Abstract
Fusarium oxysporum is the causative agent of fungal wilt disease in a variety of crops. The capacity of a fungal pathogen such as F. oxysporum f. sp. nicotianae to establish infection on its tobacco (Nicotiana tabacum) host depends in part on its capacity to evade the toxicity of tobacco defense proteins, such as osmotin. Fusarium genes that control resistance to osmotin would therefore reflect coevolutionary pressures and include genes that control mutual recognition, avoidance, and detoxification. We identified FOR (Fusarium Osmotin Resistance) genes on the basis of their ability to confer osmotin resistance to an osmotin-sensitive strain of Saccharomyces cerevisiae. FOR1 encodes a putative cell wall glycoprotein. FOR2 encodes the structural gene for glutamine:fructose-6-phosphate amidotransferase, the first and rate-limiting step in the biosynthesis of hexosamine and cell wall chitin. FOR3 encodes a homolog of SSD1, which controls cell wall composition, longevity, and virulence in S. cerevisiae. A for3 null mutation increased osmotin sensitivity of conidia and hyphae of F. oxysporum f. sp. nicotianae and also reduced cell wall β-1,3-glucan content. Together our findings show that conserved fungal genes that determine cell wall properties play a crucial role in regulating fungal susceptibility to the plant defense protein osmotin.
Studies of plant-pathogen interactions strongly suggest that under the pressure to survive, plants and pathogens continuously react to one another's defense arsenal and evolve to overcome these defenses (13). Plants recognize pathogen-associated molecular patterns, such as fungal cell wall fragments composed of chitin, glucans, oligosaccharides, or glycoprotein peptides (32). It has been established that pathogens evolved effector proteins to avoid plant surveillance mechanisms that recognize pathogen-associated molecular patterns and this in turn led to the evolution of plant surveillance mechanisms that recognize pathogen-specific effector proteins. All pathogen recognition mechanisms induce intracellular signaling that culminates in the synthesis of factors, such as antimicrobial plant proteins, that help in limiting the severity of infection (74). The antimicrobial proteins are therefore among the ultimate effectors of plant defense. There is evidence of recognition between plant antimicrobial proteins and pathogen-specific molecules (74). Therefore, pathogen mechanisms of resistance to the antimicrobial proteins and the antimicrobial proteins themselves must have coevolved. Consequently, we postulated that a screen for fungal genes that alter the sensitivity of a phytopathogen to an antifungal protein of the host plant (that is, a cognate plant defense effector) would lead to identification of genes involved in controlling pathogenicity, in controlling access of the antifungal protein to its target fungal molecules (such as genes controlling cell surface composition), and in controlling detoxification mechanisms.
The plant antifungal protein selected to test this hypothesis was osmotin. Osmotin is an antifungal protein that is overexpressed in and secreted by salt-adapted cultured tobacco (Nicotiana tabacum) cells (63). It is a member of a family of ubiquitous plant proteins, referred to as plant pathogenesis-related proteins of family 5 (PR-5), that are implicated in defense against fungi (74). Osmotin gene and protein expression is induced by biotic stresses, and overexpression of osmotin delays development of disease symptoms in transgenic plants (41, 42, 43, 84). The genetic bases of the susceptibility and resistance of Saccharomyces cerevisiae to osmotin have been explored in our laboratory (49, 50). The results show that specific interactions of osmotin with the plasma membrane are responsible for cell death signaling. However, because the cell wall governs access of osmotin to the plasma membrane, differences in cell wall composition largely account for the differential osmotin sensitivity of various S. cerevisiae strains, and specific cell wall components play a significant role in modulating osmotin toxicity (30, 31, 49, 50, 81, 82). These studies in the model nonpathogenic fungus, S. cerevisiae, support our hypothesis that a screen for genes that alter the sensitivity of a phytopathogenic fungus to an antifungal defense effector protein of the host plant will uncover genes involved in controlling access of the antifungal protein to its target fungal molecules.
Osmotin, like other plant defense antifungal proteins, has specific but broad-spectrum antifungal activity (74). One of the most osmotin-sensitive phytopathogenic fungi is Fusarium oxysporum. F. oxysporum is an ascomycete fungus, like S. cerevisiae, and has been touted as an appropriate multihost model for studying fungal virulence (53). It is a soilborne plant pathogen of economic significance, because it causes vascular wilt disease on a large variety of crop plants and produces toxic food contaminants (17, 58). In humans it also causes skin, nail, and eye disease that can become serious or life-threatening illnesses in immunocompromised patients (52, 69). F. oxysporum f. sp. lycopersici, F. oxysporum f. sp. nicotianae, and F. oxysporum f. sp. meloni, like S. cerevisiae, are quite sensitive to osmotin (1, 51; M. L. Narasimhan, unpublished data). Furthermore, it was recently shown that overexpression in F. oxysporum f. sp. nicotianae of an S. cerevisiae cell wall glycoprotein that increases the osmotin resistance of S. cerevisiae also increases the osmotin resistance of the plant pathogen and its virulence on tobacco, the osmotin-producing host plant (51). This suggested that osmotin resistance mechanisms may be conserved between S. cerevisiae and F. oxysporum and that S. cerevisiae could be used as a tool to uncover F. oxysporum genes that control osmotin sensitivity or resistance.
In the current study, we expressed an F. oxysporum f. sp. nicotianae cDNA library in the osmotin-sensitive S. cerevisiae strain BWG1-7a and selected genes for their ability to increase osmotin tolerance. We report here the identification and characterization of three FOR (Fusarium Osmotin Resistance) genes that affect the cell wall in S. cerevisiae. The product of FOR1 has homology with a putative cell surface glycoprotein; FOR2 encodes glutamine:fructose-6-phosphate amidotransferase (GFAT), an enzyme that catalyzes the first step in the biosynthetic pathway leading to amino sugar-containing macromolecules, such as glycoproteins and chitin (64); and FOR3 has high homology with S. cerevisiae SSD1, a gene that controls cell wall composition and virulence (31, 78). FOR2 and FOR3 are the functional equivalents of the corresponding S. cerevisiae genes. Our parallel analysis using two model fungi verifies the notion that cell wall proteins play a critical role in determining the sensitivity/resistance of fungi to osmotin. In addition, these results implicate that the tobacco defense protein, osmotin, can serve as an effective/useful tool in identifying genes that control cell wall composition not only in a model fungus, such as S. cerevisiae, but also in phytopathogenic fungi, such as F. oxysporum.
MATERIALS AND METHODS
Fungal strains, media, and culture conditions.
Unless specified otherwise, Saccharomyces cerevisiae strain BWG1-7a (MATa ade1-100 his4-159 leu2-3,112 ura3-52) was used throughout. S. cerevisiae strain RAY-3A (MATa his3 leu2 trp1 ura3) and its isogenic Δssd1::HIS3 derivative were kindly provided by Y. Uesono (University of Tokyo, Tokyo, Japan) and have been described elsewhere (73). YAT1588 (pir1::LEU2 pir2::HIS3 pir3::URA3) was the kind gift of A. Toh-e (University of Tokyo, Tokyo, Japan) and has been described (71). The Δgfa1::HIS3MX mutation, which replaced nucleotides 1074 to 3100 of the S. cerevisiae GFA1 (ScGFA1) open reading frame (ORF) with the HIS3 marker, was constructed using the PCR-based gene-targeting system (75) in S. cerevisiae strain W303-1A (MATa his3 leu2 trp1 ade2 ura3) (76). The S. cerevisiae strain JC1246-7A (MATa ade2 ade3 lys2-801 ura3-52 gfa1-97) (83) was the kind gift of J. F. Cannon (University of Missouri—Columbia, Columbia, MO). Standard procedures were followed for the growth and genetic manipulation of yeast (2, 60). Yeast cells were grown in either synthetic minimal medium (SD) (0.67% yeast nitrogen base without amino acids, 2% glucose, pH 6.5) with appropriate nutritional supplements or in YPD medium (1% yeast extract, 2% peptone, 2% glucose) at 28 to 30°C. Null gfa1 mutants are auxotrophic for d-glucosamine, and the gfa1 mutant was grown in media supplemented with d-glucosamine as described previously (79).
