Abstract
The human commensal yeast Candida glabrata is becoming increasingly important as an agent of nosocomial bloodstream infection. However, relatively little is known concerning the genetics and population structure of this species. We have analyzed 230 incident bloodstream isolates from previous and current population-based surveillance studies by using multilocus sequence typing (MLST). Our results show that in the U.S. cities of Atlanta, GA; Baltimore, MD; and San Francisco, CA during three time periods spanning 1992 to 2009, five populations of C. glabrata bloodstream isolates are defined by a relatively small number of sequence types. There is little genetic differentiation in the different C. glabrata populations. We also show that there has been a significant temporal shift in the prevalence of one major subtype in Atlanta. Our results support the concept that both recombination and clonality play a role in the population structure of this species.
In the most recently available survey of nosocomial bloodstream infections, Candida species were the fourth most common organism, surpassed only by Staphylococcus and Enterococcus species (24). Although Candida albicans remains the most commonly isolated Candida species worldwide, the incidence of Candida glabrata infection has been increasing steadily so that it is now the second most common cause of Candida infection in the United States (14). C. glabrata is considered a normal component of the human epithelial flora but is capable of causing serious systemic infections in susceptible hosts. This increase in the relative proportion of infections due to C. glabrata has come during the period of the introduction and prophylactic use of azole antifungal drugs (21) and may be a reflection of the decreased susceptibility of C. glabrata to these azole antifungal drugs (7, 15). Many questions regarding the epidemiology of C. glabrata infections have a direct impact on public health and still remain unanswered. Is the decreased susceptibility due to a small number of clones expanding in a population, or are all isolates capable of developing resistance to azole drugs? Are some isolates more virulent than others and therefore more prevalent in a population? Can we monitor the expansion of clonal isolates that may be more virulent or have increased drug resistance? A better understanding of the population genetics of C. glabrata may allow us to answer some of these questions.
Many DNA fingerprinting methods have been developed for the investigation of the population genetics of Candida species (19). Two of the most important aspects of a typing system are reproducibility between laboratories and the ability to archive strain types. Multilocus sequence typing (MLST) has been developed as a typing system which allows highly reproducible strain discrimination as well as the development of genotypic strain archives that can be stored digitally for both prospective and retrospective analysis of isolates (13, 22). An MLST system which utilizes six housekeeping genes on six separate chromosomes was developed for C. glabrata (4), and an online archive of sequence types (STs) was established (http://cglabrata.mlst.net). Several studies utilizing this typing system have described the molecular population structure of both regional and worldwide collections of C. glabrata isolates (4, 5, 11, 12).
During the past 2 decades, the Centers for Disease Control and Prevention (CDC) and our partners have undertaken three active, population-based surveillance studies in order to determine the incidence of candidemia, the distribution of species causing bloodstream infection, and the prevalence of antifungal drug resistance (8, 10). In each case, two major metropolitan areas were included: San Francisco, CA, and Atlanta, GA (1992 to 1993); Baltimore, MD, and the state of Connecticut (1998 to 2000); and Atlanta, GA, and Baltimore, MD (2008 to 2010). Population-based surveillance is unique in that it includes the total population of a particular geographic area and avoids the biases associated with single or select institutional studies. During each of the surveillance studies, incident bloodstream isolates from all hospitals within each defined geographic area were collected and identified to the species level. While C. glabrata isolates comprised a smaller percentage of the isolates in the 1992-to-1993 and 1998-to-2000 surveillance studies (8, 10), they represent almost a third of the isolates collected during the current surveillance (N. Iqbal and S. Lockhart, unpublished observations).
In the present work, we have characterized by MLST analysis 230 isolates of C. glabrata from five populations (excluding Connecticut) separated both geographically and temporally. This unique collection of isolates allowed an analysis of the changing population genetics of this organism. We identified 31 unique STs and showed the maintenance of a major ST both geographically and temporally that is unique to the United States. An analysis of the relatedness of specific C. glabrata populations and a strong indication for recombination within and between populations are provided.
MATERIALS AND METHODS
Isolates.
