Abstract
Npr2, a putative “nitrogen permease regulator” and homolog of the human tumor suppressor NPRL2, was found to interact with Grr1, the F-box component of the SCFGrr1 (Skp1–cullin–F-box protein complex containing Grr1) E3 ubiquitin ligase, by mass spectrometry-based multidimensional protein identification technology. Npr2 has two PEST sequences and has been previously identified among ubiquitinated proteins. Like other Grr1 targets, Npr2 is a phosphoprotein. Phosphorylated Npr2 accumulates in grr1Δ mutants, and Npr2 is stabilized in cells with inactivated proteasomes. Phosphorylation and instability depend upon the type I casein kinases (CK1) Yck1 and Yck2. Overexpression of Npr2 is detrimental to cells and is lethal in grr1Δ mutants. Npr2 is required for robust growth in defined medium containing ammonium or urea as a nitrogen source but not for growth on rich medium. npr2Δ mutants also fail to efficiently complete meiosis. Together, these data indicate that Npr2 is a phosphorylation-dependent target of the SCFGrr1 E3 ubiquitin ligase that plays a role in cell growth on some nitrogen sources.
Nitrogen, as a constituent of many biomolecules, is an essential cellular nutrient. Consequently, cells have developed sophisticated systems for nitrogen homeostasis. Glutamine is an optimal nitrogen source for yeast cells (reviewed in reference 19), whereas other amino acids, urea, and even ammonia are less efficient sources for many strains. Cells growing on those compounds as a sole source of nitrogen induce a variety of responses, including altered transport and metabolic activity, and they grow more slowly, presumably as a consequence of the increased metabolic cost.
One aspect of the response to growth on a suboptimal nitrogen source is the induction and activation of nitrogen permeases. Npr2 was originally identified along with the protein kinase Npr1, which is involved in the control of nitrogen catabolite-repressed genes, including amino acid permeases (6, 24). That association and the finding that mutations in the NPR2 gene result in poor growth when urea and proline are provided as a sole nitrogen source led to the conclusion that Npr2 (nitrogen permease regulator 2) is a nitrogen permease regulator. Although Npr2 was proposed to be a transcriptional regulator of DUR3 (24), which encodes a urea permease, a direct role in transcription has not been established. Furthermore, no direct effect of Npr2 on the permeases or their activity has been reported.
Inactivation of Npr2 has been associated with resistance to two clinically important but structurally and functionally distinct antitumor drugs, doxorubicin and cisplatin (24). Doxorubicin is a DNA-intercalating agent thought to inhibit topoisomerase II, whereas cisplatin damages DNA through the formation of platinum-DNA adducts (3). The most straightforward model based upon the current understanding of Npr2 is that the uptake of these drugs is affected by Npr2 inactivation. However, there is no evidence for decreased cisplatin or doxorubicin uptake (26). There does appear to be a modest mutator phenotype associated with Npr2 inactivation. Nevertheless, the mechanistic basis for resistance to those agents is not understood.
Despite the absence of a mechanistic understanding, interest in Npr2 has been stimulated by the finding that it is related to the amino acid sequence encoded by the human gene NPRL2/G21, originally identified as a likely candidate for the suppressor of lung cancer residing in the 3p21.3C region of the genome (16, 26). NPRL2 has since been associated with a variety of tumors, including non-small-cell lung carcinoma and renal carcinomas (29, 33). Furthermore, transduction of NPRL2 into tumor cells with deficiencies in 3p21.3c can suppress tumor formation (29). Finally, like yeast deficient in Npr2, tumor cells with 3p21.3c deficiencies exhibit resistance to cisplatin and can be resensitized by the reintroduction of NPRL2.
The role of NPRL2 in tumorigenesis provides an important impetus for understanding the function and regulation of the Nprl2 protein and therefore the yeast Npr2 protein. We have identified Npr2 among the proteins that interact with the Grr1 F-box protein, the component of the SCFGrr1 (Skp1–cullin–F-box protein complex containing Grr1) E3 ubiquitin ligase that confers specific substrate recognition. This is consistent with the finding that Npr2 has two PEST sequences and has been previously identified among ubiquitinated proteins by proteomic analysis (12, 24). Binding of Grr1 to Npr2 is independent of the F box of Grr1, the motif involved in interaction with Skp1 to form the E3 ubiquitin ligase SCFGrr1. Like all known Grr1 targets, Npr2 is a phosphoprotein that is stabilized and accumulates in its phosphorylated form in the absence of Grr1. As a consequence, overexpression of Npr2 is lethal in a grr1Δ mutant. Both phosphorylation and instability of Npr2 depend upon the type I casein kinases (CK1) Yck1 and Yck2, like several other SCFGrr1 targets involved in the regulation of nutrient permeases (22, 28). Its instability also depends upon the activity of the proteasome. Finally, Npr2 is required for robust growth in defined medium containing either ammonium or urea as a nitrogen source but not for growth on rich medium. A similar defect has been reported for npr2Δ mutants growing in proline as a nitrogen source (24). Consistent with a defect in nitrogen metabolism or sensing, npr2 mutants exhibit defects in completion of meiosis. Together, these data indicate that Npr2 is a phosphorylation-dependent target of the SCFGrr1 E3 ubiquitin ligase and plays a general role in cell growth on defined nitrogen sources.
MATERIALS AND METHODS
Yeast strains and techniques.
The yeast strains used in this study are described in Table 1. All of the strains are in the W303a background, except as noted. Cells were grown in standard culture media (YEPD = yeast extract, peptone, dextrose; SD = synthetic defined), and standard yeast genetic methods were used.
Table 1.
