Abstract
The NDT80/PhoG transcription factor family includes ScNdt80p, a key modulator of the progression of meiotic division in Saccharomyces cerevisiae. In Candida albicans, a member of this family, CaNdt80p, modulates azole sensitivity by controlling the expression of ergosterol biosynthesis genes. We previously demonstrated that CaNdt80p promoter targets, in addition to ERG genes, were significantly enriched in genes related to hyphal growth. Here, we report that CaNdt80p is indeed required for hyphal growth in response to different filament-inducing cues and for the proper expression of genes characterizing the filamentous transcriptional program. These include noteworthy genes encoding cell wall components, such as HWP1, ECE1, RBT4, and ALS3. We also show that CaNdt80p is essential for the completion of cell separation through the direct transcriptional regulation of genes encoding the chitinase Cht3p and the cell wall glucosidase Sun41p. Consistent with their hyphal defect, ndt80 mutants are avirulent in a mouse model of systemic candidiasis. Interestingly, based on functional-domain organization, CaNdt80p seems to be a unique regulator characterizing fungi from the CTG clade within the subphylum Saccharomycotina. Therefore, this study revealed a new role of the novel member of the fungal NDT80 transcription factor family as a regulator of cell separation, hyphal growth, and virulence.
Candida albicans is an opportunistic pathogen responsible for various non-life-threatening infections, such as oral thrush and vaginitis, and accounts for more than half of all Candida infections (21, 40). This pathogen is also a major cause of morbidity and mortality in bloodstream infections, especially in immunosuppressed individuals. In addition, C. albicans can colonize various biomaterials and readily forms dense biofilms that are resistant to most antifungal agents. The ability of this fungus to switch from yeast to filamentous forms (true hyphae or pseuohyphae) is a crucial determinant for host invasion and thus virulence (1). Hyphal growth can be initiated by different environmental cues, such as temperature, pH, or nutrient availability (1). Consequently, morphological switching implies a complex interplay of various sensing and signal transduction pathways, as well as transcriptional regulatory networks stimulating or repressing hyphal formation. Deciphering the molecular mechanisms that underlie this morphological switch is currently of high interest. Despite the large number of studies in recent years, the molecular determinism of C. albicans morphogenesis is still not fully understood.
The NDT80/PhoG transcription factor (TF) family includes the DNA-binding meiosis-specific protein ScNdt80p, a key modulator of the progression of the meiotic divisions in the yeast Saccharomyces cerevisiae (15, 33). This family also includes VIB-1, which is a regulator of conidiation in Neurospora crassa (41) and shares a region of similarity to PhoG, a possible non-phosphate-repressible acid phosphatase in Aspergillus nidulans (23). Structural studies revealed that this family is related to the Ig-fold family of transcription factors, which includes the human TFs p53, NF-κB, STAT, and AML-Runt and the Rel subfamilies (20, 24). All of these Ig-fold proteins bind DNA in similar manners, using loops and other features at one end of the β-sandwich. In C. albicans, a member of this family, named CaNdt80p (orf19.2119), has recently been identified as a key modulator of azole sensitivity due to its participation in the control of ergosterol biosynthesis gene expression (36). Genome-wide occupancy using chromatin immunoprecipitation coupled with high-density tiling arrays showed that, in addition to ERG genes, this TF bound a large number of gene promoters with diverse biological functions, such as cell wall, hyphal growth, carbohydrate metabolism, and the mitotic cell cycle. Additionally, de novo motif analysis of CaNdt80p-bound promoters revealed that, as in S. cerevisiae, this regulator bound to the middle sporulation element, 5′-gNCRCAAAY-3′, in C. albicans (where the lowercase letter indicates a semiconserved residue, R indicates a purine, N indicates any nucleotide, and Y indicates either a thymine or a cytosine).
The finding that CaNdt80p occupies the promoter regions of 23% of C. albicans genes suggests that the TF might control other biological processes, in addition to drug sensitivity (6, 36). Since CaNdt80p targets were significantly enriched in genes related to hyphal growth, this prompted us to study its potential role in morphological switching and host invasion. In this study, we continued to elucidate the multiple functions of CaNdt80p in C. albicans by demonstrating its central role in regulating cell separation, hyphal differentiation, and virulence. Interestingly, based on its functional-domain organization, CaNdt80p seems to be a unique TF characterizing fungi from the CTG clade within the subphylum Saccharomycotina.
MATERIALS AND METHODS
C. albicans strains, plasmids, and media.
The strains used in this study are listed in Table 1. For general propagation and maintenance conditions, the strains were cultured at 30°C in yeast-peptone-dextrose (YPD) medium supplemented with uridine (2% Bacto peptone, 1% yeast extract, 2% dextrose, and 50 μg/ml uridine, with the addition of 2% agar for solid medium). Cell growth, transformation, and DNA preparation were carried out using standard yeast procedures. For filamentation assays, cells were grown at 37°C in YPD supplemented with either 10% fetal bovine serum (Invitrogen) or 2.5 mM N-acetyl-d-glucosamine (Sigma) or in M199 medium (Sigma) buffered with 150 mM HEPES to pH 8.0. For growth under hypoxic conditions, cells were spotted on YPS (2% Bacto peptone, 1% yeast extract, 2% sucrose, 2% agar) plates and incubated in an anaerobic chamber (Oxoid; HP0011A) at 37°C. The chamber was flushed daily with nitrogen to remove oxygen and any by-products.
Table 1.