A total of 230 available incident C. glabrata bloodstream isolates from two previous population-based surveillance studies of metropolitan Atlanta, GA; Baltimore City and County, MD; and metropolitan San Francisco, CA (8, 10) and from an ongoing, population-based surveillance in metropolitan Atlanta and Baltimore City and County (2) were used in this study. For San Francisco and Atlanta from 1992 to 1993, all of the available isolates from each city were surveyed, comprising 64% of all cases of C. glabrata detected in that surveillance. For Baltimore from 1998 to 2000, 50 isolates were randomly chosen from 168 available, representing approximately 18% of all cases of C. glabrata detected during the surveillance. For Atlanta and Baltimore in 2008, all of the available isolates were chosen from Atlanta and Baltimore until more than 50 isolates from each population had been surveyed. A complete listing of strains and STs are given in Table S1 in the supplemental material.
Prior to use, all isolates were stored in glycerol at −70°C. Isolates were identified as C. glabrata by conventional biochemical means and by Luminex assay (K. Etienne and S. A. Balajee, submitted for publication) and were confirmed by positive MLST results. The closely related species Candida nivariensis and Candida bracarensis were ruled out because type isolates of these species could not be amplified with the C. glabrata MLST primer set.
DNA extraction, PCR amplification, and sequencing.
After passage of each isolate twice on Sabouraud dextrose agar plates, DNA was extracted using the Mo Bio microbial DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) according to the manufacturer's instructions. The oligonucleotide primers used for MLST analysis were those described by Dodgson and coworkers (4). PCRs were performed in a 25-μl volume containing 10 ng of genomic DNA, 0.2 μM each primer, Roche Taq DNA polymerase, and Taq PCR master mix as described by the manufacturer (Roche Diagnostics, Indianapolis, IN). Reaction conditions were as previously described for each individual primer set (4). PCR products were purified using ExoSap-It as described by the manufacturer (USB, Cleveland, OH). Sequencing reactions were performed using BigDye terminator technology (ABI, Foster City, CA) with an ABI Prism 3730 DNA sequencer. All loci were sequenced in both forward and reverse directions with the same primers as those used for the PCRs.
Data analysis.
Nucleotide sequences were determined by alignment of forward and reverse sequences by using Sequencher 4.7 software (Genecodes Inc., Ann Arbor, MI), and polymorphisms were confirmed by visual examination of the sequence traces. Sequences were then compared to the C. glabrata MLST database (http://cglabrata.mlst.net) to assign allele numbers and STs.
Population genetic analysis.
For population analysis, alleles for each strain were assigned alphabetic equivalents, and strains were grouped by populations and analyzed by Popgene v1.3 (http://www.ualberta.ca/∼fyeh/index.htm) under the criteria of haploid, codominant markers. Wright's fixation index (FST) values were calculated using the formula FST = Ht − Hs/Ht (23), and using values of Hs and Ht from Nei's analysis of gene diversity. FST values of >0.05 generally indicate little interpopulation variance, and can range from 0 for identical populations to 1 for populations sharing no alleles in common (9). Values of genetic identity and distance used Nei's unbiased measurements (17). Nei's unbiased genetic pairwise identity value calculates genetic diversity between populations across all loci simultaneously, with the assumption that differences arise due to both mutation and genetic drift. Distance values are given on a logarithmic scale, where values approaching 1 indicate complete divergence and negative numbers indicate no divergence. The likelihood-based tree was generated using a neighbor-joining algorithm in the HyPhy software package (16). Bootstrap values were calculated using the Mega 4.1 software package (20). The population-based trees were constructed using PHYLIP 3.5 (6). The Index of Association (IA) and rBarD were calculated using Multilocus 1.3b (1). Two-locus linkage disequilibrium (LD) was calculated using POPGENE v1.3 (http://www.ualberta.ca/∼fyeh).
RESULTS
General attributes of population profiles as determined by MLST.