The wild-type strain of F. oxysporum f. sp. nicotianae was obtained from G. Chilosi (Universitá della Tuscia, Viterbo, Italy). Conditions and media used for cultivating the fungus have been described (51). Potato dextrose broth (PDB) and potato dextrose agar (PDA) used for cultivating the fungus were purchased from Sigma (St. Louis, MO). The gene replacement vector pfor3::HYG was used to create a F. oxysporum Δfor3 mutant. It contained FOR3 genomic DNA (−125 bp to +3941 bp) with a hygromycin resistance cassette inserted between +697 bp and +3205 bp, replacing approximately 2.4 kb of intervening FOR3 sequence. To construct pfor3::HYG, a 0.7-kb fragment containing the 3′ part of the FOR3 ORF (+3205 bp to +3941 bp) was amplified from F. oxysporum genomic DNA by PCR using the primer pair 5′-ATCGAGTCTTGCCGACGAT-3′/5′-TGAGATCCGTCTTCAGGATC-3′ and inserted into the EcoRV site of the pSTblue-1 vector (Novagen, Madison, WI). After sequencing, to establish the fidelity of the PCR and the direction of the insert, a 1.4-kb SalI fragment of plasmid pCB1003 (Fungal Genetics Stock Center) containing a hygromycin resistance gene cassette was inserted into the SalI site on this plasmid, yielding a construct that contained the hygromycin gene cassette fused to the 5′ end of the FOR3 fragment (+3205 bp to +3941 bp). Finally, a 0.8-kb fragment of FOR3 (−125 bp to +697 bp) was amplified by PCR from F. oxysporum genomic DNA using the primer pair 5′-GCTCTAGACAGCAGCAGCAGTCTTCTCAA-3′/5′-CCGCTCGAGGGCGTTGGTTCATTGCTT-3′ and inserted as an XbaI/XhoI fragment into the corresponding restriction enzyme sites in the previous construct to yield pfor3::HYG. To disrupt FOR3, pfor3::HYG was linearized by digestion with PstI and XbaI. Linearized pfor3::HYG DNA was used to transform protoplasts of F. oxysporum f. sp. nicotianae as described previously (51), except that protoplasts were incubated at 37°C for 15 min before addition of DNA. F. oxysporum is somewhat recalcitrant to targeted gene disruption due to a high frequency of ectopic integration of the vector (16). However, a dramatic increase in the frequency of gene replacement has been reported under conditions which induce expression and chromatin remodeling of the target gene locus (25, 68). Since heat shock treatment induces SSD1 expression in yeast, it was considered likely that FOR3 would also be induced by heat shock. Therefore, F. oxysporum protoplasts were subjected to heat shock before the addition of transforming DNA, and this was indeed found to increase the frequency of homologous integration events at the FOR3 locus. Hygr transformants were purified by single-spore isolation, and disruption of FOR3 was confirmed by PCR using the primer pairs 5′ ATCGTGATCAATCACCTTCGC-3′/5′-CAAGTTATCGTGCACCAAGCA-3, 5′-ATCGTGATCAATCACCTTCGC-3′/5′-TGTATTGACCGATTCCTTGCG-3′, and 5′ ATCGTGATCAATCACCTTCGC-3′/5′-TGAGATCCGTCTTCAGGATC-3′. The PCR conditions used were 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 4 min 30 s, with a 5-s extension at every cycle, and a final elongation step at 72°C for 10 min.
Antifungal tests.
Osmotin was purified from salt-adapted tobacco cell suspension cultures (Nicotiana tabacum L. var. Wisconsin 38) to apparent homogeneity as described previously (63). The IC50 (amount of osmotin that reduces growth by 50%) of different osmotin batches was determined as described previously using S. cerevisiae strain BWG1-7a as a standard (82) and ranged between 6 and 8 μg/ml. For quantitative measurement of growth inhibition by osmotin, overnight cultures S. cerevisiae in YPD or selective media were diluted to an A600 value between 0.01 and 0.05 in the same medium, and the diluted culture (320 μl) was mixed with various concentrations of osmotin in sterile water (80 μl). Cultures were then incubated at 28 to 30°C with shaking for 20 to 22 h, and growth was measured as the absorbance at 600 nm after appropriate dilution. For measurement of growth inhibition of S. cerevisiae spheroplasts by osmotin, cells were harvested in the exponential growth phase. They were suspended in 1 M sorbitol, and cell walls were digested by treatment with lyticase (Sigma, St. Louis, MO). Spheroplasts were collected by centrifugation, washed with YPD containing 1 M sorbitol, and embedded in osmotin-supplemented YPD agar containing 1 M sorbitol. Viable counts were determined after incubation at 28 to 30°C for 2 to 3 days.
To determine the osmotin sensitivity of F. oxysporum, conidia were harvested from cultures grown on PDA plates in 2× PDB. The conidial suspension was filtered through two layers of cheesecloth to remove mycelial fragments, and conidial numbers were determined using a hemacytometer. The effect of osmotin on hyphal elongation was tested exactly as described earlier (51). The test of osmotin's effect on conidial germination was performed essentially in the same manner. Equal volumes (100 μl) of conidial suspension (1 × 106 conidia/ml) and sterile osmotin solution (or water) were mixed in wells of a 24-multiwell tissue culture plate, aliquots (10 μl) were withdrawn from each well at the end of the 16 h of the incubation period, and the concentration of germinated conidia was determined in each aliquot using a hemacytometer. Each well was sampled three times for counting, and three replicates were used for each treatment.
DNA and RNA methods.
Procedures for isolation and fractionation of F. oxysporum genomic DNA and total RNA for Southern and Northern analyses have been described previously (51). For Southern analyses, nitrocellulose membrane blots of the gel were hybridized overnight with 32P-labeled probe at 55°C in Church buffer (7% SDS–1 mM EDTA–0.5 M potassium phosphate buffer, pH 7.2). The membrane was subjected to successive washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS for 15 min, 1× SSC–0.1% SDS for 10 min, and 0.5× SSC–0.1% SDS for 10 min at room temperature (to detect FOR2 and FOR3) or 65°C (to detect FOR1) and then exposed to X-ray film at −80°C for 18 to 48 h. Probes were prepared by random primer labeling (Ready-to-go labeling system; Amersham Biosciences, Piscataway, NJ). The entire 1.2-kb EcoRI/XhoI cDNA insert of pFOR1, a 1-kb BglII/NcoI fragment, pFOR2 (base pairs 225 to 1505 of the FOR2 ORF), and a 1-kb SphI/SacI fragment of pFOR3 (base pairs 1030 to 2111 of the FOR3 ORF) were used as probes except for the Δfor3 mutation. An 822-bp fragment of FOR3 (−125 bp to +697 bp) was used for probing Southern blots for confirming the Δfor3 mutation.
The F. oxysporum cDNA library for yeast expression was constructed in p416GPD, a single-copy plasmid vector for constitutive high-level expression of the inserted gene from the glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter. F. oxysporum mRNA was isolated from total RNA using the Quick prep micro mRNA purification kit (Amersham Biosciences, Piscataway, NJ) and mRNA separator kit (Clonetech, Mountain View, CA).
cDNA synthesis was accomplished using a cDNA synthesis kit (Stratagene, La Jolla, CA). The DNA fragments were ligated between the EcoRI and XhoI restriction enzyme sites of the p416GPD vector and transformed into Escherichia coli strain XL10-Gold (Stratagene, La Jolla, CA). Approximately 107 E. coli transformants were obtained, with an average insert size of 1 kb. Plasmid DNA was extracted from pools of these E. coli transformants and used to transform cells of S. cerevisiae strain BWG1-7a using the lithium acetate method of Elble (21). Primary transformants were selected on minimal medium lacking uracil, the auxotrophy selection marker of p416GPD. Approximately 3 × 105 URA+ primary transformants were collected as 6 different pools and stored at −80°C in selective minimal medium supplemented with 15% glycerol.
The F. oxysporum cDNA library in S. cerevisiae strain BWG1-7a was screened by plating approximately 105 cells from each pool of yeast transformants on an YPD agar plate containing osmotin (30 μg/ml). The osmotin resistance phenotype of the colonies that appeared after incubation at 28 to 30°C for 2 to 3 days was confirmed by quantitative measurement of growth inhibition by osmotin. Plasmid DNA was extracted from each osmotin-resistant yeast transformant as described previously (56) and amplified by passage through E. coli. Each plasmid DNA was then used to transform S. cerevisiae strain BWG1-7a, and the osmotin resistance phenotype of the transformant was retested by quantitative measurement of growth inhibition by osmotin. Inserts of plasmids that consistently increased osmotin resistance of S. cerevisiae strain BWG1-7a were sequenced using the vector primers GPD2-5′ (5′-CTTAGTTTCGACGGATTC-3′) and Tcyc-3′ (5′-TTCGGTTAGAGCGGATGTGG-3′).
DNA was sequenced at the Recombinant DNA/Protein Resource Facility (Iowa State University, Ames, IA). DNA and protein homology searches were performed using BLAST in the Saccharomyces Genome Database (http://www.yeastgenome.org), Fusarium Group Database (http://www.broad.mit.edu), and GenBank (http://www.ncbi.nlm.nih.gov/Genbank/). Sequence alignments were performed by using the CLUSTAL W software program (70) at the Pole Bioinformatique Lyonnais Network Protein Sequence Analysis database at http://npsa-pbil.ibcp.fr/. Signal sequence prediction was made using the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/) (5). N-glycosylation sites and PHD transmembrane helix predictions were made using a PROSCAN search and the PHD secondary structure prediction method (57), respectively, at the Pole Bioinformatique Lyonnais Network Protein Sequence Analysis database at http://npsa-pbil.ibcp.fr/. Domain searches were performed using the Pfam23.0 database search (http://pfam.sanger.ac.uk/) (22). Glycosylphosphatidylinositol (GPI) anchor prediction was made using the big-PI Fungal predictor GPI Fungal Prediction Server, version 3.1 (http://mendel.imp.ac.at/) (20).
Immunoelectron microscopy.
Conidia were harvested in PDB from F. oxysporum cultures grown on PDA plates and filtered through two layers of cheesecloth to remove mycelial fragments. They were fixed and embedded as described by Mulholland et al. (48). Immunoreactions were carried out on thin sections essentially as described previously (51) with monoclonal antibodies to β-(1,3)-glucan (Biosupplies Australia, Parkville, Australia; 1:500 dilution). To establish specificity of the immunoreaction, sections were also incubated with monoclonal antibodies to β-(1,3)-glucan that had been previously adsorbed with laminarin (100 μg/ml; Sigma, St. Louis, MO) for 1 h at 4°C, as described previously (6). Rabbit anti-mouse IgG conjugated to 10-nm gold particles (1:50 dilution) was used as a secondary antibody. A Philips CM-10 Biotwin TEM (FEI Company, Hillsboro, OR) instrument was used for electron microscopy.
RESULTS
Isolation of cDNA clones.