Candida albicans strains used in the study
| Strain | Genotype | Source or reference(s) |
|---|---|---|
| BWP17 | his1/his1 ura3/ura3 arg4/arg4 | 13, 39 |
| DAY286 | his1/his1 ura3/ura3 arg4/arg4::pARG4::URA3 | 8 |
| AS31 | ndt80::HIS1/ndt80::URA3 arg4/arg4 | 36 |
| AS32 | ndt80::HIS1/ndt80::HIS1 arg4/arg4 | 36 |
| AS33 | ndt80::HIS1/ndt80::HIS1 RP10/rp10::pCIp10-NDT80 arg4/arg4 | 36 |
| AS40 | ndt80::HIS1/ndt80::HIS1 RP10/rp10::pCIpACT1-CHT3 arg4/arg4 | This study |
| AS41 | ndt80::HIS1/ndt80::HIS1 RP10/rp10::pCIpACT1-SUN41 arg4/arg4 | This study |
Cell separation defects were assessed as described previously (11), except that more than 500 cells were counted for each strain.
To overexpress CHT3 and SUN41 in the null mutant ndt80, open reading frames (ORFs) of each gene were amplified from genomic DNA using the two sets of primers Cht3F1/Cht3R1 and Sun41F1/Sun41R1 (Table 2), respectively. The PCR fragments were digested with the restriction enzymes MluI and NheI and cloned in the same sites of the CIp-ACT1-CYC vector (2). The plasmid was sequenced to confirm the integrity of the genes. Plasmids CIp-ACT1-CHT3 and CIp-ACT1-SUN41 were digested with the StuI restriction enzyme and used to transform the ndt80 mutant strain (AS32). The absence of aneuploidy was confirmed in the overexpressing mutants using comparative genome hybridization as described previously (36).
Table 2.
Primers used in the study
| Primer name | Primer sequence | Purpose |
|---|---|---|
| Cht3F1 | CGACGCGTATGCTATACTTGTTAACTATATTTTC | CHT3 overexpression |
| Cht3R1 | CTAGCTAGCATTATAGATAACCACTGTACTTGGT | |
| Sun41F1 | ATGAGATTTTCACAAGCTACTGTT | SUN41 overexpression |
| Sun41R1 | CTAGCTAGCATTATACAAGACAAAGTCAGCTTC | |
| qCht3F2 | ACTACCTCCACAGCACCAAC | qPCR |
| qCht3R2 | GTAGAAGTGGCAGGTTTAGTTG | |
| qSun41F2 | TGTGAATGGGGTGTCAAGAA | qPCR |
| qSun41R2 | AGCACCACCTCTCCAAGTGT | |
| qAct1 F1 | GAAGCCCAATCCAAAAGA | qPCR |
| qAct1 R1 | CTTCTGGAGCAACTCTCAATTC | |
| qAls3 F1 | CGGTTGCGACTGCAAAGAC | qPCR |
| qAls3 R1 | GACCAACCCAAAACAGCATTCC | |
| qHwp1 F1 | CAGTTCCACTCATGCAACCATC | qPCR |
| qHwp1 R1 | GCAATACCAATAATAGCAGCACCG | |
| qYwp1 F1 | CTG ATA TTC GTA ATG CTG GTA AAG TG | qPCR |
| qYwp1 R1 | GGA GTT TCA CCC ATT AAT CTT CTT C | |
| qEce1 F1 | CCGGCATCTCTTTTAACTGG | qPCR |
| qEce1 R1 | GAGATGGCGTTCCAGATGTT | |
| qCax4 F1 | TCAATTCATGGGATTTTTCG | qPCR |
| qCax4 R1 | CCCCGTAATTAATCCAGCAA | |
| qNrg1 F1 | TGCAACCCCAACAAACACTA | qPCR |
| qNrg1 R1 | TGACGTTGTTGATATGATGCTG | |
| qTec1 F1 | TGGTGCTTATTCACGTGTCC | qPCR |
| qTec1 R1 | GTGGTGGTCATGCCAATAGT | |
| qRBT4 F1 | CGATGCTGATGGTGGTAATG | qPCR |
| qRBT4 R1 | TTGGTCATCTGAAGGGAAGC |
Gene expression profiling.
For gene expression profiling under the yeast form, saturated overnight cultures of the wild type (wt) (BWP17) and ndt80/ndt80 strain AS31 were diluted to a starting optical density at 600 nm (OD600) of 0.1 in 50 ml fresh YPD and grown at 30°C to an OD600 of 0.8. Hyphae were induced by growing Candida cells in YPD plus 10% fetal bovine serum (FBS) at 37°C to an OD600 of 0.8. Cultures were harvested by centrifugation at 3,000 × g for 5 min, and the pellet was rapidly frozen in liquid nitrogen.
To extract RNA from cells, samples stored at −80°C were placed on ice, and RNeasy buffer RLT was added to the pellets at a buffer/pellet ratio of 10:1 (vol/vol). The pellet was allowed to thaw in the buffer while being vortexed briefly at high speed. The resuspended pellet was placed back on ice and divided into 1-ml aliquots in 2-ml screw cap microcentrifuge tubes containing 0.6 ml of 3-mm-diameter acid-washed glass beads. Samples were homogenized 5 times for 1 min each time at 4,200 rpm using a Beadbeater. The samples were placed on ice for 1 min after each homogenization step. After the homogenization, the Qiagen RNeasy protocol was followed as recommended. Total-RNA samples were eluted in RNase-free H2O. The RNA quality and integrity were assessed using an Agilent 2100 bioanalyzer.