A total of 230 incident bloodstream isolates from population-based surveillance in the United States were typed using MLST. Five populations of isolates were analyzed. Population 1 consisted of 38 isolates collected in San Francisco from 1992 to 1993; population 2 consisted of 26 isolates collected in the metro Atlanta area from 1992 to 1993 from 25 hospitals; population 3 consisted of 50 isolates collected in Baltimore City and County from 1998 to 2000 from 14 hospitals; population 4 consisted of 63 isolates collected in metro Atlanta in 2008 from 25 hospitals; population 5 consisted of 53 isolates collected in Baltimore City and County in 2008 from 15 hospitals. The six sequenced loci resulted in 3,345 combined base pairs. All of the analyzed nucleotides were within open reading frames, so no insertions or deletions were expected or detected. We observed no indications of heterozygosity for any of the six loci in any of the strains. A total of 127 nucleotide sites (3.8%) across all six genes combined were found to be polymorphic. For six isolates, STs were comprised of new combinations of previously identified alleles (4). One isolate in the 1998 Baltimore collection (CAS99-0115) was found to have both a new mutation and a different combination of polymorphisms in the NMT1 locus. This new mutation consisted of a T→C second position nonsynonymous transition (Ile→Thr) at nucleotide position 872. For the TRP1 locus, there were new combinations of previously described polymorphisms and a new mutation, an A→T second position nonsynonymous transversion (Asp→Val) at nucleotide position 427. For all populations, the most informative site was NMT1, with 18 alleles in the total collection, and the least informative site was UGP1, with only seven alleles.
MLST analysis resulted in the delineation of 31 STs from these 230 isolates. Sixteen (52%) of the STs were represented by single isolates, but this number represents only 7% of the total number of isolates. The most diverse population was that from Baltimore from 1998 to 2000, with 17 STs, and the population with the lowest ratio of STs to isolates was that from Atlanta from 1992 to 1993, with one unique ST for every 2.4 isolates genotyped (Table 1).
Table 1.
Ratio of unique STs to the number of isolates in subdivided populations and percentage of population in the three major STs
| No. of isolates (%) |
||||||
|---|---|---|---|---|---|---|
| Population | Total no. of isolates | No. of STs | Ratioa | ST16 | ST19 | ST3 |
| San Francisco 1992–1993 | 38 | 11 | 1:3.5 | 1 (3) | 10 (26) | 6 (16) |
| Atlanta 1992–1993 | 26 | 11 | 1:2.4 | 1 (4) | 4 (15) | 5 (19) |
| Baltimore 1998–2000 | 50 | 17 | 1:2.9 | 6 (12) | 8 (16) | 7 (14) |
| Atlanta 2008 | 53 | 14 | 1:3.8 | 21 (40) | 8 (15) | 15 (28) |
| Baltimore 2008 | 63 | 13 | 1:4.8 | 10 (16) | 11 (17) | 5 (8) |
| Total | 230 | 31 | 1:7.4 | 39 | 41 | 38 |
Ratio of the number of STs to the number of isolates.
We observed eight new STs that were not in the C. glabrata MLST database. One strain from the 1998 Baltimore collection, CAS99-0437, contained new alleles in five of the six loci, FKS1, NMT1, TRP1, UGP1, and URA3. In all but the TRP1 locus, the new alleles were derived from new combinations of previously characterized polymorphisms.
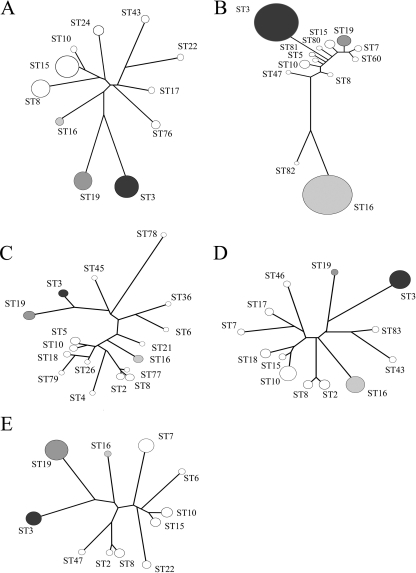
STs 16, 19, and 3 were the most abundant, and each appeared in all five of the study populations. These three STs together represented 51% of the entire study population. They represented 38% and 83% of the isolates from the 1992-to-1993 and 2008 Atlanta populations, respectively; 42% and 41% of the isolates from the 1998-to-2000 and 2008 Baltimore populations, respectively; and 45% of the isolates from the 1992-to-1993 San Francisco population (Table 1). Neighbor-joining trees were constructed for each population based on the concatenated sequences of the given genotypes (clone corrected) and are shown in Fig. 1. The frequencies of the corresponding STs are indicated by the diameters of the terminal circles. It can be seen that the overall topologies are similar, with the possible exception of the Georgia 2008 population (Fig. 1B). In this case, the tree topology is more polarized due to the inclusion of STs 80, 81, and 82, which are newly described in this study and are unique to that population.