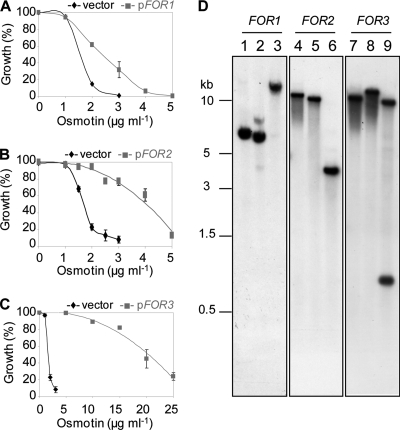
An F. oxysporum f. sp. nicotianae cDNA library was constructed in the low-copy-number yeast shuttle vector p416GPD, which allows constitutive high-level expression from yeast glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter. The cDNA library was transformed into S. cerevisiae strain BWG1-7a, an unusually osmotin-sensitive strain that has been successfully used to characterize the genetic basis of osmotin tolerance/sensitivity in yeast (31, 49, 81, 82). From a screen of about 3 × 105 primary transformants on medium containing 30 μg/ml osmotin, 52 osmotin-resistant colonies were selected. Plasmids from 15 of these 52 osmotin-resistant colonies were confirmed to confer an osmotin-resistant phenotype upon retransformation into S. cerevisiae strain BWG1-7a. These plasmids represented 7 unique inserts, all of which had significant homology to predicted cDNAs encoded in the genome of F. oxysporum f. sp. lycopersici (www.broad.mit.edu). Transformation with three plasmids, designated pFOR1, pFOR2, and pFOR3, was reproducibly associated with significant increases in the IC50 of osmotin (Fig. 1A, B, and C, respectively), and the inserted sequences appeared to be full-length cDNAs. Southern analysis of F. oxysporum f. sp. nicotianae genomic DNA, using labeled fragments of pFOR1, pFOR2, or pFOR3 inserts as a probe, showed that the corresponding FOR1, FOR2, and FOR3 genes occurred as single-copy genes in the F. oxysporum f. sp. nicotianae genome (Fig. 1D). FOR1, FOR2, and FOR3 were therefore selected for further study. Results obtained with the other genes will be presented elsewhere.
Fig. 1.
FOR1, FOR2, and FOR3 increase osmotin resistance of S. cerevisiae cells and are single-copy genes in F. oxysporum f. sp. nicotianae. (A, B, and C) Shown are osmotin sensitivities of cells of S. cerevisiae strain BWG1-7a transformed with p416GPD (vector) or the cDNA clone pFOR1 (A), pFOR2 (B), or pFOR3 (C), which were compared by estimating growth in the presence of various concentrations of osmotin. Growth of cells was measured in liquid culture in selective minimal medium supplemented with the indicated osmotin concentrations. Data were normalized to viable counts of samples without osmotin. Data are the averages ± standard errors (SE) of results from at least three experiments with duplicate samples. (D) Shown are blots of restriction enzyme-digested genomic DNA (15 μg) probed with 32P-labeled FOR1 (lanes 1, 2, and 3), FOR2 (lanes 4, 5, and 6), and FOR3 (lanes 7, 8, and 9). The restriction enzymes used were EcoRI (lanes 1, 4, and 7), PstI (lanes 2, 5, and 8), and XbaI (lanes 3, 6, and 9).
Characterization of FOR1.
The pFOR1 insert (1.5 kb) included an open reading frame of 468 nt with significant homology to hypothetical proteins encoded in the genomes of F. oxysporum f. sp. lycopersici (FOXG_05281.2; 99% identity), Fusarium graminearum (FGSG_10125.3; 86% identity), and Fusarium verticillioides (FVEG_02849; 94% identity) (see Fig. S1 in the supplemental material). FOR1 has 43% identity to S. cerevisiae SED1 (ScSED1) (YDR077w) and 24% identity to S. cerevisiae SPI1 (ScSPI1) (YER150w). However, a 43-amino-acid stretch in FOR1 is highly conserved in ScSED1 (70% identity) and ScSPI1 (60% identity) (see underline in Fig. S1). This sequence occurs as an interrupted repeat sequence in ScSED1. It is also found in hypothetical proteins of several filamentous fungi, such as Neurospora crassa (XM_95593.1; 80% identity), Coccidioides immitis (XM_001247153; 69% identity), and Chaetomium globosum (XM_001219801; 69% identity).
ScSED1(338 amino acids) is a serine- and threonine-rich, GPI-anchored, highly N- and O-glycosylated protein that accumulates as the major protein in cell walls of stationary-phase S. cerevisiae cells (8, 20, 61). Consequently, the phenotype of Δsed1 mutants is increased sensitivity to the cell wall hydrolyzing enzyme, zymolyase. SED1 is induced by stress and presumably has a role in strengthening the cell wall under stress conditions (28). ScSPI1 (149 amino acids) is also a serine- and threonine-rich, GPI-anchored, stress-induced cell wall glycoprotein (29). FOR1 is predicted to have a signal sequence and a Ser/Thr-rich 43-amino-acid stretch that is conserved in ScSED1 or ScSPI1 (see Fig. S1 in the supplemental material). Bioinformatic analyses predict that FOR1 also has N-glycosylation sites, a GPI anchor addition signal, and a 17-amino-acid C-terminal hydrophobic stretch (see Fig. S1). Thus, FOR1 is very likely to be a cell wall glycoprotein.
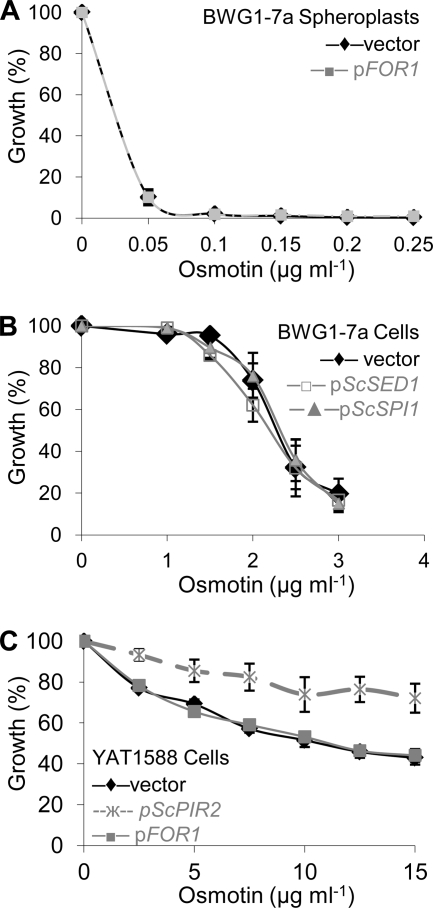
The bioinformatic analyses suggesting that FOR1 encodes a cell wall protein were supported by the observation that osmotin resistance conferred to S. cerevisiae by FOR1 requires the cell wall (Fig. 2A). Enzymatic removal of the cell wall of S. cerevisiae transformed with pFOR1 or the corresponding p416GPD vector yielded spheroplasts that were equally sensitive to osmotin. Since FOR1 is predicted to be a cell wall protein and shares a highly conserved domain with ScSED1 and ScSPI1 (see Fig. S1 in the supplemental material), we tested the effect of ScSED1 or ScSPI1 overexpression on the osmotin resistance phenotype of yeast cells. Unlike FOR1 overexpression, ScSED1 or ScSPI1 overexpression did not increase osmotin resistance (Fig. 2B). So far, the only fungal cell wall proteins known to increase osmotin resistance in S. cerevisiae are the PIR proteins (82). PIR proteins of S. cerevisiae and Candida albicans are a family of covalently linked O-glycosylated cell wall proteins that have a role in strengthening the cell wall under stress (35, 36). S. cerevisiae PIR (ScPIR) proteins were identified as osmotin resistance determinants in a screen similar to the one used to identify FOR1 (82). Although the amino acid sequences of the PIR proteins and FOR1 do not share significant homology and a characteristic repeated sequence (TAAAVSQIGDGQIQATTKT) present in PIR proteins is absent in FOR1 (see Fig. S1), it is possible that they function interchangeably in the cell wall, because PIR proteins are anchored to the cell wall (19) and bioinformatic analyses predict that FOR1 is GPI anchored (see Fig. S1). To test whether PIR proteins and FOR1 function interchangeably, we transformed FOR1 into an S. cerevisiae Δpir1 Δpir2 Δpir3 strain. It is known that the PIR family proteins are functionally redundant for conferring osmotin resistance in S. cerevisiae, so that simultaneous deletion of at least three family members is required to increase sensitivity to osmotin and, conversely, overexpression of any ScPIR gene increases osmotin resistance of an S. cerevisiae Δpir1 Δpir2 Δpir3 strain (82) (Fig. 2C). However, overexpression of FOR1 did not increase osmotin resistance of the S. cerevisiae Δpir1 Δpir2 Δpir3 strain (Fig. 2C). Thus, FOR1 cannot substitute for PIR proteins to confer osmotin resistance. However, it is worth noting that while overexpression of FOR1 does not confer resistance to a Δpir1 Δpir2 Δpir3 mutant (Fig. 2C), it increases osmotin resistance of S. cerevisiae strain BWG1-7a (Fig. 1A). S. cerevisiae strain BWG1-7a has the ssd1-d allele but contains a significant amount of PIR proteins in its cell wall, unlike the Δpir1 Δpir2 Δpir3 mutant (82, 31). Therefore, it follows that FOR1 requires cell wall-localized PIR proteins to function in osmotin resistance, and this lends further support to the notion that FOR1 is cell wall localized.
Fig. 2.