cDNA labelings and microarray hybridizations were performed as previously described (30). Briefly, 20 μg of total RNA was reverse transcribed using oligo(dT)21 in the presence of Cy3- or Cy5-dCTP (Invitrogen) and Superscript III reverse transcriptase (Invitrogen). Thereafter, the template RNA was degraded by adding 2.5 units RNase H (USB) and 1 μg RNase A (Pharmacia), followed by incubation for 15 min at 37°C. The labeled cDNAs were purified with a QIAquick PCR Purification Kit (Qiagen). Prior to hybridization, Cy3/Cy5-labeled cDNA was quantified using a NanoDrop ND-1000 UV-VIS spectrophotometer (NanoDrop) to confirm dye incorporation. Prehybridization and hybridization solutions consisted of DIG Easy Hyb solution (Roche Diagnostics, Mannheim, Germany) with 0.45% salmon sperm DNA and 0.45% yeast tRNA. The hybridization was carried out at 42°C for 20 h in a SlideBooster Hyb chamber SB 800 (Advalytix, Brunnthal, Germany) with regular microagitation of the sample. The slides were washed once in 1.0% SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.2% SDS at 42°C for 5 min; twice in 0.1% SSC, 0.2% SDS at 42°C for 5 min; and once in 0.1% SSC at 24°C for 5 min, followed by four rinses in 0.1% SSC. The chips were air dried before being scanned using a ScanArray Lite microarray scanner (Perkin Elmer). Microarray data were analyzed with GeneSpring GX v7.3 (Agilent Technologies), and genes with statistically significant changes in transcript abundance were identified using a Welch t test with a false-discovery rate (FDR) of less than 5%.
Expression analysis by qPCR.
For quantitative real-time PCR (qPCR), cDNA was synthesized from 5 μg of total RNA using the Invitrogen reverse transcription system [50 mM Tris-HCl, 75 mM KCl, 5 mM dithiothreitol (DTT), 3 mM MgCl2, 400 nM oligo(dT)15, 20 ng random octamers, 0.5 mM deoxynucleotide triphosphates (dNTPs), 200 units Superscript III reverse transcriptase]. The mixture was incubated for 60 min at 50°C. The cDNAs were then treated with 2 U of RNase H (Promega) for 20 min at 37°C, followed by heat inactivation of the enzyme at 80°C for 10 min. Aliquots were used for qPCR, which was performed using the Mx3000P QPCR System (Agilent) with the QuantiTect SYBR green PCR master mix (Qiagen). Cycling was done for 10 min at 95°C, followed by 40 cycles of 95°C for 10 s, 58°C for 15 s, and 72°C for 15 s. Samples were done in triplicate, and means were used for calculations. Fold changes were estimated by using the coding sequence of the C. albicans ACT1 ORF as a reference. Fold enrichments of the tested coding sequences were estimated using the comparative ΔΔCT method as described previously (14).
Virulence studies.
Mouse studies were carried out as previously described (27). Briefly, 8- to 12-week-old B6 mice (Jackson Laboratories, Bar Harbor, ME) were inoculated via the tail vein with 200 μl of a suspension containing 3 × 105 C. albicans cells in phosphate-buffered saline (PBS). Six mice, three females and three males, were used for each experimental group. The mice were closely monitored, and those showing ruffled fur, hunched backs, and extreme lethargy were considered moribund and were euthanized. To determine fungal loads, the kidneys from each mouse were removed aseptically and homogenized in 5 ml of PBS before being plated on YPD plates containing chloramphenicol (34 μg/ml). The number of yeast colonies per kidney was determined and log transformed. All experimental procedures involving animals were approved by the Biotechnology Research Institute Animal Care Committee, which operated under the guidelines of the Canadian Council of Animal Care.
RESULTS
The Saccharomycotina CTG clade harbors two distinct paralogs of the NDT80/PhoG-like TF family.
Screening the C. albicans genome allowed us to identify two putative proteins with a conserved NDT80/PhoG-like DNA-binding domain (DBD), corresponding to orf19.2119 (CaNdt80p) and orf19.513. Analysis of their DBDs at the amino acid level revealed that they both shared 55% similarity with the meiosis-specific transcription factor ScNdt80p of S. cerevisiae. Domain architecture analysis of the two C. albicans TFs showed that orf19.513 had a domain organization similar to that of ScNdt80p, consisting of the DBD located in the N-terminal region followed by a putative activation domain in the C-terminal region (Fig. 1A). In contrast, orf19.2119 exhibited a unique organization that was the opposite of that of orf19.513 and ScNdt80p (Fig. 1A).
Fig. 1.
The NDT80/PhoG transcription factor family across Ascomycota.ic> (A) Functional-domain organizations of NDT80/PhoG family members in Saccharomyces cerevisiae and Candida albicans. Both the DNA-binding domain (Ndt80p DBD) and putative glutamine-rich activation domain (Ndt80p AD) are represented. (B) Structural organizations of NDT80/PhoG family members across different classes of ascomycetes. The topology of the tree was based on Fitzpatrick et al. (12). The CTG clade is highlighted by a red box. WGD identifies the clade containing species in which whole-genome duplication has occurred.
Examination of ascomycete genomes using the BlastP program was performed in order to identify putative orthologs of the NDT80/PhoG TF. The results showed that all ascomycete fungi have a single putative Ndt80p displaying a domain organization similar to those of ScNdt80p and orf19.513. The sole exception was the monophyletic CTG clade within the Saccharomycotina, containing organisms that translate CTG as serine instead of leucine and that harbor an additional gene homolog with a domain organization similar to that of orf19.2119 (Fig. 1B).
Deletion of CaNDT80 affects cell separation.
By combining genome-wide location and transcriptional profiling, we have previously revealed a key role of CaNdt80p in modulating azole sensitivity through the regulation of the expression of ergosterol (ERG) biosynthesis genes (36). In addition to ERG gene promoters, Ndt80p was found to bind a large number of gene promoters, demonstrating that the regulator might operate in other biological processes. In order to further analyze other potential cellular functions controlled by CaNdt80p, we monitored the yeast growth morphology of cells missing both ndt80 alleles when cultured in YPD at 30°C. As shown in Fig. 2A, microscopic observation revealed that ndt80 cells showed altered cell morphology corresponding to a defect in cell separation, as well as abnormal cell size, compared to both wt and revertant strains. ndt80 mutants consist of relatively swollen yeast cells forming chains connected by septa, as visualized by calcofluor white staining, along with a significant increase in the percentages of chains of cells with 3 or 4, 5 or 6, or more than 6 cells (Fig. 2B).
