Abstract
There are currently intensive global research efforts aimed at increasing and modifying the accumulation of lipids, alcohols, hydrocarbons, polysaccharides, and other energy storage compounds in photosynthetic organisms, yeast, and bacteria through genetic engineering. Many improvements have been realized, including increased lipid and carbohydrate production, improved H2 yields, and the diversion of central metabolic intermediates into fungible biofuels. Photosynthetic microorganisms are attracting considerable interest within these efforts due to their relatively high photosynthetic conversion efficiencies, diverse metabolic capabilities, superior growth rates, and ability to store or secrete energy-rich hydrocarbons. Relative to cyanobacteria, eukaryotic microalgae possess several unique metabolic attributes of relevance to biofuel production, including the accumulation of significant quantities of triacylglycerol; the synthesis of storage starch (amylopectin and amylose), which is similar to that found in higher plants; and the ability to efficiently couple photosynthetic electron transport to H2 production. Although the application of genetic engineering to improve energy production phenotypes in eukaryotic microalgae is in its infancy, significant advances in the development of genetic manipulation tools have recently been achieved with microalgal model systems and are being used to manipulate central carbon metabolism in these organisms. It is likely that many of these advances can be extended to industrially relevant organisms. This review is focused on potential avenues of genetic engineering that may be undertaken in order to improve microalgae as a biofuel platform for the production of biohydrogen, starch-derived alcohols, diesel fuel surrogates, and/or alkanes.
Interest in a variety of renewable biofuels has been rejuvenated due to the instability of petroleum fuel costs, the reality of peak oil in the near future, a reliance on unstable foreign petroleum resources, and the dangers of increasing atmospheric CO2 levels. Photosynthetic algae, both microalgae and macroalgae (i.e., seaweeds), have been of considerable interest as a possible biofuel resource for decades (165). Several species have biomass production rates that can surpass those of terrestrial plants (41), and many eukaryotic microalgae have the ability to store significant amounts of energy-rich compounds, such as triacylglycerol (TAG) and starch, that can be utilized for the production of several distinct biofuels, including biodiesel and ethanol. It is believed that a large portion of crude oil is of microalgal origin, with diatoms being especially likely candidates, considering their lipid profiles and productivity (153). If ancient algae are responsible for creating substantial crude oil deposits, it is clear that investigation of the potential of living microalgae to produce biofuels should be a priority. Microalgae are especially attractive as a source of fuel from an environmental standpoint because they consume carbon dioxide and can be grown on marginal land, using waste or salt water (41). In addition, it may be possible to leverage the metabolic pathways of microalgae to produce a wide variety of biofuels (Fig. 1). In contrast to corn-based ethanol or soy/palm-based biodiesel, biofuels derived from microalgal feedstocks will not directly compete with the resources necessary for agricultural food production if inorganic constituents can be recycled and saltwater-based cultivation systems are developed.
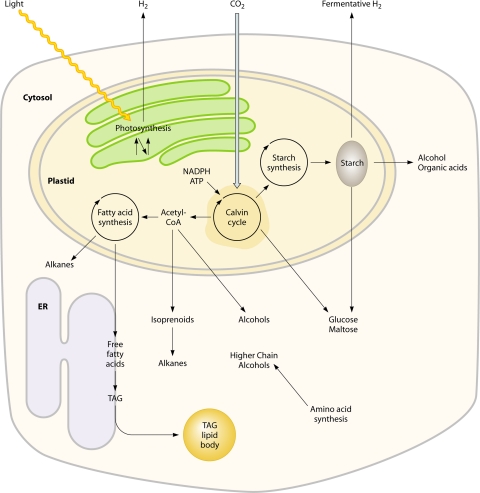
Fig. 1.
Microalgal metabolic pathways that can be leveraged for biofuel production. ER, endoplasmic reticulum.
However, several technical barriers need to be overcome before microalgae can be used as an economically viable biofuel feedstock (139). These include developing low-energy methods to harvest microalgal cells, difficulties in consistently producing biomass at a large scale in highly variable outdoor conditions, the presence of invasive species in large-scale ponds, low light penetrance in dense microalgal cultures, the lack of cost-effective bioenergy carrier extraction techniques, and the potentially poor cold flow properties of most microalga-derived biodiesel. To advance the utilization of microalgae in biofuel production, it is important to engineer solutions to optimize the productivity of any microalgal cultivation system and undertake bioprospecting efforts to identify strains with as many desirable biofuel traits as possible. Over 40,000 species of algae have been described, and this is likely only a small fraction of the total number of available species (75). The U.S. Department of Energy's Aquatic Species Program analyzed approximately 3,000 different microalgae for their potential to produce biofuels, and numerous additional species have subsequently been investigated (165). Although these efforts demonstrated that many species of microalgae have properties that are desirable for biofuel production, most have drawbacks that have prevented the emergence of an economically viable algal biofuel industry. It is postulated that a light-harvesting footprint of at least 20,000 square miles will be required to satisfy most of the current U.S. transportation fuel demand (41). Therefore, even modest improvements in photon conversion efficiencies will dramatically reduce the land area and cost required to produce biofuels. Consequently, continued bioprospecting efforts and the development and engineering of select microalgal strains are required to improve the yields of bioenergy carriers. Current commercial agriculture crops have been cultivated for thousands of years, with desired traits selected over time. It stands to reason that with microalgae, it would be beneficial to use genetic engineering in an attempt to bypass such a lengthy selection process. However, despite the recent advances in biotechnological approaches, the full potential of genetic engineering in some microalgal species, particularly diploid diatoms, can be fully realized only if conventional breeding methods become firmly established, thereby allowing useful traits or mutations to be easily combined (5, 24, 25). Since the topic of microalgal sexual breeding is beyond the scope of this review, we will instead focus on genetic engineering approaches that could be utilized in the industry's efforts to improve microalgae as a source of biofuels.
GENETIC ENGINEERING OF MICROALGAE
Significant advances in microalgal genomics have been achieved during the last decade. Expressed sequence tag (EST) databases have been established; nuclear, mitochondrial, and chloroplast genomes from several microalgae have been sequenced; and several more are being sequenced. Historically, the green alga Chlamydomonas reinhardtii has been the focus of most molecular and genetic phycological research. Therefore, most of the tools for the expression of transgenes and gene knockdown have been developed for and are specific for this species. However, tools are now also being rapidly developed for diatoms and other algae that are of greater interest for industrial applications.
Microalgal genomes.
Access to microalgal genome sequences that are of interest for academic or industrial applications greatly facilitates genetic manipulation, and the availability of rapid large-scale sequencing technology represents a revolution in microalga research. Several nuclear genome sequencing projects have now been completed, including those for C. reinhardtii (116, 171), Phaeodactylum tricornutum (15), Thalassiosira pseudonana (6), Cyanidioschyzon merolae (109), Ostreococcus lucimarinus (135), Ostreococcus tauri (36), and Micromonas pusilla (201). Currently, ongoing microalgal genome sequencing projects include those for Fragilariopsis cylindrus, Pseudo-nitzschia, Thalassiosira rotula, Botryococcus braunii, Chlorella vulgaris, Dunaliella salina, Micromonas pusilla, Galdieria sulphuraria, Porphyra purpurea, Volvox carteri, and Aureococcus anophageferrens (100). In addition, there are several completed and ongoing efforts to sequence plastid and mitochondrial genomes, as well as dynamic transcriptomes from many different microalgae (4, 9, 29, 66, 73, 101, 105, 130, 133, 149, 161, 169, 171, 196, 198).
Methods for transformation and expression.
Successful genetic transformation has been reported for the green (Chlorophyta), red (Rhodophyta), and brown (Phaeophyta) algae; diatoms; euglenids; and dinoflagellates (2, 3, 21–23, 26, 30, 37, 42, 44, 45, 49, 54–56, 58, 65, 69, 76, 83, 85, 87, 88, 92–95, 108, 119, 121, 123, 147, 148, 150, 151, 163, 170, 180, 181, 183, 187, 210, 211, 214, 217). More than 30 different strains of microalgae have been transformed successfully to date. In many cases, transformation resulted in stable expression of transgenes, from either the nucleus or the plastid, but in some cases only transient expression was observed. Methods developed primarily with C. reinhardtii (for a recent review by Eichler-Stahlberg et al., see reference 48) demonstrate that the stability of expression can be improved through proper codon usage, the use of strong endogenous promoters, and inclusion of species-specific 5′, 3′, and intron sequences. The efficiency of transformation seems to be strongly species dependent, and the method of transformation has to be carefully selected and optimized for each microalga. A variety of transformation methods have been used to transfer DNA into microalgal cells, including agitation in the presence of glass beads or silicon carbide whiskers (44, 87, 119), electroporation (21, 22, 26, 108, 170, 181, 184), biolistic microparticle bombardment (2, 45, 49, 51, 52, 81, 83, 88, 183, 187, 210, 211), and Agrobacterium tumefaciens-mediated gene transfer (23, 93).
Efficient isolation of genetic transformants is greatly facilitated by the use of selection markers, including antibiotic resistance and/or fluorescent/biochemical markers. Several different antibiotic resistance genes have been used successfully for microalgal transformant selection, including bleomycin (2, 52, 56, 104, 210), spectinomycin (19, 42), streptomycin (42), paromomycin (81, 173), nourseothricin (210), G418 (45, 148, 210), hygromycin (12), chloramphenicol (184), and others. Due to the fact that many microalgae are resistant to a wide range of antibiotics, the actual number of antibiotics that work with a specific strain may be much more limited. In addition, antibiotics like nourseothricin and G418 are much less effective in salt-containing media and are not ideal for use with marine algae (210). Other markers that have been used include luciferase (51, 55, 83), β-glucuronidase (22, 23, 26, 49, 51, 92), β-galactosidase (58, 85, 151), and green fluorescent protein (GFP) (23, 50, 54, 56, 148, 210).
Transgene expression and protein localization in the chloroplast is needed for the proper function of many metabolic genes of interest for biofuel production. In C. reinhardtii, it is possible to achieve transformation of the chloroplast through homologous recombination (for a review by Marín-Navarro et al., see reference 106). While chloroplast transformation has not been demonstrated with diatoms, several publications have used plastid targeting sequences to translocate proteins to the chloroplast (3, 65).
Nuclear transformation of microalgae generally results in the random integration of transgenes. While this may be suitable for transgene expression or for random mutagenesis screens, it makes it difficult to delete specific target genes. Some progress in homologous recombination has been made with the nuclear genome of C. reinhardtii, but the efficiency remains low (217). Homologous recombination has also been reported for the red microalga C. merolae (121). Another option for gene inactivation is the use of RNA silencing to knock down gene expression; the mechanisms for RNA silencing have been studied with microalgae, and RNA silencing has been used successfully with both C. reinhardtii and P. tricornutum (16, 37, 122, 123, 214). Recent improvements in gene knockdown strategies include the development of high-throughput artificial-micro-RNA (armiRNA) techniques for C. reinhardtii that are reportedly more specific and stable than traditional RNA interference (RNAi) approaches (123, 214).
Members of the chlorophyte group that have been transformed include C. reinhardtii, which has been transformed using a variety of methods (44, 87, 88, 93, 170); Chlorella ellipsoidea (22, 83); Chlorella saccharophila (108); C. vulgaris (30, 69); Haematococcus pluvialis (177, 187); V. carteri (81, 163); Chlorella sorokiniana (30); Chlorella kessleri (49); Ulva lactuca (76); Dunaliella viridis (180); and D. salina (181, 183). Heterokontophytes that have reportedly been transformed include Nannochloropsis oculata (21); diatoms such as T. pseudonana (147), P. tricornutum (2, 210, 211), Navicula saprophila (45), Cylindrotheca fusiformis (52, 148), Cyclotella cryptica (45), and Thalassiosira weissflogii (51); and phaeophytes, such as Laminaria japonica (150) and Undaria pinnatifada (151). Rhodophytes, such as C. merolae (121), Porphyra yezoensis (23), Porphyra miniata (92), Kappaphycus alvarezii (94), Gracilaria changii (58), and Porphyridium sp. (95), have also been transformed. Dinoflagellates that have been transformed include Amphidinium sp. and Symbiodinium microadriaticum (119). The only euglenid that has been transformed to date is Euglena gracilis (42).
GENETIC ENGINEERING OF THE LIPID METABOLISM
Understanding microalgal lipid metabolism is of great interest for the ultimate production of diesel fuel surrogates. Both the quantity and the quality of diesel precursors from a specific strain are closely linked to how lipid metabolism is controlled. Lipid biosynthesis and catabolism, as well as pathways that modify the length and saturation of fatty acids, have not been as thoroughly investigated for algae as they have for terrestrial plants. However, many of the genes involved in lipid metabolism in terrestrial plants have homologs in the sequenced microalgal genomes. Therefore, it is probable that at least some of the transgenic strategies that have been used to modify the lipid content in higher plants will also be effective with microalgae.
Lipid biosynthesis.
