt52 and Ypt53 could substitute for Vps21 in membrane binding. But we also found that the triple mutant Δvps21 Δypt52 Δypt53 still showed membrane binding (data not shown).
In addition to Δvps21, a whole collection of pertinent mutants (as detailed in Table 2) was examined for their effects on Vps8 membrane association in flotation experiments. Especially, we focused on the mutants of the class C/HOPS core complex (26) and the CORVET subunit mutant Δvps3 (Fig. 1C). Vps8 was still found at the membrane in these mutants; however, the Vps8 proportion in the membrane fraction was slightly reduced compared to the wild type. Thus, although Vps8 membrane association is not absolutely dependent on the HOPS core components, the presence of a functional CORVET complex seems to further stabilize Vps8 at the membrane. Alternatively, the Vps8 membrane association is destabilized due to the trafficking block induced in the mutants. We also considered the possibility that Vps8 may be held at the membrane by two different interactions mediated by Vps21 and HOPS. Therefore, a flotation experiment was performed with a Δvps11 Δvps21 double mutant. However, Vps8 was still found in the membrane fraction in this mutant (Fig. 1B).
Table 2.
Mutants tested for Vps8 membrane associationa
| Protein | Mutant(s) | Function |
|---|---|---|
| Arf1 | arf1-13, arf1-16, arf1-18 | Small GTPase, regulation of coated vesicle formation at Golgi apparatus |
| Chc1 | chc1 tsf | Clathrin heavy chain |
| Pep12 | Δpep12, pep12 tsf | t-SNARE at late endosome |
| Pik1 | pik1-63, pik1-83 | Phosphatidylinositol-4-kinase, PI-4-P formation at Golgi apparatus |
| SacI | 2μm-SAC1 (overexpression) | Phosphatidylinositol-phosphatase |
| Sec18 | sec18-1 | N-Ethylmaleimide-sensitive fusion protein, dissociation of cis-SNARE complexes |
| Vps3 | Δvps3 | Subunit of the CORVET tethering complex |
| Vps11/Pep5 | Δvps11 | Subunit of the class C/HOPS core complex |
| Vps16 | Δvps16 | Subunit of the class C/HOPS core complex |
| Vps18/Pep3 | Δvps18 | Subunit of the class C/HOPS core complex |
| Vps19/Pep7/Vac1 | Δvps19 | Multivalent adapter protein for vacuolar protein sorting |
| Vps21 | Δvps21, vps21 S21N, VPS21 Q66L | Rab GTPase at late endosome, Rab5 homologue |
| Vps33 | Δvps33 | Subunit of the class C/HOPS core complex |
| Vps34 | Δvps34 | Phosphatidylinositol-3-kinase |
| Vps45 | vps45 1-3 tsf | Sec1/Munc18 (SM) protein, functional interaction with Pep12 |
| Ypt52 | Δypt52, Δypt52 Δypt53 | Homologue of Vps21 |
| Δvps21 Δypt52 Δypt53 | ||
| Ypt53 | Δypt53, Δypt52 Δypt53 | Homologue of Vps21 |
| Δvps21 Δypt52 Δypt53 |
Transformed with pRK827; membrane associations were tested by flotation.
So far, we have not been able to identify any mutant that completely abolishes the Vps8 membrane association. The most likely interpretation of these data therefore is that Vps8 has an intrinsic ability to bind to membranes. We wanted to address this point by testing the lipid binding properties of purified Vps8. However, due to aggregation problems with Vps8 expressed in Escherichia coli, we were unable to resolve this issue.
Mapping of an N-terminal membrane binding domain in Vps8.
Using C-terminal truncations, we tried to map the membrane binding domain in Vps8. Again, the deletions were examined for membrane binding by flotation (Fig. 2). A small N-terminal fragment comprising 253 amino acids was still able to bind to membranes. But, the membrane association was lost upon further shortening to 68 N-terminal amino acids. Furthermore, a fragment spanning amino acids 164 to 339 did bind to membranes. From this analysis, we conclude that a membrane binding domain is contained in Vps8 between amino acids 164 and 253.
Fig. 2.