Fig. 1.
Neighbor-joining trees for each population in this study showing the abundance of each genotype in the population. (A) Atlanta, 1992; (B) Atlanta, 2008; (C) Baltimore, 1998; (D) Baltimore, 2008; (E) San Francisco, 1992. Circles represent the percentage of each population of a given ST. Major STs ST3, ST16, and ST19 are shaded in gray or black.
Temporal changes in population structure.
Because candidemia surveillance took place in both Atlanta and Baltimore over two different time periods, 15 years apart for Atlanta and 10 years apart for Baltimore, temporal changes in the C. glabrata populations could be observed. The single statistically significant change that was observed between current and past populations was an increase in ST16 in Atlanta during the 2008 surveillance compared to the earlier (1992) surveillance period (P = 0.013; Fisher's exact test). ST16 represented approximately 40% of the 2008 Atlanta survey compared to only 4% from 1992 to 1993. An increase was also observed for ST3, although it was not statistically significant.
The temporal changes in genetic diversity in the populations between the previous and current surveillance studies are primarily due to the gain and loss of STs represented by single isolates. In Baltimore from 1998 to 2000, there were 17 STs comprised of 55 different combined alleles. In 2008, nine of these STs were lost, and an additional five were gained. However, there was a net loss of 11 alleles in the population. In Atlanta from 1992 to 1993, there were 11 STs comprised of 45 alleles. In 2008, the number of isolates in the population had more than doubled, but the number of STs climbed to only 14, with a loss of five STs, a gain of eight STs, and a net loss of four alleles.
Geographic changes in population structure.
Only two STs represented by more than one isolate were unique to a particular region. Six isolates from Baltimore were ST18, and two isolates from Atlanta were ST24. There were no STs which were unique to San Francisco. A total of 12 and 8 STs were found to be unique to the Baltimore and Atlanta populations, respectively. Further analysis revealed that for all six loci, there were 30 and 13 alleles that were unique to the Baltimore and Atlanta populations, respectively, and that these alleles were evenly divided across the loci. Locus NMT1 had the highest number, with 11 unshared alleles. In addition, we observed that there were <20 individual polymorphisms that were unshared between groups and that these were divided approximately evenly between the two populations.
Measures of genetic distance between populations.
In order to assess genetic diversity among the populations, Wright's fixation index (FST) and Nei's genetic distance (DN) values were calculated for each locus for all pairwise combinations of the five populations (Table 2). FST measures the diversity between populations at an individual locus. The two population pairs that differ geographically but not temporally, San Francisco and Atlanta from 1992 to 1993 and Atlanta and Baltimore 2008, showed little difference in their population structure, as indicated by their low FST values, and had average FST values that were lower than the overall mean (Table 2). The two temporally isolated Baltimore populations had the lowest FST values of all the pairwise comparisons, an indication that there was little genetic diversity between the two populations when measured at the level of individual loci. For the two temporally separated Atlanta populations, the FST values were higher than the overall average, which may be a reflection of the shift in major STs within these populations. The pairwise populations with the highest mean FST values in all of the comparisons were San Francisco in 1992 compared to Atlanta in 2008, which reflected the large temporal and geographic distance between these two populations. Overall, however, very few of the FST values were found to be significantly above 0.05, normally considered to be the cutoff between little and moderate differentiation (9), indicating that the allelic identity between populations was higher than the identity within populations.
Table 2.