In recent years, many of the genes involved in lipid synthesis have been subjected to both knockout and overexpression in order to clarify their importance in lipid accumulation and to establish strategies to increase the lipid content in the oleaginous seeds of higher plants, such as Arabidopsis thaliana, soy bean (Glycine max), and rapeseed (Brassica napus). See Fig. 2 for a simplified overview of lipid biosynthesis pathways. Several of these transgenic overexpression strategies have resulted in the increased production of triacylglycerols in seeds and in other plant tissues. Ohlrogge and Jaworski have proposed that the fatty acid supply helps determine the regulation of oil synthesis (134); therefore, some efforts have been made to increase the expression of enzymes that are involved in the pathways of fatty acid synthesis. One early committing step in fatty acid synthesis is the conversion of acetyl-coenzyme A (CoA) to malonyl-CoA, catalyzed by acetyl-CoA carboxylase (ACCase), which is considered the first committed step in fatty acid biosynthesis in many organisms. However, several attempts to utilize ACCase overexpression to increase lipid content in various systems have been somewhat disappointing. Dunahay et al. overexpressed native ACCase in the diatom C. cryptica (45). Despite a 2- or 3-fold increase in ACCase activity, no increased lipid production could be observed (165). ACCase from A. thaliana has been overexpressed in B. napus and Solanum tuberosum (potato) (89, 157). Overexpression of ACCase in the oleaginous seeds of B. napus resulted in a minor increase in seed lipid content of about 6% (384 mg g−1 and 408 mg g−1 dry weight for wild-type [WT] and transgenic ACCase rapeseed lines, respectively). Interestingly, the effect of ACCase overexpression in potato tubers, a tissue that normally is very starch rich and lipid poor, resulted in a 5-fold increase in TAG content (from 0.0116 to 0.0580 mg g−1 fresh weight). It may be that ACCase levels are a limiting step in lipid biosynthesis mainly in cells that normally do not store large amounts of lipid. Another attempt to increase expression of a protein involved in fatty acid synthesis, 3-ketoacyl-acyl-carrier protein synthase III (KASIII), was not successful in increasing lipid production. KASIII from spinach (Spinacia oleracea) or Cuphea hookeriana was expressed in tobacco (Nicotiana tabacum), A. thaliana, and B. napus, resulting in either no change or reduced seed oil content (33).
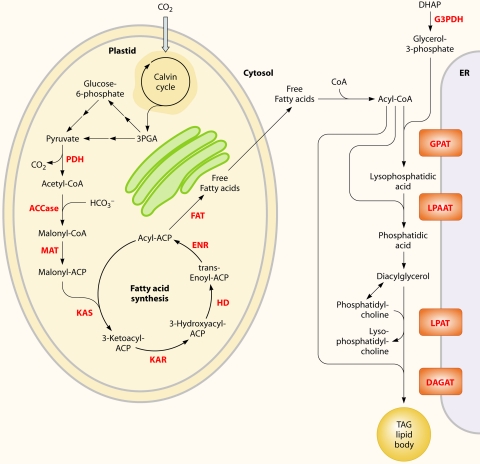
Fig. 2.
Simplified overview of the metabolites and representative pathways in microalgal lipid biosynthesis shown in black and enzymes shown in red. Free fatty acids are synthesized in the chloroplast, while TAGs may be assembled at the ER. ACCase, acetyl-CoA carboxylase; ACP, acyl carrier protein; CoA, coenzyme A; DAGAT, diacylglycerol acyltransferase; DHAP, dihydroxyacetone phosphate; ENR, enoyl-ACP reductase; FAT, fatty acyl-ACP thioesterase; G3PDH, gycerol-3-phosphate dehydrogenase; GPAT, glycerol-3-phosphate acyltransferase; HD, 3-hydroxyacyl-ACP dehydratase; KAR, 3-ketoacyl-ACP reductase; KAS, 3-ketoacyl-ACP synthase; LPAAT, lyso-phosphatidic acid acyltransferase; LPAT, lyso-phosphatidylcholine acyltransferase; MAT, malonyl-CoA:ACP transacylase; PDH, pyruvate dehydrogenase complex; TAG, triacylglycerols.
While increasing the expression of genes involved in fatty acid synthesis has had small successes, with regard to increasing the total amount of seed oils, some interesting results have been achieved through the overexpression of genes involved in TAG assembly. One of the most successful attempts to increase the amount of seed lipid is the overexpression of a cytosolic yeast, glycerol-3-phosphate dehydrogenase (G3PDH), in the seeds of B. napus, which resulted in a 40% increase in lipid content (191). G3PDH catalyzes the formation of glycerol-3-phosphate, which is needed for TAG formation. This interesting result suggests that genes involved in TAG assembly are of importance for total seed oil production. This is further supported by several other studies in which overexpression of TAG assembly genes resulted in increases in seed oil content. For example, overexpression of glycerol-3-phosphate acyltransferase, lysophosphatidic acid acyltransferase, or diacylglycerol acyltransferase (DAGAT) all result in significant increases in plant lipid production (79, 80, 96, 186, 215, 218). Due to the fact that enzymes such as these seem to be good candidates for overexpression strategies with the goal of increasing storage lipid content, an attempt has also been made to use directed evolution to increase the efficiency of one of these enzymes, DAGAT (172).
Another possible approach to increasing the cellular lipid content is blocking metabolic pathways that lead to the accumulation of energy-rich storage compounds, such as starch. For example, two different starch-deficient strains of C. reinhardtii, the sta6 and sta7 mutants, have disruptions in the ADP-glucose pyrophosphorylase or isoamylase genes, respectively (10, 124, 144, 146, 209). Wang et al. (197), as well as unpublished results from our laboratory, have shown that these mutants accumulate increased levels of TAG during nitrogen deprivation. Another starchless mutant of Chlorella pyrenoidosa has also been shown to have elevated polyunsaturated fatty acid content (154).
In addition to what has been accomplished with higher plants, successful modifications have also been achieved with bacteria and yeast to increase and/or modify their lipid content. Due to the ease of genetic engineering with Escherichia coli and Saccharomyces cerevisiae, these modifications include quite comprehensive modulations of entire metabolic pathways, with the simultaneous overexpression or deletion of several key enzymes. Such modifications are, of course, much harder to achieve with microalgae, but they should be attainable with organisms with established protocols for genetic transformation and available selectable markers. One example of a comprehensive modification of E. coli, which resulted in a 20-fold increase in free fatty acid production, entailed overexpression of the lipid biosynthesis genes encoding acetyl-CoA carboxylase, an endogenous thioesterase, and a plant thioesterase, as well as knocking out a gene product involved in β-oxidation of fatty acids, acyl-CoA synthetase (encoded by fadD) (102). Of particular interest in this study is the substantial increase in free fatty acid production that was due to the expression of the two thioesterases. With E. coli it has been shown that long-chain fatty acids can inhibit fatty acid synthesis and that this inhibition can be released by expression of specific thioesterases (84, 193).
Lipid catabolism.
A complementary strategy to increase lipid accumulation is to decrease lipid catabolism. In the case of lipid biosynthesis, most of what we know regarding successful strategies to decrease lipid catabolism comes from studies of higher plants and yeast. Genes involved in the activation of both TAG and free fatty acids, as well as genes directly involved in β-oxidation of fatty acids, have been inactivated, sometimes resulting in increased cellular lipid content. To circumvent the current lack of efficient homologous recombination in microalgae, gene inactivation would have to be achieved either through random mutagenesis or through the use of RNA silencing (37, 123, 214). Due to the fact that cells rely on the β-oxidation of fatty acids for cellular energy under certain physiological conditions, knocking out lipid catabolism genes not only may result in increased lipid storage but also could have deleterious effects on cellular growth and proliferation. For example, inactivation of the peroxisomal long-chain acyl-CoA synthetase (LACS) isozymes, LACS6 and LACS7, in A. thaliana inhibits seed lipid breakdown, which increased oil content. However, proper seedling development was also inhibited without the addition of an external carbon source (57). Similar results were achieved through the inactivation of 3-ketoacyl-CoA thiolase (KAT2) in A. thaliana (59). Another potential problem with strategies that involve inhibition of lipid catabolism is that enzymes with overlapping functions exist for many of the steps of β-oxidation, making it difficult to completely abolish these functions. An example is the short-chain acyl-CoA oxidase enzymes ACX3 and ACX4 in A. thaliana. Single mutants of ACX3 or ACX4 have normal lipid breakdown and seedling development, while double mutants are nonviable, putatively due to complete elimination of short-chain acyl-CoA oxidase activity (159).
During diel light-dark cycles, many microalgae initiate TAG storage during the day and deplete those stores at night to support cellular ATP demands and/or cell division. Consequently, inhibition of β-oxidation would prevent the loss of TAG during the night, but most likely at the cost of reduced growth. This strategy, therefore, may not be beneficial for microalgae grown in outdoor open ponds, but it may be a valid strategy to increase lipid production in microalgae grown in photobioreactors with exogenous carbon sources and/or continuous light.
In some studies, inhibition of lipid oxidation has caused unexpected phenotypes. Several publications have shown that knocking out genes involved in β-oxidation in S. cerevisiae not only can lead to increased amounts of intracellular free fatty acids but also results in extracellular fatty acid secretion in some instances (120, 132, 162). The lipid catabolism genes that have been implicated in fatty acid secretion in S. cerevisiae include acyl-CoA oxidase and several acyl-CoA synthetases (see below).
Modification of lipid characteristics.
In addition to engineering microalgae for the increased production of lipids, it is also reasonable to attempt to increase the quality of the lipids, with regard to suitability as a diesel fuel feedstock. The carbon chain length and degree of unsaturation of the fatty acids in each microalgal species can affect the cold flow and oxidative stability properties of a biodiesel fuel which is derived from this feedstock. Typically, most microalgal fatty acids have a chain length between 14 and 20; major species are often 16:1, 16:0, and 18:1. Ideal fatty acids for diesel production should be 12:0 and 14:0. The chain lengths of fatty acids are determined by acyl-ACP thioesterases, which release the fatty acid chain from the fatty acid synthase. There are several acyl-ACP thioesterases from a variety of organisms that are specific for certain fatty acid chain lengths, and transgenic overexpression of thioesterases can be used to change fatty acid chain length. Expression of a 12:0-biased thioesterase from Umbellularia californica in both A. thaliana and E. coli drastically changed the lipid profiles in these organisms, with a great increase in the production of lauric acid (193, 194). Similarly, a 14:0-biased thioesterase from Cinnamomum camphorum was expressed in A. thaliana and E. coli, greatly increasing the production of myristic acid (208). Both of these thioesterases are obviously interesting candidates for transgenic overexpression in microalgae since their activities could improve the suitability of microalga-derived diesel feedstock.
Fatty acids of even shorter chain lengths can also be used for production of gasoline and jet fuel. It is possible to use hydrocracking to break down longer hydrocarbons into shorter chain lengths that are more suitable as feedstocks for gasoline or jet fuel, but it may also be possible to reduce production costs through genetically engineering microalgae to directly produce these shorter chain lengths. Transgenic overexpression of an 8:0- and 10:0-biased thioesterase from C. hookeriana in canola has had some success in increasing the production of short chain fatty acids (32). Interestingly, combined overexpression of the 8:0/10:0-biased thioesterase and a ketoacyl ACP synthase (KAS), both from C. hookeriana, increases 8 to 10 fatty acids by an additional 30 to 40% (31).
The potential for modification of lipid content in microalgae.
It is reasonable to believe that some of the strategies that result in increased oil seed content in terrestrial plants may be able to increase the lipid content of microalgal cells as well. Many microalgae do not produce large amounts of storage lipids during logarithmic growth. Instead, when they encounter environmental stress, such as a lack of nitrogen, they slow down their proliferation and start producing energy storage products, such as lipids and/or starch (75). It will be interesting to see how overexpression of lipid synthesis pathway genes will affect microalgal proliferation. It may be that increased lipid synthesis will result in a reduction of cell division. In such a case, overexpression of lipid synthesis genes may still be beneficial if they can be controlled by an inducible promoter that can be activated once the microalgal cells have grown to a high density and have entered stationary phase. Examples of inducible promoters in algae include copper-responsive elements in C. reinhardtii (152) and a nitrate-responsive promoter in diatoms (148). Inhibiting lipid catabolism may also cause problems with proliferation and biomass productivity since microalgae often rely on catabolic pathways to provide energy and precursors for cell division.
DIRECT BIOLOGICAL SYNTHESIS OF BIOFUELS
Industrial methods for the production of biofuels using energy-rich carbon storage products, such as sugars and lipids, are well established and are currently being used on a large scale in the production of bioethanol from corn grain and biodiesel from oil seed crops. However, it might be possible to introduce biological pathways in microalgal cells that allow for the direct production of fuel products that require very little processing before distribution and use. Several biological pathways have been described for the production of fatty acid esters, alkanes, and alcohols. However, the introduction of metabolic pathways for the direct production of fuels faces many challenges. The product yields for pathways that lead to the accumulation of compounds that are not necessarily useful for the cell are unlikely to be economically viable without comprehensive engineering of many aspects of microalgal metabolism. In addition, many types of fuel products have the potential to be toxic, and tolerant species of microalgae may have to be generated.
Fatty acid ester production.
Triacylglycerols can be used for the production of biodiesel through the creation of fatty acid esters. Microalgal lipids can also be used to produce a “green” or renewable diesel through the process of hydrotreating. However, these transformations require additional energy carriers (e.g., methanol or hydrogen) and chemical processing, which increases the cost of biofuel production. Every production step that can be transferred to biological pathways will likely improve the overall economics. An interesting example is the in vivo conversion of fatty acids to fuel by the simultaneous overexpression of the ethanol production genes from Zymomonas mobilis and the wax ester synthase/acyl-CoA-diacylglycerol acyltransferase (WS/DGAT) gene from the Acinetobacter baylyi strain ADP1 in E. coli, which resulted in the synthesis of fatty acid ethyl esters that could be used directly as biodiesel (86). Although the ethyl ester yield from this manipulation was not overly impressive for E. coli, it will be interesting to see if higher productivities can be achieved and what effect fatty acid ester accumulation has on microalgal growth.
Straight-chain alkanes.