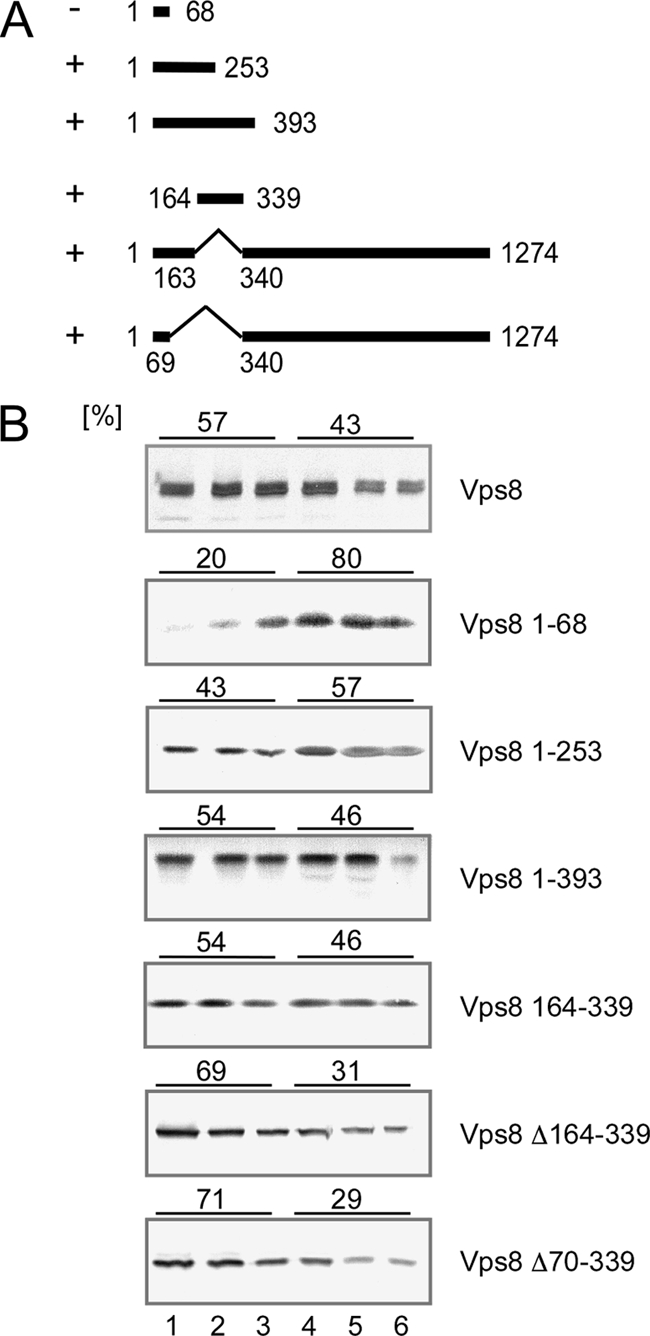
Mapping of the N-terminal membrane binding region in Vps8. (A) The range of Vps8 amino acids encoded by the different constructs is indicated by solid lines and marked with the corresponding sequence positions. +, membrane association; −, no membrane association. All Vps8 constructs were 13myc tagged at the C terminus. (B) Flotation analysis was performed as described for Fig. 1. Cell extracts were prepared from RKY1843 (Δvps8) transformed with the following various plasmids (from top to bottom): pRK827 (VPS8-13myc), pRK1000 (VPS8 1–68), pRK990 (VPS8 1–253), pRK992 (VPS8 1–393), pRK1015 (VPS8 164–339), pRK1038 (VPS8 Δ164–339), and pRK1052 (VPS8 Δ70–339). The amounts of Vps8 (percentages) in the top three fractions (membrane fraction) and the lower three fractions (soluble proteins) are given on top of the panels.
When we deleted the mapped membrane binding region from Vps8 (Δ164–339 or Δ70–339), the resulting proteins were still membrane bound. This indicates that there are additional regions in Vps8 that mediate membrane binding.
In fact, all Vps8 deletion mutants examined in this study (except for the short N-terminal peptide) were still membrane bound (data not shown). This indicates that there are multiple membrane binding regions in Vps8.
Mapping of Vps21 and HOPS core complex binding domains in Vps8.
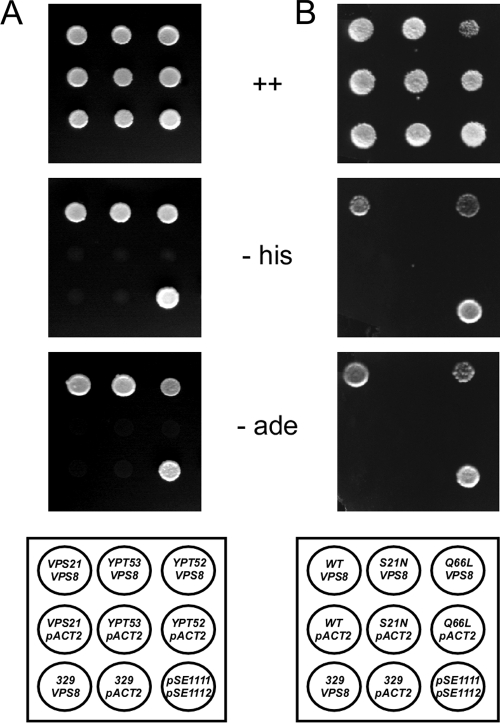
It has been claimed that Vps21 functionally interacts with Vps8 (13). Although we could not detect an effect of Δvps21 on Vps8 membrane binding, Vps21 could still affect other aspects of Vps8 function. Therefore, we examined Vps8 for Vps21 binding by using the yeast two-hybrid system. Fusions between Vps8 and the Gal4 activation domain (Gal4AD-Vps8) and between Vps21 and the Gal4 DNA binding domain (Gal4DB-Vps21) were expressed in a two-hybrid tester strain. Interaction of the two fusion partners is reflected by activation of the GAL UAS-dependent HIS3 and ADE2 reporter cassettes. When the Gal4AD-Vps8 fusion was expressed together with the Gal4DB-Vps21 fusion, activation of the HIS3 and ADE2 reporter cassettes was observed (Fig. 3A). No activation was observed with the single constructs. This indicates that Vps21 indeed interacts with Vps8. A similar interaction was also observed with the Vps21 homologues Ypt52 and Ypt53. We also tried to verify this interaction by coimmunoprecipitation (co-IP). However, we failed to detect a specific co-IP signal between Vps8-13myc and Vps21 (data not shown). We further tested whether the nucleotide state of the Vps21 GTPase had an influence on Vps8 binding. To this end, the dominant negative Vps21 S21N mutant, locked in the GDP-bound form, and the permanently active Vps21 Q66L mutant, locked in the GTP-bound form, were examined for Vps8 two-hybrid interactions (Fig. 3B). We found that Vps21 was only able to interact with Vps8 in its GTP-bound (Q66L) form. A similar result has been reported recently elsewhere (19).