FST and Nei's distance values for subdivided populationsa
|
FST for: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Population | nb | FKS | LEU2 | NMT | TRP | UGP | URA3 | Mean FST | DNc |
| Atlanta (1992) vs San Francisco (1992) | 64 | 0.0205 | 0.0285 | 0.0159 | 0.0154 | 0.0104 | 0.0339 | 0.0207 | 0.0983 |
| Atlanta (1992) vs Baltimore (1998) | 76 | 0.0110 | 0.0150 | 0.0064 | 0.0043 | 0.0144 | 0.0071 | 0.0097 | 0.0182 |
| Atlanta (1992) vs Atlanta (2008) | 89 | 0.0404 | 0.0618 | 0.0366 | 0.0458 | 0.0512 | 0.0584 | 0.0490 | 0.3260 |
| Atlanta (1992) vs Baltimore (2008) | 79 | 0.0178 | 0.0327 | 0.0132 | 0.0119 | 0.0195 | 0.0239 | 0.0198 | 0.1081 |
| San Francisco (1992) vs Baltimore (1998) | 88 | 0.0186 | 0.0092 | 0.0185 | 0.0163 | 0.0298 | 0.0286 | 0.0201 | 0.1095 |
| San Francisco (1992) vs Atlanta (2008) | 101 | 0.0392 | 0.0589 | 0.0407 | 0.0510 | 0.0602 | 0.0585 | 0.0514 | 0.3372 |
| San Francisco (1992) vs Baltimore (2008) | 91 | 0.0194 | 0.0070 | 0.0195 | 0.0200 | 0.0277 | 0.0252 | 0.0197 | 0.1066 |
| Baltimore (1998) vs Atlanta (2008) | 113 | 0.0193 | 0.0412 | 0.0277 | 0.0339 | 0.0402 | 0.0338 | 0.0327 | 0.2020 |
| Baltimore (1998) vs Baltimore (2008) | 103 | 0.0024 | 0.0051 | 0.0049 | 0.0044 | 0.0039 | 0.0066 | 0.0045 | −0.0030 |
| Atlanta (2008) vs Baltimore (2008) | 116 | 0.0172 | 0.0366 | 0.0212 | 0.0256 | 0.0263 | 0.0218 | 0.0248 | 0.1346 |
| All populations | 230 | 0.0327 | 0.0457 | 0.0325 | 0.0360 | 0.0447 | 0.0466 | 0.0397 | |
FST = (Ht − Hs)/Ht.
n = population size.
DN, Nei's unbiased measure of genetic distance.
Nei's genetic identity values were calculated pairwise among all populations. Again, the values suggested that when all loci are considered simultaneously, the five populations are relatively undifferentiated. As similarly seen using FST values, the 1992-to-1993 Atlanta population was the most divergent from the other populations, the largest value was for the populations with the greatest geographic and temporal isolation (San Francisco 1992 and Atlanta 2008), and the two Baltimore populations were the most similar (Table 2).
Analysis of recombination.
To determine the extent of clonality and recombination in the different populations, we used three different tests of linkage disequilibrium (LD): two measures of association (IA and rBarD) and a two-locus LD test by the use of all pairwise allelic combinations. Since clonal reproduction can mask the effects of recombination, we prepared two data sets. One included all strains of each ST, and the other included the clone-corrected data of each ST from which identical genotypes were removed (haplotypes only).
For the two-locus LD test, all five populations and the total combined population gave similar results. Considering that each locus had between four and eight alleles, there were approximately 1,300 total pairwise comparisons. As seen in Table 3, a large majority of combinations showed significant LD (P < 0.05), thus contradicting the null hypothesis of recombination (23).
Table 3.
Multilocus LD analyses in each of the haploid populations
| Population | No. of isolates | No. of haplotypes | Value for all isolates |
LDa | Value for haplotypes only |
||
|---|---|---|---|---|---|---|---|
| IA | rDc | IA | rDc | ||||
| Atlanta 1992 | 26 | 11 | 2.83b | 0.580b | 114 | 0.311 | 0.075 |
| San Francisco 1992 | 38 | 12 | 2.95b | 0.602b | 153 | 1.32b | 0.287b |
| Baltimore 1998 | 50 | 17 | 2.80b | 0.566b | 188 | 1.10b | 0.233b |
| Atlanta 2008 | 63 | 15 | 3.40b | 0.683b | 205 | 1.57b | 0.326b |
| Baltimore 2008 | 53 | 14 | 3.17b | 0.641b | 172 | 1.08b | 0.229b |
| All isolates | 230 | 31 | 3.01b | 0.608b | 485 | 1.87b | 0.385b |
Normalized LD. The number of significant (P < 0.05) linkage disequilibria from all possible pairwise combinations of all alleles.
P < 0.01.
rD, rBarD.
In the association tests, the Index of Association (IA) and rBarD are expected to be zero if populations are freely recombining and greater than zero if there is association between alleles (clonality). The rBarD statistic takes into consideration the number of loci tested and is considered a more robust measure of association (1). For the uncorrected populations, both IA and rBarD tests rejected the null hypothesis of recombination in all five cases as well as the total isolates considered to be a single population. For the clone-corrected populations, all but the 1992 Atlanta population could be rejected at a P value of <0.01. The 1992 Atlanta population had a probability for both IA and rBarD of 0.05.