Short- and medium-chain alkanes have the potential to be used directly as transportation fuel. Since alkanes can be derived from fatty acids, microalgae that are good lipid producers could perhaps be genetically transformed to produce alkanes. This conversion relies on the serial transformation of fatty acids to aldehydes and then to alkanes. The last step is thought to be catalyzed by a decarbonylase enzyme; however, no functional decarbonylase enzyme has been cloned to date, and the actual mechanism for the conversion of aldehyde to alkane remains to be found. Interestingly, a suggested decarbonylase enzyme involved in alkane production has been studied with the green microalga B. braunii, which has the ability to produce very-long-chain alkanes (35). Strains of B. braunii differ in which long-chain hydrocarbons are synthesized, with strain A producing very-long-chain dienes and trienes, while strain B produces very-long-chain triterpenoid hydrocarbons (117). Decarbonylase activity has also been found in the leaves of the pea Pisum sativum (20, 164, 192), and several possible decarbonylases that are thought to be involved in wax formation, including Cer1 and Cer22, have been found in A. thaliana (1, 155). The alkanes that are generated by these putative decarbonylases all have very-long-chain lengths (>22 carbons) and will require further processing for fuel production. A possible example of long-chain-alkane (14- to 22-carbon) production has been reported for the bacterium Vibrio furnissii (137); however, a more recent study disputed these claims (195). Production of shorter-chain-length alkanes that are suitable for direct use as fuel remains an existing goal, and further research is needed to clarify how alkanes are generated and to reveal the precise enzymes involved.
Ethanol, butanol, isopropanol, and other longer-chain alcohols.
Ethanol for biofuels is currently produced from the fermentations of food starches or cellulose-derived sugars. Algal starches have been shown to be fermentable by yeast (129), but an approach to directly couple ethanol production to photosynthetic carbon fixation in situ may be preferred. Many microalgae have fermentative metabolic pathways to ethanol, but to couple ethanol production to photoautotrophic metabolism will require changes in regulatory pathways or the insertion of new metabolic pathways. With cyanobacteria, the creation of a pathway for ethanol biosynthesis has been demonstrated, with the insertion of pyruvate decarboxylase and alcohol dehydrogenase from the ethanologenic bacterium Z. mobilis (34, 40). This pathway produces ethanol during photoautotrophic growth and could be incorporated into algae; however, these enzymes are not optimized for performance in oxic conditions and may need to be configured for eukaryotic systems.
As a fuel, ethanol has a lower energy density than gasoline and poor storage properties. Longer-chain alcohols C3 to C5 have higher energy densities that are similar to those of gasoline and are easier to store and transport than ethanol. Recently, many exciting advances toward the biological production of C3 up to C8 alcohols have been achieved. Isopropanol and butanol are naturally produced by bacteria of the genus Clostridium, and production has been industrialized using Clostridium acetobutylicum. Because C. acetobutylicum has a low growth rate and is somewhat difficult to genetically engineer, attempts have been made to express the production pathways for isopropanol and butanol in the more user-friendly host E. coli. For isopropanol production, several combinations of up to five genes from various species of Clostridium were overexpressed in E. coli, resulting in the production of 4.9 g/liter of isopropanol (67). In a similar fashion, six genes encoding the entire pathway for butanol production were transferred from C. acetobutylicum into E. coli by Atsumi et al. (7). With optimization, the overexpression of the butanol production pathway resulted in 1-butanol production of approximately 140 mg/liter. Interestingly, yields were greatly improved by the deletion of 5 host genes that compete with the 1-butanol pathway for acetyl-CoA and NADH, resulting in the production of 550 mg/liter (7). A further increase in production was achieved through the expression of the entire pathway from a single plasmid, resulting in production of 1.2 g/liter 1-butanol (78). Production of biofuels through transgenic overexpression of entire production pathways can cause problems for the host organism when the nonnative enzymes interfere with the host's normal metabolism. An alternative synthetic pathway for the production of butanol utilized the endogenous keto acid pathways for amino acid synthesis. These ubiquitous pathways normally produce amino acids through 2-keto acid precursors. Atsumi et al. proved that it is possible to divert some of the 2-keto acid intermediates from amino acid production into alcohol production, especially that of isobutanol, which was produced at titers up to 22 g/liter (8). This was achieved through the simultaneous transgenic overexpression of a 2-keto-acid decarboxylase and an alcohol dehydrogenase. Using similar approaches, it is possible to obtain longer-chain alcohols as well, and up to C8 alcohols have been synthesized (for a review by Connor and Liao, see reference 28). Some of the 2-keto acid intermediates, such as 2-ketobutyrate, are conserved in microalgae, and it is therefore reasonable to believe that a similar approach to production of alcohols in algae is possible.
Isoprenoids.
Isoprenoids, also known as terpenoids, represent an incredibly diverse group of natural compounds, with more than 40,000 different molecules. In microalgae, isoprenoids are synthesized via the methylerythritol (MEP) pathway using glyceraldehydes-3-phosphate and pyruvate to generate the basic building blocks of isoprenoid biosynthesis, isopentyl diphosphate (IPP), and dimethylallyl diphosphate (DMAPP). Molecules that could potentially work as gasoline substitutes, including isopentenol, have been produced by E. coli using isoprenoid biosynthesis pathways. Two enzymes from Bacillus subtilis that utilize IPP and DMAPP for the biosynthesis of isopentenol were overexpressed in E. coli, resulting in production of 112 mg/liter isopentenol (107, 200). Other compounds that could replace diesel and jet fuel can also be produced through isoprenoid pathways (for recent reviews by Fortman et al. and Lee et al., see references 53 and 98, respectively).
Feasibility of direct biological synthesis of fuels.
There are many examples of successful pathway manipulation to generate compounds that can be used as fuels. Most of these come from the manipulation of E. coli, in which the overexpression and deletion of entire metabolic pathways are feasible. With advances in genetic transformation methods and increased knowledge regarding expression systems in microalgae, comprehensive modifications, such as those performed with E. coli, may be attempted. It is reasonable to believe that some of the biochemical pathways in microalgae could be leveraged for the direct production of fuels. Considering the reported yields from the various pathways that have been utilized so far, the production of isobutanol through the keto acid pathways should be carefully considered for microalgal systems.
SECRETION OF TRIACYLGLYCEROL, ALKANES, FREE FATTY ACIDS, AND WAX ESTERS
It is believed that among the most costly downstream processing steps in fuel production using microalgal feedstocks are the harvesting/dewatering steps and the extraction of fuel precursors from the biomass. Based on currently achievable productivities, most microalgae will not grow to a density higher than a few grams of biomass per liter of water. While there are several possible low-cost solutions to concentrating the biomass, including settling and flocculation, these methods are slow and the resulting biomass may still require further dewatering. Alternative methods to concentrate algal biomass include centrifugation and filtration, which are faster, but they are also typically much more expensive and energy intensive. In addition, many microalgal species have a very tough outer cell wall that makes extraction of fuel feedstocks difficult, thereby requiring the use of harsh lysis conditions. One possible solution is to manipulate the biology of microalgal cells to allow for the secretion of fuels or feedstocks directly into the growth medium. There are in fact several pathways in nature that lead to secretion of hydrophobic compounds, including TAGs, free fatty acids, alkanes, and wax esters.
Secretion of free fatty acids in yeast.
As mentioned in the section on lipid catabolism, inactivation of genes involved in β-oxidation has been shown to result in fatty acid secretion in some instances. These genes were identified to have a function in fatty acid secretion through the use of a screening method wherein mutated yeast colonies were overlaid with an agar containing a fatty acid auxotrophic yeast strain that requires free fatty acids in order to proliferate. Mutant colonies that secrete fatty acids were thereby identified by the formation of a halo in the overlaid agar (120, 132). Similar screening methods could be utilized to identify microalgae that have the ability to secrete fatty acids. In one of the yeast studies, random mutagenesis resulted in the secretion of TAGs. Unfortunately, neither the genes involved nor the mechanism has been described (132). S. cerevisiae has five genes with fatty acyl-CoA synthetase activity, including those encoding FAA1 and FAA4. Combined inactivation of FAA1 and FAA4, or FAA1 together with acyl-CoA oxidase activity, results in a buildup of intracellular free fatty acids and secretion of free fatty acids. Importantly, the highest levels of fatty acid secretion also seemed to be associated with reduced proliferation (120). This kind of secretion was found to take place mainly during logarithmic growth, whereas the cells started importing free fatty acids in stationary phase (162). The actual mechanisms for TAG and/or free fatty acid secretion in S. cerevisiae in these cases are not known. It is possible that any manipulation that allows yeast cells to accumulate high levels of intracellular free fatty acids will result in secretion of free fatty acids, and it may be possible to reproduce this kind of secretion in microalgae. A similar form of fatty acid secretion has been achieved by Synthetic Genomics with cyanobacteria (158). To improve the rates of secretion and to allow for the secretion of other types of bioenergy carriers, it could be beneficial to investigate some of the efficient mechanisms that exist for the secretion of fatty acids and related compounds in other organisms, which could be transferred to microalgae.
Mechanisms for secretion of lipids and related compounds.
There are several examples of established pathways for the secretion of lipophilic compounds. These include the secretion of TAG-containing very-low-density lipid (VLDL) vesicles from hepatocytes, TAG-containing vesicles from mammary glands, and the ATP-binding cassette (ABC) transporter-mediated export of plant waxes, which consist of many types of hydrocarbons. In addition to cellular export pathways, there are also known pathways for intracellular transport of fatty acids between organelles, including import of fatty acids into mitochondria and peroxisomes for β-oxidation, and it may be possible to utilize such pathways for the export of lipids. Several key genes are known for these pathways, and transgenic expression of ABC transporters has been used to enable drug transport, resulting in resistance. However, the successful transgenic expression and utilization of lipid secretion pathways to secrete molecules suitable for biofuel production remain largely to be demonstrated.
Even though many of the genes that are involved in secretion have been identified, the exact mechanisms are generally not known. For example, the secretion of VLDL from hepatocytes has been shown to be affected both by deletion and overexpression of a wide range of genes, such as those encoding apolipoprotein E (ApoE), microsomal triglyceride transfer protein (MTP), triacylglycerol hydrolase (TGH), and arylacetamide deacetylase (AADA) (62, 99, 189, 190). Overexpression of these genes in hepatocytes that already have the capacity to secrete VLDL results in increased secretion; however, it is not known which genes would be needed to enable VLDL secretion from cell types that do not normally secrete VLDL. In a similar fashion, several genes have been identified that are involved in milk TAG vesicle secretion. These genes include those that encode adipophilin (ADPH), xanthine oxidoreductase (XOR), and butyrophilin (BTN) (for a review by McManaman et al., see reference 111). But again, the exact genes that would be needed to enable a functional secretion pathway in another cell type are not known. With further research, it should become clear whether the transfer of these very efficient TAG-secreting pathways is feasible.
What is perhaps a more straightforward approach to enabling secretion of lipids from microalgae is the use of ABC transporters. ABC transporters mediate the export of plant waxes consisting of a multitude of compounds that are derived from very-long-chain fatty acids, including alkanes, ketones, alcohols, aldehydes, alkyl esters, and fatty acids. In A. thaliana there are over 120 different ABC transporters, and in addition to transporting compounds that are related to very-long-chain fatty acids, they are also responsible for the transport of molecules that are produced through the isoprenoid synthesis pathway, including terpenoids and other compounds that are of interest for biofuel production. As with VLDL and milk TAG secretion, the exact mechanisms are not known, but the ABC transporter systems may rely on fewer components, and transgenic expression of ABC transporters has resulted in transport of a variety of compounds, including kanamycin, cholesterol, and sterols (82, 115, 205). Of particular interest are ABC transporters that have been shown to have the ability to transport plant wax components that are derived from very-long-chain fatty acids. These transporters include the A. thaliana Desperado/AtWBC11 transporter and the Cer5/AtWBC12 transporter, both of which have been shown to be important for exporting wax to the epidermis (136, 140). Long-chain fatty acids are imported into the peroxisome for β-oxidation by ABC transporters, and inactivation of the ABC transporter Ped3p or Pxa1 in A. thaliana inhibits peroxisomal uptake of long-chain fatty acids (70, 216). One interesting characteristic of ABC transporters is their promiscuous gating properties. For example, Cer5/AtWBC12 facilitates the export of very-long-chain aldehydes, ketones, alcohols, alkanes, and perhaps fatty acids (140). While wax transporters have not been shown to export products that are derived from medium-chain fatty acids, it would perhaps be possible to use random mutagenesis or directed evolution to generate mutant ABC transporters that have the ability to secrete short- and medium-chain fatty acids.
In summary, lipid secretion is an attractive alternative to harvesting algal biomass that could potentially lower the cost of producing microalga-derived biofuels. However, the current knowledge of secretion pathways is still rather limited and therefore may not necessarily be easily transferred to microalgal cells. In addition, a secretion strategy may not be the best solution when a significant number of contaminating microorganisms are present in the cultivation system. The secretion of the fuel intermediates into the culture medium would provide these microorganisms with a rich source of nutrient, thereby reducing product yields. It is currently possible to induce secretion of free fatty acids from yeast and cyanobacteria, but it remains to be demonstrated at a scale significant for biofuel production from microalgal feedstocks. Since there are several highly efficient pathways for lipid secretion in nature that are being explored with various model organisms, research efforts should be aimed at transferring these pathways into organisms that are good lipid producers.
GENETIC MODIFICATION OF CARBOHYDRATE METABOLISM
Carbohydrates can be metabolized into a variety of biofuels, including ethanol, butanol, H2, lipids, and/or methane. Glucans are stored in microalgae in a variety of ways. The phyla Chlorophyta, Dinophyta, Glaucophyta, and Rhodophyta store glucans in linear α-1,4 and branched α-1,6 glycosidic linkages (10). In Heterokontophyta, Phaeophyceae, and Bacillariophyceae, water-soluble granules of laminarin and chrysolaminarin are synthesized, which are made up of β-1,3 linkages with branching at the C-2 and C-6 positions of glucose (74). In green algae, starch is synthesized and stored within the chloroplast, while it is stored in the cytoplasm in Dinophyta, Glaucophyta, and Rhodophyta and in the periplastidial space in Cryptophyceae (38, 39).
Genetic strategies for increasing glucan storage.