Yeast strains used in this study
| Strain | Background | Relevant genotype | Source |
|---|---|---|---|
| K699 | W303 | MATaura3-1his3-11,15leu2-3,111trp1-1ade2can1-100 | 31 |
| NSY228 | W303 | MATagrr1::KanMX2pCUP1-FLAG-GRR1 | This study |
| CWY1960 | W303 | MATapCUP1-FLAG-GRR1 | This study |
| CWY1556 | W303 | MATaGAL1-3 × HA-NPR2::HIS3 | This study |
| CWY1951 | W303 | MATaGAL1-3 × HA-NPR2::HIS3pCUP1-FLAG-GRR1 | This study |
| CWY1538 | W303 | MATaNPR2-3 × HA::HIS3 | This study |
| CWY1553 | W303 | MATagrr1::KanMX2NPR2-3 × HA::HIS3 | This study |
| NSY213 | W303 | MATagrr1::KanMX2HIS3::GAL1-3 × HA-NPR2 | This study |
| CWY1910 | W303 | MATaHIS3LEU2URA3 | This study |
| CWY1793 | W303 | MATaHIS3::GAL1-3 × HA-NPR2LEU2URA3 | This study |
| CWY1913 | W303 | MATagrr1::KanMX2LEU2URA3HIS3 | This study |
| CWY1795 | W303 | MATaURA3::GAL1-NPR2::3 × HA::HIS3grr1::KanMX2LEU2 | This study |
| NSY199 | W303 | MATanpr2::KanMX2 | This study |
| NSY195 | W303 | MATa/a | This study |
| NSY203 | W303 | MATa/anpr2::KanMX2/npr2::KanMX2 | This study |
| CWY1584 | S288C | MATaNPR2-3 × HA::HIS3leu2ura3-52 | This study |
| CWY1585 | S288C | MATaHIS3::GAL1-3 × HA-NPR2leu2ura3-52 | This study |
| CWY1586 | S288C | MATaNPR2-3 × HA::HIS3leu2ura3-52yck1-Δ1yck2-2ts | This study |
| CWY1587 | S288C | MATaHIS3::GAL1-3 × HA-NPR2leu2ura3-52yck1-Δ1yck2-2ts | This study |
| CWY1999 | BY4741 | his3Δ1leu2Δ0met15Δ0pdr5Δ::KanMX4HIS3::GAL1-3 × HA-NPR2 | This study |
The carboxy- and amino-terminal epitope-tagged proteins were generated via chromosomal integration of PCR-amplified fragments (18). Deletion mutants were constructed using PCR-based methods (18, 30).
FLAG purification.
The purification of FLAG-tagged Grr1 was done essentially as described previously (8). Cells were grown in uracil-deficient medium, and the expression of FLAG-GRR1 was induced for 4 h by the addition of 100 mM copper sulfate. The cells were harvested, washed in ice-cold water, and ground in liquid nitrogen. The proteins were extracted in purification buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 10% glycerol, 0.2% NP-40, 1× lambda phosphatase, adjusted to pH 8.0). FLAG-tagged protein was affinity purified under native conditions. Washes were done with purification buffer without glycerol, and elution was done by competition with the FLAG peptide (200-mg/ml final concentration) for 2 h at room temperature. The resulting proteins were precipitated by trichloroacetic acid to proceed on further analysis.
Multidimensional protein identification technology (MudPIT) analysis was carried out as described previously (21).
Protein preparation, coimmunoprecipitation, and phosphatase treatments.
Cells were harvested by centrifugation, and the pellets were stored at −80°C.
Protein extracts were prepared in lysis buffer (50 mM Tris-HCl [pH 7.5], 0.1% NP-40, 250 mM NaCl) containing phosphatase inhibitors (10 mM NaPPi, 5 mM EGTA, 0.1 mM orthovanadate) and protease inhibitors (100 mM phenylmethylsulfonyl fluoride, 1 mg of leupeptin/ml, 1 mg of aprotinin/ml). Lysis buffer extraction was performed by lysing cells at 4°C with glass beads (four times for 40 s) in a FastPrep FP120 apparatus. The protein extracts were collected after 15 min of centrifugation at 10,000 × g at 4°C.
Coimmunoprecipitations and the phosphatase treatments were performed as described previously (28). Briefly, cells were grown to mid-logarithmic phase in rich galactose medium containing 100 mM copper sulfate to induce both promoters and the proteins were extracted in lysis buffer as described above for protein preparation. Npr2-HA was immunoprecipitated with anti-hemagglutinin (anti-HA) monoclonal antibodies covalently bound to protein A-Sepharose. The proteins were immunoblotted with mouse 12CA5 anti-HA monoclonal antibodies or anti-FLAG antibodies (Sigma), respectively. Whole-cell extracts represent 100 μg of protein, whereas 1 mg of whole-cell extract was used for each immunoprecipitation.
Real-time RT-PCRs.
Real-time reverse transcription (RT)-PCRs were carried out as described previously (7).
Cell growth assays.
Strains were grown overnight in the respective media and diluted in the morning at the indicated optical density at 600 nm in liquid culture at 30°C. Measurements were taken every 2 h. The SD media were composed of 2% glucose, 0.17% yeast nitrogen base without amino acids and ammonium sulfate, and the indicated source of nitrogen (100 mM urea, 0.1 mM urea, or 100 mM ammonium sulfate) along with adenine and uracil.
Sporulation.
The strain of interest was grown overnight as a liquid culture in YEPD and plated for 2 to 3 days at 30°C on potassium acetate. The spore membranes were digested for 5 min at 30°C in 0.5 mg/ml Zymolyase T-100 on ice in 2 M sorbitol. The resulting asci were microdissected on YEPD medium.
RESULTS
Npr2 interacts with the Grr1 F-box protein.
In the interest of identifying proteins that interact with Grr1 as either substrates or regulators, we applied mass spectrometry-based MudPIT. Initial attempts to tag Grr1 with the tandem affinity purification epitope were judged to be unsuccessful based upon the similarity of the phenotype of cells expressing the tagged gene to mutants with compromised Grr1 function (data not shown). Consequently, we constructed GRR1 tagged with the FLAG epitope at the amino terminus and expressed it from the copper-inducible CUP1 promoter on plasmid pRS416 (15). Grr1 accumulating from that promoter was expressed at a level approximately three- to fourfold higher than the wild-type level (data not shown). The FLAG-Grr1 protein was expressed in a grr1Δ mutant strain growing in rich glucose-containing medium with the addition of copper and Grr1-containing protein complexes purified by adsorption to anti-FLAG epitope monoclonal antibody, followed by elution with the FLAG peptide. FLAG-Grr1 was highly enriched in that fraction (data not shown). The resulting protein complexes were analyzed by MudPIT analysis essentially as described previously (8, 15).