Fig. 2.
Ndt80p is required for cell separation completion. (A) Images of wt (DAY286), ndt80/ndt80 mutant (AS31), and revertant ndt80/ndt80/NDT80 (AS33) strains showing the cell separation defect. The cells were stained with calcofluor white. (B) Quantification of the cell separation defect. Shown are the percentages of chains of cells containing 1 or 2, 3 or 4, 5 or 6, or more than 6 cells.
CaNdt80p is required for the transcriptional activation of cell separation genes.
To gain insight into the underlying molecular mechanism leading to the cell separation defect, we examined transcriptional differences between wt and ndt80 cells growing in YPD at 30°C using whole-genome microarrays. Using a statistical-significance analysis with an estimated false-discovery rate of 5%, in addition to a cutoff of 1.5-fold, we identified 111 genes that require CaNdt80p for their proper expression, including 68 upregulated genes and 43 downregulated genes (see Table S1 in the supplemental material).
Our previous genome-wide location study demonstrated that, under yeast-promoting growth conditions, CaNdt80p binds 1,446 gene promoters (36). By cross-referencing these data with the list of Ndt80p transcriptionally dependent genes, we were able to identify candidate CaNdt80p direct target promoters whose genes are transcriptionally regulated during the yeast growth phase (Fig. 3A). Gene expression analysis indicated that some CaNdt80p direct targets were activated (46 genes), whereas others were repressed (25 genes), in the ndt80 mutant. This finding suggests that the TF functions as both an activator and a repressor of gene expression.
Fig. 3.
Ndt80p directly regulates genes implicated in cell separation. (A) Relationship between Ndt80p-bound genes and genes showing altered expression in the ndt80 mutant (AS31). (B) Ndt80p promoter occupancies of the endochitinase Cht3p and the glucosidase Sun41p as determined by Sellam et al. (36). The positions of the middle sporulation element motifs are shown.
Interestingly, among the genes that the ndt80 mutants failed to activate, we found the putative cell wall glycosidase gene SUN41, which is required for cell separation in C. albicans (11, 16). Additionally, we were interested in the Cht3p chitinase, which showed an average reduction in transcript abundance of 4.5-fold in ndt80/ndt80 mutants, although these levels of repression were not consistent enough for it to pass the threshold of statistical significance in our 4 replicates. Both the SUN41 and CHT3 gene promoters are bound by CaNdt80p (Fig. 3B), and their inactivation leads to a cell separation defect similar to that observed in ndt80 mutants. This suggests that CaNdt80p could directly control the expression of genes implicated in cell separation completion and that the cell separation defect in ndt80 mutants is the consequence of CHT3 and/or SUN41 depletion.
The ndt80 cell separation phenotype is suppressed by overexpression of CHT3 or SUN41.
In order to confirm that the cell separation defect observed in ndt80 is related to the depletion of at least one of these two cell wall degradation genes, we sought to investigate if overexpression of either CHT3 or SUN41 could restore the wt phenotype in ndt80 mutants. For this purpose, the CHT3 and SUN41 ORFs were placed under the control of the CaACT1 promoter and expressed in the ndt80 background. Overexpression of these two genes was confirmed using quantitative real-time PCR, and the results demonstrated that the expression levels of CHT3 and SUN41 were significantly augmented compared to the wt strain (Fig. 4A). We then quantified the cell separation defect by counting the cells per chain in the overexpression mutants. As shown in Fig. 4B, the ndt80 cell separation phenotype was reverted by overexpressing either CHT3 or SUN41. Indeed, cells were found predominantly as single cells or as mother-daughter cells in overexpression mutants, similar to what was observed in the wt and revertant strains. This supports the idea that the cell separation defect in ndt80 cells can be attributed to the depletion of CHT3 and SUN41.
Fig. 4.
The ndt80 cell separation defect is reverted by the overexpression of CHT3 or SUN41. (A) Average transcript levels of CHT3 and SUN41 in overexpression mutants relative to the wt strain (DAY286). For each gene, two clones (clones C1 and C20 for the ndt80/Act::SUN41 strain and clones C1 and C11 for the ndt80/Act::CHT3 strain) were evaluated. The reported values are the means of two independent experiments. (B) Evaluation of the percentages of chains of cells with 1 or 2, 3 or 4, 5 or 6, or more than 6 cells in overexpressing strains.
CaNdt80p is essential for hyphal growth in response to different filament-inducing conditions.
The ndt80 mutants were also tested for the ability to form hyphae under different environmental conditions. Wt cells grown in liquid medium containing serum or N-acetylglucosamine revealed vigorous filamentation (Fig. 5A and B). In contrast, the ndt80 mutants differentiated into short and swollen elongated cells with a high frequency of lateral branches in the presence of the serum (Fig. 5A).
Fig. 5.
CaNdt80p is essential for hyphal growth in response to different filament-inducing conditions. (A and B) Cell morphologies of wt (DAY286), ndt80/ndt80 mutant (AS31), and revertant ndt80/ndt80/NDT80 (AS33) strains on liquid media supplemented with fetal bovine serum (A) or N-acetyl-d-glucosamine (B). (C and D) Colony morphologies of wt, ndt80, and revertant strains after 5 days of growth on solid media under hypoxic conditions (C) or in M199 medium at pH 8 (D).
Similar results were obtained under hypoxic conditions and at pH 8. Indeed, while the wt and revertant strains showed abundant filamentation at the edges of colonies, ndt80 developed smooth colonies with no mycelial growth even after prolonged incubation (Fig. 5C and D). This filamentation defect of ndt80 mutants was also observed under other filament-inducing conditions, namely, RPMI and spider media (data not shown).