The role of FOR1 in osmotin resistance. (A) Overexpression of FOR1 does not increase osmotin resistance of yeast spheroplasts. Shown are osmotin sensitivities of spheroplasts of S. cerevisiae strain BWG1-7a transformed with p416GPD (vector) or FOR1 in the same vector (pFOR1). Growth of spheroplasts was measured by embedding 500 spheroplasts in YPD agar supplemented with 1 M sorbitol and the indicated concentration of osmotin and determining the number of viable colonies after 3 days of incubation at 30°C. Data were normalized to viable counts of samples without osmotin. Data are the averages ± standard deviations of results from at least two experiments with duplicate samples. (B) Overexpression of ScSED1 and ScSPI1 does not increase osmotin resistance of yeast cells. Cells of S. cerevisiae strain BWG1-7a transformed with p416GPD (vector), as well as p416GPD containing ScSED1 (pScSED1) or ScSPI1 (pScSPI1) inserts, were grown in liquid culture in selective minimal medium supplemented with the indicated osmotin concentrations. Growth was measured as the absorbance at 600 nm and normalized to the absorbance of control cultures grown without osmotin. Data are the averages ± SE of results from at least two experiments with three samples. (C) The function of FOR1 is distinct from that of the ScPIR genes. Cells of S. cerevisiae strain YAT1588 (Δpir1 Δpir2 Δpir3) transformed with p416GPD (vector), pFOR1, or ScPIR2 in the same vector (pScPIR2) were grown in liquid culture in selective minimal medium supplemented with the indicated osmotin concentrations. Growth was measured as the absorbance at 600 nm and normalized to the absorbance of control cultures grown without osmotin. Data are the averages ± SE of results from at least two experiments with three samples.
Characterization of FOR2.
The sequence of the FOR2 gene revealed an open reading frame of 2,100 nucleotides (nt), encoding a protein of 699 amino acids with homology to GFA (see Fig. S2 in the supplemental material), the gene encoding glutamine:fructose-6-phosphate amidotransferase (GFAT). GFAT has been identified from bacterial, fungal, plant, and mammalian sources (3, 18, 46, 77). It catalyzes the formation of glucosamine-6-phosphate, the first step in the biosynthetic pathway leading to amino sugar-containing macromolecules, such as glycoproteins and chitin (64). A single locus with significant sequence identity to FOR2 exists in the genomes of F. oxysporum f. sp. lycopersici (FOXG_00190.2; 100% identity), F. graminearum (FGSG_01199.3; 94% identity), and F. verticillioides (FVEG_01326.3; 97% identity) (data not shown). FOR2 has significant but lower homology with GFAT of S. cerevisiae (60% identity), C. albicans (60% identity), Escherichia coli (36% identity), and Homo sapiens (52% identity) (see Fig. S2; also data not shown). However, GFAT-specific domains were particularly conserved in FOR2 (data not shown; also see Fig. S2). FOR2 has a glutamine amidotransferase class 2 domain (amino acids 1 to 199) and two sugar isomerase (SIS) domains, structures that are contained in all GFAT enzymes (see Fig. S2). SIS domains are found in many phosphosugar isomerases and phosphosugar binding proteins (4). These sequence comparisons/features strongly suggest that FOR2 is the structural gene for the F. oxysporum glutamine:fructose-6-phosphate amidotransferase.
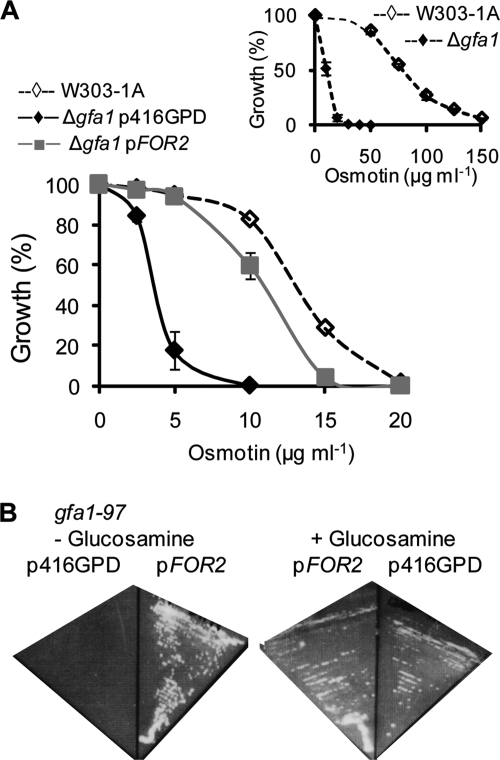
A null mutation in ScGFA1, the gene encoding ScGFAT, increases osmotin sensitivity of the osmotin-tolerant S. cerevisiae strain W303-1A (Fig. 3A, inset). Transformation of the osmotin-sensitive W303-1A Δgfa1 mutant with pFOR2, which expresses FOR2 from the constitutive GPD promoter in the p416GPD vector, restores osmotin resistance comparable to that of the wild-type S. cerevisiae strain W303-1A (Fig. 3A). Having established that FOR2 can replace the function of ScGFA1 in osmotin resistance, we next examined whether FOR2 functions in glucosamine biosynthesis. The S. cerevisiae strain JC1246-7A (gfa1-97) has a point mutation in GFA1 that confers a temperature-sensitive glucosamine-auxotrophic phenotype (83). Thus, transformants of this strain that carry the vector p416GPD are glucosamine auxotrophs at 37°C (Fig. 3B). However, transformants carrying pFOR2 revert to glucosamine prototrophy at 37°C, indicating that FOR2 functions in glucosamine biosynthesis, as expected from its sequence homology with GFAT from various sources. Collectively, these data establish that FOR2 is a structural gene for glutamine:fructose-6-phosphate amidotransferase.
Fig. 3.
FOR2 is the functional homolog of ScGFA1, the gene for GFAT. (A) Cells of S. cerevisiae strain W303-1A and an isogenic Δgfa1::HIS3MX strain transformed with vector (Δgfa1p416GPD) or FOR2 in the same vector (Δgfa1pFOR2) were grown in liquid culture in selective minimal medium supplemented with the indicated osmotin concentrations. Growth was measured as the absorbance at 600 nm and normalized to the absorbance of control cultures grown without osmotin. Data are the averages ± SE of results from two experiments with three independent transformants for each strain. Inset: relative osmotin sensitivities of S. cerevisiae strain W303-1A and the isogenic Δgfa1::HIS3MX strain (Δgfa1) in YPD medium in a similar assay are shown. Data are the averages ± SE of results from two independent experiments. (B) Shown are plates of S. cerevisiae strain JC1246-7A (gfa1-97) transformed with p416GPD or pFOR2, grown for 3 days at 37°C in selective minimal medium without (−) or with (+) glucosamine (1 mg/ml) supplement.
Characterization of FOR3.
The pFOR3 insert includes an open reading frame of 3,954 nt, encoding a protein of 1,318 amino acids that has significant sequence similarity to SSD1 of various fungi (see Fig. S3 in the supplemental material). Hypothetical proteins with high similarity to FOR3 are encoded by single loci in the genomes of F. oxysporum f. sp. lycopersici (FOXG_01798.2; 90% identity), F. verticillioides (FVEG_08172.3; 89% identity), and F. graminearum (FGSG_07009.3; 87% identity) (see Fig. S3; also data not shown). FOR3 has significant but lower identity with SSD1 of other fungi: Colletotrichum lagenarium (76% identity), Magnaporthe grisea (78% identity), C. albicans (44% identity), and S. cerevisiae (40% identity) (see Fig. S3; also data not shown). Furthermore, the C-terminal RNase II homology domain that is essential for ScSSD1 function is conserved in FOR3 (see Fig. S3) (73, 80).
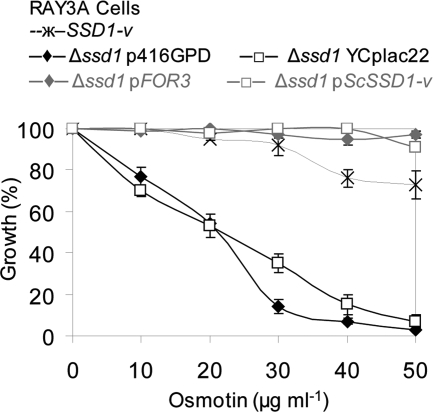
ScSSD1 is the major determinant of osmotin resistance in S. cerevisiae, with the natural polymorphism of SSD1 in strains of S. cerevisiae accounting for most of their differential sensitivities to osmotin (31). The SSD1-v allele, which encodes a 160-kDa protein, can suppress mutations in genes that regulate cellular integrity, cell cycle progression, growth at high temperatures, differentiation, and life span, whereas the SSD1-d allele, which encodes an 83-kDa C-terminally truncated version of the protein, does not have these suppressor functions (33, 34, 39, 45, 65, 72). The osmotin-sensitive phenotype of S. cerevisiae strain BWG1-7a, which has the SSD1-d allele, can be suppressed by the SSD1-v allele (31). It is interesting to note that FOR3, which is the structural equivalent of the SSD1-v allele in S. cerevisiae, produces the greatest increase in osmotin resistance of S. cerevisiae strain BWG1-7a of the three FOR genes isolated (see Fig. S3 in the supplemental material) (Fig. 1C). The functional equivalence of FOR3 and ScSSD1 was further confirmed by comparing the osmotin sensitivities of an S. cerevisiae Δssd1 strain (Ray3A Δssd1) transformed with plasmids expressing or not expressing either FOR3 or ScSSD1-v (Fig. 4). The Δssd1 transformants expressing FOR3 or ScSSD1-v were resistant to osmotin, just like the wild-type strain Ray3A (SSD1-v), while the vector transformants remained sensitive to osmotin, as expected (31). These results demonstrate the functional equivalence of FOR3 and ScSSD1 in S. cerevisiae.
Fig. 4.
FOR3 is the functional homolog of ScSSD1. Shown is the relative growth of cells of isogenic S. cerevisiae strains RAY3A (SSD1-v) and RAY3AΔssd1::HIS3MX transformed with the vector p416GPD (Δssd1p416GPD), FOR3 in p416GPD (Δssd1pFOR3), vector YCplac22 (Δssd1YCplac22), or ScSSD1-v in YCplac22 (Δssd1pScSSD1-v) in minimal selective medium supplemented with the indicated osmotin concentrations. Values are the averages ± SE of results from two independent experiments and are normalized to the A600 of control cultures without osmotin.