Combined analysis of the populations.
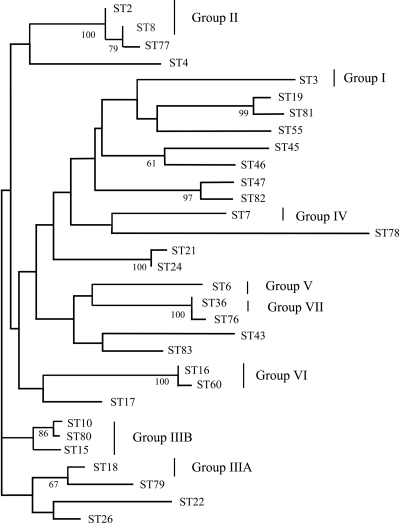
A neighbor-joining tree was constructed to show the genetic relationship among the 31 observed STs in our combined populations (Fig. 2). All seven of the groups defined by Dodgson and coworkers (4, 5) from two global population studies of C. glabrata isolates were identified in our collection, and the isolates themselves showed a high degree of diversity. Bootstrap values were high among paired isolates but dropped off considerably when larger groups were compared (data not shown). Our analysis supports previous findings (4) that group III partitions into subgroups A and B and that group II is better resolved than are groups I and IV (4).
Fig. 2.
Neighbor-joining dendrogram for the 31 STs identified in this study. Major groups previously identified (4) are indicated. Horizontal bar indicates percent divergence. Bootstrap values above 60% are indicated at nodes (replicates = 1,000).
DISCUSSION
The present study was undertaken as part of a long-term prospective surveillance to determine species distribution and drug resistance profiles in hospital-associated Candida bloodstream infections (8, 10). Isolates studied were all incident bloodstream isolates collected from residents of each surveillance area. From a total of 230 isolates, representing the sequencing of ∼7.7 × 105 base pairs, we observed only two new mutations, both nonsynonymous polymorphisms. We described six additional isolates as new STs resulting from new combinations of existing alleles.
A number of interesting observations have resulted from this analysis. For these selected U.S. isolates, the uncovering of new polymorphisms (by definition, new alleles) may be increasingly proportional to the number of isolates examined, suggesting a finite number of alleles in the population. The definition of an allele as a discrete set of polymorphisms as described by Dodgson et al. (4) is strongly supported, with little evidence of homoplasy in the creation of allelic combinations. There is evidence from our analysis for recombination in this species. Because alleles and STs are shared among all populations in this study, this implies that recombination has, or is, occurring among our defined populations. Earlier studies have suggested that such recombination is sexual (meiotic) recombination (3, 5). As in earlier work (4), our data supports the concept that these six unlinked loci are representative of the genome as a whole and reflect the underlying mechanisms creating diversity and differentiation in this species.
We have also provided evidence that there is a clonal component to the population, such as the increasing proportion of ST16 in Atlanta and the overall abundance both temporally and geographically of ST16, ST19, and ST3. The temporal stability of some STs suggests that at least some isolates with identical STs may be related by descent. More specifically, the isolates in the major STs may be clonally related. Both the IA and linkage association analyses support this concept, even when populations were clone corrected. Our findings are in agreement with other published work (4) and are supported by the knowledge that these MLST loci are physically unlinked (18). Additionally, we have reanalyzed IA and rBarD for a subpopulation of 20 isolates containing groups I, IV, V, VI, and VII. As seen in Fig. 2, this large middle group of STs are lacking in bootstrap support for the branch nodes. The IA and rBarD values for the clone-corrected subpopulation were found to be 0.598 and 0.135, respectively. Although still below the level of statistical significance for recombination (P < 0.02), these values are less than those for the the combined population as a whole (Table 3). We interpret this as suggesting that some subpopulations may be recombining at low, but variable, rates.