The rate-limiting step of starch synthesis (see Fig. 3 for an overview of starch metabolism) is the ADP-glucose pyrophosphorylase (AGPase)-catalyzed reaction of glucose 1-phosphate with ATP, resulting in ADP-glucose and pyrophosphate (176). AGPase is typically a heterotetramer composed of large regulatory and small catalytic subunits and is allosterically activated by 3-phosphoglyceric acid (3-PGA), linking starch synthesis to photosynthesis (209). Polysaccharides are often found surrounding the pyrenoid in microalgae, likely because it is a source of 3-PGA (10).
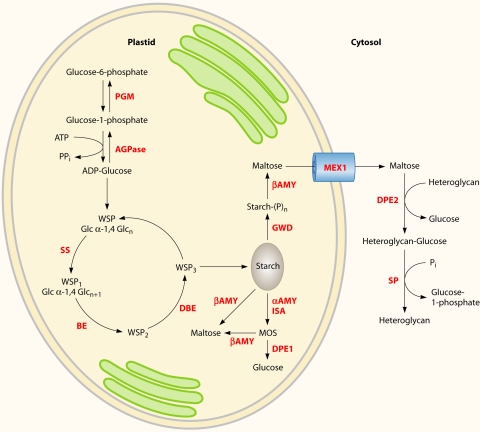
Fig. 3.
Starch metabolism in green microalgae. The metabolites and simplified representative pathways in microalgal starch metabolism are shown in black, and enzymes are shown in red. Glucans are added to the water soluble polysaccharide (WSP) by α-1,4 glycosidic linkages (WSP1) until a branching enzyme highly branches the ends (WSP2). Some of these branches are trimmed (WSP3), and this process is repeated until a starch granule is formed. Phosphorolytic [Starch-(P)n] and hydrolytic degradation pathways are shown. αAMY, α-amylase; AGPase, ADP-glucose pyrophosphorylase; βAMY, β-amylases; BE, branching enzymes; DBE, debranching enzymes; DPE, disproportionating enzyme (1 and 2) α-1,4 glucanotransferase; Glc, glucose; GWD, glucan-water dikinases; ISA, isoamylases; MEX1, maltose transporter; MOS, malto-oligosaccharides; PGM, plastidial phosphoglucomutase; P, phosphate; Pi, inorganic phosphate; PPi, pyrophosphate; SP, starch phosphorylases; SS, starch synthases.
Much work has been done on the catalytic and allosteric properties of AGPases in crop plants to increase starch production, with mixed results (174). Designer AGPases, such as those encoded by glgC16 from E. coli (176) or the recombinant rev6 (63), that have successfully increased starch content in other plants should be expressed in microalgae, preferably in a background with no native AGPase activity, such as the C. reinhardtii sta1 or sta6 mutant. An alternative approach for increasing microalgal starch would be to introduce starch-synthesizing enzymes into the cytosol. The cytosol would give more physical space for the starch granules to accumulate (174). A problem for cytosolic starch synthesis could be that because the AGPase is far from the pyrenoid and thus 3-PGA, it may not be activated. This problem could be circumvented by the introduction of an AGPase that does not require 3-PGA, such as the Mos(1-198)/SH2 AGPase, which still has activity even without the presence of an activator (14).
Decreasing starch degradation in microalgae.
The precise mechanisms of starch catabolism in green microalgae are largely unknown (10) but are more widely understood for A. thaliana (175). Starch can be degraded by hydrolytic and/or phosphorolytic mechanisms. Hydrolytic starch degradation requires an enzyme capable of hydrolyzing semicrystalline glucans at the surface of the insoluble starch granule. In A. thaliana, α-amylase (AMY3) is thought to participate in starch degradation, and a homologous protein is found in C. reinhardtii (175). Interestingly, starch can be degraded even when all three α-amylases in A. thaliana have been knocked out, indicating alternative mechanisms for starch degradation (207). Plastidial starch degradation is stimulated by phosphorylation of glucose residues at the root of amylopectin by glucan-water dikinases (GWD). GWD catalyzes the transfer of β-phosphate in ATP to the C-6 position of the glucans in amylopectin (156). The C-3 position in the glucan can also be phosphorylated by the phosphoglucan water dikinases (PWD), and both of these phosphorylations are thought to help disrupt the crystalline structure of the starch granules to allow glucan-metabolizing enzymes access (212). In A. thaliana, the disruption of the GWD (sex1 phenotype) results in starch levels that are four to six times higher than those in wild-type leaves (206), while disruptions in PWD result in a less severe starch excess phenotype (156). These phosphorylation steps are critical for starch degradation and are excellent gene knockout targets for a starch accumulation phenotype in microalgae.
Secretion strategies and soluble sugars.
Many microalgae have the native ability to secrete fixed carbon products. Mannitol, arabinose, glutamic acid, proline, glycerol, lysine, aspartic acid, and various polysaccharides have been reported to be secreted (71). Ankistrodesmus densus secretes polysaccharides when exposed to light, even during stationary phase (138). Although little is currently known about these secretion events, a further understanding of their regulation and mechanism could potentially be leveraged for continuous biofuel production from secreted saccharides.
The production of soluble sugars may be preferred over polysaccharides because soluble sugars are smaller and easier to process, in addition to likely being more amenable for engineered secretion because many transporters have been described. Maltose, a product of starch degradation in the chloroplast, is transported to the cytosol in green microalgae and land plants by the maltose export protein (MEX1) (38). This protein facilitates bidirectional diffusion and could be leveraged to export maltose out of the cell. Although sucrose has largely been unexplored with microalgae, evidence exists that some of the enzymes involved in sucrose metabolism, such as the sucrose synthetase and sucrose phosphate synthetase, are present (46). The synthesis of sucrose could be exploited by sucrose transporters, such as SUC1 and SUC2 found in A. thaliana, for extracellular excretion. S. cerevisiae cells transformed with SUC1 and SUC2 have been shown to transport sucrose and some maltose across their plasma membranes (160).
In addition to exporting soluble sugars, intracellular sugar accumulation is also a desirable microalgal biofuel trait. Maltose is metabolized in the cytosol by a glucosyltransferase, DPE2, but when it is knocked out in Arabidopsis, it results in a 30-fold increase of maltose in the leaves, enough to cause maltose exportation to the roots, where the concentration is doubled. In addition, when the MEX1 transporter is knocked out there is at least 40 times more maltose in the leaves of the Arabidopsis mutant than in the wild type (103). In sugar cane, an increase in total sugar production has been accomplished by the transgenic expression of a sucrose isomerase from a bacterium. This isomerase converts sucrose to isomaltulose, a nonplant metabolite, and as a result, the total sugar levels of isomaltulose and sucrose are twice as high as those of control plants. This may imply that there is a signaling system that gives negative feedback when sucrose levels reach a certain level, but when sucrose is converted to isomaltulose, the level of sucrose is not detected, allowing for higher levels of total sugar accumulation (202). In microalgae, maltose could be converted to other isoforms that may be silent to the native metabolic regulatory system, which could result in an increase in total sugar content. For example, a glucosyltransferase from a bacterium converting maltose to trehalose (131) could be expressed as a potential strategy to increase total sugar content. A potential side effect of increased trehalose is its ability to induce starch synthesis and AGPase expression, which has been shown with Arabidopsis (199).
Mutant considerations.
Mutants that synthesize less starch or have a reduced capacity to degrade starch often have reduced growth rates (175). A. thaliana mutants grown in a diurnal cycle that cannot synthesize or degrade starch grow more slowly than the wild type (17, 18, 213). An A. thaliana plastidial phosphoglucomutase mutant (corresponding to STA5 in C. reinhardtii) had more of a reduced growth rate during short daylight periods compared to long daylight periods, but at continuous light its growth rates were equal (17). These considerations may become issues when microalgae are grown for biofuels and are subject to diurnal light cycles.
MICROALGAL HYDROGEN PRODUCTION
Many eukaryotic microalgae and cyanobacteria have evolved in ecosystems that become depleted of O2, especially during the night, and diverse fermentation metabolisms that can be leveraged in renewable bioenergy strategies are present in these organisms (64, 118, 125). Of particular interest is the ability of many green microalgae to produce H2; however, it should be noted that additional fermentation metabolites, including organic acids and alcohols, are also secreted by many species during anoxia (118, 143). Hydrogen metabolism has been studied extensively with C. reinhardtii (61, 68, 72), resulting in significant advances in both our fundamental understanding of H2 metabolism in this organism (43, 110, 143) and in improvements in overall H2 yields (90, 91, 114). Hydrogenases are classified according to the metal ions at their active sites, and the [NiFe] and [FeFe] hydrogenases are capable of the reversible reduction of protons to H2. These two enzyme classes are phylogenetically distinct, and interestingly, only the [FeFe] hydrogenases have been described in eukaryotic microalgae; whereas, only the [NiFe] hydrogenases have been reported for cyanobacteria. The [FeFe] hydrogenases in many green microalgae are able to effectively couple to the photosynthetic electron transport chain at the level of ferredoxin, providing the means to generate H2 directly from water oxidation. However, all microalgal [FeFe] hydrogenases characterized to date are particularly O2 sensitive, and H2 photoproduction is only transiently observed prior to the accumulation of O2 to inhibitory levels under nutrient-replete conditions (27, 178). In 2000, Melis and coworkers described the use of sulfur deprivation (203), which decreases photosynthetic activity, as an effective means of sustaining H2 photoproduction when respiration is able to consume all of the photosynthetic O2 produced by the cells (114). Recently, it was demonstrated that alginate-immobilized cultures of nutrient-deprived C. reinhardtii could sustain H2 photoproduction even in the presence of an oxygenated atmosphere (90). Genetic techniques have been applied with the aim of increasing H2 photoproduction activity by decreasing light-harvesting antenna size, inhibiting state transitions, and hydrogenase engineering (11, 60, 68, 112). In combination with physiological and biochemical approaches, these studies have rapidly advanced our understanding of H2 metabolism and enzyme maturation (13, 145) in green microalgae, and numerous strategies are emerging to further advance our ability to optimize H2 production in eukaryotic phototrophs.
IMPROVED GROWTH CAPACITY THROUGH INCREASED STRESS TOLERANCE OR INCREASED PHOTOSYNTHETIC EFFICIENCY
The production of any biofuel is dependent on the efficiency of the metabolic pathways that lead to accumulation of storage compounds, such as lipids and starch, as well as on the ability of microalgae to rapidly produce large amounts of biomass. Experiments with small- and large-scale microalgal photobioreactors and molecular research in photosynthetic efficiency have revealed several factors that can limit biomass accumulation. These include stress factors, such as salt concentration, temperature, pH, and light intensity. Depending on the design of the cultivation facilities, it is possible to control these factors to a certain degree through engineering and manipulation of the growth environment, but these manipulations add to the cost of growing microalgae. Therefore, it would be of great benefit to develop genetic strategies to increase the cellular tolerance to a variety of stress factors.
High-light stress.
One important consideration is the intensity of light at which a certain strain of microalga reaches its maximum growth rate; this intensity, which corresponds to the maximum photosynthetic efficiency, is usually around 200 to 400 μmol photons m−2 s−1 for most species. Light intensities above the maximum photosynthetic efficiency actually reduce the growth rate, a phenomenon known as photoinhibition. Photosynthetically active radiation intensities from sunlight can exceed 2,000 μmol photons m−2 s−1 during midday (113). Consequently, most microalgae will not grow at maximum efficiency during most of the day. Microalgae are considered great model organisms to study photosynthetic efficiency, and several attempts have been made to improve the photosynthetic efficiency and/or reduce the effects of photoinhibition on microalgal growth. Much of this work has been focused on reducing the size of the chlorophyll antenna or lowering the number of light-harvesting complexes to minimize the absorption of sunlight by individual chloroplasts (97, 126–128, 141, 142, 188). This approach may seem counterintuitive, but this strategy may have two positive effects; first, it permits higher light penetration in high-density cultures, and second, it can allow a higher maximum rate of photosynthesis due to the fact that the cells are less likely to be subjected to photoinhibition since their light-harvesting complexes absorb less light (for an excellent review of the subject by Melis, see reference 113). Earlier research relied on random mutagenesis strategies to generate mutants with fewer or smaller chlorophyll antennae, but a recent publication efficiently used an RNAi-based strategy to knock down both LHCI and LHCII in C. reinhardtii (126). This strategy can most likely be applied to many different microalgae more easily than a random mutagenesis approach. It seems clear that manipulation of light-harvesting complexes can lead to increased biomass productivity under high light in controlled laboratory conditions. However, it remains to be seen how well these mutants will perform in larger-scale cultures with more varied conditions and perhaps with competition from wild invasive microalgal species. In one study of algal antenna mutants, no improvement in productivity was observed with outdoor ponds (77). However, they also did not observe any improved productivity in laboratory cultures. With more research, it should become clear whether the current approach can be successfully applied to increase biomass production.
Other stress factors.
High light is not the only environmental variable that can cause stress and inhibit microalgal growth. Salt, pH, temperature, and other stimuli can also cause stress to microalgal cultures. Many genes have been identified that are important for withstanding stress conditions. These include genes that are directly involved in scavenging reactive oxygen, such as those encoding glutathione peroxidase and ascorbate peroxidase (168, 182, 204) as well as enzymes that catalyze the production of osmolytes, such as mannitol and ononitol (166, 167, 185), and interestingly, an ATP synthase subunit that is involved in the regulation of intracellular ATP levels and stress tolerance (179). The antistress properties of these genes isolated from bacteria as well as a marine stress-tolerant microalga (Chlamydomonas sp. W80) were demonstrated through transgenic overexpression in several different systems, including tobacco and E. coli, which resulted in increased resistance to several different stressful stimuli, including high salt and low temperature. It is likely that similar improvements can be achieved with microalgae. An additional benefit of increasing stress tolerance is the possibility of growing select microalgae under extreme conditions that limit the proliferation of invasive species.