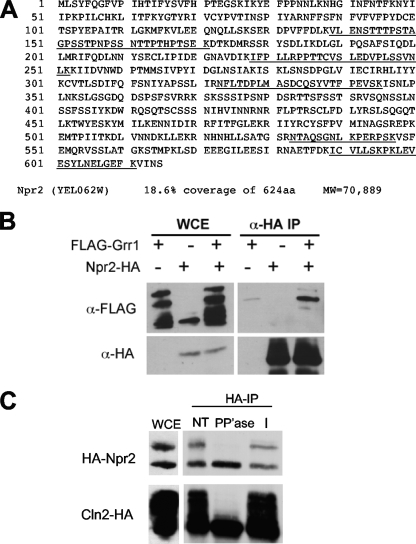
MudPIT analysis of Grr1 complexes led to the identification of a number of proteins that were specific based upon comparison to FLAG antibody precipitation from wild-type cells expressing only untagged proteins (15) and to a database of nonspecific proteins purified from yeast cells using anti-FLAG beads (13). Among the specific peptides identified by MudPIT, six unique peptides out of a total of seven peptides had sequences derived from Npr2, a putative negative regulator of nitrogen permeases (Fig. 1A). Together, those peptides represent 18.6% of the 71-kDa protein. Purification of FLAG-Grr1 with mutations in the F box that eliminate Skp1-interacting residues also yielded Npr2 peptides, suggesting that the interaction was directly with Grr1 and not other subunits of the SCF complex (data not shown). In addition, we recovered peptides derived by MudPIT analysis of Npr2 from cells growing in rich medium containing raffinose as a carbon source.
Fig. 1.
Npr2 interacts with Grr1. (A) Npr2 peptides identified by FLAG-Grr1 MudPIT analysis. Grr1 was tagged with a single FLAG epitope at the amino terminus and expressed in a grr1Δ mutant strain from a centromeric plasmid under the control of the copper-inducible CUP1 promoter (NSY228). The tryptic peptides derived from Npr2 by mass spectrometry-based MudPIT analysis are underlined in the amino acid sequence. (B) Npr2 associates stably with Grr1. Cells expressing GAL-HA-NPR2 (CWY1556), CUP1-FLAG-GRR1 (CWY1960), or both (CWY1951) were analyzed by immunoprecipitation (IP) with anti-HA or anti-FLAG antibodies and evaluated by immunoblotting as indicated. (C) Npr2 is a phosphoprotein. Npr2-HA immune complexes were treated with lambda phosphatase (PP'ase), left untreated (NT), or treated with phosphatase plus phosphatase inhibitor (I) and visualized by immunoblotting. The slower-migrating form of Npr2 immunodetected was specifically sensitive to phosphatase. A parallel treatment of the phosphoprotein Cln2-HA immune complexes is presented as a control. WCE, whole-cell extract.
To confirm that the interaction detected by MudPIT could also be observed by other methods, we expressed FLAG-Grr1 in a strain in which Npr2 was tagged with the HA epitope and subjected lysates of that strain to coimmunoprecipitation using anti-HA antibody. We could not detect that interaction by coimmunoprecipitation when Npr2-HA was expressed from its own promoter. However, when we constructed an allele of NPR2 tagged with a triple HA epitope at its amino terminus under the control of the yeast GAL1 promoter at the endogenous locus, we could detect a specific interaction with FLAG-Grr1 (Fig. 1B). We conclude that the abundance or avidity of the Npr2-Grr1 complex is low in wild-type cells.
Npr2 is a moderately unstable phosphoprotein.
To evaluate the behavior of the protein, we analyzed the protein in strains expressing Npr2-HA from the endogenous Npr2 locus. Npr2 in protein extracts from those strains migrated as a higher-mobility form of about 70 kDa and a lower-mobility species (Fig. 1C). Our ability to resolve these two species was very sensitive to the ionic conditions in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Although some variability in the ratio of the two species of Npr2 was observed between experiments and in cells grown on a variety of carbon sources, including glucose and galactose, those changes were relatively subtle (data not shown). Because lower-mobility species often represent phosphoproteins, we treated immune complexes precipitated with anti-HA antibodies with lambda phosphatase and evaluated the mobility of the treated protein relative to that of both untreated samples and samples that were treated with phosphatase in the presence of phosphatase inhibitors. The phosphatase-treated sample migrated as a single species comparable to the higher-mobility species in the untreated sample (Fig. 1C). This indicated that, like other targets of Grr1, Npr2 is a phosphoprotein and is consistent with the finding that phosphorylated proteins interact with Grr1 via the leucine-rich repeat that makes up its protein-protein interaction domain (14; our unpublished data).
The role of Grr1 as the F-box component of an SCF E3 ubiquitin ligase immediately suggested that the interaction might be a consequence of the targeting of phosphorylated Npr2 for ubiquitin-mediated degradation by SCFGrr1. We therefore predicted that Npr2 should be unstable. To evaluate that possibility, we expressed HA-Npr2 from the GAL1 promoter at the endogenous locus under inducing conditions and then added glucose to repress GAL1-directed gene expression (Fig. 2C). Samples were taken over a 6-hour time course for analysis of HA-Npr2 abundance by immunoblotting. We noticed that the abundance of both species of Npr2 is increased in those cells relative to that of Npr2 expressed from its own promoter and that the bulk of the accumulation is in the unphosphorylated species. After repression with glucose, the abundance of Npr2 protein decreased steadily over the time course (Fig. 2C, top left). These data indicate that Npr2 is a moderately unstable protein.
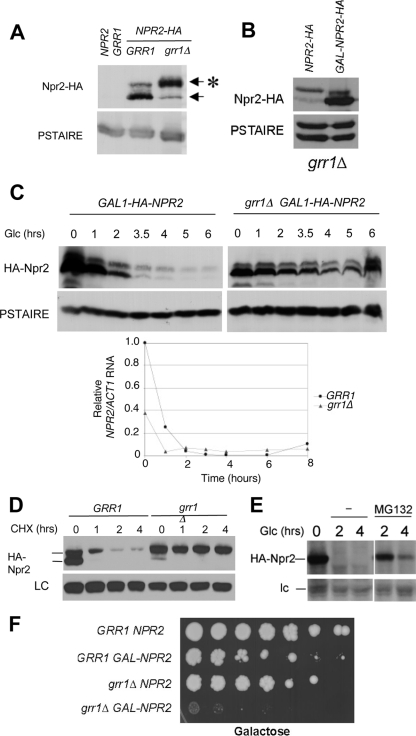
Fig. 2.