Fig. 3.
Vps8-Vps21 interaction. Interactions were tested by two-hybrid analysis. Strain PJ69-4A was transformed with two plasmids coding for Gal4-DNA binding domain (DB) and Gal4 activation domain (AD) fusions. Equal amounts of cells were spotted onto minimal plates (from top to bottom) containing adenine and histidine (++), adenine (−his), or histidine (−ade). Plates were scored after 3 days at 30°C. Plasmid combinations spotted are indicated in the lower panels (from left to right). (A) pRK1002 (GAL-DB-VPS21) + pRK979 (GAL-AD-VPS8); pRK1003 (GAL-DB-YPT53) + pRK979; pRK1024 (GAL-DB-YPT52) + pRK979; pRK1002 + pACT2; pRK1003 + pACT2; pRK1024 + pACT2; pRK329 + pRK979; pRK329 + pACT2; pSE1111 + pSE1112 (positive controls) (14). (B) pRK1002 + pRK979; pRK1004 (GAL-DB-VPS21 S21N); pRK1005 (GAL-DB-VPS21 Q66L); pRK1002 + pACT2; pRK1004 + pACT2; pRK1005 + pACT2; pRK329 + pRK979; pRK329 + pACT2; pSE1111 + pSE1112.
In addition to Vps21, Vps8 also associates with the core components of the HOPS complex (20, 32). We were interested in mapping the binding domains for Vps21 and the HOPS core complex in Vps8. To this end, various deletions were introduced into Vps8 (Fig. 4), and the deletion constructs were examined for Vps21 and HOPS core complex interactions by two-hybrid analysis and by coimmunoprecipitation experiments.
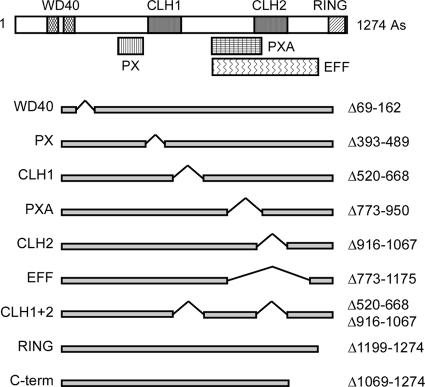
Fig. 4.
Vps8 deletions. Putative motifs (top) and the positions of the deletions (bottom) within the Vps8 protein sequence are indicated. The names of putative sequence motifs (as originally defined in the SMART database [http://smart.embl-heidelberg.de/]) are listed on the left; the range of deleted amino acids is listed on the right. The single-copy plasmids encoding the C-terminally 13myc-tagged Vps8 deletion variants are (from top to bottom): pRK999 (ΔWD40), pRK1006 (ΔPX), pRK868 (ΔCLH1), pRK1007 (ΔPXA), pRK863 (ΔCLH2), pRK1008 (ΔEFF), pRK864 (ΔCLH1+2), pRK858 (ΔRING), and pRK1054 (ΔC-term).
By database inspection, we looked for conserved motifs in the Vps8 protein sequence, as a starting point for our deletion analysis. At present, only three conserved motifs in Vps8 are listed with high confidence in the SMART database (http://smart.embl-heidelberg.de/): two N-terminal WD40 repeats and a C-terminal RING-H2 domain. When we started this analysis, two putative clathrin heavy chain repeats (CLH) were also displayed, but these are no longer listed in the current version of the database, presumably because their scores are less significant than the required threshold. Yet, these motifs could still be meaningful, since for Vps41, which appears to be functionally equivalent to Vps8, such a motif is predicted with high confidence.
In addition to the confidently predicted domains, a whole list of putative domains in Vps8 was presented in the SMART database with scores below the selected threshold. It is well possible that some of them are meaningful. For our deletion study, we selected putative motifs related to membrane binding (PX and PXA) and motifs related to GTPase function (FH2, RasGEFN, RhoGEF, RhoGAP, ARF-GAP, and Sec7). The deletion comprising the motifs related to GTPase function was called an effector domain (EFF) deletion. Although we are well aware that these motifs may not be significant, for simplicity we still chose to use these motif designations to name our deletion constructs. The deletions introduced into Vps8 are depicted in Fig. 4.