From a population standpoint, we have observed slight differences in the overall abundance of individual STs among geographic groups, primarily among minor STs, although frequency shifts in the major STs were also found. This is consistent with other observations which have shown FST values between cities and countries to be generally smaller than those between continents (3). Our data showing that the C. glabrata population in three large U.S. cities consists of a relatively small number of major STs are consistent with the previous study of U.S. isolates, in which the same and other major STs were observed (4). For example, STs 3 and 10 have been shown to be major STs worldwide (4). Some of the major STs in other collections do not appear in ours, while one of the major STs observed in this study, ST16, appears to be restricted to the United States (4, 11, 12). In addition, we observed a large temporal increase in the frequency of ST16 in Atlanta. Taken collectively, this suggests that STs may vary, or drift, between major and minor types over distance and time. The abundance of a particular ST in a geographic locale may be a reflection of the adaptation of a specific ST to a geographic niche, while the general drift in the temporal and geographic populations may reflect the ability of isolates of C. glabrata to adapt to new environments.
The low FST and genetic distance values among the populations are, in part, a reflection of the relative abundance in each population of the three major STs, ST19, ST16, and ST3, which account for an average of 50% of the isolates across all populations. The amount of diversity within a population ranged from one unique ST for every 2.4 isolates in Atlanta from 1992 to 1993 to one unique ST for every 4.8 patients in Baltimore in 2008. Two other studies of ST diversity of C. glabrata isolates have been recently published. Odds and coworkers (12) identified 27 STs from 50 patients in a 1-year study of Candida isolates from Scotland, for a ratio of one unique ST for every 1.9 isolates. Lin and coworkers (11) identified 15 STs from 37 patients in a single hospital in Taiwan over 2 years, for a ratio of one unique ST for every 2.5 isolates. Ratios from both of these previous studies indicate a higher degree of diversity in these populations in terms of STs than we have found. However, ST is not indicative of overall allelic diversity within a population. Because neither dendrograms nor individual STs were provided in either of the previous studies, it is not possible to tell whether the ST diversity is a reflection of overall diversity or whether it reflects a diverse population of closely related isolates. The populations observed in the other studies were also not bound by the constraints of case patient residency in a defined geographic boundary.
There is an intriguing correlation between genetic distance measures and incidence rates in the Atlanta and Baltimore populations. When the two Baltimore populations were compared temporally, they were shown to be highly similar genetically. The incidence rate of C. glabrata in Baltimore from 1998 to 2000 was 6.6/100,000 (8). Preliminary analysis showed that this rate dropped only slightly in 2008 to 6.3/100,000 (2). In contrast, the largest amount of genetic diversity was seen between the two Atlanta populations when they were compared temporally. The incidence rate of C. glabrata in Atlanta from 1992 to 1993 was only 0.96/100,000 (10). Preliminary analysis showed that in 2008 the incidence rate in Atlanta was 3.9/100,000 (2). It is interesting to note that the large change in the C. glabrata population structure in Atlanta also correlated with an increased incidence rate. While there are multiple factors that contribute to an increased incidence rate, it is reasonable to speculate that changes in the population structure of the organism, perhaps to better fill an available niche, played a contributing role. Knowledge of the population genetic structure of the various C. glabrata populations within the ongoing candidemia surveillance will contribute significantly to our further analysis of incidence rates, patient outcomes, and antifungal resistance within these populations. Research to better understand these relationships is ongoing.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the significant contributions of the following members of the CDC Candidemia Surveillance Group: Angela Ahlquist, Monica Farley, Lee Harrison, Wendy Baughman, Betsy Siegel, Rosemary Hollick, Kizee Etienne, Eszter Deak, Joyce Peterson, Naureen Iqbal, Lauren Smith, and Tom Chiller. We thank the staff of all the institutions that contributed isolates to this study. We thank Arun Balajee for critical reading of the manuscript. We also acknowledge the members of the DFBMD core sequencing laboratory at the Centers for Disease Control and Prevention for their technical assistance.
The findings and conclusions of this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 26 February 2010.