Npr2 is a phosphoprotein that is stabilized in a grr1Δ mutant. (A) Npr2 accumulates in a hyperphosphorylated form in grr1Δ mutants. Wild-type (CWY1538) or grr1Δ mutant (CWY1553) cells expressing NPR2-HA from the endogenous locus (or an untagged control strain, K699) were grown in YEPD to mid-logarithmic phase and immunoblotted for Npr2-HA with 12CA5 anti-HA monoclonal antibodies. Anti-PSTAIRE antibodies were used for the loading control. The asterisk denotes phosphorylated Npr2-HA. (B) Npr2 expressed from the GAL1 promoter in grr1Δ mutant cells primarily accumulates the higher-mobility species. NPR2-HA was expressed under the control of its endogenous promoter or from the GAL1 promoter in grr1Δ mutant cells by continuous growth on galactose, followed by immunoblotting for Npr2-HA with 12CA5 anti-HA monoclonal antibodies. (C) Npr2 protein expressed from the GAL1 promoter is stabilized in a grr1Δ mutant. (Top) NPR2-HA was expressed under the control of the repressible GAL1 promoter by growing cells of either the wild-type background (CWY1556) or the grr1Δ mutant background (NSY213) in 2% galactose to the mid-logarithmic phase. The GAL1 promoter was repressed by addition of 4% glucose (Glc). Cells were harvested after glucose addition at the times indicated, and the abundance of Npr2 was monitored by immunoblotting compared to the loading control. (Bottom) NPR2 gene expression in grr1Δ mutant cells. To confirm the transcriptional repression of NPR2 by glucose in the cells analyzed in the top half of panel C, RNA was isolated from the same cells and analyzed by real-time RT-PCR. Note that the relatively low abundance of NPR2 mRNA at 0 min in the grr1Δ mutant strain was reflected in the relatively low abundance of the Npr2 protein detected in the same sample. (D) Npr2 protein expressed from the endogenous locus is stabilized in a grr1Δ mutant. Strains expressing NPR2-HA from the endogenous locus under the control of its own promoter in the wild-type and grr1Δ mutant backgrounds were treated with cycloheximide (CHX; 100 μg/ml) at 0 min, and then samples were taken at the times indicated and analyzed by immunoblotting for Npr2-HA protein or an endogenous 50-kDa anti-HA-reactive protein as a loading control (LC). (E) Npr2 protein is stabilized by inhibition of the proteasome. Cells carrying a pdr5Δ mutation to promote drug sensitivity and expressing NPR2-HA from the GAL1 promoter (CWY1999) were grown on galactose for 1 h. Glucose was added to repress NPR2-HA expression, 50 μM MG132 was added to one half of the culture, and the other half was left untreated. The abundance of the Npr2-HA protein was determined by immunoblotting at 2 and 4 h following glucose addition. lc, loading control. (F) The accumulation of Npr2 protein is toxic for the cells. Wild-type cells (CWY1910 and CWY1753) and grr1Δ mutant cells (CWY1913 and CWY 1795) either overexpressing Npr2 from the GAL1 promoter or expressing a wild-type level of Npr2, respectively, were plated as a series of 1:5 dilutions on rich galactose medium to induce GAL1-NPR2. The extent of colony growth was recorded after 5 days at 30°C.
Npr2 is stabilized in a grr1Δ mutant.
SCFGrr1 is specifically involved in the ubiquitylation and degradation of a subset of phosphorylated proteins. Consequently, we evaluated the effect of inactivation of Grr1 on the abundance and modification state of Npr2. We expressed HA-tagged Npr2 from its own promoter in grr1Δ mutant cells and analyzed the state of the Npr2 protein (Fig. 2A). The abundance of the phosphorylated species of Npr2 accumulated to a substantially higher level than the unphosphorylated species in the grr1Δ mutant, and in comparison to wild-type cells grown under the same conditions, the phosphorylated form was much more abundant. Nevertheless, the overall level of the protein was only slightly higher in the grr1Δ mutant than in wild-type cells.
To evaluate the effect of inactivation of Grr1 on Npr2 turnover, we introduced the GAL-HA-NPR2 construct into a grr1Δ mutant strain and monitored the abundance of HA-Npr2 over a time course of 6 h following repression of the GAL1 promoter by glucose (Fig. 2C, top). Unlike Npr2 expressed from its own promoter, the protein accumulated from the GAL1 promoter in the grr1Δ mutant accumulates primarily in the higher-mobility species, suggesting that the capacity to phosphorylate Npr2 is limiting when the protein is overexpressed. That becomes more obvious when the protein overexpressed from the GAL1 promoter in grr1Δ mutant cells is directly compared to protein expressed in the same strain from the wild-type promoter (Fig. 2B). The abundance of Npr2 protein in the mutant cells is only slightly lower after 3.5 h following repression of the GAL1 promoter and easily detectable after 6 h of repression. That is despite the elimination of NPR2 transcript accumulated from the GAL1 promoter within 1 h (Fig. 2C, bottom). In contrast, Npr2 is largely lost from wild-type cells after 3.5 h. Based upon these results, we conclude that Grr1 is required for the instability of Npr2 protein, consistent with the hypothesis that Npr2 is a target for the SCFGrr1 ubiquitin ligase.
To evaluate whether inactivation of Grr1 affects turnover of Npr2 when it is expressed from its own promoter at the endogenous NPR2 locus, we treated wild-type and grr1Δ mutant strains expressing NPR2-HA with cycloheximide to inhibit new protein synthesis and then monitored the abundance and modification state of Npr2-HA over a time course of 4 h (Fig. 2D). Both low- and high-mobility forms of Npr2-HA were lost rapidly from the wild-type cells. However, the lower-mobility Npr2 protein, which was most abundant at the time of cycloheximide addition in the grr1Δ mutant cells, remained stable throughout the time course. The higher-mobility species disappeared rapidly in both cases, suggesting that it was converted to the lower-mobility species by phosphorylation.