CaNdt80p is required for the activation of hypha-specific genes.
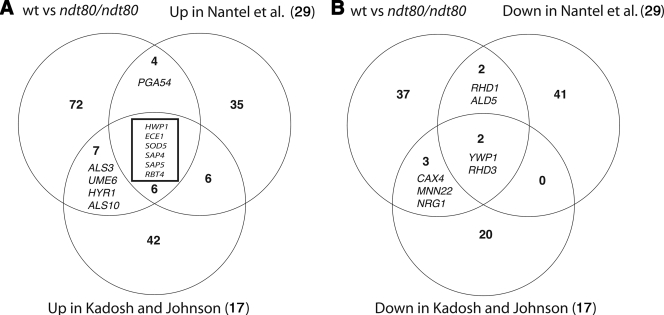
To gain insight into the role of CaNdt80p in mediating hyphal growth in C. albicans, we used microarrays to compare the transcriptomes of ndt80 mutant and wt cells grown in the presence of serum at 37°C. Using a statistical-significance analysis with an estimated false-discovery rate of 5%, in addition to a stringent cutoff of 3-fold, we found that ndt80 mutant cells had a severe gene expression defect, as they failed to fully activate 82 genes and to repress 41 genes (see Table S2 in the supplemental material). As illustrated in Fig. 6, the ndt80 mutant was unable to upregulate genes that had been previously characterized as being activated during the yeast-to-hypha switch, including genes encoding cell wall components (HWP1, ECE1, RBT4, ALS3, ALS10, and HYR1) and a superoxide dismutase (SOD5), as well as two secreted aspartyl proteinases (SAP4 and SAP5). Quantitative real-time PCR was used to confirm the expression defects of some of these genes specifying the yeast-to-hypha transition (Table 3) .
Fig. 6.
CaNdt80p is required for the activation of hypha-specific genes. Shown are the overlaps between genes differentially regulated during the yeast-to-hypha switch as determined by Nantel et al. (29) and Kadosh et al. (17) and genes downregulated (A) or upregulated (B) in a wt-versus-ndt80 comparison.
Table 3.
qPCR analysis of genes identified as differentially expressed by microarray experiments in wt-versus-ndt80 comparison under hypha-promoting conditions
| Gene | ORF | Microarraya | qPCRa,b |
|---|---|---|---|
| ALS3 | orf19.1816 | 0.03 | 5.08E−03 ± 0.0 |
| HWP1 | orf19.1321 | 0.08 | 8.73E−03 ± 0.0 |
| ECE1 | orf19.3374 | 0.03 | 2.16E−02 ± 0.2 |
| YWP1 | orf19.3618 | 30.59 | 867.06 ± 0.4 |
| CAX4 | orf19.3682 | 5.10 | 19.42 ± 3.2 |
| NRG1 | orf19.7150 | 3.87 | 7.16 ± 0.1 |
| TEC1 | orf19.5908 | 0.20 | 0.23 ± 0.1 |
| RBT4 | orf19.6202 | 0.02 | 0.34 ± 0.1 |
Average fold change.
Each value is the mean ± standard deviation of two independent experiments, each with three replicates.
Interestingly, activation of two TFs required for the positive regulation of hypha-specific genes, Ume6p and Tec1p, was found to be dependent on CaNdt80p. Additionally, ndt80 mutants failed to downregulate genes repressed during hypha formation, including YWP1, CAX4, MNN22, RHD1, RHD3, ALD5, and the transcriptional repressor NRG1 (Fig. 6). This clearly demonstrates that CaNdt80p is required for both the activation and the repression of genes characterizing the morphogenetic transcriptional program in C. albicans.
CaNdt80p is required for full virulence in a mouse model.
The ability of C. albicans to switch from yeast to hyphae is critical for host invasion and virulence. Since ndt80 mutants were unable to form hyphae in response to different filament-inducing conditions, we investigated if the TF is required for C. albicans virulence by using a mouse model. The C. albicans strain DAY286 (wt), an ndt80 mutant strain, and a revertant were tested in a mouse model for systemic infection by intravenous injection in the tail vein. As shown in Fig. 7A, while 70% of the mice infected with the wt and revertant strains became moribund within 12 days postinfection, none of the ndt80 strain-infected mice showed any clinical signs of advanced infection, such as ruffled fur, hunched back, or extreme lethargy, until the end of the experiment (day 21). Fungal loads from kidney tissues were also examined. As shown in Fig. 7B, the fungal loads were significantly lower in ndt80 strain-infected mice than in mice challenged with the wt or revertant strain. Taken together, these findings demonstrate that Ndt80p is a critical determinant of C. albicans pathogenicity.
Fig. 7.
Ndt80p is required for full virulence in a B6 mouse model. (A) Survival of mice infected with ndt80 mutant, ndt80 revertant, and wt parental strains. Mice were inoculated by tail vein injection, and survival was measured over a 21-day period. (B) The kidney fungal load was determined 7 days after injection. The error bars indicate standard deviations.
DISCUSSION
CaNdt80p is a novel transcriptional regulator unique to the Saccharomycotina CTG clade.
In the present study, we have shown that CaNdt80p, encoded by orf19.2119, has a unique functional domain organization distinct from that of ScNdt80p. However, another member of the Ndt80/PhoG TF family, encoded by orf19.513, exists in C. albicans, and in addition to significant sequence similarity, this TF showed exactly the same domain organization as the meiosis-specific TF ScNdt80p. This suggests that orf19.513 is the “true” C. albicans ortholog of ScNdt80p. Nevertheless, we have decided to continue using the common name CaNDT80 to define orf19.2119, as this gene has already been the subject of several publications by us and others and we have been unable to find any growth conditions that would result in the transcription of the orf19.513 gene (results not shown).