FOR3 controls cell wall composition and osmotin sensitivity in F. oxysporum.
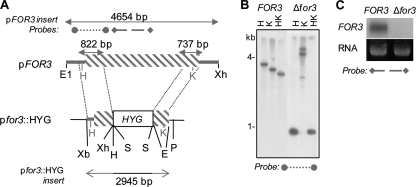
To investigate the function of FOR3 in F. oxysporum, a gene disruption vector, pfor3::HYG, was constructed in which 2,466 bp of the FOR3 coding region was replaced by the hygromycin resistance cassette from pCB1003 (Fungal Genetics Stock Center) (Fig. 5A). Protoplasts of F. oxysporum f. sp. nicotianae were transformed with linearized pfor3::HYG, and hygromycin-resistant colonies were screened by PCR for the presence of the insertion in the FOR3 gene. Among single-spore isolates of several independent PCR-positive transformants, one transformant (Δfor3) exhibited a banding pattern on Southern blots that was consistent with a homologous recombination event and replacement of the FOR3 gene (Fig. 5B). Northern analysis confirmed the absence of the FOR3 transcript in the mutant (Fig. 5C). The Δfor3 strain was therefore used for further study.
Fig. 5.
Disruption of the FOR3 gene in F. oxysporum f. sp. nicotianae. (A) Schematic view of the FOR3 gene disruption strategy. The DNA inserts of pFOR3 and pfor3::HYG are shown. The FOR3 coding region is indicated by the gray striped box and noncoding regions by the gray line. The vector sequences are indicated by the black line. Restriction sites on the cloning vector are indicated in black, and those in FOR3 are indicated in gray. The sizes of cloned DNA fragments (double-headed arrows), as well as the probes used for Southern (dotted line) or Northern (dashed line) blot analyses, are indicated. Restriction site abbreviations are as follows: E, EcoRV; E1, EcoRI; H, HindIII; K, KpnI; P, PstI; S, SalI; Xb, XbaI; Xh, XhoI. (B) Southern blot of restriction-digested genomic DNA of the wild-type (FOR3) and Δfor3 mutant strains. Restriction site abbreviations are as in panel A. (C) Northern blot of total RNA of the wild-type (FOR3) and Δfor3 mutant strains with a FOR3 probe (FOR3) and the corresponding ethidium bromide signal (RNA) shown.
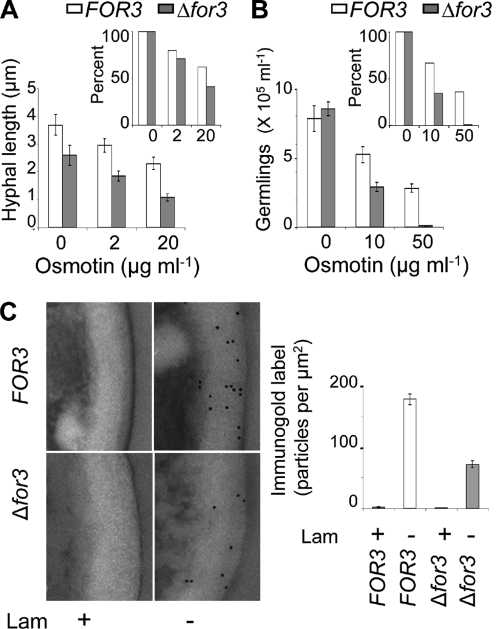
To ascertain the effect of the Δfor3 mutation on osmotin sensitivity, conidia from the Δfor3 and wild-type strains of F. oxysporum f. sp. nicotianae were incubated in growth medium for 16 h in the presence of various concentrations of osmotin. In both strains, all the conidia germinated at the lower concentrations of osmotin used, and the lengths of the emergent hyphae were affected inversely in relation to the concentration of osmotin. At higher concentrations of osmotin, only a fraction of the conidia were able to germinate. Thus, the effect of osmotin on hyphal length and ability to germinate were quantified separately (Fig. 6A and 6B). By analysis of variance (ANOVA) at the level of α = 0.05, the P value of the hyphal lengths of the wild type and Δfor3 mutant in the absence of osmotin was <0.05, indicating that the Δfor3 mutant had significantly shorter hyphae even in the absence of osmotin (Fig. 6A). The average hyphal lengths after osmotin treatment were therefore normalized to the average hyphal lengths of control samples without added osmotin. Figure 6A shows that the inhibition of hyphal elongation by osmotin was greater in the Δfor3 mutant than in the wild-type strain of F. oxysporum f. sp. nicotianae (about 1.5-fold difference at 20 μg/ml osmotin; Fig. 6A, inset). The effect of the Δfor3 mutation on the ability of conidia to germinate (or hyphal emergence), measured as the number of germlings obtained after 16 h of incubation of conidia in growth medium, is shown in Fig. 6B. By ANOVA at the level of α = 0.05, the P value of the number of germlings of the wild type and Δfor3 mutant in the absence of osmotin was >0.05, indicating no significant effect of the Δfor3 mutation on the ability of conidia to germinate. Osmotin significantly inhibited the ability of conidia to germinate in both wild-type and Δfor3 strains, but the inhibitory effect was much more pronounced in the Δfor3 mutant. In order to visualize the results more clearly, the average number of germlings observed in osmotin-treated samples was normalized to the average number of germlings observed in the sample without osmotin and expressed as a percentage (Fig. 6B, inset). The results clearly demonstrated that the inhibition of conidial germination (or hyphal emergence) by osmotin was greater in the Δfor3 mutant than in the wild-type strain (about a 33-fold difference at 50 μg/ml osmotin; Fig. 6B, inset). These data show that a null mutation in FOR3 results in increased sensitivity to osmotin in F. oxysporum.
Fig. 6.
Disruption of the FOR3 gene increases osmotin sensitivity and alters cell wall composition. (A) Inhibition of hyphal elongation by osmotin. Hyphal lengths of germlings of the wild type (FOR3) and Δfor3 mutant strain (Δfor3) after 16 h of incubation with the indicated osmotin concentration are shown. The inset shows the same data normalized to the average hyphal length in the absence of osmotin. Each value is the mean of three different observations ± SE. (B) Inhibition of conidial germination by osmotin. Conidia (106/ml) were incubated with the indicated osmotin concentrations, and the concentration of germinated conidia of the wild type (FOR3) or the Δfor3 mutant strain (Δfor3) after 16 h of incubation is shown. The inset shows the same data normalized to the average concentration of germlings in the absence of osmotin. Values are the means of three different observations ± SE. (C) Immunocytochemical measurement of cell wall β-1,3-glucan content. Shown are ultrathin sections of conidia of the wild-type (FOR3) and Δfor3 mutant strains that were incubated with β-1,3-glucan antibody preadsorbed (+) or not (−) with laminarin (Lam). Secondary antibodies were conjugated to 10-nm gold particles, which appear as small black dots at sites of positive reaction. The graph shows the average number of gold particles on the cell walls in these samples ± SE (n = 10 cells).
In S. cerevisiae, SSD1 controls cell wall composition (31, 78). To determine whether FOR3, the homolog of SSD1 in F. oxysporum, regulates cell wall composition, conidia of the wild-type and Δfor3 strains of F. oxysporum f. sp. nicotianae were subjected to immunocytochemical analysis in ultrathin sections with an antibody that specifically recognizes linear β-1,3-glucan. β-1,3-Glucan occurs specifically in fungal cell walls, and accordingly immunogold labeling was observed only in the cell wall of both strains (Fig. 6C). Preadsorption of the antibody with laminarin, a linear β-1,3-glucan, completely abolished labeling, confirming that the immunoreaction was specifically detecting linear β-1,3-glucan. Quantitative measurement of immunogold labeling as the average densities of the gold particles per μm2 showed that the cell wall content of linear β-1,3-glucan is reduced in the Δfor3 mutant to 40% of that of the wild-type strain (Fig. 6C). Taken together, these data demonstrate that FOR3 controls osmotin sensitivity as well as cell wall composition in F. oxysporum and is both the structural and functional equivalent of S. cerevisiae SSD1.
DISCUSSION
We have identified FOR1, FOR2, and FOR3 as three F. oxysporum genes that increase osmotin resistance in a heterologous fungal species. Interestingly, all three genes affect the cell wall of S. cerevisiae. The cell walls of S. cerevisiae and other ascomycetous fungi are similar and contain mainly β-1,3-glucans, glycoproteins, and chitin, along with smaller amounts of α-1,3-glucans or β-1,6-glucans (15, 38, 59). It is therefore not surprising that some osmotin resistance determinants are conserved between ascomycetous fungi.
FOR3.
Several lines of evidence suggest that FOR3 is the structural and functional homolog of ScSSD1 (Fig. 4, 5, and 6). First, FOR3 has significant similarity with SSD1 of other fungi (see Fig. S3 in the supplemental material). Second, overexpression of FOR3 confers osmotin resistance to the S. cerevisiae BWG1-7a strain, which carries a truncated ssd1-d allele (Fig. 1). Third, FOR3 complements the osmotin sensitivity phenotype of an S. cerevisiae Δssd1 mutant (Fig. 4). Like FOR3, SSD1 orthologs from other fungi show considerable sequence similarity with the full-length S. cerevisiae SSD1-v allele and can complement phenotypes of S. cerevisiae strains carrying the truncated ssd1-d allele or null Δssd1 mutations. C. albicans SSD1 can suppress synthetic lethality of swi4 ssd1-d mutations in S. cerevisiae that are also suppressed by the ScSSD1-v allele (12). SSD1 of the cucumber anthracnose fungus Colletotrichum lagenarium can suppress the caffeine sensitivity phenotype of an S. cerevisiae Δssd1 mutant (67). Fourth, null mutation of FOR3 reduced the β-1,3-glucan content of the cell wall and increased osmotin sensitivity of F. oxysporum f. sp. nicotianae. Likewise, null mutation of ScSSD1, the major osmotin resistance determinant of S. cerevisiae, results in a slight deficiency of β-glucans accompanied by an increase in mannan and chitin in both laboratory and feral strains (31, 78). It is not surprising that FOR3 was isolated as the major osmotin resistance determinant in our screen, since the S. cerevisiae strain BWG1-7a that was transformed with the F. oxysporum cDNA library has the defective truncated ssd1-d allele (31). This finding is consistent with our previous work indicating SSD1 as the major osmotin resistance determinant in S. cerevisiae (31).