CONCLUSION
Microalgae are an extremely diverse group of organisms, many of which possess novel metabolic features that can be exploited for the production of renewable biofuels. These include (i) high photosynthetic conversion efficiencies, (ii) rapid biomass production rates, (iii) the capacity to produce a wide variety of biofuel feedstocks, and (iv) the ability to thrive in diverse ecosystems. Although microalgae have long been considered a promising platform for the production of biofuels, earlier studies concluded that the economics of microalgal biofuel production needed to be significantly improved. In contrast to these previous efforts, we are now equipped with a wide variety of new genetic tools, genome sequences, and high-throughput analytical techniques that will allow scientists to analyze and manipulate metabolic pathways with unprecedented precision. Promising advances in metabolic engineering allow for not only the increased production of endogenous carbon storage compounds, such as TAGs and starch, but also the direct production, and perhaps secretion, of designer hydrocarbons that may be used directly as fuels. The application of these modern metabolic engineering tools in photosynthetic microalgae has the potential to create important sources of renewable fuel that will not compete with food production or require fresh water and arable land.
ACKNOWLEDGMENTS
We acknowledge support from the Air Force Office of Scientific Research grant FA9550-05-1-0365 and the Office of Biological and Environmental Research, GTL program, Office of Science, U.S. Department of Energy.
Footnotes
Published ahead of print on 5 February 2010.
REFERENCES
- 1.Aarts M. G. M., Keijzer C. J., Stiekema W. J., Pereira A. 1995. Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7:2115–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apt K. E., Grossman A. R., Kroth-Pancic P. G. 1996. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 252:572–579 [DOI] [PubMed] [Google Scholar]
- 3.Apt K. E., Zaslavkaia L., Lippmeier J. C., Lang M., Kilian O., Wetherbee R., Grossman A. R., Kroth P. G. 2002. In vivo characterization of diatom multipartite plastid targeting signals. J. Cell Sci. 115:4061–4069 [DOI] [PubMed] [Google Scholar]
- 4.Archibald J. M., Rogers M. B., Toop M., Ishida K., Keeling P. J. 2003. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc. Natl. Acad. Sci. U. S. A. 100:7678–7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armbrust E. 1999. Identification of a new gene family expressed during the onset of sexual reproduction in the centric diatom Thalassiosira weissflogii. Appl. Environ. Microbiol. 65:3121–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armbrust E. V., Berges J. A., Bowler C., Green B. R., Martinez D., Putnam N. H., Zhou S., Allen A. E., Apt K. E., Bechner M., Brzezinski M. A., Chaal B. K., Chiovitti A., Davis A. K., Demarest M. S., Detter J. C., Glavina T., Goodstein D., Hadi M. Z., Hellsten U., Hildebrand M., Jenkins B. D., Jurka J., Kapitonov V. V., Kroger N., Lau W. W., Lane T. W., Larimer F. W., Lippmeier J. C., Lucas S., Medina M., Montsant A., Obornik M., Parker M. S., Palenik B., Pazour G. J., Richardson P. M., Rynearson T. A., Saito M. A., Schwartz D. C., Thamatrakoln K., Valentin K., Vardi A., Wilkerson F. P., Rokhsar D. S. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79–86 [DOI] [PubMed] [Google Scholar]
- 7.Atsumi S., Cann A. F., Connor M. R., Shen C. R., Smith K. M., Brynildsen M. P., Chou K. J. Y., Hanai T., Liao J. C. 2008. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10:305–311 [DOI] [PubMed] [Google Scholar]
- 8.Atsumi S., Hanai T., Liao J. C. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89 [DOI] [PubMed] [Google Scholar]
- 9.Bachvaroff T. R., Concepcion G. T., Rogers C. R., Herman E. M., Delwiche C. F. 2004. Dinoflagellate expressed sequence tag data indicate massive transfer of chloroplast genes to the nuclear genome. Protist 155:65–78 [DOI] [PubMed] [Google Scholar]
- 10.Ball S. G., Deschamps P. 2009. Starch metabolism, p. 1–40InHarris E. H., Stern D. B. (ed.), The Chlamydomonas sourcebook, second edition: organellar and metabolic processes , vol. 2, Academic Press, Oxford, England [Google Scholar]
- 11.Beckmann J., Lehr F., Finazzi G., Hankamer B., Posten C., Wobbe L., Kruse O. 2009. Improvement of light to biomass conversion by de-regulation of light-harvesting protein translation in Chlamydomonas reinhardtii. J. Biotechnol. 142:70–77 [DOI] [PubMed] [Google Scholar]
- 12.Berthold P., Schmitt R., Mages W. 2002. An engineered Streptomyces hygroscopicus aph 7 gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153:401–412 [DOI] [PubMed] [Google Scholar]
- 13.Bock A., King P. W., Blokesch M., Posewitz M. C. 2006. Maturation of hydrogenases. Adv. Microb. Physiol. 51:1–71 [DOI] [PubMed] [Google Scholar]
- 14.Boehlein S. K., Shaw J. R., Stewart J. D., Hannah L. C. 2009. Characterization of an autonomously activated plant ADP-glucose pyrophosphorylase. Plant Physiol. 149:318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowler C., Allen A. E., Badger J. H., Grimwood J., Jabbari K., Kuo A., Maheswari U., Martens C., Maumus F., Otillar R. P., Rayko E., Salamov A., Vandepoele K., Beszteri B., Gruber A., Heijde M., Katinka M., Mock T., Valentin K., Verret F., Berges J. A., Brownlee C., Cadoret J. P., Chiovitti A., Choi C. J., Coesel S., De Martino A., Detter J. C., Durkin C., Falciatore A., Fournet J., Haruta M., Huysman M. J., Jenkins B. D., Jiroutova K., Jorgensen R. E., Joubert Y., Kaplan A., Kroger N., Kroth P. G., La Roche J., Lindquist E., Lommer M., Martin-Jezequel V., Lopez P. J., Lucas S., Mangogna M., McGinnis K., Medlin L. K., Montsant A., Oudot-Le Secq M. P., Napoli C., Obornik M., Parker M. S., Petit J. L., Porcel B. M., Poulsen N., Robison M., Rychlewski L., Rynearson T. A., Schmutz J., Shapiro H., Siaut M., Stanley M., Sussman M. R., Taylor A. R., Vardi A., von Dassow P., Vyverman W., Willis A., Wyrwicz L. S., Rokhsar D. S., Weissenbach J., Armbrust E. V., Green B. R., Van de Peer Y., Grigoriev I. V. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244 [DOI] [PubMed] [Google Scholar]
- 16.Casas-Mollano J. A., Rohr J., Kim E. J., Balassa E., van Dijk K., Cerutti H. 2008. Diversification of the core RNA interference machinery in Chlamydomonas reinhardtii and the role of DCL1 in transposon silencing. Genetics 179:69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caspar T., Huber S. C., Somerville C. 1985. Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 79:11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caspar T., Lin T.-P., Kakefuda G., Benbow L., Preiss J., Somerville C. 1991. Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol. 95:1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerutti H., Johnson A. M., Gillham N. W., Boynton J. E. 1997. A eubacterial gene conferring spectinomycin resistance on Chlamydomonas reinhardtii: integration into the nuclear genome and gene expression. Genetics 145:97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheesbrough T. M., Kolattukudy P. E. 1984. Alkane biosynthesis by decarbonylation of aldehydes catalyzed by a particulate preparation from Pisum sativum. Proc. Natl. Acad. Sci. U. S. A. 81:6613–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H. L., Li S. S., Huang R., Tsai H. J. 2008. Conditional production of a functional fish growth hormone in the transgenic line of Nannochloropsis oculata (Eustigmatophyceae). J. Phycol. 44:768–776 [DOI] [PubMed] [Google Scholar]
- 22.Chen Y., Wang Y., Sun Y., Zhang L., Li W. 2001. Highly efficient expression of rabbit neutrophil peptide-1 gene in Chlorella ellipsoidea cells. Curr. Genet. 39:365–370 [DOI] [PubMed] [Google Scholar]
- 23.Cheney D., Metz B., Stiller J. 2001. Agrobacterium-mediated genetic transformation in the macroscopic marine red alga Porphyra yezoensis. J. Phycol. 37Suppl.:11 [Google Scholar]
- 24.Chepurnov V. A., Mann D. G., Sabbe K., Vyverman W. 2004. Experimental studies on sexual reproduction in diatoms. Int. Rev. Cytol. 237:91–154 [DOI] [PubMed] [Google Scholar]
- 25.Chepurnov V. A., Mann D. G., von Dassow P., Vanormelingen P., Gillard J., Inzé D., Sabbe K., Vyverman W. 2008. In search of new tractable diatoms for experimental biology. Bioessays 30:692–702 [DOI] [PubMed] [Google Scholar]
- 26.Chow K. C., Tung W. L. 1999. Electrotransformation of Chlorella vulgaris. Plant Cell Rep. 18:778–780 [Google Scholar]
- 27.Cohen J., Kim K., Posewitz M., Ghirardi M. L., Schulten K., Seibert M., King P. 2005. Molecular dynamics and experimental investigation of H(2) and O(2) diffusion in [Fe]-hydrogenase. Biochem. Soc. Trans. 33:80–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connor M. R., Liao J. C. 2009. Microbial production of advanced transportation fuels in non-natural hosts. Curr. Opin. Biotechnol. 20:307–315 [DOI] [PubMed] [Google Scholar]
- 29.Crépineau F., Roscoe T., Kaas R., Kloareg B., Boyen C. 2000. Characterisation of complementary DNAs from the expressed sequence tag analysis of life cycle stages of Laminaria digitata (Phaeophyceae). Plant Mol. Biol. 43:503–513 [DOI] [PubMed] [Google Scholar]
- 30.Dawson H. N., Burlingame R., Cannons A. C. 1997. Stable transformation of Chlorella: rescue of nitrate reductase-deficient mutants with the nitrate reductase gene. Curr. Microbiol. 35:356–362 [DOI] [PubMed] [Google Scholar]
- 31.Dehesh K., Edwards P., Fillatti J., Slabaugh M., Byrne J. 1998. KAS IV: a 3-ketoacyl-ACP synthase from Cuphea sp. is a medium chain specific condensing enzyme. Plant J. 15:383–390 [DOI] [PubMed] [Google Scholar]
- 32.Dehesh K., Jones A., Knutzon D. S., Voelker T. A. 1996. Production of high levels of 8:0 and 10:0 fatty acids in transgenic canola by overexpression of Ch FatB 2, a thioesterase cDNA from Cuphea hookeriana. Plant J. 9:167–172 [DOI] [PubMed] [Google Scholar]
- 33.Dehesh K., Tai H., Edwards P., Byrne J., Jaworski J. G. 2001. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 125:1103–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng M.-D., Coleman J. R. 1999. Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol. 65:523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennis M., Kolattukudy P. E. 1992. A cobalt-porphyrin enzyme converts a fatty aldehyde to a hydrocarbon and CO. Proc. Natl. Acad. Sci. U. S. A. 89:5306–5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derelle E., Ferraz C., Rombauts S., Rouze P., Worden A. Z., Robbens S., Partensky F., Degroeve S., Echeynie S., Cooke R., Saeys Y., Wuyts J., Jabbari K., Bowler C., Panaud O., Piegu B., Ball S. G., Ral J. P., Bouget F. Y., Piganeau G., De Baets B., Picard A., Delseny M., Demaille J., Van de Peer Y., Moreau H. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. U. S. A. 103:11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Riso V., Raniello R., Maumus F., Rogato A., Bowler C., Falciatore A. 2009. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 37:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deschamps P., Haferkamp I., d'Hulst C., Neuhaus H. E., Ball S. G. 2008. The relocation of starch metabolism to chloroplasts: when, why and how. Trends Plant Sci. 13:574–582 [DOI] [PubMed] [Google Scholar]
- 39.Deschamps P., Haferkamp I., Dauvillee D., Haebel S., Steup M., Buleon A., Putaux J.-L., Colleoni C., d'Hulst C., Plancke C., Gould S., Maier U., Neuhaus H. E., Ball S. 2006. Nature of the periplastidial pathway of starch synthesis in the cryptophyte Guillardia theta. Eukaryot. Cell 5:954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dexter J., Fu P. 2009. Metabolic engineering of cyanobacteria for ethanol production. Energy Environ. Sci. 2:857–864 [Google Scholar]
- 41.Dismukes G. C., Carrieri D., Bennette N., Ananyev G. M., Posewitz M. C. 2008. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 19:235–240 [DOI] [PubMed] [Google Scholar]
- 42.Doetsch N. A., Favreau M. R., Kuscuoglu N., Thompson M. D., Hallick R. B. 2001. Chloroplast transformation in Euglena gracilis: splicing of a group III twintron transcribed from a transgenic psbK operon. Curr. Genet. 39:49–60 [DOI] [PubMed] [Google Scholar]
- 43.Dubini A., Mus F., Seibert M., Grossman A. R., Posewitz M. C. 2009. Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J. Biol. Chem. 284:7201–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunahay T. G. 1993. Transformation of Chlamydomonas reinhardtii with silicon carbide whiskers. Biotechniques 15:452–455,457–458,460 [PubMed] [Google Scholar]
- 45.Dunahay T. G., Jarvis E. E., Roessler P. G. 1995. Genetic transformation of the diatoms Cyclotella cryptica and Navicula saprophila. J. Phycol. 31:1004–1011 [Google Scholar]
- 46.Duran W. R., Pontis H. G. 1977. Sucrose metabolism in green algae I. The presence of sucrose synthetase and sucrose phosphate synthetase. Mol. Cell Biochem. 16:149–152 [DOI] [PubMed] [Google Scholar]
- 47. Reference deleted.