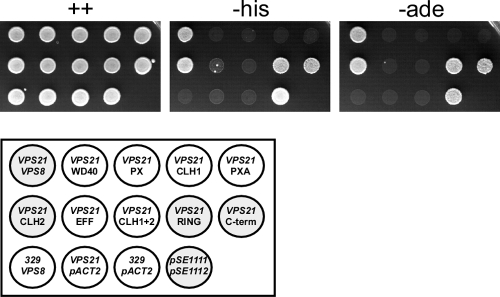
The interaction between Vps8 deletion mutants and Vps21 was again examined by two-hybrid analysis (Fig. 5). Gal4DB-Vps21 was expressed together with Gal4AD-Vps8 fusions in the two-hybrid tester strain, and induction levels of the HIS3 and ADE2 cassettes were monitored by growth on −His and −Ade plates. Three deletion mutants, Vps8ΔCLH2, Vps8ΔRING, and Vps8ΔC-term, were still able to interact with Vps21, while all other deletion mutants did not show an interaction. The three deletions which still allowed binding are located in the C-terminal part of Vps8. Thus, the N-terminal three-quarters of the protein appear to be important for Vps21 binding.
Fig. 5.
Vps21 interaction with Vps8 deletion variants. Interactions were tested by two-hybrid analysis. Strain PJ69-4A was transformed with pRK1002 (GAL-DB-VPS21) and a second plasmid coding for one of several different Gal-AD-Vps8Δ fusions. Equal amounts of cells were spotted onto minimal plates containing adenine and histidine (++), only adenine (−his), or only histidine (−ade). Plates were scored after 3 days at 30°C. The transformants contained the following VPS8 plasmids (from left to right): pRK979 (VPS8), pRK1047 (ΔWD40), pRK1033 (ΔPX), pRK1051 (ΔCLH1), pRK1034 (ΔPXA), pRK1045 (ΔCLH2), pRK1035 (ΔEFF), pRK1032 (ΔCLH1+2), pRK1030 (ΔRING), pRK1101 (ΔC-term). Positive controls were pRK329 (GAL-DB vector)/pRK979 (VPS8), pRK1002 (VPS21)/pACT2 (GAL-AD vector), pRK329 + pACT2, and pSE1111 + pSE1112.
The interaction between Vps8 deletions mutants and the HOPS core complex was examined by coimmunoprecipitation (Fig. 6). The 13myc-tagged Vps8 variants were immunoprecipitated from cell extracts with anti-myc antibodies, and the immunoprecipitates were examined for the presence of the 3HA-tagged HOPS subunit Vps11 by Western blotting. For most of the deletion variants, a Vps11 signal was obtained. Interestingly, most of the deletions that abrogated Vps21 binding (WD40, PX, CLH1, and PXA) did not affect Vps11 binding. Only one deletion, the ΔC-term deletion, completely prevented Vps11 binding. The Vps11 signal was reduced with the ΔCLH2 and ΔEFF deletions and was very faint with the ΔCLH1+2 double deletion. Thus, deletions in the C-terminal part of Vps8 seem to affect HOPS binding.
Fig. 6.
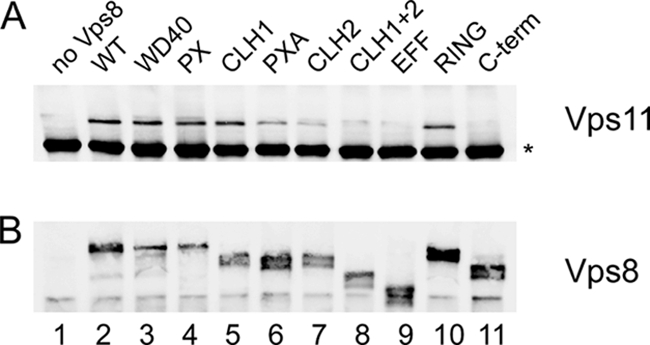
HOPS core complex interaction with Vps8 deletion variants. The interaction between the 13myc-tagged Vps8 deletion variants and 3HA-tagged Vps11 was examined by coimmunoprecipitation. Strain RKY2046 (Δvps8 VPS11-3HA) was transformed with the following plasmids (lanes indicated in parentheses): YCplac33 (vector) (1), pRK827 (VPS8-13myc) (2); pRK999 (ΔWD40) (3), pRK1006 (ΔPX) (4), pRK868 (ΔCLH1) (5), pRK1007 (ΔPXA) (6), pRK863 (ΔCLH2) (7), pRK864 (ΔCLH1+2) (8), pRK1008 (ΔEFF) (9), pRK858 (ΔRING) (10), and pRK1054 (ΔC-term) (11). Vps8-13myc was precipitated from cell extracts with anti-myc antibody (9E10; Covance), and the immunoprecipitates were examined for the presence of Vps11-3HA by Western blotting (WB) with anti-HA antibodies (HA.11; Covance) (A) and Vps8-13myc with anti-myc antibodies (B). *, background band.