REFERENCES
- 1.Agapow P.-M., Burt A. 2001. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1:101–102 [Google Scholar]
- 2.Ahlquist A., Farley M. M., Harrison L. H., Baughman W., Siegel B., Hollick R., Lockhart S. R., Magill S. S., Chiller T. 2009. Epidemiology of candidemia in metropolitan Atlanta and Baltimore City and County: preliminary results of population-based active, laboratory surveillance—2008-2010, abstr. M-1241. Abstr. 49th Intersci. Conf.Antimicrob. Agents Chemother., San Francisco, CA [Google Scholar]
- 3.Brisse S., Pannier C., Angoulvant A., deMeeus T., Diancourt L., Faure O., Muller H., Peman J., Viviani M. A., Grillot R., Dujon B., Fairhead C., Hennequin C. 2009. Uneven distribution of mating types among genotypes of Candida glabrata isolates from clinical samples. Eukaryot. Cell 8:287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dodgson A. R., Pujol C., Denning D. W., Soll D. R., Fox A. J. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J. Clin. Microbiol. 41:5709–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodgson A. R., Pujol C., Pfaller M. A., Denning D. W., Soll D. R. 2005. Evidence for recombination in Candida glabrata. Fung. Genet. Biol. 42:233–243 [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein J. 1973. Maximum likelihood and minimum-step methods for estimating evolutionary trees from data on discrete characters. Syst. Zool. 22:240–249 [Google Scholar]
- 7.Fidel P. L., Jr., Vazquez J. A., Sobel J. D. 1999. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12:80–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajjeh R. A., Sofair N. A., Harrison L. H., Lyon G. M., Arthington-Skaggs B. A., Mirza S. A., Phelan M., Morgan J., Lee-Yang W., Ciblak M. A., Benjamin L. E., Sanza L. T., Huie S., Yeo S. F., Brandt M. E., Warnock D. W. 2004. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J. Clin. Microbiol. 42:1519–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartl D. L., Clark A. G. 1997. Principles of population genetics, 3rd ed.Sinauer Associates,Inc., Sunderland, MA [Google Scholar]
- 10.Kao A. S., Brandt M. E., Pruitt W. R., Conn L. A., Perkins B. A., Stevens D. S., Baughman W. S., Reingold A. L., Rothrock G. A., Pfaller M. A., Pinner R. W., Hajjeh R. A. 1999. The epidemiology of candidemia in two United States cities: results of a population-based active surveillance. Clin. Infect. Dis. 29:1164–1170 [DOI] [PubMed] [Google Scholar]
- 11.Lin C. Y., Chen Y. C., Lo H. J., Chen K. W., Li S. Y. 2007. Assessment of Candida glabrata strain relatedness by pulsed-field gel electrophoresis and multilocus sequence typing. J. Clin. Microbiol. 45:2452–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odds F. C., Hanson M. F., Davidson A. D., Jacobsen M. D., Wright P., Whyte J. A., Gow N. A., Jones B. L. 2007. One year prospective survey of Candida bloodstream infections in Scotland. J. Med. Microbiol. 56:1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odds F. C., Jacobsen M. D. 2008. Multilocis sequence typing of pathogenic Candida species. Eukaryot. Cell 7:1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaller M. A., Diekema D. J. 2007. The epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaller M. A., Diekema D. J., Gibbs D. L., Newell V. A., Meis J. F., Gould I. M., Fu W., Colombo A. L., Rodriguez-Noriega E., and the Global Antifungal Surveillance Group 2007. Results from the ARTEMIS DISK Global Antifungal Surveillance study, 1997 to 2005: an 8.5-year analysis of susceptibilities of Candida species and other yeast species to fluconazole and voriconazole determined by CLSI standardized disk diffusion testing. J. Clin. Microbiol. 45:1735–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pond S. L., Frost S. D., Muse S. V. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679 [DOI] [PubMed] [Google Scholar]
- 17.Raymond M., Rousset F. 1995. GENEPOP (version 1.3): population genetics software for exact tests and ecumenicism. J. Hered. 86:248–249 [Google Scholar]
- 18.Sherman D. J., Martin T., Nikolski M., Cayla C., Souciet J. L., Durrens P., and the Genolevures Consortium 2009. Genolevures: protein families and synteny among complete hemiascomycetous yeast proteomes and genomes. Nucleic Acids Res. 37:D550–D554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soll D. R. 2000. The ins and outs of DNA fingerprinting the infectious fungi. Clin. Microbiol. Rev. 13:332–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 21.Trick W. E., Fridkin S. K., Edwards J. R., Hajjeh R. A., Gaynes R. P., et al. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35:627–630 [DOI] [PubMed] [Google Scholar]
- 22.Urwin R., Maiden C. J. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479–487 [DOI] [PubMed] [Google Scholar]
- 23.Weir B. S. 1996. Genetic data analysis II. Sinauer, Sunderland, MA [Google Scholar]
- 24.Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., Edmond M. B. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.