- 48.Eichler-Stahlberg A., Weisheit W., Ruecker O., Heitzer M. 2009. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 229:873–883 [DOI] [PubMed] [Google Scholar]
- 49.El-Sheekh M. M. 1999. Stable transformation of the intact cells of Chlorella kessleri with high velocity microprojectiles. Biol. Plant. 42:209–216 [Google Scholar]
- 50.Ender F., Godl K., Wenzl S., Sumper M. 2002. Evidence for autocatalytic cross-linking of hydroxyproline-rich glycoproteins during extracellular matrix assembly in Volvox. Plant Cell 14:1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falciatore A., Casotti R., Leblanc C., Abrescia C., Bowler C. 1999. Transformation of nonselectable reporter genes in marine diatoms. Mar. Biotechnol. 1:239–251 [DOI] [PubMed] [Google Scholar]
- 52.Fischer H., Robl I., Sumper M., Kröger N. 1999. Targeting and covalent modification of cell wall and membrane proteins heterologously expressed in the diatom Cylindrotheca fusiformis (Bacillariophyceae). J. Phycol. 35:113–120 [Google Scholar]
- 53.Fortman J. L., Chhabra S., Mukhopadhyay A., Chou H., Lee T. S., Steen E., Keasling J. D. 2008. Biofuel alternatives to ethanol: pumping the microbial well. Trends Biotechnol. 26:375–381 [DOI] [PubMed] [Google Scholar]
- 54.Franklin S., Ngo B., Efuet E., Mayfield S. P. 2002. Development of a GFP reporter gene for Chlamydomonas. Plant J. 30:733–744 [DOI] [PubMed] [Google Scholar]
- 55.Fuhrmann M., Hausherr A., Ferbitz L., Schödl T., Heitzer M., Hegemann P. 2004. Monitoring dynamic expression of nuclear genes in Chlamydomonas reinhardtii by using a synthetic luciferase reporter gene. Plant Mol. Biol. 55:869–881 [DOI] [PubMed] [Google Scholar]
- 56.Fuhrmann M., Oertel W., Hegemann P. 1999. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 19:353–362 [DOI] [PubMed] [Google Scholar]
- 57.Fulda M., Schnurr J., Abbadi A., Heinz E. 2004. Peroxisomal acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16:394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gan S. Y., Qin S., Othman R. Y., Yu D., Phang S. M. 2003. Transient expression of lacZ in particle bombarded Gracilaria changii (Gracilariales, Rhodophyta). J. Appl. Phycol. 15:345–349 [Google Scholar]
- 59.Germain V., Rylott E. L., Larson T. R., Sherson S. M., Bechtold N., Carde J. P., Bryce J. H., Graham I. A., Smith S. M. 2001. Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid beta-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J. 28:1–12 [DOI] [PubMed] [Google Scholar]
- 60.Ghirardi M. L., Dubini A., Yu J., Maness P.-C. 2009. Photobiological hydrogen-producing systems. Chem. Soc. Rev. 38:52–61 [DOI] [PubMed] [Google Scholar]
- 61.Ghirardi M. L., Posewitz M. C., Maness P. C., Dubini A., Yu J., Seibert M. 2007. Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu. Rev. Plant Biol. 58:71–91 [DOI] [PubMed] [Google Scholar]
- 62.Gibbons G. F., Islam K., Pease R. J. 2000. Mobilisation of triacylglycerol stores. Biochim. Biophys. Acta 1483:37–57 [DOI] [PubMed] [Google Scholar]
- 63.Giroux M. J., Shaw J., Barry G., Cobb B. G., Greene T., Okita T., Hannah L. C. 1996. A single mutation that increases maize seed weight. Proc. Natl. Acad. Sci. U. S. A. 93:5824–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grossman A. R., Croft M., Gladyshev V. N., Merchant S. S., Posewitz M. C., Prochnik S., Spalding M. H. 2007. Novel metabolism in Chlamydomonas through the lens of genomics. Curr. Opin. Plant Biol. 10:190–198 [DOI] [PubMed] [Google Scholar]
- 65.Gruber A., Vugrinec S., Hempel F., Gould S. B., Maier U. G., Kroth P. G. 2007. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol. Biol. 64:519–530 [DOI] [PubMed] [Google Scholar]
- 66.Hackett J. D., Scheetz T. E., Yoon H. S., Soares M. B., Bonaldo M. F., Casavant T. L., Bhattacharya D. 2005. Insights into a dinoflagellate genome through expressed sequence tag analysis. BMC Genomics 6:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanai T., Atsumi S., Liao J. C. 2007. Engineered synthetic pathway for isopropanol production in Escherichia coli. Appl. Environ. Microbiol. 73:7814–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hankamer B., Lehr F., Rupprecht J., Mussgnug J. H., Posten C., Kruse O. 2007. Photosynthetic biomass and H2 production by green algae: from bioengineering to bioreactor scale-up. Physiol. Plant. 131:10–21 [DOI] [PubMed] [Google Scholar]
- 69.Hawkins R. L., Nakamura M. 1999. Expression of human growth hormone by the eukaryotic alga, Chlorella. Curr. Microbiol. 38:335–341 [DOI] [PubMed] [Google Scholar]
- 70.Hayashi M., Nito K., Takei-Hoshi R., Yagi M., Kondo M., Suenaga A., Yamaya T., Nishimura M. 2002. Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid beta-oxidation. Plant Cell Physiol. 43:1. [DOI] [PubMed] [Google Scholar]
- 71.Hellebust J. A. 1965. Excretion of some organic compounds by marine phytoplankton. Limnol. Oceanogr. 10:192–206 [Google Scholar]
- 72.Hemschemeier A., Melis A., Happe T. 2009. Analytical approaches to photobiological hydrogen production in unicellular green algae. Photosynth. Res. 102:523–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Henry I. M., Wilkinson M. D., Hernandez J. M., Schwarz-Sommer Z., Grotewold E., Mandoli D. F. 2004. Comparison of ESTs from juvenile and adult phases of the giant unicellular green alga Acetabularia acetabulum. BMC Plant Biol. 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hirokawa Y., Fujiwara S., Suzuki M., Akiyama T., Sakamoto M., Kobayashi S., Tsuzuki M. 2008. Structural and physiological studies on the storage β-polyglucan of haptophyte Pleurochrysis haptonemofera. Planta 227:589–599 [DOI] [PubMed] [Google Scholar]
- 75.Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., Darzins A. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54:621–639 [DOI] [PubMed] [Google Scholar]
- 76.Huang X., Weber J. C., Hinson T. K., Mathieson A. C., Minocha S. C. 1996. Transient expression of the GUS reporter gene in the protoplasts and partially digested cells of Ulva lactuca L.(Chlorophyta). Bot. Mar. 39:467–474 [Google Scholar]
- 77.Huesemann M. H., Hausmann T. S., Bartha R., Aksoy M., Weissman J. C., Benemann J. R. 2009. Biomass productivities in wild type and pigment mutant of Cyclotella sp. (diatom). Appl. Biochem. Biotechnol. 157:507–526 [DOI] [PubMed] [Google Scholar]
- 78.Inui M., Suda M., Kimura S., Yasuda K., Suzuki H., Toda H., Yamamoto S., Okino S., Suzuki N., Yukawa H. 2008. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl. Microbiol. Biotechnol. 77:1305–1316 [DOI] [PubMed] [Google Scholar]
- 79.Jain R. K., Coffey M., Lai K., Kumar A., MacKenzie S. L. 2000. Enhancement of seed oil content by expression of glycerol-3-phosphate acyltransferase genes. Biochem. Soc. Trans. 28:959–960 [PubMed] [Google Scholar]
- 80.Jako C., Kumar A., Wei Y., Zou J., Barton D. L., Giblin E. M., Covello P. S., Taylor D. C. 2001. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 126:861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jakobiak T., Mages W., Scharf B., Babinger P., Stark K., Schmitt R. 2004. The bacterial paromomycin resistance gene, aphH, as a dominant selectable marker in Volvox carteri. Protist 155:381–393 [DOI] [PubMed] [Google Scholar]
- 82.Janvilisri T., Venter H., Shahi S., Reuter G., Balakrishnan L., van Veen H. W. 2003. Sterol transport by the human breast cancer resistance protein (ABCG2) expressed in Lactococcus lactis. J. Biol. Chem. 278:20645–20651 [DOI] [PubMed] [Google Scholar]
- 83.Jarvis E. E., Brown L. M. 1991. Transient expression of firefly luciferase in protoplasts of the green alga Chlorella ellipsoidea. Curr. Genet. 19:317–321 [Google Scholar]
- 84.Jiang P., Cronan J. E., Jr 1994. Inhibition of fatty acid synthesis in Escherichia coli in the absence of phospholipid synthesis and release of inhibition by thioesterase action. J. Bacteriol. 176:2814–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang P., Qin S., Tseng C. K. 2003. Expression of the lacZ reporter gene in sporophytes of the seaweed Laminaria japonica (Phaeophyceae) by gametophyte-targeted transformation. Plant Cell Rep. 21:1211–1216 [DOI] [PubMed] [Google Scholar]
- 86.Kalscheuer R., Stolting T., Steinbuchel A. 2006. Microdiesel: Escherichia coli engineered for fuel production. Microbiology 152:2529–2536 [DOI] [PubMed] [Google Scholar]
- 87.Kindle K. L. 1990. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 87:1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kindle K. L., Schnell R. A., Fernandez E., Lefebvre P. A. 1989. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 109:2589–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klaus D., Ohlrogge J. B., Neuhaus H. E., Dörmann P. 2004. Increased fatty acid production in potato by engineering of acetyl-CoA carboxylase. Planta 219:389–396 [DOI] [PubMed] [Google Scholar]
- 90.Kosourov S. N., Seibert M. 2009. Hydrogen photoproduction by nutrient-deprived Chlamydomonas reinhardtii cells immobilized within thin alginate films under aerobic and anaerobic conditions. Biotechnol. Bioeng. 102:50–58 [DOI] [PubMed] [Google Scholar]
- 91.Kruse O., Rupprecht J., Bader K. P., Thomas-Hall S., Schenk P. M., Finazzi G., Hankamer B. 2005. Improved photobiological H2 production in engineered green algal cells. J. Biol. Chem. 280:34170–34177 [DOI] [PubMed] [Google Scholar]
- 92.Kubler J. E., Minocha S. C., Mathieson A. C. 1994. Transient expression of the GUS reporter gene in protoplasts of Porphyra miniata (Rhodophyta). J. Mar. Biotechnol. 1:165–169 [Google Scholar]
- 93.Kumar S. V., Misquitta R. W., Reddy V. S., Rao B. J., Rajam M. V. 2004. Genetic transformation of the green alga—Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 166:731–738 [Google Scholar]
- 94.Kurtzman A. M., Cheney D. P. 1991. Direct gene transfer and transient expression in a marine red alga using the biolistic method. J. Phycol. 27Suppl.:42 [Google Scholar]
- 95.Lapidot M., Raveh D., Sivan A., Arad S. M., Shapira M. 2002. Stable chloroplast transformation of the unicellular red alga Porphyridium species. Plant Physiol. 129:7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lardizabal K., Effertz R., Levering C., Mai J., Pedroso M. C., Jury T., Aasen E., Gruys K., Bennett K. 2008. Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol. 148:89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee J. W., Mets L., Greenbaum E. 2002. Improvement of photosynthetic CO2 fixation at high light intensity through reduction of chlorophyll antenna size. Appl. Biochem. Biotechnol. 98:37–48 [DOI] [PubMed] [Google Scholar]
- 98.Lee S. K., Chou H., Ham T. S., Lee T. S., Keasling J. D. 2008. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19:556–563 [DOI] [PubMed] [Google Scholar]
- 99.Lehner R., Vance D. E. 1999. Cloning and expression of a cDNA encoding a hepatic microsomal lipase that mobilizes stored triacylglycerol. Biochem. J. 343:1–10 [PMC free article] [PubMed] [Google Scholar]
- 100.Liolios K., Mavromatis K., Tavernarakis N., Kyrpides N. C. 2008. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 36:D475–D479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lluisma A. O., Ragan M. A. 1997. Expressed sequence tags (ESTs) from the marine red alga Gracilaria gracilis. J. Appl. Phycol. 9:287–293 [Google Scholar]
- 102.Lu X., Vora H., Khosla C. 2008. Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab. Eng. 10:333–339 [DOI] [PubMed] [Google Scholar]
- 103.Lu Y., Steichen J. M., Weise S. E., Sharkey T. D. 2006. Cellular and organ level localization of maltose in maltose-excess Arabidopsis mutants. Planta 224:935–943 [DOI] [PubMed] [Google Scholar]
- 104.Lumbreras V., Stevens D. R., Purton S. 1998. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14:441–447 [Google Scholar]
- 105.Maheswari U., Montsant A., Goll J., Krishnasamy S., Rajyashri K. R., Patell V. M., Bowler C. 2005. The diatom EST database. Nucleic Acids Res. 33:D344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Marín-Navarro J., Manuell A. L., Wu J., Mayfield S. P. 2007. Chloroplast translation regulation. Photosynth. Res. 94:359–374 [DOI] [PubMed] [Google Scholar]
- 107.Martin V. J. J., Pitera D. J., Withers S. T., Newman J. D., Keasling J. D. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21:796–802 [DOI] [PubMed] [Google Scholar]
- 108.Maruyama M., Horáková I., Honda H., Xing X., Shiragami N., Unno H. 1994. Introduction of foreign DNA into Chlorella saccharophila by electroporation. Biotechnol. Tech. 8:821–826 [Google Scholar]