FOR2.
FOR2 encodes a functional glutamine:fructose-6-phosphate amidotransferase (GFAT) since it was able to complement both the osmotin sensitivity and glucosamine auxotrophy phenotypes of S. cerevisiae gfa1 mutants (Fig. 3; see also Fig. S2 in the supplemental material). GFAT is the highly conserved first and rate-limiting enzyme in the highly conserved biosynthetic pathway for UDP-N-acetylglucosamine, which is the donor of N-acetylglucosamine monomer units incorporated into glycoproteins and chitin (64). In Aspergillus nidulans, an increase in cell wall chitin content is associated with increased osmotin resistance (14). In S. cerevisiae, it has been shown that the availability of donor monomer units for chitin biosynthesis is normally limiting (10). The expression level of GFA1 is therefore a major determinant of cell wall chitin content (40). Therefore, it is plausible that increased cell wall chitin content contributes to the osmotin resistance associated with overexpression of FOR2 in S. cerevisiae. In S. cerevisiae, F. oxysporum, A. niger, and Penicillium chrysogenum, GFAT is induced by cell wall stressors such as calcofluor white (7, 24, 40, 55). The induced expression of GFAT is accompanied by an increase in cell wall chitin content, suggesting that GFAT content could be a major factor limiting chitin content of the cell walls of many ascomycete fungi (55). Therefore, it is very likely that FOR2 will be an osmotin resistance determinant in F. oxysporum, just as GFA1 controls osmotin resistance in S. cerevisiae.
GFA1 and SSD1 are conserved master controllers of cell wall strength and osmotin resistance.
The cell walls of S. cerevisiae and other ascomycetous fungi are similarly organized (15, 37). The inner layer of the cell wall of S. cerevisiae contains chitin and alkali-insoluble cross-linked glucan and thereby makes a major contribution to the mechanical strength of the cell wall (9, 27, 37, 38, 54). SSD1 and GFA1 control the structure and composition of this layer. SSD1 controls the content of alkali-insoluble cross-linked glucan, and GFA1 controls chitin content (30, 40). In addition, SSD1 controls deposition of PIR glycoproteins that are known to be cross-linked to cell wall β-1,3-glucan fibrils of the inner wall (31, 36). Cell wall and electron microscopy analyses show that Δssd1 mutants of S. cerevisiae have weak walls and altered composition, which are suggestive of cross-linking defects (31, 54). Null SSD1 mutants of S. cerevisiae also exhibit increased sensitivity to cell wall perturbing agents (66). Deletion of orthologs of SSD1 in other fungi is also shown to weaken the cell wall. For instance, deletion of the SSD1 ortholog in the human pathogenic yeast Cryptococcus neoformans results in increased sensitivity to cell wall-degrading enzymes (26). Null ssd1 mutants of C. lagenarium and Magnaporthe grisea have a lower rate of hyphal growth and increased sensitivity to caffeine, which is an indicator of cell wall weakening (67). In fact, we also observed that hyphal growth was retarded in the Δfor3 mutant of F. oxysporum compared to the wild-type strain even in the absence of osmotin (Fig. 6A). Deletion of the gene encoding GFAT in S. cerevisiae and A. niger results in glucosamine insufficiency and is therefore lethal (55, 83). A temperature-sensitive mutant of S. cerevisiae GFA1 fails to grow normally when glucosamine is limiting unless osmotic stabilizers are added to the medium, indicating that GFA1 is a major contributor to cell wall strength (83). Conversely, increasing gene dosage of SSD1 and GFA1 compensates for cell wall weakening in S. cerevisiae (33, 34, 40, 44) and also confers resistance to osmotin (82). All these data provide further evidence that cell wall weakening is closely associated with increased sensitivity to osmotin. Genes such as GFA1 and SSD1, which are master controllers of cell wall strength, are therefore conserved fungal osmotin resistance determinants.
FOR1 and the divergent relationship between cell wall glycoprotein structure and function.
Cell wall glycoproteins of ascomycetous fungi form a layer surrounding the core β-glucan and chitin-containing layer (37). Due to the bulk of the carbohydrate moieties involved in N glycosylation and the presence of disulfide bridges, the cell wall glycoproteins determine permeability of the cell wall to macromolecules (38). Cell wall glycoproteins also have a limited role in maintaining and strengthening the cell wall (15). We have shown that heterologous expression of ScPIR2 in F. oxysporum targets the protein to the F. oxysporum cell wall and increases osmotin resistance, suggesting that cell wall targeting mechanisms are conserved between these fungi (51). However, FOR1 presents an example of a divergent structure-function relationship of glycoprotein between fungi. Although FOR1 is predicted to be a cell wall protein similar to ScSED1/ScSPI1 based on sequence analysis (see Fig. S1 in the supplemental material) and is unable to confer osmotin resistance to S. cerevisiae if cell walls are removed (Fig. 2A), neither ScSED1 nor ScSPI1 was able to confer osmotin resistance to the osmotin-sensitive yeast strain BWG1-7a as FOR1 was (Fig. 2B). Additionally, while FOR1 cooperates with ScPIR proteins, it cannot replace their function in increasing osmotin resistance of S. cerevisiae (Fig. 2C). We can only speculate about the mechanism by which FOR1 increases osmotin resistance in S. cerevisiae. It does not contain the signature DGQJQ sequence (J is any hydrophobic amino acid) that is essential for cross-linking of PIR glycoproteins of S. cerevisiae to β-1,3-glucan and thereby strengthening the cell wall (11, 36, 37, 47). However, FOR1 has a predicted GPI anchor sequence for covalent linkage to cell wall β-1,6-glucan (see Fig. S1) (37). Glycoproteins that are anchored by the remnant of a GPI anchor to cell wall β-glucan are known to contribute to cell wall strengthening (28, 61, 62). FOR1 could also be bound to other cell wall proteins by disulfide bonds or noncovalent linkages, mechanisms that are known to anchor proteins in the S. cerevisiae cell wall (37). The existence of putative N-glycosylation sites, a Ser/Thr-rich domain containing multiple cysteine residues, and a C-terminal domain with weak homology to collagen domains (see Fig. S1; collagen domains are annotated as Pfam PF01391 and can form triple helices and multimers of these helices) suggests that FOR1 could exist in the cell wall as high-molecular-weight bulky oligomers of an N- and O-glycosylated monomer. The expression level of PIR proteins correlates inversely with cell wall permeability in S. cerevisiae (37). Therefore, on the basis of the data in Fig. 2C, we can consider the possibility that the mechanism by which FOR1 increases osmotin resistance in yeast consists of FOR1 aggregates reducing permeability of the cell wall to osmotin when the inner cell wall layer is already rendered somewhat impermeable by cross-linking of β-1,3-glucan with PIR proteins, a process that neither mandates nor excludes covalent linking of FOR1 to the cell wall.
Conservation of osmotin resistance determinants between fungi.
Together, our results validate our hypothesis that osmotin resistance mechanisms may be conserved between S. cerevisiae and F. oxysporum to a great extent and that S. cerevisiae could be used as a tool to uncover F. oxysporum genes that control osmotin sensitivity or resistance. Most importantly, our results show that resistance of fungi to the plant defense protein osmotin is determined by the impermeability of their cell walls to this protein. Consequently, genes such as GFA1 and SSD1 that control deposition of multiple cell wall constituents are major, and conserved, fungal osmotin resistance determinants. Individual cell wall glycoproteins can make small contributions to osmotin resistance and are not conserved osmotin resistance determinants.
Effect of for3 mutation on pathogenicity.
Fungal cell walls are unique and are therefore the targets of antifungals of medical and agronomic importance. GFAT is an essential and ubiquitous enzyme and is therefore less attractive as an antifungal target than SSD1, which is unique to fungi. It has been shown that deletion of ScSSD1 orthologs of the cucumber anthracnose fungus C. lagenarium and the rice blast fungus M. grisea impaired their ability to initially establish infection on their hosts (67). Compared with the wild-type strains, the Δssd1 mutants of C. lagenarium and M. grisea were shown to have weaker cell walls and were also able to induce host defenses more strongly, which accounted for their reduced infectivity. Similarly, null ssd1 mutants of C. albicans were found to have decreased virulence (23). The null ssd1 mutants of C. albicans were more sensitive to host defense antimicrobial peptides than the isogenic wild-type strain. Since the null for3 mutation depleted cell wall β-1,3-glucan, which could weaken the wall, and increased sensitivity to the tobacco defense protein osmotin, we expected the Δfor3 mutant of F. oxysporum f. sp. nicotianae to also be impaired in its ability to cause disease symptoms in the host tobacco plant. However, we were unable to observe significant differences in disease symptom development between soil-grown tobacco plants inoculated by the root dip method with the wild-type strain of F. oxysporum or the Δfor3 mutant (see Fig. S4 in the supplemental material). Wheeler et al. (78) observed that SSD1 knockout mutants of feral strains of S. cerevisiae had altered cell surface properties and changes in cell wall composition, were more-potent elicitors of a proinflammatory response in cultured macrophages, and were also more virulent in mouse models of infection. Our results, when considered with the reports of Wheeler et al. (78) and Tanaka et al. (67), suggest that cell surface changes induced by inactivation of SSD1 affect pathogenicity in a unique manner for every pathogen-host combination. The results support the conclusion that SSD1 is not a good antifungal target.
