Abstract
Acanthamoeba castellanii is a facultative pathogen that has a two-stage life cycle comprising the vegetatively growing trophozoite stage and the dormant cyst stage. Cysts are formed when the cell encounters unfavorable conditions, such as environmental stress or food deprivation. Due to their rigid double-layered wall, Acanthamoeba cysts are highly resistant to antiamoebic drugs. This is problematic as cysts can survive initially successful chemotherapeutic treatment and cause relapse of the disease. We studied the Acanthamoeba encystment process by using two-dimensional gel electrophoresis (2DE) and found that most changes in the protein content occur early in the process. Truncated actin isoforms were found to abound in the encysting cell, and the levels of translation elongation factor 2 (EF2) were sharply decreased, indicating that the rate of protein synthesis must be low at this stage. In the advanced stage of encystment, however, EF2 levels and the trophozoite proteome were partly restored. The protease inhibitors PMSF (phenylmethylsulfonyl fluoride) and E64d [(2S,3S)-trans-epoxysuccinyl-l-leucylamido-3-methylbutane ethyl ester] inhibited the onset of encystment, whereas the protein synthesis inhibitor cycloheximide was ineffective. Changes in the protein profile, similar to those of encysting cells, could be observed with trophozoite homogenates incubated at room temperature for several hours. Interestingly, these changes could be inhibited significantly by cysteine protease inhibitors but not by inhibitors against other proteases. Taken together, we conclude that the encystment process in A. castellanii is of a bipartite nature consisting of an initial phase of autolysis and protein degradation and an advanced stage of restoration accompanied by the expression of encystment-specific genes.
The bacteriovorous Acanthamoeba spp. occur ubiquitously in the environment (27) and have a two-stage life cycle consisting of the replicating and feeding trophozoite stage and the dormant, double-walled, cyst stage (16). Cysts are formed in order to survive in an inhospitable environment and are able to persist in a wide variety of habitats (4, 17). Indeed, the ubiquity of Acanthamoeba is made possible by the extreme resistance of the cyst against desiccation, temperature changes, chemicals, radiation, and prolonged starvation. Also, various antiamoebic agents, such as benzalkonium chloride and propamidine isethionate, have no effect on cysts (9, 13, 29). Since acanthamoebae are facultative pathogens that can cause Acanthamoeba keratitis (AK) and granulomatous amoebic encephalitis (GAE), encystment is also of medical relevance (16). An often occurring complication in the treatment of AK is the presence of viable cysts that remain in the corneal stroma after initial successful therapy, as these can eventually excyst again and lead to recurrent infections (23).
According to Weisman (31), the encystment process comprises three phases: induction, wall synthesis, and dormancy. During the induction phase, trophozoites begin to lose their amoeboid appearance and become round. The first wall that is formed gives rise to the exocyst; this wall is 0.3 to 0.5 μm thick and consists mostly of acid-insoluble proteins. The endocyst is formed after the appearance of a well-defined layer whose major component is cellulose (31). Cell wall synthesis is usually accompanied by a decrease in cytoplasmic mass of approximately 80% through a gradual dehydration of the amoeba, thereby causing retraction of the protoplast from the cell wall (2). Rather early, autolysosomes appear and remain in the cytoplasm throughout the whole encystment process. In light of these dramatic changes in the cell's physiology, it is surprising that the encysting cell can stop and revert the process until 15 h after induction (30). Afterwards, however, cells become committed to the completion of the encystment process.
At the molecular level, a number of factors involved in the encystment process have been characterized thus far. For example, cyst-specific protein 21 (Csp21) is a cyst wall protein found in group II acanthamoebae and was reported to be synthesized approximately 12 h after induction (6). The expression of the respective gene is repressed under normal growth conditions via one or more repressor elements between the TATA box and nucleotide (nt) +63 (3). Furthermore, encystment requires serine protease activity (5, 20) and autophagy proteins (22), all of which are suggested to be involved in autolytic processes, and glycogen phosphorylase, which is necessary for the breakdown of glycogen (14). The glucose-1-phosphate that is thereby liberated is subsequently used for the buildup of cellulose in the cyst wall.