The presence of an ScNdt80p-like TF in all ascomycetes suggests an important function of this regulator (Fig. 1B). Surprisingly, orf19.2119-like TFs were found exclusively in fungi belonging to the monophylic CTG clade of Saccharomycotina. In addition to C. albicans, this group contains a large number of closely related pathogenic yeasts, such as Candida parapsilosis, Candida tropicalis, Candida lusitaniae, and Candida guilliermondii. Recently, Butler et al. (3) revealed significant expansions of the cell wall, secreted aspartase, and transporter gene families in pathogenic species of the CTG clade, suggesting adaptations associated with pathogenesis. Such large-scale amplification of a gene family and its contributions to promoting virulence have also been attributed to other ascomycete and basidiomycete species (34, 37). Gene duplication and the expansion of multiple gene families are considered to be major forces in evolution by allowing functional innovation. These observations argue that the presence of two members of the Ndt80/PhoG TF family in C. albicans, with highly similar DBDs, is most likely the result of gene duplication events from a common ancestor. Taking into consideration the role of CaNdt80p in virulence, the appearance or the retention of this TF in the Candida clade could be related to its critical role in adaptation to its mammalian host environment.
CaNdt80p is a general transcriptional regulator acting as both an activator and a repressor.
Using the global and unbiased approach of chromatin immunoprecipitation (ChIP)-Chip assays, we have demonstrated that CaNdt80p occupies a large number of promoter regions representing 23% of C. albicans genes (36). In addition, we have clearly demonstrated that CaNdt80p is required for the modulation of different biological processes, such as cell separation, hyphal growth, virulence, and azole sensitivity. Taken together, these results suggest that CaNdt80p is a multifunctional general transcriptional regulator. Gene expression analysis indicated that some direct targets of CaNdt80p were upregulated whereas others were repressed in ndt80 mutants. These findings suggest that this TF plays bifunctional roles as a repressor and an activator. Interestingly, in addition to structural and functional similarities between the CaNdt80p and p53 family members (20), p53 also has positive or negative effects on the activities of its target promoters (31).
Intriguingly, CaNdt80p was found to bind a large number of gene promoters that did not show any significant changes in gene expression under yeast-promoting conditions when the TF was absent. This phenomenon seems to be common in human TFs, where changing the level of a TF alters the expression levels of only 1 to 10% of its known target genes (10). It seems, therefore, that only a small proportion of the binding sites for a factor might be functional in a given cell type and that their functionality could be determined by cell-specific partners that need to be recruited for transcriptional activation or repression. Alternatively, TF networks may be fairly robust and able to compensate for a missing regulator. In our study, this can be illustrated by the example of the promoter region of NRG1, which exhibits significant Ndt80p binding in cells growing as yeasts. However, no NRG1 transcript level alteration was observed in ndt80 mutants grown under the same conditions. On the other hand, transcriptional profiling in the presence of serum revealed that the ndt80 mutants failed to downregulate NRG1 in response to serum. Assuming that Ndt80p binds to the NRG1 promoter under hypha-promoting conditions, this suggests that the repression of this transcriptional repressor requires a hypha-specific partner.
Ndt80p is a new regulator of cell separation in C. albicans.
In S. cerevisiae, cell separation occurs during the G1 phase and is achieved by the degradation of septal components, composed essentially of chitin, which holds the mother and daughter cells together after cytokinesis (4, 9). Degradation of septal materials is accomplished by the endochitinase Cts1p, which is responsible for the lysis of the primary septum at the neck (19). In addition to chitinase, glucanases, such as Eng1p, Scw11p, and Sun4p, play a complementary role and are required for dissolution of the secondary septum and/or the surrounding cell wall materials holding the mother and daughter cells together (25, 42). In addition to the tight spatial regulation of hydrolytic enzymes, strict temporal regulation during the cell cycle is also required (42). In both S. cerevesiae and fission yeast, this is achieved by the transcription factor Ace2p, which activates the expression of the chitinase CTS1 and other glucanases specifically in the G1 phase (9). In C. albicans, deleting CaACE2 results in a dramatic defect in cell separation, as well as attenuated virulence (18). Additionally, it was shown that CaAce2p is required for the transcriptional activation of the chitinase Cht3p and other glucanases, highlighting an evolutionarily conserved role of this TF in fungi (18, 26). Recent genome-wide investigations using DNA microarrays have revealed four successive waves of genes that are expressed periodically during the C. albicans cell cycle (7). Among these waves, ACE2 was found to peak periodically during the G2/M transition and to activate cell separation genes (CHT3, DSE1, SCW4, SCW11, and ENG1), which therefore are transcribed periodically at the M/G1 transition. Interrogation of the Candida Cell Cycle database (http://www.bri.nrc.ca/candida/cycle/) for cycling transcripts revealed that NDT80 was not considered a periodic gene; however, a slight peak was observed exactly at the G2/M transition in synchrony with ACE2. Based on this observation, the cell separation role of Ndt80p might be cell cycle regulated, as it is for Ace2p.
In this work, we have reported a novel function of Ndt80p in controlling cell separation in C. albicans through direct transcriptional activation of genes encoding the endochitinase Cht3p and the β-glucanase Sun41p. It is therefore likely that cell separation completion in this pathogen is the result of contributions from both Ace2p and Ndt80p gene targets. While both Ndt80p and Ace2p are required for cell separation during mitotic exit, the overlap between the targets of these two TFs consists of only two genes, CHT3 and SUN41 (26). Ndt80p was not found to be required for transcriptional activation of other Ace2p cell separation targets, such as DSE1, DSE4, and SCW11. Considering that Ndt80p was not found in the promoter region of ACE2 (36) and is not required for its proper expression, in addition to the fact that NDT80 expression was not altered in ace2 mutants, the two TFs might operate independently in two distinct transcriptional regulatory networks to control cell separation.