Supplementary Material
ACKNOWLEDGMENTS
The services of the Life Sciences Microscopy Center, Purdue University, are gratefully acknowledged. We thank J. F. Cannon, A. Toh-e, Y. Uesono, and G. Chilosi for materials provided.
This work was supported by NSF grant MCB 035049 and by KAUST.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 26 February 2010.
REFERENCES
- 1.Abad L. R., Paino D'Urzo M., Liu D., Narasimhan M. L., Reuveni M., Zhu J. K., et al. 1996. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 118:11–23 [Google Scholar]
- 2.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Sedman J. G., Smith J. A., Struhl K. (ed.). 1988. Current protocols in molecular biology, vol. 1 and 2 John Wiley and Sons, New York, NY [Google Scholar]
- 3.Baev N., Endre G., Petrovics G., Banfalvi Z., Kondorosi A. 1991. Six nodulation genes of nod box locus 4 in Rhizobium meliloti are involved in nodulation signal production: nodM codes for D-glucosamine synthetase. Mol. Gen. Genet. 228:113–124 [DOI] [PubMed] [Google Scholar]
- 4.Bateman A. 1999. The SIS domain: a phosphosugar-binding domain. Trends Biochem. Sci. 24:94–95 [DOI] [PubMed] [Google Scholar]
- 5.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 6.Benhamou N., Ouellette G. B. 1986. Ultrastructural localization of glycoconjugates in the fungus Ascocalyx abietina, the Scleroderris canker agent of conifers, using lectin-gold complexes. J. Histochem. Cytochem. 34:855–867 [DOI] [PubMed] [Google Scholar]
- 7.Boorsma A., de Nobel H., ter Riet B., Bargmann B., Brul S., Hellingwerf K. J., Klis F. M. 2004. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast 21:413–427 [DOI] [PubMed] [Google Scholar]
- 8.Bowen S., Wheals A. E. 2004. Incorporation of Sed1p into the cell wall of Saccharomyces cerevisiae involves KRE6. FEMS Yeast Res. 4:731–735 [DOI] [PubMed] [Google Scholar]
- 9.Bowman S. M., Free S. J. 2006. The structure and synthesis of the fungal cell wall. Bioessays 28:799–808 [DOI] [PubMed] [Google Scholar]
- 10.Bulik D. A., Olczak M., Lucero H. A., Osmond B. C., Robbins P. W., Specht C. A. 2003. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell 2:886–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castillo L., Martinez A. I., Garcerá A., Elorza M. V., Valentín E., Sentandreu R. 2003. Functional analysis of the cysteine residues and the repetitive sequence of Saccharomyces cerevisiae Pir4/Cis3: the repetitive sequence is needed for binding to the cell wall beta-1,3-glucan. Yeast 20:973–983 [DOI] [PubMed] [Google Scholar]
- 12.Chen C. Y., Rosamond J. 1998. Candida albicans SSD1 can suppress multiple mutations in Saccharomyces cerevisiae. Microbiology 144:2941–2950 [DOI] [PubMed] [Google Scholar]
- 13.Chisholm S. T., Coaker G., Day B., Staskawicz B. J. 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124:803–814 [DOI] [PubMed] [Google Scholar]
- 14.Coca M. A., Damsz B., Yun D. J., Hasegawa P. M., Bressan R. A., Narasimhan M. L. 2000. Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant J. 22:61–69 [DOI] [PubMed] [Google Scholar]
- 15.De Groot P. W., Ram A. F., Klis F. M. 2005. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol. 42:657–675 [DOI] [PubMed] [Google Scholar]
- 16.Di Pietro A., Roncero M. I. G. 1998. Cloning, expression and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant Microbe Interact. 11:91–98 [DOI] [PubMed] [Google Scholar]
- 17.Di Pietro A., Madrid M. P., Caracuel Z., Delgado-Jarana J., Roncero M. I. 2003. Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4:315–325 [DOI] [PubMed] [Google Scholar]
- 18.Dutka-Malen S., Mazodier P., Badet B. 1988. Molecular cloning and overexpression of the glucosamine synthetase gene from Escherichia coli. Biochimie 70:287–290 [DOI] [PubMed] [Google Scholar]
- 19.Ecker M., Deutzmann R., Lehle L., Mrsa V., Tanner W. 2006. Pir proteins of Saccharomyces cerevisiae are attached to beta-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 281:11523–11529 [DOI] [PubMed] [Google Scholar]
- 20.Eisenhaber B., Schneider G., Wildpaner M., Eisenhaber F. 2004. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. J. Mol. Biol. 337:243–253 [DOI] [PubMed] [Google Scholar]
- 21.Elble R. 1992. A simple and efficient procedure for transformation of yeasts. Biotechniques 13:18–20 [PubMed] [Google Scholar]
- 22.Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut J. S., Hotz H. R., et al. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gank K. D., Yeaman M. R., Kojima S., Yount N. Y., Park H., Edwards J. E., Jr., Filler S. G., Fu Y. 2008. SSD1 is integral to host defense peptide resistance in Candida albicans. Eukaryot. Cell 7:1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García R., Bermejo C., Grau C., Pérez R., Rodríguez-Peña J. M., Francois J., Nombela C., Arroyo J. 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279:15183–15195 [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Maceira F. I., Di Pietro A., Roncero M. I. G. 2000. Cloning and disruption of pgx4 encoding an in planta expressed exopolygalaturonase from Fusarium oxysporum. Mol. Plant Microbe Interact. 13:359–365 [DOI] [PubMed] [Google Scholar]
- 26.Gerik K. J., Donlin M. J., Soto C. E., Banks A. M., Banks I. R., Maligie M. A., Selitrennikoff C. P., Lodge J. K. 2005. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58:393–408 [DOI] [PubMed] [Google Scholar]
- 27.Gill S., Singh B., Sunder S. 2005. Isolation of 4-aminopyridine resistant mutants affecting alkali-insoluble glucan content of cell walls in Saccharomyces cerevisiae. Indian J. Exp. Biol. 43:897–901 [PubMed] [Google Scholar]
- 28.Hagen I., Ecker M., Lagorce A., Francois J. M., Sestak S., Rachel R., Grossman G., Hauser N. C., Hoheisel J. D., Tanner W., Strahl S. 2004. Sed1p and Srl1p are required to compensate for cell wall instability in Saccharomyces cerevisiae mutants defective in multiple GPI-anchored mannoproteins. Mol. Microbiol. 52:1413–1425 [DOI] [PubMed] [Google Scholar]
- 29.Hamada K., Fukuchi S., Arisawa M., Baba M., Kitada K. 1998. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol. Gen. Genet. 258:53–59 [DOI] [PubMed] [Google Scholar]
- 30.Ibeas J. I., Lee H., Damsz B., Prasad D. T., Pardo J. M., Hasegawa P. M., Bressan R. A., Narasimhan M. L. 2000. Fungal cell wall phosphomannans facilitate the toxic activity of a plant PR-5 protein. Plant J. 23:375–383 [DOI] [PubMed] [Google Scholar]
- 31.Ibeas J. I., Yun D.-J., Damsz B., Narasimhan M. L., Uesono Y., Ribas J. C., et al. 2001. Resistance to the plant PR-5 protein osmotin in the model fungus Saccharomyces cerevisiae is mediated by the regulatory effects of SSD1 on cell wall composition. Plant J. 25:271–280 [DOI] [PubMed] [Google Scholar]
- 32.Jones J. D., Dangl J. L. 2006. The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
- 33.Kaeberlein M., Guarente L. 2002. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics 160:83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaeberlein M., Andalis A. A., Liszt G. B., Fink G. R., Guarente L. 2004. Saccharomyces cerevisiae SSD1-V confers longevity by a Sir2p independent mechanism. Genetics 166:1661–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapteyn J. C., Hoyer L. L., Hecht J. E., Müller W. H., Andel A., Verkleij A. J., et al. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 35:601–611 [DOI] [PubMed] [Google Scholar]
- 36.Kapteyn J. C., Van Egmond P., Sievi E., Van Den Ende H., Makarow M., Klis F. M. 1999. The contribution of the O-glycosylated protein Pir2p/Hsp150 to the construction of the yeast cell wall in wild-type cells and beta 1,6-glucan-deficient mutants. Mol. Microbiol. 31:1835–1844 [DOI] [PubMed] [Google Scholar]
- 37.Klis F. M., Boorsma A., De Groot P. W. 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23:185–202 [DOI] [PubMed] [Google Scholar]
- 38.Klis F. M., Mol P., Hellingwerf K., Brul S. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26:239–256 [DOI] [PubMed] [Google Scholar]
- 39.Kurischko C., Weiss G., Ottey M., Luca F. C. 2005. A role for the Saccharomyces cerevisiae regulation of Ace2 and polarized morphogenesis signaling network in cell integrity. Genetics 171:443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lagorce A., Le Berre-Anton V., Aguilar-Uscanga B., Martin-Yken H., Dagkessamanskaia A., François J. 2002. Involvement of GFA1, which encodes glutamine-fructose-6-phosphate amidotransferase, in the activation of the chitin synthesis pathway in response to cell-wall defects in Saccharomyces cerevisiae. Eur. J. Biochem. 269:1697–1707 [DOI] [PubMed] [Google Scholar]
- 41.LaRosa P. C., Chen Z., Nelson D. E., Singh N. K., Hasegawa P. M., Bressan R. A. 1992. Osmotin gene expression is posttranscriptionally regulated. Plant Physiol. 100:409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li R., Wu N., Fan Y., Song B. 1999. Transgenic potato plants expressing osmotin gene inhibits fungal development in inoculated leaves. Chin. J. Biotechnol. 15:71–75 [PubMed] [Google Scholar]