So, an inverse pattern of interactions was observed for Vps21 and the HOPS core complex: while the N-terminal three-quarters of Vps8 appear to be important for Vps21 binding, the C-terminal quarter appears to be important for binding to the HOPS core complex.
All deletion mutants were also tested for membrane association in flotation experiments. But again, none of the deletions had an effect on membrane association (data not shown).
Phenotypes of VPS8 deletions.
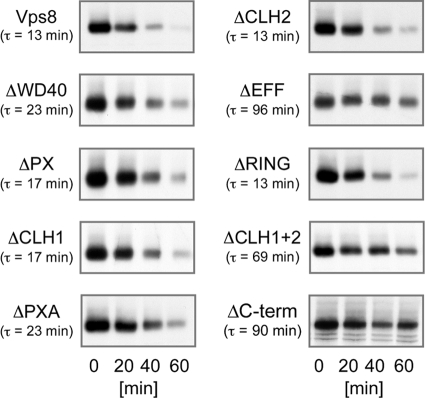
VPS8 mutants are defective in sorting of biosynthetic and endocytic cargoes to the yeast vacuole. To study the effects of our Vps8 deletions on vacuolar protein sorting, we examined the transport of the endocytic cargo protein Ste6, the yeast a-factor transporter. The short-lived Ste6 protein (half-life, 13 min) is strongly stabilized in a Δvps8 strain. The Ste6 half-life was determined by pulse-chase experiments in a Δvps8 strain transformed with single-copy plasmids expressing the different Vps8 deletion mutants (as detailed in Fig. 4) and with a multicopy plasmid expressing Ste6-3HA (Fig. 7). Some deletions had a strong effect on Ste6 half-life, and some had only a moderate effect or no effect at all. A strong effect was observed with those deletions that affect HOPS binding (ΔEFF, ΔCHL1+2, and ΔC-term), while only a moderate effect was seen with the deletions that affect Vps21 binding (ΔWD40, ΔPX, ΔCHL1, and ΔPXA). The deletions that affect neither Vps21 nor HOPS binding had no effect on the Ste6 half-life (ΔCLH2 and ΔRING). So, there appears to be a correlation between the binding properties of the Vps8 mutants and the observed Ste6 phenotype. While loss of HOPS core complex binding leads to a severe defect in Ste6 turnover, loss of Vps21 binding has only modest effects on Ste6.
Fig. 7.
Effects of VPS8 deletions on Ste6 trafficking. RKY1843 (Δvps8) was transformed with the 2μm plasmid pRK906 expressing Ste6-3HA and with a second plasmid expressing one of the 13myc-tagged Vps8 deletion variants. The Ste6 half-life was determined in a pulse-chase experiment. Cells were labeled for 15 min with [35S]methionine and then chased with cold methionine. Cell extracts were prepared at the indicated time intervals, and Ste6 was immunoprecipitated from the extracts with anti-Ste6 antibodies. The immunoprecipitates were analyzed by SDS-PAGE and autoradiography. Signals were quantified with ImageJ. Calculated half-lives (τ) are indicated. The VPS8 plasmids are as follows (from top to bottom): left panels, pRK827 (VPS8-13myc), pRK999 (ΔWD40), pRK1006 (ΔPX), pRK868 (ΔCLH1), and pRK1007 (ΔPXA); right panels, pRK863 (ΔCLH2), pRK1008 (ΔEFF), pRK858 (ΔRING), pRK864 (ΔCLH1+2), and pRK1054 (ΔC-term).
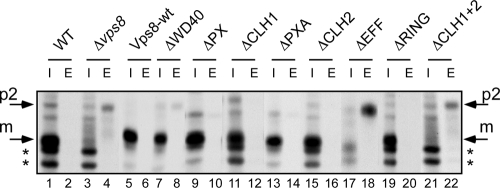
To exclude that the effects of the VPS8 deletions are restricted to Ste6 turnover, we also examined the trafficking of another protein, CPY, which passes through endosomes on its way to the vacuole. By passing from one compartment to the other, CPY receives different modifications that can be distinguished by gel mobility. In the endoplasmic reticulum, it is core glycosylated to the p1 form, in the Golgi complex it is converted into the slower-migrating, outer chain glycosylated p2 form, and in the vacuole the mature m form is finally generated by proteolytic cleavage of the precursor. In vacuolar protein sorting (vps) mutants, which are defective in endosomal transport, p2-CPY is missorted to the culture medium. To detect missorting, CPY was immunoprecipitated from cell extracts and from culture supernatants. CPY sorting was examined in a pulse-chase experiment (Fig. 8). After a 40-min chase period, CPY was completely converted to the m-CPY form in the wild-type strain, which is exclusively found in the internal fraction. In the VPS8 deletion strain, m-CPY could not be detected in the internal fraction, while p2-CPY could now be precipitated from the culture medium. A similar phenotype was observed for the mutants that abrogate HOPS binding (ΔEFF and ΔCLH1+2). Most of the mutants, however, that abolished the Vps21 interaction showed a wild-type pattern (m-CPY internal and no p2-CPY in the external fraction). Only with the ΔWD40 mutant could some p2-CPY be detected in the external fraction, while most of CPY was still found internal in its mature form. Thus, the VPS8 deletions affect CPY sorting in a way that is similar to Ste6 trafficking.