- 109.Matsuzaki M., Misumi O., Shin-i T., Maruyama S., Takahara M., Miyagishima S.-Y., Mori T., Nishida K., Yagisawa F., Nishida K., Yoshida Y., Nishimura Y., Nakao S., Kobayashi T., Momoyama Y., Higashiyama T., Minoda A., Sano M., Nomoto H., Oishi K., Hayashi H., Ohta F., Nishizaka S., Haga S., Miura S., Morishita T., Kabeya Y., Terasawa K., Suzuki Y., Ishii Y., Asakawa S., Takano H., Ohta N., Kuroiwa H., Tanaka K., Shimizu N., Sugano S., Sato N., Nozaki H., Ogasawara N., Kohara Y., Kuroiwa T. 2004. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653–657 [DOI] [PubMed] [Google Scholar]
- 110.Matthew T., Zhou W., Rupprecht J., Lim L., Thomas-Hall S. R., Doebbe A., Kruse O., Hankamer B., Marx U. C., Smith S. M., Schenk P. M. 2009. The metabolome of Chlamydomonas reinhardtii following induction of anaerobic H2 production by sulfur depletion. J. Biol. Chem. 284:23415–23425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McManaman J. L., Russell T. D., Schaack J., Orlicky D. J., Robenek H. 2007. Molecular determinants of milk lipid secretion. J. Mammary Gland Biol. Neoplasia 12:259–268 [DOI] [PubMed] [Google Scholar]
- 112.Melis A. 2007. Photosynthetic H2 metabolism in Chlamydomonas reinhardtii (unicellular green algae). Planta 226:1075–1086 [DOI] [PubMed] [Google Scholar]
- 113.Melis A. 2009. Solar energy conversion efficiencies in photosynthesis: minimizing the chlorophyll antennae to maximize efficiency. Plant Sci. 177:272–280 [Google Scholar]
- 114.Melis A., Zhang L., Forestier M., Ghirardi M. L., Seibert M. 2000. Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122:127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mentewab A., Stewart C. N. 2005. Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat. Biotechnol. 23:1177–1180 [DOI] [PubMed] [Google Scholar]
- 116.Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Marechal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent. R., Dutcher S., Fernandez E., Fukuzawa H., Gonzalez-Ballester D., Gonzalez-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riano-Pachon D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martinez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Metzger P., Largeau C. 2005. Botryococcus braunii: a rich source for hydrocarbons and related ether lipids. Appl. Microbiol. Biotechnol. 66:486–496 [DOI] [PubMed] [Google Scholar]
- 118.Meuser J. E., Ananyev G., Wittig L. E., Kosourov S., Ghirardi M. L., Seibert M., Dismukes G. C., Posewitz M. C. 2009. Phenotypic diversity of hydrogen production in chlorophycean algae reflects distinct anaerobic metabolisms. J. Biotechnol. 142:21–30 [DOI] [PubMed] [Google Scholar]
- 119.Michael R., Miller D. J. 1998. Genetic transformation of dinoflagellates (Amphidinium and Symbiodinium): expression of GUS in microalgae using heterologous promoter constructs. Plant J. 13:427–435 [Google Scholar]
- 120.Michinaka Y., Shimauchi T., Aki T., Nakajima T., Kawamoto S., Shigeta S., Suzuki O., Ono K. 2003. Extracellular secretion of free fatty acids by disruption of a fatty acyl-CoA synthetase gene in Saccharomyces cerevisiae. J. Biosci. Bioeng. 95:435–440 [DOI] [PubMed] [Google Scholar]
- 121.Minoda A., Sakagami R., Yagisawa F., Kuroiwa T., Tanaka K. 2004. Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 45:667–671 [DOI] [PubMed] [Google Scholar]
- 122.Moellering E. R., Benning C. 13November2009. RNAi silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell doi:10.1128/EC.00203-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Molnar A., Bassett A., Thuenemann E., Schwach F., Karkare S., Ossowski S., Weigel D., Baulcombe D. 2009. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58:165–174 [DOI] [PubMed] [Google Scholar]
- 124.Mouille G., Maddelein M. L., Libessart N., Talaga P., Decq A., Delrue B., Ball S. 1996. Preamylopectin processing: a mandatory step for starch biosynthesis in plants. Plant Cell 8:1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mus F., Dubini A., Seibert M., Posewitz M. C., Grossman A. R. 2007. Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282:25475–25486 [DOI] [PubMed] [Google Scholar]
- 126.Mussgnug J. H., Thomas-Hall S., Rupprecht J., Foo A., Klassen V., McDowall A., Schenk P. M., Kruse O., Hankamer B. 2007. Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion. Plant Biotechnol. J. 5:802–814 [DOI] [PubMed] [Google Scholar]
- 127.Nakajima Y., Tsuzuki M., Ueda R. 2001. Improved productivity by reduction of the content of light-harvesting pigment in Chlamydomonas perigranulata. J. Appl. Phycol. 13:95–101 [Google Scholar]
- 128.Nakajima Y., Ueda R. 1997. Improvement of photosynthesis in dense microalgal suspension by reduction of light harvesting pigments. J. Appl. Phycol. 9:503–510 [Google Scholar]
- 129.Nguyen M. T., Choi S. P., Lee J., Lee J. H., Sim S. J. 2009. Hydrothermal acid pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. J. Microbiol. Biotechnol. 19:161–166 [DOI] [PubMed] [Google Scholar]
- 130.Nikaido I., Asamizu E., Nakajima M., Nakamura Y., Saga N., Tabata S. 2000. Generation of 10,154 expressed sequence tags from a leafy gametophyte of a marine red alga, Porphyra yezoensis. DNA Res. 7:223–227 [DOI] [PubMed] [Google Scholar]
- 131.Nishimoto T., Nakano M., Ikegami S., Chaen H., Fukuda S., Sugimoto T., Kurimoto M., Tsujisaka Y. 1995. Existence of a novel enzyme converting maltose into trehalose. Biosci. Biotechnol. Biochem. 59:2189–2190 [Google Scholar]
- 132.Nojima Y., Kibayashi A., Matsuzaki H., Hatano T., Fukui S. 1999. Isolation and characterization of triacylglycerol-secreting mutant strain from yeast, Saccharomyces cerevisiae. J. Gen. Appl. Microbiol. 45:1–6 [DOI] [PubMed] [Google Scholar]
- 133.O'Brien E. A., Koski L. B., Zhang Y., Yang L. S., Wang E., Gray M. W., Burger G., Lang B. F. 2007. TBestDB: a taxonomically broad database of expressed sequence tags (ESTs). Nucleic Acids Res. 35:D445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ohlrogge J. B., Jaworski J. G. 1997. Regulation of fatty acid synthesis. Annu. Rev. Plant Biol. 48:109–136 [DOI] [PubMed] [Google Scholar]
- 135.Palenik B., Grimwood J., Aerts A., Rouze P., Salamov A., Putnam N., Dupont C., Jorgensen R., Derelle E., Rombauts S., Zhou K., Otillar R., Merchant S. S., Podell S., Gaasterland T., Napoli C., Gendler K., Manuell A., Tai V., Vallon O., Piganeau G., Jancek S., Heijde M., Jabbari K., Bowler C., Lohr M., Robbens S., Werner G., Dubchak I., Pazour G. J., Ren Q., Paulsen I., Delwiche C., Schmutz J., Rokhsar D., Van de Peer Y., Moreau H., Grigoriev I. V. 2007. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U. S. A. 104:7705–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Panikashvili D., Savaldi-Goldstein S., Mandel T., Yifhar T., Franke R. B., Hofer R., Schreiber L., Chory J., Aharoni A. 2007. The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol. 145:1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Park M. O. 2005. New pathway for long-chain n-alkane synthesis via 1-alcohol in Vibrio furnissii M1. J. Bacteriol. 187:1426–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Paulsen B. S., Aslaksen T., Freire-Nordi C. S., Vieira A. A. H. 1998. Extracellular polysaccharides from Ankistrodesmus densus (Chlorophyceae). J. Phycol. 34:638–641 [Google Scholar]
- 139.Pienkos P. T., Darzins A. 2009. The promise and challenges of microalgal-derived biofuels. Biofuels Bioprod. Bioref. 3:431–440 [Google Scholar]
- 140.Pighin J. A., Zheng H., Balakshin L. J., Goodman I. P., Western T. L., Jetter R., Kunst L., Samuels A. L. 2004. Plant cuticular lipid export requires an ABC transporter. Science 306:702–704 [DOI] [PubMed] [Google Scholar]
- 141.Polle J. E. W., Kanakagiri S., Jin E. S., Masuda T., Melis A. 2002. Truncated chlorophyll antenna size of the photosystems—a practical method to improve microalgal productivity and hydrogen production in mass culture. Int. J. Hydrogen Energy 27:1257–1264 [Google Scholar]
- 142.Polle J. E. W., Kanakagiri S. D., Melis A. 2003. tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta 217:49–59 [DOI] [PubMed] [Google Scholar]
- 143.Posewitz M. C., Dubini A., Meuser J. E., Seibert M., Ghirardi M. L. 2009. Hydrogenases, hydrogen production, and anoxia, p. 217–246InHarris E. H., Stern D. B. (ed.), The Chlamydomonas sourcebook, 2nd ed., vol. 2 Organellar and metabolic processes Academic Press, Oxford, United Kingdom [Google Scholar]
- 144.Posewitz M. C., King P. W., Smolinski S. L., Smith R. D., Ginley A. R., Ghirardi M. L., Seibert M. 2005. Identification of genes required for hydrogenase activity in Chlamydomonas reinhardtii. Biochem. Soc. Trans. 33:102–103 [DOI] [PubMed] [Google Scholar]
- 145.Posewitz M. C., King P. W., Smolinski S. L., Zhang L., Seibert M., Ghirardi M. L. 2004. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J. Biol. Chem. 279:25711–25720 [DOI] [PubMed] [Google Scholar]
- 146.Posewitz M. C., Smolinski S. L., Kanakagiri S., Melis A., Seibert M., Ghirardi M. L. 2004. Hydrogen photoproduction is attenuated by disruption of an isoamylase gene in Chlamydomonas reinhardtii. Plant Cell 16:2151–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Poulsen N., Chesley P. M., Kröger N. 2006. Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae). J. Phycol. 42:1059–1065 [Google Scholar]
- 148.Poulsen N., Kroger N. 2005. A new molecular tool for transgenic diatoms. FEBS J. 272:3413–3423 [DOI] [PubMed] [Google Scholar]
- 149.Pruitt K. D., Tatusova T., Maglott D. R. 2005. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33:D501–D504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Qin S., Sun G. Q., Jiang P., Zou L. H., Wu Y., Tseng C. K. 1999. Review of genetic engineering of Laminaria japonica (Laminariales, Phaeophyta) in China. Hydrobiologia 398:469–472 [Google Scholar]
- 151.Qin S., Yu D. Z., Jiang P., Teng C. Y., Zeng C. K. 2003. Stable expression of lacZ reporter gene in seaweed Undaria pinnatifida. High Technol. Lett. 13:87–89 [Google Scholar]
- 152.Quinn J. M., Merchant S. 1995. Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7:623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ramachandra T. V., Mahapatra D. M., Karthick B., Gordon R. 2009. Milking diatoms for sustainable energy: biochemical engineering versus gasoline-secreting diatom solar panels. Ind. Eng. Chem. Res. 48:8769–8788 [Google Scholar]
- 154.Ramazanov A., Ramazanov Z. 2006. Isolation and characterization of a starchless mutant of Chlorella pyrenoidosa STL-PI with a high growth rate, and high protein and polyunsaturated fatty acid content. Phycol. Res. 54:255–259 [Google Scholar]
- 155.Rashotte A. M., Jenks M. A., Ross A. S., Feldmann K. A. 2004. Novel eceriferum mutants in Arabidopsis thaliana. Planta 219:5–13 [DOI] [PubMed] [Google Scholar]
- 156.Ritte G., Heydenreich M., Mahlow S., Haebel S., Kötting O., Steup M. 2006. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 580:4872–4876 [DOI] [PubMed] [Google Scholar]
- 157.Roesler K., Shintani D., Savage L., Boddupalli S., Ohlrogge J. 1997. Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol. 113:75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Roessler P. G., Chen Y., Liu B., Dodge C. N. December2009. Secretion of fatty acids by photosynthetic microorganisms. U.S. patent 20090298143 [Google Scholar]
- 159.Rylott E. L., Rogers C. A., Gilday A. D., Edgell T., Larson T. R., Graham I. A. 2003. Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid beta-oxidation is essential for embryo development. J. Biol. Chem. 278:21370–21377 [DOI] [PubMed] [Google Scholar]
- 160.Sauer N., Stolz J. 1994. SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine-tagged protein. Plant J. 6:67–77 [DOI] [PubMed] [Google Scholar]
- 161.Scala S., Carels N., Falciatore A., Chiusano M. L., Bowler C. 2002. Genome properties of the diatom Phaeodactylum tricornutum. Plant Physiol. 129:993–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Scharnewski M., Pongdontri P., Mora G., Hoppert M., Fulda M. 2008. Mutants of Saccharomyces cerevisiae deficient in acyl-CoA synthetases secrete fatty acids due to interrupted fatty acid recycling. FEBS J. 275:2765–2778 [DOI] [PubMed] [Google Scholar]
- 163.Schiedlmeier B., Schmitt R., Müller W., Kirk M. M., Gruber H., Mages W., Kirk D. L. 1994. Nuclear transformation of Volvox carteri. Proc. Natl. Acad. Sci. U. S. A. 91:5080–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Schneider-Belhaddad F., Kolattukudy P. 2000. Solubilization, partial purification, and characterization of a fatty aldehyde decarbonylase from a higher plant, Pisum sativum. Arch. Biochem. Biophys. 377:341–349 [DOI] [PubMed] [Google Scholar]
- 165.Sheehan J., Dunahay T., Benemann J., Roessler P. 1998. A look back at the US Department of Energy's Aquatic Species Program—biodiesel from algae. Report no. NREL/TP-580-24190 National Renewable Energy Laboratory, Golden, Colorado [Google Scholar]