- 43.Liu D., Raghothama K. G., Hasegawa P. M., Bressan R. A. 1994. Osmotin overexpression in potato delays development of disease symptoms. Proc. Natl. Acad. Sci. U. S. A. 91:1888–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martín H., Castellanos M. C., Cenamor R., Sánchez M., Molina M., Nombela C. 1996. Molecular and functional characterization of a mutant allele of the mitogen-activated protein-kinase gene SLT2(MPK1) rescued from yeast autolytic mutants. Curr. Genet. 29:516–522 [DOI] [PubMed] [Google Scholar]
- 45.McDonald H. B., Helfant A. H., Mahony E. M., Khosla S. K., Goetsch L. 2002. Mutational analysis reveals a role for the C terminus of the proteasome subunit Rpt4p in spindle pole body duplication in Saccharomyces cerevisiae. Genetics 162:705–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKnight G. L., Mudri S. L., Mathewes S. L., Traxinger R. R., Marshall S., Sheppard P. O., O'Hara P. J. 1992. Molecular cloning, cDNA sequence, and bacterial expression of human glutamine:fructose-6-phosphate amidotransferase. J. Biol. Chem. 267:25208–25212 [PubMed] [Google Scholar]
- 47.Mrsa V., Tanner W. 1999. Role of NaOH-extractable cell wall proteins Ccw5p, Ccw6p, Ccw7p and Ccw8p (members of the Pir protein family) in stability of the Saccharomyces cerevisiae cell wall. Yeast 15:813–820 [DOI] [PubMed] [Google Scholar]
- 48.Mulholland J., Preuss D., Moon A., Wong A., Drubin D., Botstein D. 1994. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J. Cell Biol. 125:381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narasimhan M. L., Coca M. A., Jin J., Yamauchi T., Ito Y., Kadowaki T., et al. 2005. Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol. Cell 17:171–180 [DOI] [PubMed] [Google Scholar]
- 50.Narasimhan M. L., Damsz B., Coca M. A., Ibeas J. I., Yun D. J., Pardo J. M., Hasegawa P. M., Bressan R. A. 2001. A plant defense response effector induces microbial apoptosis. Mol. Cell 8:921–930 [DOI] [PubMed] [Google Scholar]
- 51.Narasimhan M. L., Lee H., Damsz B., Singh N. K., Ibeas J. I., Matsumoto T. K., Woloshuk C. P., Bressan R. A. 2003. Overexpression of a cell wall glycoprotein in Fusarium oxysporum increases virulence and resistance to a plant PR-5 protein. Plant J. 36:390–400 [DOI] [PubMed] [Google Scholar]
- 52.Nucci M., Anaissie E. 2007. Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20:695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ortoneda M., Guarro J., Madrid M. P., Caracuel Z., Roncero M. I., Mayayo E., Di Pietro A. 2004. Fusarium oxysporum as a multihost model for the genetic dissection of fungal virulence in plants and mammals. Infect. Immun. 72:1760–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popolo L., Gilardelli D., Bonfante P., Vai M. 1997. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1delta mutant of Saccharomyces cerevisiae. J. Bacteriol. 179:463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ram A. F., Arentshorst M., Damveld R. A., van Kuyk P. A., Klis F. M., van den Hondel C. A. 2004. The cell wall stress response in Aspergillus niger involves increased expression of the glutamine:fructose-6-phosphate amidotransferase-encoding gene (gfaA) and increased deposition of chitin in the cell wall. Microbiology 150:3315–3326 [DOI] [PubMed] [Google Scholar]
- 56.Robzyk K., Kassir Y. 1992. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 20:3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rost B., Sander C. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584–599 [DOI] [PubMed] [Google Scholar]
- 58.Saremi H., Okhovvat S. M. 2006. Mycotoxin producing Fusarium species associated with plant disease on potato, wheat, corn and animal diseases in northwest Iran. Commun. Agric. Appl. Biol. Sci. 71:1175–1185 [PubMed] [Google Scholar]
- 59.Schoffelmeer E. A., Klis F. M., Sietsma J. H., Cornelissen B. J. 1999. The cell wall of Fusarium oxysporum. Fungal Genet. Biol. 27:275–282 [DOI] [PubMed] [Google Scholar]
- 60.Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:12–17 [DOI] [PubMed] [Google Scholar]
- 61.Shimoi H., Kitagaki H., Ohmori H., Iimura Y., Ito K. 1998. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J. Bacteriol. 180:3381–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simões T., Teixeira M. C., Fernandes A. R., Sá-Correia I. 2003. Adaptation of Saccharomyces cerevisiae to the herbicide 2,4-dichlorophenoxyacetic acid, mediated by Msn2p- and Msn4p-regulated genes: important role of SPI1. Appl. Environ. Microbiol. 69:4019–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh N. K., Bracker C. A., Hasegawa P. M., Handa A. K., Buckel S., Hermodson M. A., et al. 1987. Characterization of osmotin: a thaumatin-like protein associated with osmotic adaptation in plant cells. Plant Physiol. 85:529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith R. J., Milewski S., Brown A. J., Gooday G. W. 1996. Isolation and characterization of the GFA1 gene encoding the glutamine:fructose-6-phosphate amidotransferase of Candida albicans. J. Bacteriol. 178:2320–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sutton A., Immanuel D., Arndt K. T. 1991. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 11:2133–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi T., Shimoi H., Ito K. 2001. Identification of genes required for growth under ethanol stress using transposon mutagenesis in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:1112–1119 [DOI] [PubMed] [Google Scholar]
- 67.Tanaka S., Yamada K., Yabumoto K., Fujii S., Huser A., Tsuji G., et al. 2007. Saccharomyces cerevisiae SSD1 orthologues are essential for host infection by the ascomycete plant pathogens Colletotrichum lagenarium and Magnaporthe grisea. Mol. Microbiol. 64:1332–1349 [DOI] [PubMed] [Google Scholar]
- 68.Thomas B. J., Rothstein R. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619–630 [DOI] [PubMed] [Google Scholar]
- 69.Thomas P. A. 2003. Fungal infections of the cornea. Eye 17:852–862 [DOI] [PubMed] [Google Scholar]
- 70.Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toh-e A., Yasunaga S., Nisogi H., Tanaka K., Oguchi T., Matsui Y. 1993. Three yeast genes, PIR1, PIR2 and PIR3, containing internal tandem repeats, are related to each other, and PIR1 and PIR2 are required for tolerance to heat shock. Yeast 9:481–494 [DOI] [PubMed] [Google Scholar]
- 72.Uesono Y., Fujita A., Toh-e A., Kikuchi Y. 1994. The MCS1/SSD1/SRK1/SSL1 gene is involved in stable maintenance of the chromosome in yeast. Gene 143:135–138 [DOI] [PubMed] [Google Scholar]
- 73.Uesono Y., Toh-e A., Kikuchi Y. 1997. Ssd1p of Saccharomyces cerevisiae associates with RNA. J. Biol. Chem. 272:16103–16109 [DOI] [PubMed] [Google Scholar]
- 74.Veronese P., Ruiz M. T., Coca M. A., Hernandez-Lopez A., Lee H., Ibeas J. I., et al. 2003. In defense against pathogens.Both plant sentinels and foot soldiers need to know the enemy. Plant Physiol. 131:1580–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wach A., Brachat A., Rebischung C., Steiner S., Pokorni K., te Heesen S., Philippsen P. 1998. Methods Enzymol. 26:67–81 [Google Scholar]
- 76.Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. 1989. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58:409–419 [DOI] [PubMed] [Google Scholar]
- 77.Watzele G., Tanner W. 1989. Cloning of the glutamine:fructose-6 phosphate amidotransferase gene from yeast. Pheromonal regulation of its transcription. J. Biol. Chem. 264:8753–8758 [PubMed] [Google Scholar]
- 78.Wheeler R. T., Kupiec M., Magnelli P., Abeijon C., Fink G. R. 2003. A Saccharomyces cerevisiae mutant with increased virulence. Proc. Natl. Acad. Sci. U. S. A. 100:2766–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whelan W. L., Ballou C. E. 1975. Sporulation in D-glucosamine auxotrophs of Saccharomyces cerevisiae: meiosis with defective ascospore wall formation. J. Bacteriol. 124:1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilson R. B., Brenner A. A., White T. B., Engler M. J., Gaughran J. P., Tatchell K. 1991. The Saccharomyces cerevisiae SRK1 gene, a suppressor of bcy1 and ins1, may be involved in protein phosphatase function. Mol. Cell. Biol. 11:3369–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yun D. J., Ibeas J. I., Lee H., Coca M. A., Narasimhan M. L., Uesono Y., et al. 1998. Osmotin, a plant antifungal protein, subverts signal transduction to enhance fungal cell susceptibility. Mol. Cell 1:807–817 [DOI] [PubMed] [Google Scholar]
- 82.Yun D. J., Zhao Y., Pardo J. M., Narasimhan M. L., Damsz B., Lee H., et al. 1997. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proc. Natl. Acad. Sci. U. S. A. 94:7082–7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng J., Khalil M., Cannon J. F. 2000. Glc7p protein phosphatase inhibits expression of glutamine-fructose-6-phosphate transaminase from GFA1. J. Biol. Chem. 275:18070–18078 [DOI] [PubMed] [Google Scholar]
- 84.Zhu B., Chen T. H., Li P. H. 1996. Analysis of late-blight disease resistance and freezing tolerance in transgenic potato plants expressing sense and antisense genes for an osmotin-like protein. Planta 198:70–77 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.