The dependence upon Grr1 for instability suggests that Npr2 is degraded via the ubiquitin proteasome pathway. To test that hypothesis, the degradation of Npr2 expressed from the GAL1 promoter was evaluated following glucose repression in pdr5Δ mutant cells that were treated with the proteasome inhibitor MG132 (Fig. 2E). The pdr5Δ mutation potentiates the sensitivity of cells to MG132 (17). Treatment of cells with MG132 dramatically decreased the rate of loss of Npr2, which persisted in the treated cells for more than 4 h following glucose repression. Because the gel conditions used for this experiment did not resolve the two species of Npr2 in the pdr5Δ mutant, we are uncertain whether the phosphorylation state is affected. Nevertheless, we can conclude that the activity of the proteasome is required for Npr2 instability.
Overexpression of Npr2 from the GAL1 promoter is modestly toxic for wild-type cells, causing slow growth and lethality (Fig. 2F and data not shown). The basis for this toxicity is unknown. However, that toxicity is dramatically increased in a grr1Δ mutant background. Thus, at least under conditions of overexpression (Fig. 2B), Grr1 is required to keep Npr2 below a lethal level. This is consistent with a role for Grr1 in the inactivation of Npr2 via degradation.
The instability of Npr2 depends upon CK1.
Grr1 is required for nutrient signaling by both amino acids and glucose. Mth1 is an established target of SCFGrr1 that must be degraded for extracellular glucose to induce the expression of hexose permease genes (10, 22, 28). Targeting of Mth1 by SCFGrr1 requires phosphorylation of Mth1 by CK1 encoded by the YCK1 and YCK2 genes. The target of Grr1 required for signaling by amino acids via the SPS pathway leading to induction of amino acid and peptide permeases is as yet unknown (1, 2). However, activation of that pathway also requires CK1 (1, 28). Because Npr2 has been proposed to participate in the control of nutrient permeases, we hypothesized that its turnover might also require CK1 activity.
To test that hypothesis, we first evaluated the phosphorylation of Npr2 in cells deficient in CK1 activity by tagging Npr2 with HA at its endogenous locus in a yck1Δ yck2-ts strain (Fig. 3A). Whereas Npr2-HA expressed in a wild-type strain was distributed between the phosphorylated and unphosphorylated species, the protein was almost entirely in the higher-mobility unphosphorylated form when expressed in the yck mutant.
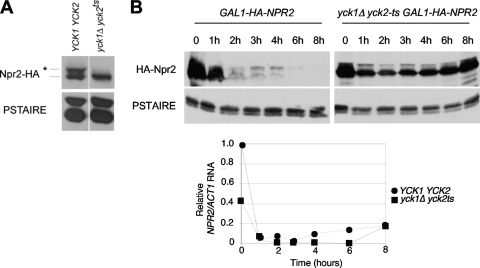
Fig. 3.
Npr2 phosphorylation and stability are dependent upon CK1. (A) Npr2 phosphorylation depends upon CK1. Npr2 was expressed from its own promoter in either wild-type cells (CWY1584) or yck1Δ yck2-ts mutant cells (CWY1586) and detected by immunoblotting with anti-HA antibody. Anti-PSTAIRE antibodies were used to detect the loading control. (B) The instability of Npr2 depends upon CK1. (Top) Npr2 was expressed under the control of the repressible GAL1 promoter integrated into the wild-type background (CWY1585) and the yck1Δ yck2-ts mutant background (CWY1587). The cells were grown in 2% galactose to the mid-logarithmic phase and shifted to 37°C for 2 h to inactivate casein kinase. The GAL1 promoter was then shut off, and the cells were harvested at the times indicated. The abundance of the Npr2 protein was monitored by immunoblotting and compared to the PSTAIRE loading control. (Bottom) GAL1-NPR2 expression in yck1Δ yck2-ts mutant cells. The transcriptional repression of NPR2 by the GAL1 promoter was confirmed by real-time RT-PCR as described above. As in the grr1Δ mutant background, the maximum expression of Npr2 is much lower in the yck1Δ yck2-ts mutant background at time zero than in the equivalent wild-type background.
Next, to evaluate the effect of inactivating CK1 on the stability of Npr2, we constructed a yck1Δ yck2-ts strain containing GAL-HA-NPR2 at the endogenous NPR2 locus. CK1 is inactive in that strain at 37°C, the restrictive temperature for the yck2-ts allele. yck1Δ yck2-ts mutant and wild-type cultures, both carrying the GAL-HA-NPR2 construct, were grown at the permissive temperature in the presence of galactose to induce the GAL1 promoter and then shifted to the restrictive temperature for 30 min prior to the addition of glucose. Cells were then monitored for the abundance of HA-Npr2 and NPR2 RNA over a time course of 8 h following glucose addition (Fig. 3B). A dramatic stabilization of HA-Npr2 was observed in the yck1Δ yck2-ts mutant at the restrictive temperature, whereas, as in prior experiments, the loss of HA-Npr2 was nearly complete within 2 h in the wild-type cells (Fig. 3B, top). Again, the abundance of the NPR2 transcript was lower in galactose-induced mutant cells and repressed by glucose nearly as efficiently as in wild-type cells (Fig. 3B, bottom). Finally, the abundance of the lower-mobility, hyperphosphorylated form of Npr2 was dramatically diminished in the mutant cells, suggesting that Npr2, like Mth1, is a target for phosphorylation by CK1 and that, like that of Mth1, phosphorylation is required for SCFGrr1-dependent degradation.
Npr2 is required for robust growth on some defined nitrogen sources.