Fig. 8.
Effects of VPS8 deletions on CPY sorting. Cells were labeled for 5 min with [35S]methionine and then chased with cold methionine for 40 min. CPY was immunoprecipitated from cell extracts (I) and from the culture medium (E) with anti-CPY antibodies. The immunoprecipitates were analyzed by SDS-PAGE and autoradiography. Lanes: 1 and 2, JD52 (3–22) RKY1843 (Δvps8) transformed with plasmids expressing 13myc-tagged Vps8 deletion variants; 3 and 4, YCplac33; 5 and 6, pRK827; 7 and 8, pRK999; 9 and 10, pRK1006; 11 and 12, pRK868; 13 and 14, pRK1007; 15 and 16, pRK863; 17 and 18, pRK1008; 19 and 20, pRK858; 21 and 22, pRK864. Arrows show positions of the p2 and m forms; asterisks indicate background bands.
DISCUSSION
In this report, membrane association of the class D Vps function Vps8 and the interaction with its binding partners Vps21 and the HOPS core complex were examined.
Based on the fact that Vps8 can be sedimented by centrifugation, it has been concluded that Vps8 is membrane associated (5, 13). Based on flotation experiments, we now present conclusive evidence that this notion is indeed correct. Membrane binding was not absolutely dependent on Vps21 or on the class C/HOPS core complex. In Δvps21 and in class C/HOPS core complex single mutants, as well as in the Δvps21 Δvps11 double mutant, Vps8 was still associated with membranes.
In a previous report, it was claimed that the Rab GTPase Vps21, the yeast homologue of mammalian Rab5, recruits Vps8 to membranes (13). This notion, however, is not supported by our experiments. Differences in the methods used may be responsible for the divergent results. In the study by Horazdovsky et al. the yeast cells were subjected to a number of stress conditions (high osmolarity, high pH, starvation, and cell wall stress) before extract preparation. Yeast spheroplasts were generated and subjected to a pulse-chase regimen followed by immunoprecipitation of Vps8 after an extended chase period. Our experiments, however, were performed in a different way. Cell extracts prepared from exponentially growing cells by rapid glass bead lysis were immediately fractionated by flotation without further treatment. Thus, our experiment more closely reflects the steady-state distribution of Vps8. Indeed, Vps8 membrane association was lost when we used the Horazdovsky et al. conditions for extract preparation (unpublished data).
Since neither Vps21 nor HOPS appears to be essential for Vps8 membrane recruitment under steady-state conditions, we looked for other potential Vps8 receptors. Vps8 is part of the so-called CORVET tethering complex, which shares core components with the HOPS complex. HOPS binds to the Rab GTPase Ypt7 and to the vacuolar SNARE complex, which appears to contribute to anchoring HOPS to the vacuolar membrane. Therefore, components of the endosomal SNARE complex (Pep12) and associated factors like Vps45 involved in the same trafficking step as Vps8 were obvious candidates for potential Vps8 receptors. However, membrane association was also unaffected by deletion of PEP12 and inactivation of Vps45.
So far, we have failed to identify a mutant that completely abolishes Vps8 membrane binding. We therefore believe that Vps8 has an intrinsic ability to bind to membranes. Our data further suggest that this Vps8 membrane association is more stable in the context of a functional CORVET complex. Several components of the endocytic machinery have been shown to bind directly to membranes, mostly through interactions with phosphatidyl inositol phosphates (29). Notably, HOPS also has an affinity for phosphoinositides (31). PI-3-P appears to play a prominent role as a receptor in the endocytic pathway, but PI-3-P does not appear to be essential for Vps8 membrane binding, since Vps8 was still at the membrane in the PI-3 kinase mutant Δvps34, which is devoid of PI-3-P (27). Also, lowering the PI-4-P level by inactivation of the PI-4 kinase Pik1 or by overexpression of the PI-4-P-specific phosphatase SacI (10) did not affect Vps8 membrane association.
No obvious membrane binding motifs could be found in the Vps8 sequence. Thus, Vps8 may contain novel binding motifs. Vps8 could bind to membranes in a way analogous to Bet3, the membrane binding component of the TRAPP tethering complex (24). The crystal structure of mouse bet3 reveals a flat, wide membrane-interacting surface decorated with positively charged residues (15). Charge inversion mutations on this surface affect TRAPP localization and cause trafficking defects. In support of an extended membrane binding surface, our data indicate that Vps8 makes multiple contacts with the membrane. This is suggested by the finding that deletion of the mapped N-terminal membrane binding region (amino acids 164 to 253) did not abolish membrane binding. Furthermore, we found that portions (quarters) of Vps8 were all membrane associated (unpublished data). So, there must be at least four independent membrane binding areas in Vps8.