In the search for additional factors, there have been several successful attempts in the past years to screen encysting Acanthamoeba castellanii for genes specifically expressed during encystment at the mRNA level (19, 21) as well as at the protein level (1, 24). However, there is still a lack of information on the extent of cellular reorganization during the encystment process at the protein level. In this study, we therefore aimed to monitor the encystment process in PAT06, a new clinical isolate of A. castellanii (10), by using two-dimensional gel electrophoresis (2DE) and to analyze the developmental and molecular processes at the proteomic level.
MATERIALS AND METHODS
Chemicals.
PMSF (phenylmethylsulfonyl fluoride), E64 (l-trans-3-carboxyoxiran-2-carbonyl-l-leucylagmatine), E64d [(2S,3S)-trans-epoxysuccinyl-l-leucylamido-3-methylbutane ethyl ester], AEBSF (4-(2-aminoethyl)benzenesulfonyl fluoride), pepstatin A, phenanthroline, cycloheximide, and Triton X-100 were all purchased from Sigma-Aldrich.
Selected strain and culture conditions.
A strain that can encyst quickly and in a synchronized fashion had to be selected in order to be able to observe definite developmental stages. We chose the new clinical A. castellanii isolate, PAT06 (morphological group II, genotype T4), because in a previous study this strain was found to encyst synchronously when incubated in starvation medium, whereas the type strain NEFF had lost much of its encystment capacity (10). PAT06 encystment was induced by placing cells in Tris-buffered starvation medium (pH 9) (6). Normal culture of trophozoites was done axenically in PYG (peptone-yeast extract-glucose) medium as described previously (10).
Induction of encystment.
The cultures were harvested by centrifugation (800 × g) at room temperature (RT). Cell pellets were washed twice in Tris-buffered starvation medium (pH 9) (6), taken up in 25-ml portions of starvation medium, and incubated in 300-ml tissue culture flasks at RT for the time periods indicated.
Processing of samples for two-dimensional gel electrophoresis (2DE).
The cells (approximately 107) were harvested at RT by centrifugation at 800 × g (5 min), and the resulting cell pellets were subsequently washed in two centrifugation steps (800 × g at RT, 5 min) in 1× phosphate-buffered saline (PBS) at the same speed. The cells were resuspended in 2 ml of 10% trichloroacetic acid (TCA), 20% ultrapure water, and 70% acetone and incubated at −20°C for at least 1 h in order to deactivate proteases and kinases, to remove salts, and to precipitate cellular proteins. Afterwards, the samples were centrifuged at 20,000 × g at 4°C, and the resulting pellets were washed twice by resuspending the pellet in a mixture of acetone and double-distilled water (ddH2O) (9:1), followed by centrifugation at 20,000 × g (at 4°C for 20 min). Finally, the samples were resolubilized in an appropriate amount of sample buffer (12), and insoluble material was removed by centrifugation at 20,000 × g (at 20°C for 20 min).
Two-dimensional gel electrophoresis (2DE).
Two-dimensional gel electrophoresis (2DE) was performed according to the protocol described for Entamoeba histolytica (12). Gel images were evaluated using Melanie 4 software (Genebio). For isoelectric focusing in 17-cm IPG (immobilized pH gradient) strips, 500-μg amounts of protein were loaded. The second dimension was run in 20- by 20-cm gels in a Protean II xi cell (Bio-Rad).
Homogenization of cells and removal of the large organelle fraction.
After the cells were harvested, approximately 107 cells were resuspended in 600 μl of 1× PBS and broken up in a Dounce homogenizer. Protease inhibitors were added at the concentrations given. After removal of intact cells (500 × g, 3 min), the cell lysate was either used immediately for assays or centrifuged at 20,000 × g for 20 min in order to remove the large organelle fraction. All centrifugation steps were performed at 4°C.