Ndt80p plays a central role in hyphal development and virulence in C. albicans.
In C. albicans, the role of transcriptional regulators of hyphal growth has been the subject of numerous investigations. In this work, we have enriched the repertoire of C. albicans filamentation TFs by demonstrating the critical function of CaNdt80p in hyphal development. In C. albicans, the yeast-to-hypha transition is triggered by various environmental stimuli, such as serum, neutral pH, high temperature, nutrient starvation, and CO2 (1). Deletion of CaNDT80 abolished the ability of C. albicans to undergo the yeast-to-hypha transition in response to a large set of hypha-inducing conditions. Since sensing of filamentation stimuli operates through a variety of distinct sensing/signalization pathways, Ndt80p is thus thought to act as a critical downstream effector promoting hypha formation in response to different signals conveyed by different upstream pathways.
Transcriptional profiling revealed that the hyphal defect of ndt80 mutants was correlated with the inability to activate hypha-specific genes, such as HWP1, ECE1, RBT4, ALS3, HYR1, SAP4, and SAP5. Additionally, given the fact that Ndt80p can act as a repressor, the hyphal defect of the ndt80 mutants could also be the consequence of the nonrepression of yeast-specific genes, such as YWP1, CAX4, MNN22, RHD1, RHD3, and ALD5. Based on our transcriptional-profiling data, the molecular basis of the hyphal defects of the ndt80 mutants can be explained in three possible ways. (i) ndt80 cells fail to repress the transcriptional repressor Nrg1p. Taking into account the critical role of Nrg1p in hypha-specific gene regulation (17), together with the occupancy of the NRG1 promoter by Ndt80p (36), C. albicans filamentous growth mediated by CaNdt80p might be an outcome of the release of Nrg1p repression at hypha-specific gene promoters. (ii) ndt80 cells fail to activate the expression of genes encoding the transcriptional activators Ume6p and Tec1p. In our previous study, the UME6 and TEC1 promoters were shown to be bound by Ndt80p, and we have shown here that UME6 and TEC1 activation in the presence of serum requires Ndt80p. Thus, CaNdt80p acts upstream of Ume6p and Tec1p, which, in turn, are implicated in activating the filamentation transcriptional program. (iii) ndt80 cells fail to activate key hypha-specific genes. In addition to the indirect-regulation models, we cannot rule out the possibility that CaNdt80p directly activates part of the hypha transcriptional responses, since it was found to bind the promoters of many hypha-specific genes, such as ECE1, ALS3, ALS1, HWP1, HYR1, and RBT4.
The ability of C. albicans to undergo morphological switching is a critical pathogenicity determinant. In fact, hyphal differentiation facilitates the invasion of host tissues and also helps C. albicans to escape from phagocytosis (22, 35). The loss of virulence of ndt80 mutants is most likely attributable to the key role of Ndt80p in controlling hyphal growth. However, the ability of this TF to activate other virulence-related functions, such as adhesion (HWP1, ALS1, ALS3, and ALS10) (5, 32, 38) or extracellular proteolytic activity (SAP4 and SAP5) (28), cannot be ruled out.
Supplementary Material
ACKNOWLEDGMENTS
Thanks are due to members of the BRI Microarray Laboratory and the BRI Animal Facility, especially Khairul Islam, Jean-Sébastien Deneault, Mario Mercier, and Jessy Tremblay, for technical assistance.
This work was supported by grants from the Canadian Institute of Health Research (CIHR) to A.N. (MOP-42516). C.A. was supported by an Alexander Graham Bell CGS-NSERC scholarship.
This is NRC publication number 50681.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 22 January 2010.
REFERENCES
- 1.Biswas S., Van Dijck P., Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackwell C., Russell C. L., Argimon S., Brown A. J., Brown J. D. 2003. Protein A-tagging for purification of native macromolecular complexes from Candida albicans. Yeast 20:1235–1241 [DOI] [PubMed] [Google Scholar]
- 3.Butler G., Rasmussen M. D., Lin M. F., Santos M. A., Sakthikumar S., Munro C. A., Rheinbay E., Grabherr M., Forche A., Reedy J. L., Agrafioti I., Arnaud M. B., Bates S., Brown A. J., Brunke S., Costanzo M. C., Fitzpatrick D. A., de Groot P. W., Harris D., Hoyer L. L., Hube B., Klis F. M., Kodira C., Lennard N., Logue M. E., Martin R., Neiman A. M., Nikolaou E., Quail M. A., Quinn J., Santos M. C., Schmitzberger F. F., Sherlock G., Shah P., Silverstein K. A., Skrzypek M. S., Soll D., Staggs R., Stansfield I., Stumpf M. P., Sudbery P. E., Srikantha T., Zeng Q., Berman J., Berriman M., Heitman J., Gow N. A., Lorenz M. C., Birren B. W., Kellis M., Cuomo C. A. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabib E., Roberts R., Bowers B. 1982. Synthesis of the yeast cell wall and its regulation. Annu. Rev. Biochem. 51:763–793 [DOI] [PubMed] [Google Scholar]
- 5.Chaffin W. L. 2008. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72:495–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C. G., Yang Y. L., Shih H. I., Su C. L., Lo H. J. 2004. CaNdt80 is involved in drug resistance in Candida albicans by regulating CDR1. Antimicrob. Agents Chemother. 48:4505–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cote P., Hogues H., Whiteway M. 2009. Transcriptional analysis of the Candida albicans cell cycle. Mol. Biol. Cell 20:3363–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis D. A., Bruno V. M., Loza L., Filler S. G., Mitchell A. P. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dohrmann P. R., Butler G., Tamai K., Dorland S., Greene J. R., Thiele D. J., Stillman D. J. 1992. Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 6:93–104 [DOI] [PubMed] [Google Scholar]