It has been claimed that Vps8 functionally interacts with Vps21 (13), but physical association had not been examined until recently. By two-hybrid analysis, we have shown that Vps8 interacts with Vps21 in its GTP-bound form. By deletion analysis, we found that the N-terminal three-quarters of Vps8 are important for Vps21 binding, while the C-terminal quarter appears to be important for binding to the HOPS core complex. While the manuscript was under review, a study was published reporting a Vps21-Vps8 two-hybrid interaction (19). Although the initial mapping experiments from that study are compatible with our data, Markgraf et al. came to a completely opposite conclusion as to the localization of the Vps21 and class C/HOPS binding regions. However, we think that their data were obviously not properly interpreted.
What is the significance of this Vps8-Vps21 interaction? Rab/tethering complex interactions are of central importance for vesicle docking and fusion. Yet, other subunits of CORVET and HOPS appear to be more important in this context. For instance, the two equivalent subunits, Vps3 in CORVET and Vps39/Vam6 in HOPS, have been shown to display GEF activity toward the respective Rabs, Vps21 and Ypt7 (20, 35). In addition, whole tethering complexes bind to active Rab-GTP, i.e., act as Rab effectors (6, 20, 31). In contrast, the Vps8-Vps21 interaction does not appear to be absolutely required for endocytic trafficking. Vps8 deletions that abolish Vps21 binding only had a modest effect on Ste6 trafficking and CPY sorting, while deletions that abolish interaction with the class C/HOPS core complex caused severe trafficking defects. Thus, the Vps8-Vps21 interaction probably serves an auxiliary function. For instance, it could play a role in removing the tethers from the membrane after fusion in preparation for a new cycle of membrane fusion.
In contrast to our findings, a central role for the Vps21-Vps8 interaction in endocytic trafficking has been postulated in a recent report (19). This was concluded from the finding that overexpression of Vps21 as well as Vps8 leads to a clustering of late endosomes and multivesicular bodies. Both Vps21 and Vps8, among other Vps proteins, could be detected in these clusters. However, the functional significance of these structures for endocytic trafficking is unclear, since trafficking of endosomal cargo proteins like Ste3 and CPS was apparently not impaired by this clustering. This rather supports our contention that the Vps21-Vps8 interaction under normal conditions is not essential for targeting endosomal cargo proteins to the vacuole.
At present, Vps8 is viewed as a subunit of the distinct 700-kDa CORVET complex (20). However, some of our data are more compatible with Vps8 being part of some higher-order structure. For instance, Vps8 displayed an unusual fractionation behavior. After detergent or high-salt extraction, Vps8 was still largely insoluble (unpublished data). In this respect Vps8 resembles the TRAPP complex, which also proved to be resistant to detergent extraction (23). It is tempting to speculate that the CORVET subunit Vps8 performs a similar function as its cognate HOPS subunit, Vps41. It has been demonstrated that Vps41 forms homotypic, hexameric complexes (7). Vps41 has two functions: in addition to its role in homotypic vacuole fusion as part of the HOPS complex, it is also involved in formation of vesicles at the trans-Golgi network (TGN) as part of the so-called ALP pathway, a direct transport route from the TGN to the vacuole (22). It has been suggested that Vps41 may form coats at TGN membranes. Vps8 could perform a similar function at early endosomes.
ACKNOWLEDGMENTS
We thank Aki Nakano, Greg Payne, and Jeremy Thorner for sending us yeast strains. We are also grateful to Karin Krapka for her assistance with some of the experiments.
This work was supported by DFG grant Ko 963/4-2 to R.K.
Footnotes
Published ahead of print on 19 February 2010.