Protein identification.
Spots of interest were manually excised from Coomassie brilliant blue-stained gels for tryptic digestion and subsequent mass spectrometry (MS) analysis. Peptide mass fingerprinting (PMF) and postsource decay (PSD) fragmentation experiments were performed on a matrix-assisted laser desorption ionization (MALDI) reflectron time-of-flight (TOF) instrument (AXIMA-CFRplus; Shimadzu Biotech Kratos Analytical) as previously described (15). For protein identification, measured monoisotopic m/z values were submitted after removal of matrix and enzyme-derived contaminations (8) to an in-house Mascot server (26) searching protein and DNA sequences from public databases (NCBInr 9/26/2009 [9,775,507 sequences; 3,338,911,485 residues; taxonomy: other eukaryota containing 190,336 sequences and eucaryota containing 3,566,107 sequences]). A protein was considered identified if the scores of database searches clearly exceeded the algorithm's significance threshold (P < 0.05) for PMF and/or PSD data.
Strain accession number.
Acanthamoeba castellanii strain PAT06 was deposited at the ATCC and is available under accession number NRS-10465 (preliminary number).
RESULTS
Most of the changes in the protein profile of encysting cells occur early in the encystment process.
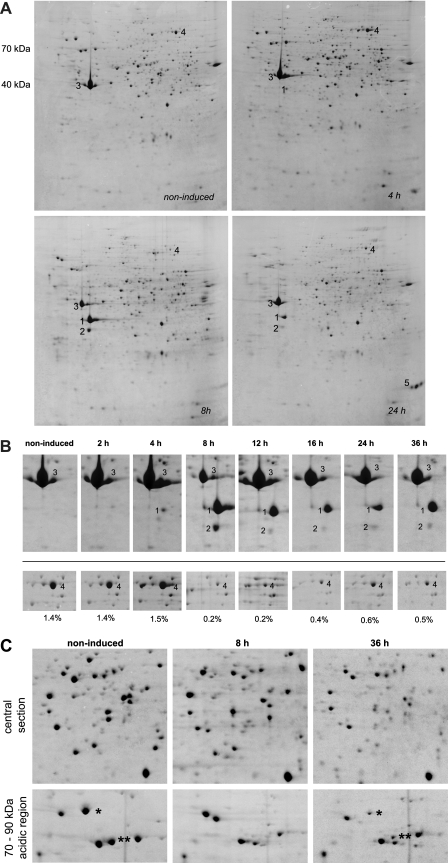
At defined time points after induction of encystment (2 h, 4 h, 8 h, 16 h, 24 h, and 36 h), the cultures were harvested, lysed, and prepared for two-dimensional gel electrophoresis (2DE). Although hardly any differences could be observed in the protein profile of cells 4 h after induction compared to noninduced trophozoites (Fig. 1A), one novel protein spot was detected (spot 1) at approximately 35 kDa and pI 6. After 8 h, the protein profile had already changed profoundly (Fig. 1A). Spot 1 had become the most prominent protein visualized, with a second spot (spot 2, at about 30 kDa and pI 6) appearing at a slightly lower molecular mass. Conspicuously, the levels of the protein above the two novel spots had decreased (spot 3, at about 40 kDa and pI 6), suggesting that spots 1 and 2 were degradation products of this protein, rather than newly synthesized protein. Spot 1 remained present throughout the time interval studied after its appearance (Fig. 1B), whereas the intensity of spot 2 decreased again after 12 h (Fig. 1B). After their isolation from 2D gels for protein identification, spots 1 to 3 were firmly identified as actin (Table 1). In the case of spot 3, the peptide mass fingerprints (PMF) gave a Mascot score for A. castellanii actin just very slightly below the threshold of statistical significance, but the homology to actin from the choanoflagellate Monosiga brevicollis was clearly statistically significant (Table 1). Moreover, the homology of the identified actin and the protein sequence of A. castellanii actin is above 93%.
Fig. 1.