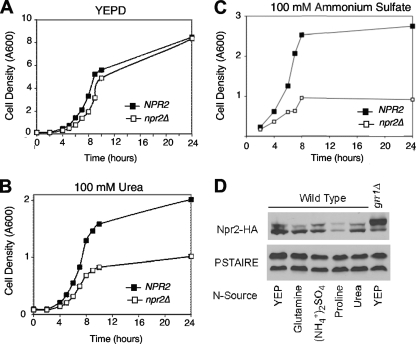
Npr2 has been proposed to play a role in urea uptake, and inactivation of NPR2 has been reported to increase the expression of DUR3, which encodes a urea permease (24). Although the effect on DUR3 expression is not observed in our strain background (data not shown), we have found that npr2Δ affects the capacity of cells to grow on urea as a sole nitrogen source (Fig. 4). Whereas the growth of npr2Δ mutant cells inoculated into rich medium (YEPD, a complex, amino acid-containing medium) is unaffected with regard to growth rate or maximal density (Fig. 4A), the same cells grow substantially more slowly than wild-type cells on medium containing 100 mM urea as a nitrogen source (Fig. 4B).
Fig. 4.
Growth of Npr2 mutant cells in low nitrogen or poor nitrogen sources. (A, B, and C) npr2Δ mutants are compromised for growth on urea as a nitrogen source. The CWY701 and NSY199 strains were grown to the mid-log phase in either rich YEPD or SD medium containing either 100 mM urea or 100 mM ammonium sulfate as a nitrogen source and then diluted to the density indicated and grown for 28 h. Cell density was determined at the times indicated. (D) Npr2 protein abundance and modification in various nitrogen sources. A wild-type strain expressing NPR2-3 × HA (CWY1538) was grown to the mid-log phase in either YEPD or SD medium containing 10 mM glutamate, 10 mM proline, 100 mM ammonium sulfate, or 100 mM urea. Cells were harvested, and the abundance of Npr2 in protein extracts was monitored by immunoblotting and compared to the PSTAIRE loading control.
The deleterious effect of inactivation of Npr2 on growth on urea is consistent with the reported role of Npr2 in urea uptake (24). However, we considered the possibility that it was the consequence of a more general effect of npr2Δ on the efficiency of nitrogen uptake or metabolism. We asked whether the npr2Δ mutation resulted in a similar effect on growth rate and maximal density when cells are grown in 100 mM ammonium as a nitrogen source (Fig. 4C). Similar to the effect of npr2Δ mutants growing on urea, the cells growing on ammonium grew slower and to a lower density than cells growing on rich medium (Fig. 4A). This suggests that the utilization of ammonium and urea as a nitrogen source is compromised in the absence of functional Npr2. Thus, Npr2 is likely to be required for the utilization of nitrogen derived from diverse sources at a point downstream of ammonium assimilation (19). Although these observations are consistent with a defect in the assimilation of these forms of nitrogen, a more likely explanation was recently advanced by Neklesa and Davis (23), who reported that Npr2 is required to inactivate TORC1 and thereby adapt to nitrogen sources other than glutamine, a “nonrepressing” nitrogen source.
Finally, we analyzed the abundance and modification state of the Npr2 protein in a number of nitrogen sources, including yeast extract-peptone and glutamate, which are expected not to be repressing and others thought to be repressing (Fig. 4D). We found no substantial differences in the relative modification state of Npr2 between those conditions. However, there was a significant reduction in the abundance of the Npr2 protein in cells growing on proline, which also supported the slowest growth rate of this group of nitrogen sources.
Npr2 is required for meiosis and sporulation.
The apparent defect in nitrogen uptake or metabolism suggested to us that Npr2 might be important for the proper regulation of commitment to meiosis. To evaluate that possibility, we incubated diploid wild-type and npr2Δ/npr2Δ mutant cells in sporulation medium, which is deficient in nitrogen and low in carbon, and determined the number of cells that were competent to complete meiosis and sporulation (Table 2). We found that although nearly 50% of the wild-type cells could enter meiosis and were at least partially proficient in sporulation, less than 13% of the npr2Δ/npr2Δ mutant cells could do so. Furthermore, less than 5% of the cells that entered meiosis went on to form complete tetrads. This is compared to the ability of nearly half of the wild-type cells that entered meiosis to form tetrads in the wild-type culture. We conclude that the Npr2-deficient cells cannot appropriately respond to nitrogen starvation and enter or complete meiosis. The extent of this defect may vary between strains, as a wild-type level of sporulation of npr2Δ mutants was observed in the SK1 background (20) whereas a defect in overall sporulation similar to that observed here was observed in a separate study (23).
Table 2.
Sporulation of wild-type and npr2Δ/npr2Δ diploid cellsa
| Phenotype |
NPR2/NPR2 |
npr2Δ/npr2Δ |
||
|---|---|---|---|---|
| No. of cells | % of cells | No. of cells | % of cells | |
| Nonsporulated | 557 | 53.5 | 897 | 86.8 |
| Monads to triads | 259 | 24.8 | 130 | 12.6 |
| Tetrads | 225 | 21.6 | 6 | 0.6 |
The sporulation assay was carried out using wild-type diploid cells (NSY203) and npr2Δ/npr2Δ diploid cells (NSY203) as described in Materials and Methods. More than 1,000 sporulation products for each strain were visually examined and the percentage in each class is reported.
DISCUSSION
One of the well-characterized roles of the SCFGrr1 E3 ubiquitin ligase is to promote nutrient uptake by regulating nutrient permease gene expression. Here we describe a previously unrecognized target, Npr2, identified based upon a physical interaction with Grr1 observed by mass spectrometry-based MudPIT analysis. Although Npr2 is a relatively stable protein, its turnover is dependent upon Grr1. Previous genome-wide approaches identified Npr2 as a ubiquitinated protein but failed to reveal its regulation by Grr1, presumably because of its relatively low abundance (4, 12). As is typical of SCFGrr1 substrates, Npr2 is a phosphoprotein. Both its phosphorylation and instability are dependent upon CK1, a common feature of Grr1-dependent targets involved in nutrient regulation (1, 22, 28). Accumulation of hyperphosphorylated Npr2 is associated with inactivation of Grr1.