- 10.Farnham P. J. 2009. Insights from genomic profiling of transcription factors. Nat. Rev. Genet 10:605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firon A., Aubert S., Iraqui I., Guadagnini S., Goyard S., Prevost M. C., Janbon G., d'Enfert C. 2007. The SUN41 and SUN42 genes are essential for cell separation in Candida albicans. Mol. Microbiol. 66:1256–1275 [DOI] [PubMed] [Google Scholar]
- 12.Fitzpatrick D. A., Logue M. E., Stajich J. E., Butler G. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 6:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 14.Guillemette T., Sellam A., Simoneau P. 2004. Analysis of a nonribosomal peptide synthetase gene from Alternaria brassicae and flanking genomic sequences. Curr. Genet. 45:214–224 [DOI] [PubMed] [Google Scholar]
- 15.Hepworth S. R., Friesen H., Segall J. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiller E., Heine S., Brunner H., Rupp S. 2007. Candida albicans Sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation. Eukaryot. Cell 6:2056–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadosh D., Johnson A. D. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly M. T., MacCallum D. M., Clancy S. D., Odds F. C., Brown A. J., Butler G. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol. Microbiol. 53:969–983 [DOI] [PubMed] [Google Scholar]
- 19.Kuranda M. J., Robbins P. W. 1991. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 266:19758–19767 [PubMed] [Google Scholar]
- 20.Lamoureux J. S., Stuart D., Tsang R., Wu C., Glover J. N. 2002. Structure of the sporulation-specific transcription factor Ndt80 bound to DNA. EMBO J. 21:5721–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leroy O., Gangneux J. P., Montravers P., Mira J. P., Gouin F., Sollet J. P., Carlet J., Reynes J., Rosenheim M., Regnier B., Lortholary O. 2009. Epidemiology, management, and risk factors for death of invasive Candida infections in critical care: a multicenter, prospective, observational study in France (2005-2006). Crit. Care Med. 37:1612–1618 [DOI] [PubMed] [Google Scholar]
- 22.Lo H. J., Kohler J. R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G. R. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 23.MacRae W. D., Buxton F. P., Sibley S., Garven S., Gwynne D. I., Arst H. N., Jr., Davies R. W. 1993. Characterization of an Aspergillus nidulans genomic DNA fragment conferring phosphate-non-repressible acid-phosphatase activity. Gene 130:247–251 [DOI] [PubMed] [Google Scholar]
- 24.Montano S. P., Cote M. L., Fingerman I., Pierce M., Vershon A. K., Georgiadis M. M. 2002. Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast. Proc. Natl. Acad. Sci. U. S. A. 99:14041–14046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouassite M., Camougrand N., Schwob E., Demaison G., Laclau M., Guerin M. 2000. The ′SUN′ family: yeast SUN4/SCW3 is involved in cell septation. Yeast 16:905–919 [DOI] [PubMed] [Google Scholar]
- 26.Mulhern S. M., Logue M. E., Butler G. 2006. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 5:2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullick A., Elias M., Picard S., Bourget L., Jovcevski O., Gauthier S., Tuite A., Harakidas P., Bihun C., Massie B., Gros P. 2004. Dysregulated inflammatory response to Candida albicans in a C5-deficient mouse strain. Infect. Immun. 72:5868–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naglik J. R., Challacombe S. J., Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67:400–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nantel A., Dignard D., Bachewich C., Harcus D., Marcil A., Bouin A. P., Sensen C. W., Hogues H., van het Hoog M., Gordon P., Rigby T., Benoit F., Tessier D. C., Thomas D. Y., Whiteway M. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nantel A., Rigby T., Hogues H., Whiteway M. 2006. Microarrays for studying pathogenicity in Candida albicans, p. 181–210InKavanagh K.(ed.), Medical mycology: cellular and molecular techniques Wiley Press, Hoboken, NJ [Google Scholar]
- 31.Oren M. 2003. Decision making by p53: life, death and cancer. Cell Death Differ. 10:431–442 [DOI] [PubMed] [Google Scholar]
- 32.Phan Q. T., Myers C. L., Fu Y., Sheppard D. C., Yeaman M. R., Welch W. H., Ibrahim A. S., Edwards J. E., Jr., Filler S. G. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce M., Benjamin K. R., Montano S. P., Georgiadis M. M., Winter E., Vershon A. K. 2003. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol. Cell. Biol. 23:4814–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Powell A. J., Conant G. C., Brown D. E., Carbone I., Dean R. A. 2008. Altered patterns of gene duplication and differential gene gain and loss in fungal pathogens. BMC Genomics 9:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocha C. R., Schroppel K., Harcus D., Marcil A., Dignard D., Taylor B. N., Thomas D. Y., Whiteway M., Leberer E. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sellam A., Tebbji F., Nantel A. 2009. Role of Ndt80p in sterol metabolism regulation and azole resistance in Candida albicans. Eukaryot. Cell 8:1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soanes D. M., Alam I., Cornell M., Wong H. M., Hedeler C., Paton N. W., Rattray M., Hubbard S. J., Oliver S. G., Talbot N. J. 2008. Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS One 3:e2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staab J. F., Bradway S. D., Fidel P. L., Sundstrom P. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535–1538 [DOI] [PubMed] [Google Scholar]
- 39.Wilson R. B., Davis D., Mitchell A. P. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., Edmond M. B. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 41.Xiang Q., Glass N. L. 2002. Identification of vib-1, a locus involved in vegetative incompatibility mediated by het-c in Neurospora crassa. Genetics 162:89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeong F. M. 2005. Severing all ties between mother and daughter: cell separation in budding yeast. Mol. Microbiol. 55:1325–1331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.