Course of encystment in A. castellanii PAT06. (A) The cells were induced to encyst in starvation medium and harvested at the time points given. Subsequently, samples were prepared for 2DE. Spot 1, actin isoform; spot 2, another actin isoform; spot 3, actin; spot 4, translation elongation factor 2 (EF2); spot 5, unidentified encystment specific protein. The 2D gels (12.5% polyacrylamide) have a pI range of 3 (left) to 10 (right). The molecular masses are indicated next to the top left gel. (B) Sections from 2DE gels, including more time points. The numbers refer to the same spots or proteins as in panel A. The proportion of EF2 of the total protein visualized on the 2D gels is given below the respective images. (C) Two sections from 2D gels corresponding to noninduced cells and cells 8 h and 36 h after induction of encystment. In the 70- to 90-kDa region in the acidic part of the gel, several trophozoite proteins are marked by asterisks; these proteins disappear 8 h after induction and reappear later in encystment.
Table 1.
Proteins identified in this study
| Spot identification | Protein | Database | Database entry | Mascot score for PMFa | No. of matched peptides/total no. of peptides | Sequence coverage (%) | Mascot score for PSDa |
|---|---|---|---|---|---|---|---|
| Spot 1 | Actin | NCBInr/other eucaryota | gi‖223855 actin (Acanthamoeba castellanii) | 83 (>65) | 10/32 | 24 | NAb |
| Spot 2 | Actin | NCBInr/other eucaryota | gi‖223855 actin (Acanthamoeba castellanii) | 145 (>65) | 12/24 | 37 | NA |
| Spot 3 | Actin | NCBInr/other eucaryota | gi‖16554294 actin (Monosiga brevicollis) | 77 (>65) | 8/21 | 19 | NA |
| NCBInr/other eucaryota | gi‖223855 actin (Acanthamoeba castellanii) | 64 (>65) | 7/21 | 16 | NA | ||
| Spot 4 | Elongation factor 2 | NCBInr/eucaryota | Elongation factor 2 (Dictyostelium discoideum) | 77 (>78) | 11/24 | 12 | NA |
| NCBInr/other eucaryota | gi‖464158 elongation factor 2 (Entamoeba histolytica) | NA | NA | NA | 47 (>40) FYAFGR |
Mascot scores (threshold value for significant protein identification) for peptide mass fingerprints (PMF) and the postsource decay (PSD)-derived peptide sequence of spot 4 are given.
NA, not applicable.
In addition to the appearance of these novel protein spots, we also noted that the levels of a protein in the higher-molecular-mass range (spot 4, at about 90 kDa and pI 8.5) had sharply decreased at 8 h after induction (0.2% of total cellular protein) compared to the trophozoite stage (1.4% of total cellular protein) (Fig. 1A and B). This protein was identified as translation elongation factor 2 (EF2) on the basis of its significant sequence homology to Entamoeba histolytica EF2 and a close to significant PMF Mascot score for Dictyostelium discoideum EF2 (Table 1). The steep decrease in the levels of expression of EF2 during early encystment suggests that the rate of protein synthesis must be very low at this stage.
At 24 h after induction (Fig. 1A), a prominent protein in the lower-molecular-mass range and basic pI range was readily detected (spot 5, at about 20 kDa and pI 9). As this encystment-specific protein was still absent 16 h after induction (data not shown), it must be synthesized sometime between 16 and 24 h after induction. Unfortunately, we did not find a statistically significant match for this protein in the databases, but it is interesting to note that its molecular mass matches well to that of Csp21 (20,187 Da and pI 9.06), a cyst-specific protein localizing to the cyst wall in A. castellanii NEFF (6). Interestingly, the level of expression of EF2 (spot 4) had partly recovered by 24 h (0.6% of total protein) and 36 h (0.5% of total protein) after induction compared to the level 8 h after induction (Fig. 1B), presumably because the synthesis of novel encystment-specific proteins, e.g., spot 5, is required in the advanced stage of encystment. However, proteins that had been present in the trophozoite but were absent 8 h after induction had also been resynthesized (Fig. 1C). In general, however, the levels of expression of most proteins had decreased sharply during encystment (Fig. 1C).