Npr2 is required for efficient growth on some defined nitrogen sources and has been proposed to be a regulator of nitrogen permeases (24). Although the precise role of Npr2 is unclear, it was originally identified based upon a defect of the mutant in growth on urea and proline as a nitrogen source, a property shared with Npr1, a protein kinase involved in the regulation of nitrogen permeases (5, 9, 27). We have shown the phenotype of the npr2 mutant to include slow growth and compromised mass accumulation when it is grown on several nonoptimal nitrogen sources. These observations are consistent with a recent report from Neklesa and Davis (23) showing that a complex of Npr2 and Npr3 plays an inhibitory role upstream of TORC1, an important regulator of protein synthesis and amino acid uptake. Genetic analysis performed in the context of that study suggests that Npr2 acts to restrict TORC1 function in the absence of glutamine, an optimal nitrogen source, thereby promoting the expression of nitrogen metabolite-repressible genes and decreasing the rate of ribosome biosynthesis. The metabolism of glutamine is unaffected by inactivation of Npr2. We hypothesized that the defect in growth on ammonia might be a consequence of misregulation of ammonium uptake rather than in its metabolism. In fact, inhibition of TOR by rapamycin has been reported to induce expression of the major ammonium permease encoded by MEP2 (6). However, we find that regulation of MEP2 expression is intact in the npr2Δ mutant (data not shown), suggesting that any defect must occur either at the level of the permease itself or in some other aspect of ammonium assimilation.
Npr2 is a relatively stable protein with a half-life of greater than an entire cell cycle. However, Npr2 turnover is dependent upon Grr1 and, like that of several other targets of SCFGrr1, Npr2 turnover depends upon CK1. The long half-life of Npr2 is unexpected for targets of ubiquitin-mediated degradation but might reflect our failure to identify conditions that favor degradation. For example, Mth1 appears to be rather stable in cells growing on nonglucose carbon sources but is rapidly degraded via phosphorylation-dependent ubiquitination by SCFGrr1 when glucose is added to the growth medium, which, in turn, induces glucose permease gene expression (10, 22, 28). A similar phenomenon involving Grr1 may accelerate Npr2 turnover. Although Npr2 appears to be dispensable in cells growing in rich medium and therefore might be expected to be degraded under those conditions, neither the phosphorylation state nor accumulation of Npr2 appears to be strongly affected by the nitrogen source. We do know that hyperaccumulation of Npr2 is detrimental to cells and that it becomes lethal when Grr1 is inactivated. However, we do not know whether similar hyperaccumulation occurs under specific conditions in wild-type cells. Instead, Grr1 may be involved in a homeostatic mechanism regulating the overall abundance of Npr2 protein.
The requirement of casein kinase for the phosphorylation and instability of Npr2 is shared with several other Grr1 targets involved in the regulation of nutrient permease gene expression. In the case of Mth1, a corepressor of HXT genes that is inactivated by glucose, phosphorylation by Yck1/2 is a prerequisite for recognition by SCFGrr1 (22, 28). The target in the pathway leading to the activation of amino acid permeases is currently unknown. Both glucose and amino acid permeases are induced when nutrient uptake is desirable. Although our data point to Npr2 as a target of Yck1/2, neither the importance of turnover nor the stimulus leading to that turnover is understood.
Loss of Npr2 function has been reported to lead to resistance to the genotoxic agents doxorubicin, a topoisomerase II inhibitor, and cisplatin, which induces DNA damage by forming platinum-DNA adducts, two clinically important chemotherapeutics (11, 26). That phenotype is shared with mutants deficient in the Sky1 serine-rich protein-specific kinase. Resistance to a broad range of other compounds is not observed, and there is no evidence that this is related to a defect in uptake. Although the mechanism by which either of these genes confers resistance to the drugs is unknown, NPR2 and SKY2 are members of the same epistasis group, indicating that they function in the same pathway. This is consistent with the observation that both npr2Δ and sky1Δ mutants have a mild mutator phenotype (25, 26). Sky1 activity has been associated with mismatch repair defects and other aberrations of DNA metabolism. Although the NPR2 SKY2 epistasis could indicate a pathway in which Npr2 is phosphorylated by the Sky2 kinase, it seems unlikely because Npr2 lacks consensus sites for Sky2 (32) and because we have shown that the majority of Npr2 phosphorylation is lost in the absence of CK1 activity, eliminating the need to invoke the involvement of another kinase. Whether CK1 or Grr1 contributes to the role of Npr2 in modulating sensitivity to genotoxic agents is not known.
The closest human homolog of Npr2 is the tumor suppressor encoded by NPRL2 (16, 26). Inactivation of that protein, like that of its yeast counterpart, leads to cisplatin resistance, rendering cells refractory to this common cancer chemotherapeutic agent (29). Also, like its yeast ortholog, Nprl2 has been shown to form a complex with Nprl3, although its role as a TOR pathway regulator has not been established (23). Despite the fact that the physiological roles of Npr2 and Nprl2 are unknown, the shared phenotype of cisplatin resistance suggests that those roles at least partially overlap. Similarly, since the regulation of the Nprl2 protein has yet to be determined, it is not known whether it is a target for phosphorylation or ubiquitin-mediated proteolysis. However, based upon the similarity between these two proteins and the phenotypes of mutants in them, we suggest that Nprl2 may be a target for SCF-mediated ubiquitylation and subsequent turnover. If so, aberrant regulation of Nprl2 abundance could be important in the development and treatment of human cancer.
ACKNOWLEDGMENTS
We acknowledge excellent technical assistance by Tatyana Kalashnikova, comments on the manuscript from Rob de Bruin, and discussion and comments from members of the TSRI Cell Cycle Group. We thank the laboratories of Steve Reed and Mark Johnston for strains. We also thank anonymous reviewers for insightful comments that improved the presentation and interpretation of this report.
This work was supported by United States Public Health Service grants GM043487 and GM059441 to C.W. and grant P4 1RR11823 from the National Center for Research Resources of the National Institutes of Health to T. N. Davis.