REFERENCES
- 1.Bagnat M., Keranen S., Shevchenko A., Simons K. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. U. S. A. 97:3254–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becherer K. A., Rieder S. E., Emr S. D., Jones E. W. 1996. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell 7:579–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant N. J., Stevens T. H. 1998. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol. Mol. Biol. Rev. 62:230–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burd C. G., Peterson M., Cowles C. R., Emr S. D. 1997. A novel Sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol. Biol. Cell 8:1089–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y. J., Stevens T. H. 1996. The VPS8 gene is required for localization and trafficking of the CPY sorting receptor in Saccharomyces cerevisiae. Eur. J. Cell Biol. 70:289–297 [PubMed] [Google Scholar]
- 6.Collins K. M., Thorngren N. L., Fratti R. A., Wickner W. T. 2005. Sec17p and HOPS, in distinct SNARE complexes, mediate SNARE complex disruption or assembly for fusion. EMBO J. 24:1775–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darsow T., Katzmann D. J., Cowles C. R., Emr S. D. 2001. Vps41p function in the alkaline phosphatase pathway requires homo-oligomerization and interaction with AP-3 through two distinct domains. Mol. Biol. Cell 12:37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerrard S. R., Bryant N. J., Stevens T. H. 2000. VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol. Biol. Cell 11:613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosshans B. L., Ortiz D., Novick P. 2006. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U. S. A. 103:11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hama H., Schnieders E. A., Thorner J., Takemoto J. Y., DeWald D. B. 1999. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274:34294–34300 [DOI] [PubMed] [Google Scholar]
- 11.Hama H., Tall G. G., Horazdovsky B. F. 1999. Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J. Biol. Chem. 274:15284–15291 [DOI] [PubMed] [Google Scholar]
- 12.Horazdovsky B. F., Busch G. R., Emr S. D. 1994. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 13:1297–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horazdovsky B. F., Cowles C. R., Mustol P., Holmes M., Emr S. D. 1996. A novel RING finger protein, Vps8p, functionally interacts with the small GTPase, Vps21p, to facilitate soluble vacuolar protein localization. J. Biol. Chem. 271:33607–33615 [DOI] [PubMed] [Google Scholar]
- 14.James P., Halladay J., Craig E. A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y. G., Sohn E. J., Seo J., Lee K. J., Lee H. S., Hwang I., Whiteway M., Sacher M., Oh B. H. 2005. Crystal structure of bet3 reveals a novel mechanism for Golgi localization of tethering factor TRAPP. Nat. Struct. Mol. Biol. 12:38–45 [DOI] [PubMed] [Google Scholar]
- 16.Lemmon S. K., Traub L. M. 2000. Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 12:457–466 [DOI] [PubMed] [Google Scholar]
- 17.Lewis M. J., Pelham H. R. 2002. A new yeast endosomal SNARE related to mammalian syntaxin 8. Traffic 3:922–929 [DOI] [PubMed] [Google Scholar]
- 18.Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 19.Markgraf D. F., Ahnert F., Arlt H., Mari M., Peplowska K., Epp N., Griffith J., Reggiori F., Ungermann C. 2009. The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol. Biol. Cell 20:5276–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peplowska K., Markgraf D. F., Ostrowicz C. W., Bange G., Ungermann C. 2007. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev. Cell 12:739–750 [DOI] [PubMed] [Google Scholar]
- 21.Raymond C. K., Howald S. I., Vater C. A., Stevens T. H. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehling P., Darsow T., Katzmann D. J., Emr S. D. 1999. Formation of AP-3 transport intermediates requires Vps41 function. Nat. Cell Biol. 1:346–353 [DOI] [PubMed] [Google Scholar]
- 23.Sacher M., Barrowman J., Schieltz D., Yates J. R., III, Ferro-Novick S. 2000. Identification and characterization of five new subunits of TRAPP. Eur. J. Cell Biol. 79:71–80 [DOI] [PubMed] [Google Scholar]
- 24.Sacher M., Jiang Y., Barrowman J., Scarpa A., Burston J., Zhang L., Schieltz D., Yates J. R., III, Abeliovich H., Ferro-Novick S. 1998. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 17:2494–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sapperstein S. K., Walter D. M., Grosvenor A. R., Heuser J. E., Waters M. G. 1995. p115 is a general vesicular transport factor related to the yeast endoplasmic reticulum to Golgi transport factor Uso1p. Proc. Natl. Acad. Sci. U. S. A. 92:522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato T. K., Rehling P., Peterson M. R., Emr S. D. 2000. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell 6:661–671 [DOI] [PubMed] [Google Scholar]
- 27.Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., Emr S. D. 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260:88–91 [DOI] [PubMed] [Google Scholar]
- 28.Seals D. F., Eitzen G., Margolis N., Wickner W. T., Price A. 2000. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. U. S. A. 97:9402–9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonsen A., Wurmser A. E., Emr S. D., Stenmark H. 2001. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13:485–492 [DOI] [PubMed] [Google Scholar]
- 30.Singer-Krüger B., Stenmark H., Dusterhoft A., Philippsen P., Yoo J. S., Gallwitz D., Zerial M. 1994. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J. Cell Biol. 125:283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stroupe C., Collins K. M., Fratti R. A., Wickner W. 2006. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 25:1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian S., Woolford C. A., Jones E. W. 2004. The Sec1/Munc18 protein, Vps33p, functions at the endosome and the vacuole of Saccharomyces cerevisiae. Mol. Biol. Cell 15:2593–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sztul E., Lupashin V. 2006. Role of tethering factors in secretory membrane traffic. Am. J. Physiol. Cell Physiol. 290:C11–C26 [DOI] [PubMed] [Google Scholar]
- 34.Weiß P., Huppert S., Kölling R. 2009. Analysis of the dual function of the ESCRT-III protein Snf7 in endocytic trafficking and in gene expression. Biochem. J. 424:89–97 [DOI] [PubMed] [Google Scholar]
- 35.Wurmser A. E., Sato T. K., Emr S. D. 2000. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151:551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yahara N., Ueda T., Sato K., Nakano A. 2001. Multiple roles of Arf1 GTPase in the yeast exocytic and endocytic pathways. Mol. Biol. Cell 12:221–238 [DOI] [PMC free article] [PubMed] [Google Scholar]