- 166.Shen B., Jensen R. G., Bohnert H. J. 1997. Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to chloroplasts. Plant Physiol. 113:1177–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sheveleva E., Chmara W., Bohnert H. J., Jensen R. G. 1997. Increased salt and drought tolerance by d-ononitol production in transgenic Nicotiana tabacum L. Plant Physiol. 115:1211–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shigeoka S., Ishikawa T., Tamoi M., Miyagawa Y., Takeda T., Yabuta Y., Yoshimura K. 2002. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53:1305–1319 [PubMed] [Google Scholar]
- 169.Shimko N., Liu L., Lang B. F., Burger G. 2001. GOBASE: the organelle genome database. Nucleic Acids Res. 29:128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shimogawara K., Fujiwara S., Grossman A., Usuda H. 1998. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148:1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shrager J., Hauser C., Chang C. W., Harris E. H., Davies J., McDermott J., Tamse R., Zhang Z., Grossman A. R. 2003. Chlamydomonas reinhardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol. 131:401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Siloto R. M. P., Truksa M., Brownfield D., Good A. G., Weselake R. J. 2009. Directed evolution of acyl-CoA: diacylglycerol acyltransferase: development and characterization of Brassica napus DGAT1 mutagenized libraries. Plant Physiol. Biochem. 47:456–461 [DOI] [PubMed] [Google Scholar]
- 173.Sizova I., Fuhrmann M., Hegemann P. 2001. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277:221–229 [DOI] [PubMed] [Google Scholar]
- 174.Smith A. M. 2008. Prospects for increasing starch and sucrose yields for bioethanol production. Plant J. 54:546–558 [DOI] [PubMed] [Google Scholar]
- 175.Smith A. M., Zeeman S. C., Smith S. M. 2005. Starch degradation. Annu. Rev. Plant Biol. 56:73–98 [DOI] [PubMed] [Google Scholar]
- 176.Stark D. M., Timmerman K. P., Barry G. F., Preiss J., Kishore G. M. 1992. Regulation of the amount of starch in plant tissues by ADP glucose pyrophosphorylase. Science 258:287–292 [DOI] [PubMed] [Google Scholar]
- 177.Steinbrenner J., Sandmann G. 2006. Transformation of the green alga Haematococcus pluvialis with a phytoene desaturase for accelerated astaxanthin biosynthesis. Appl. Environ. Microbiol. 72:7477–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Stripp S. T., Goldet G., Brandmayr C., Sanganas O., Vincent K. A., Haumann M., Armstrong F. A., Happe T. 2009. How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc. Natl. Acad. Sci. U. S. A. 106:17331–17336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Suda Y., Yoshikawa T., Okuda Y., Tsunemoto M., Tanaka S., Ikeda K., Miyasaka H., Watanabe M., Sasaki K., Harada K. 2009. Isolation and characterization of a novel antistress gene from Chlamydomonas sp. W80. J. Biosci. Bioeng. 107:352–354 [DOI] [PubMed] [Google Scholar]
- 180.Sun Y., Gao X., Li Q., Zhang Q., Xu Z. 2006. Functional complementation of a nitrate reductase defective mutant of a green alga Dunaliella viridis by introducing the nitrate reductase gene. Gene 377:140–149 [DOI] [PubMed] [Google Scholar]
- 181.Sun Y., Yang Z., Gao X., Li Q., Zhang Q., Xu Z. 2005. Expression of foreign genes in Dunaliella by electroporation. Mol. Biotechnol. 30:185–192 [DOI] [PubMed] [Google Scholar]
- 182.Takeda T., Miyao K., Tamoi M., Kanaboshi H., Miyasaka H., Shigeoka S. 2003. Molecular characterization of glutathione peroxidase-like protein in halotolerant Chlamydomonas sp. W80. Physiol. Plant. 117:467–475 [DOI] [PubMed] [Google Scholar]
- 183.Tan C., Qin S., Zhang Q., Jiang P., Zhao F. 2005. Establishment of a micro-particle bombardment transformation system for Dunaliella salina. J. Microbiol. 43:361–365 [PubMed] [Google Scholar]
- 184.Tang D. K., Qiao S. Y., Wu M. 1995. Insertion mutagenesis of Chlamydomonas reinhardtii by electroporation and heterologous DNA. Biochem. Mol. Biol. Int. 36:1025–1035 [PubMed] [Google Scholar]
- 185.Tarczynski M. C., Jensen R. G., Bohnert H. J. 1993. Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science 259:508–510 [DOI] [PubMed] [Google Scholar]
- 186.Taylor D. C., Katavic V., Zou J., MacKenzie S. L., Keller W. A., An J., Friesen W., Barton D. L., Pedersen K. K., Michael Giblin E. 2002. Field testing of transgenic rapeseed cv. Hero transformed with a yeast sn-2 acyltransferase results in increased oil content, erucic acid content and seed yield. Mol. Breed. 8:317–322 [Google Scholar]
- 187.Teng C., Qin S., Liu J., Yu D., Liang C., Tseng C. 2002. Transient expression of lacZ in bombarded unicellular green alga Haematococcus pluvialis. J. Appl. Phycol. 14:497–500 [Google Scholar]
- 188.Tetali S. D., Mitra M., Melis A. 2007. Development of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii is regulated by the novel Tla1 gene. Planta 225:813–829 [DOI] [PubMed] [Google Scholar]
- 189.Tietge U. J. F., Bakillah A., Maugeais C., Tsukamoto K., Hussain M., Rader D. J. 1999. Hepatic overexpression of microsomal triglyceride transfer protein (MTP) results in increased in vivo secretion of VLDL triglycerides and apolipoprotein B. J. Lipid Res. 40:2134–2139 [PubMed] [Google Scholar]
- 190.Tietge U. J. F., Maugeais C., Cain W., Grass D., Glick J. M., de Beer F. C., Rader D. J. 2000. Overexpression of secretory phospholipase A2 causes rapid catabolism and altered tissue uptake of high density lipoprotein cholesteryl ester and apolipoprotein A-I. J. Biol. Chem. 275:10077–10084 [DOI] [PubMed] [Google Scholar]
- 191.Vigeolas H., Waldeck P., Zank T., Geigenberger P. 2007. Increasing seed oil content in oil-seed rape (Brassica napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol. J. 5:431–441 [DOI] [PubMed] [Google Scholar]
- 192.Vioque J., Kolattukudy P. E. 1997. Resolution and purification of an aldehyde-generating and an alcohol-generating fatty acyl-CoA reductase from pea leaves (Pisum sativum L.). Arch. Biochem. Biophys. 340:64–72 [DOI] [PubMed] [Google Scholar]
- 193.Voelker T. A., Davies H. M. 1994. Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J. Bacteriol. 176:7320–7327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Voelker T. A., Worrell A. C., Anderson L., Bleibaum J., Fan C., Hawkins D. J., Radke S. E., Davies H. M. 1992. Fatty acid biosynthesis redirected to medium chains in transgenic oilseed plants. Science 257:72–74 [DOI] [PubMed] [Google Scholar]
- 195.Wackett L. P., Frias J. A., Seffernick J. L., Sukovich D. J., Cameron S. M. 2007. Genomic and biochemical studies demonstrating the absence of an alkane-producing phenotype in Vibrio furnissii M1. Appl. Environ. Microbiol. 73:7192–7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wahlund T. M., Hadaegh A. R., Clark R., Nguyen B., Fanelli M., Read B. A. 2004. Analysis of expressed sequence tags from calcifying cells of marine coccolithophorid (Emiliania huxleyi). Mar. Biotechnol. 6:278–290 [DOI] [PubMed] [Google Scholar]
- 197.Wang Z. T., Ullrich N., Joo S., Waffenschmidt S., Goodenough U. 30October2009. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starch-less Chlamydomonas reinhardtii. Eukaryot. Cell doi:10.1128/EC.00272-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Weber A., Oesterhelt C., Gross W., Bräutigam A., Imboden L., Krassovskaya I., Linka N., Truchina J., Schneidereit J., Voll H. 2004. EST-analysis of the thermo-acidophilic red microalga Galdieriasulphuraria reveals potential for lipid A biosynthesis and unveils the pathway of carbon export from rhodoplasts. Plant Mol. Biol. 55:17–32 [DOI] [PubMed] [Google Scholar]
- 199.Wingler A., Fritzius T., Wiemken A., Boller T., Aeschbacher R. A. 2000. Trehalose induces the ADP-gucose pyrophosphorylase gene, ApL3, and starch synthesis in Arabidopsis. Plant Physiol. 124:105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Withers S. T., Gottlieb S. S., Lieu B., Newman J. D., Keasling J. D. 2007. Identification of isopentenol biosynthetic genes from Bacillus subtilis by a screening method based on isoprenoid precursor toxicity. Appl. Environ. Microbiol. 73:6277–6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Worden A. Z., Lee J. H., Mock T., Rouze P., Simmons M. P., Aerts A. L., Allen A. E., Cuvelier M. L., Derelle E., Everett M. V., Foulon E., Grimwood J., Gundlach H., Henrissat B., Napoli C., McDonald S. M., Parker M. S., Rombauts S., Salamov A., Von Dassow P., Badger J. H., Coutinho P. M., Demir E., Dubchak I., Gentemann C., Eikrem W., Gready J. E., John U., Lanier W., Lindquist E. A., Lucas S., Mayer K. F., Moreau H., Not F., Otillar R., Panaud O., Pangilinan J., Paulsen I., Piegu B., Poliakov A., Robbens S., Schmutz J., Toulza E., Wyss T., Zelensky A., Zhou K., Armbrust E. V., Bhattacharya D., Goodenough U. W., Van de Peer Y., Grigoriev I. V. 2009. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324:268–272 [DOI] [PubMed] [Google Scholar]
- 202.Wu L., Birch R. G. 2007. Doubled sugar content in sugarcane plants modified to produce a sucrose isomer. Plant Biotechnol. J. 5:109–117 [DOI] [PubMed] [Google Scholar]
- 203.Wykoff D. D., Davies J. P., Melis A., Grossman A. R. 1998. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117:129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yoshimura K., Miyao K., Gaber A., Takeda T., Kanaboshi H., Miyasaka H., Shigeoka S. 2004. Enhancement of stress tolerance in transgenic tobacco plants overexpressing Chlamydomonas glutathione peroxidase in chloroplasts or cytosol. Plant J. 37:21–33 [DOI] [PubMed] [Google Scholar]
- 205.Yu L., Gupta S., Xu F., Liverman A. D. B., Moschetta A., Mangelsdorf D. J., Repa J. J., Hobbs H. H., Cohen J. C. 2005. Expression of ABCG5 and ABCG8 is required for regulation of biliary cholesterol secretion. J. Biol. Chem. 280:8742–8747 [DOI] [PubMed] [Google Scholar]
- 206.Yu T.-S., Kofler H., Hausler R. E., Hille D., Flugge U.-I., Zeeman S. C., Smith A. M., Kossmann J., Lloyd J., Ritte G., Steup M., Lue W.-L., Chen J., Weber A. 2001. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13:1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yu T.-S., Zeeman S. C., Thorneycroft D., Fulton D. C., Dunstan H., Lue W.-L., Hegemann B. R., Tung S.-Y., Umemoto T., Chapple A., Tsai D.-L., Wang S.-M., Smith A. M., Chen J., Smith S. M. 2005. α-Amylase is not required for breakdown of transitory starch in Arabidopsis leaves. J. Biol. Chem. 280:9773–9779 [DOI] [PubMed] [Google Scholar]
- 208.Yuan L., Voelker T. A., Hawkins D. J. 1995. Modification of the substrate specificity of an acyl-acyl carrier protein thioesterase by protein engineering. Proc. Natl. Acad. Sci. U. S. A. 92:10639–10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zabawinski C., Van Den Koornhuyse N., D'Hulst C., Schlichting R., Giersch C., Delrue B., Lacroix J.-M., Preiss J., Ball S. 2001. Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-glucose pyrophosphorylase. J. Bacteriol. 183:1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zaslavskaia L. A., Lippmeier J. C., Kroth P. G., Grossman A. R., Apt K. E. 2000. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J. Phycol. 36:379–386 [Google Scholar]
- 211.Zaslavskaia L. A., Lippmeier J. C., Shih C., Ehrhardt D., Grossman A. R., Apt K. E. 2001. Trophic conversion of an obligate photoautotrophic organism through metabolic engineering. Science 292:2073–2075 [DOI] [PubMed] [Google Scholar]
- 212.Zeeman S. C., Delatte T., Messerli G., Umhang M., Stettler M., Mettler T., Streb S., Reinhold H., Kotting O. 2007. Starch breakdown: recent discoveries suggest distinct pathways and novel mechanisms. Funct. Plant Biol. 34:465–473 [DOI] [PubMed] [Google Scholar]
- 213.Zeeman S. C., Northrop F., Smith A. M., Rees T. A. 1998. A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J. 15:357–365 [DOI] [PubMed] [Google Scholar]
- 214.Zhao T., Wang W., Bai X., Qi Y. 2009. Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 58:157–164 [DOI] [PubMed] [Google Scholar]
- 215.Zheng P., Allen W. B., Roesler K., Williams M. E., Zhang S., Li J., Glassman K., Ranch J., Nubel D., Solawetz W. 2008. A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat. Genet. 40:367–372 [DOI] [PubMed] [Google Scholar]
- 216.Zolman B. K., Silva I. D., Bartel B. 2001. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 127:1266–1278 [PMC free article] [PubMed] [Google Scholar]
- 217.Zorin B., Lu Y., Sizova I., Hegemann P. 2009. Nuclear gene targeting in Chlamydomonas as exemplified by disruption of the PHOT gene. Gene 432:91–96 [DOI] [PubMed] [Google Scholar]
- 218.Zou J., Katavic V., Giblin E. M., Barton D. L., MacKenzie S. L., Keller W. A., Hu X., Taylor D. C. 1997. Modification of seed oil content and acyl composition in the Brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell 9:909–923 [DOI] [PMC free article] [PubMed] [Google Scholar]