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Abdel-Sater F., El Bakkoury M., Urrestarazu A., Vissers S., Andre B. 2004. Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol. Cell. Biol. 24:9771–9785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andréasson C., Heessen S., Ljungdahl P. O. 2006. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 20:1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baruah H., Barry C. G., Bierbach U. 2004. Platinum-intercalator conjugates: from DNA-targeted cisplatin derivatives to adenine binding complexes as potential modulators of gene regulation. Curr. Top. Med. Chem. 4:1537–1549 [DOI] [PubMed] [Google Scholar]
- 4.Benanti J. A., Cheung S. K., Brady M. C., Toczyski D. P. 2007. A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat. Cell Biol. 9:1184–1191 [DOI] [PubMed] [Google Scholar]
- 5.Boeckstaens M., Andre B., Marini A. M. 2007. The yeast ammonium transport protein Mep2 and its positive regulator, the Npr1 kinase, play an important role in normal and pseudohyphal growth on various nitrogen media through retrieval of excreted ammonium. Mol. Microbiol. 64:534–546 [DOI] [PubMed] [Google Scholar]
- 6.Cardenas M. E., Cutler N. S., Lorenz M. C., Di Como C. J., Heitman J. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bruin R. A., Kalashnikova T. I., Chahwan C., McDonald W. H., Wohlschlegel J., Yates J., Russell P., Wittenberg C. 2006. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23:483–496 [DOI] [PubMed] [Google Scholar]
- 8.de Bruin R. A., McDonald W. H., Kalashnikova T. I., Yates J., Wittenberg C. 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117:887–898 [DOI] [PubMed] [Google Scholar]
- 9.De Craene J. O., Soetens O., Andre B. 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276:43939–43948 [DOI] [PubMed] [Google Scholar]
- 10.Flick K. M., Spielewoy N., Kalashnikova T. I., Guaderrama M., Zhu Q., Chang H. C., Wittenberg C. 2003. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 14:3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuertesa M. A., Castillab J., Alonsoa C., Perez J. M. 2003. Cisplatin biochemical mechanism of action: from cytotoxicity to induction of cell death through interconnections between apoptotic and necrotic pathways. Curr. Med. Chem. 10:257–266 [DOI] [PubMed] [Google Scholar]
- 12.Hitchcock A. L., Auld K., Gygi S. P., Silver P. A. 2003. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. U. S. A. 100:12735–12740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sorensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180–183 [DOI] [PubMed] [Google Scholar]
- 14.Hsiung Y. G., Chang H. C., Pellequer J. L., La Valle R., Lanker S., Wittenberg C. 2001. F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol. Cell. Biol. 21:2506–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kesti T., McDonald W. H., Yates J. R., III, Wittenberg C. 2004. Cell cycle-dependent phosphorylation of the DNA polymerase epsilon subunit, Dpb2, by the Cdc28 cyclin-dependent protein kinase. J. Biol. Chem. 279:14245–14255 [DOI] [PubMed] [Google Scholar]
- 16.Li J., Wang F., Haraldson K., Protopopov A., Duh F. M., Geil L., Kuzmin I., Minna J. D., Stanbridge E., Braga E., Kashuba V. I., Klein G., Lerman M. I., Zabarovsky E. R. 2004. Functional characterization of the candidate tumor suppressor gene NPRL2/G21 located in 3p21.3C. Cancer Res. 64:6438–6443 [DOI] [PubMed] [Google Scholar]
- 17.Liu C., Apodaca J., Davis L. E., Rao H. 2007. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques 42:158, 160–162 [DOI] [PubMed] [Google Scholar]
- 18.Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 19.Magasanik B., Kaiser C. A. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1–18 [DOI] [PubMed] [Google Scholar]
- 20.Marston A. L., Tham W. H., Shah H., Amon A. 2004. A genome-wide screen identifies genes required for centromeric cohesion. Science 303:1367–1370 [DOI] [PubMed] [Google Scholar]
- 21.McDonald W. H., Yates J. R., III 2002. Shotgun proteomics and biomarker discovery. Dis. Markers 18:99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriya H., Johnston M. 2004. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. U. S. A. 101:1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neklesa T. K., Davis R. W. 2009. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 5:e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rousselet G., Simon M., Ripoche P., Buhler J. M. 1995. A second nitrogen permease regulator in Saccharomyces cerevisiae. FEBS Lett. 359:215–219 [DOI] [PubMed] [Google Scholar]
- 25.Schenk P. W., Boersma A. W., Brok M., Burger H., Stoter G., Nooter K. 2002. Inactivation of the Saccharomyces cerevisiae SKY1 gene induces a specific modification of the yeast anticancer drug sensitivity profile accompanied by a mutator phenotype. Mol. Pharmacol. 61:659–666 [DOI] [PubMed] [Google Scholar]
- 26.Schenk P. W., Brok M., Boersma A. W., Brandsma J. A., Den Dulk H., Burger H., Stoter G., Brouwer J., Nooter K. 2003. Anticancer drug resistance induced by disruption of the Saccharomyces cerevisiae NPR2 gene: a novel component involved in cisplatin- and doxorubicin-provoked cell kill. Mol. Pharmacol. 64:259–268 [DOI] [PubMed] [Google Scholar]
- 27.Soetens O., De Craene J. O., Andre B. 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276:43949–43957 [DOI] [PubMed] [Google Scholar]
- 28.Spielewoy N., Flick K., Kalashnikova T. I., Walker J. R., Wittenberg C. 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 24:8994–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueda K., Kawashima H., Ohtani S., Deng W. G., Ravoori M., Bankson J., Gao B., Girard L., Minna J. D., Roth J. A., Kundra V., Ji L. 2006. The 3p21.3 tumor suppressor NPRL2 plays an important role in cisplatin-induced resistance in human non-small-cell lung cancer cells. Cancer Res. 66:9682–9690 [DOI] [PubMed] [Google Scholar]
- 30.Wach A., Brachat A., Pohlmann R., Philippsen P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808 [DOI] [PubMed] [Google Scholar]
- 31.Willems A. R., Lanker S., Patton E. E., Craig K. L., Nason T. F., Mathias N., Kobayashi R., Wittenberg C., Tyers M. 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86:453–463 [DOI] [PubMed] [Google Scholar]
- 32.Yun C. Y., Fu X. D. 2000. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol. 150:707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zabarovsky E. R., Lerman M. I., Minna J. D. 2002. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene 21:6915–6935 [DOI] [PubMed] [Google Scholar]