In total, 42 proteins of interest were isolated from gels for identification, but with the exception of actin (spots 1 to 3) and EF2 (spot 4) (Table 1), unambiguous mass spectrometric/bioinformatic identification was up to now not successful due the still very limited A. castellanii genome information available and the often poorly pronounced similarities to homologues from organisms whose genomes have been sequenced.
The early phase of encystment can be completely inhibited by the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF).
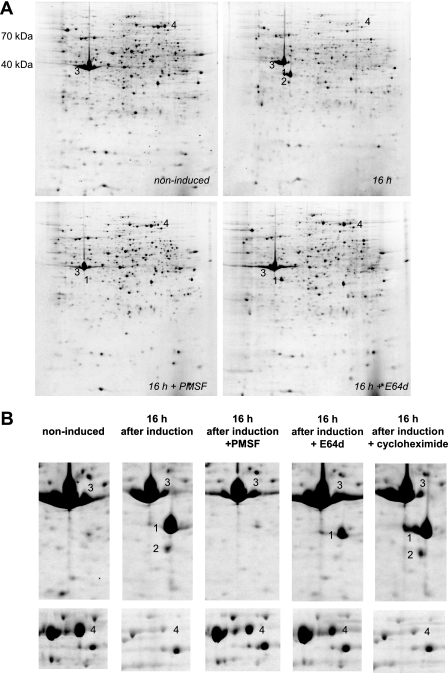
We had observed that novel actin isoforms appearing early during encystment could be degradation products of actin (Fig. 1A and B). Since it had been previously reported that the inhibition of serine proteases with PMSF blocks the encystment process in Acanthamoeba (5, 20), we aimed to assess the effect of PMSF on the 2DE profiles of encysting cells.
Although the addition of 1 mM PMSF to the starvation medium did not prevent the rounding up of cells, as typically seen during encystment, it totally abolished all alterations previously observed in the 2DE profiles of encysting cells (Fig. 2A and B). In fact, even after overnight incubation (16 h) in starvation medium containing 1 mM PMSF, the 2DE profile of the cells was practically indistinguishable from that of noninduced trophozoites. Also, 100 μM E64d, the membrane-permeable variant of the cysteine protease inhibitor E64 (l-trans-3-carboxyoxiran-2-carbonyl-l-leucylagmatine), slowed down encystment compared to the control culture, but inhibition was incomplete (Fig. 2A and B). This was probably due to the lower concentration of E64d used compared to the concentration of PMSF, which might have been insufficient to inhibit all cysteine protease activity during the time interval studied. Indeed, PMSF was ineffective at a concentration of 100 μM (not shown). However, because of the very high price of E64d, the experiment was not repeated at a concentration of 1 mM.
Fig. 2.
The protease inhibitor PMSF prevents cells from starting the encystment process. (A) The cells were placed in encystment medium either without further additive or with the protease inhibitor PMSF (1 mM) or E64d (100 μM). The numbers refer to the same spots or proteins as in Fig. 1. The 2D gels (12.5% polyacrylamide) have a pI range of 3 (left) to 10 (right). The molecular masses are indicated next to the top left gel. (B) Close-up of actin and EF2 in noninduced cells, encysting cells without further additive, and encysting cells with either PMSF (1 mM), E64d (100 μM), or the protein synthesis inhibitor cycloheximide (100 μM). The numbers refer to the same spots or proteins as in Fig. 1.
Strikingly, the addition of the protein synthesis inhibitor cycloheximide at 100 μM had no observable effect on the early phase of encystment (Fig. 2B), although this compound is clearly effective in strain PAT06 and inhibited cell growth when we applied it at this concentration (not shown).
The proteolytic processes observed during the early phase of encystment are mediated by proteases already present in the trophozoite.
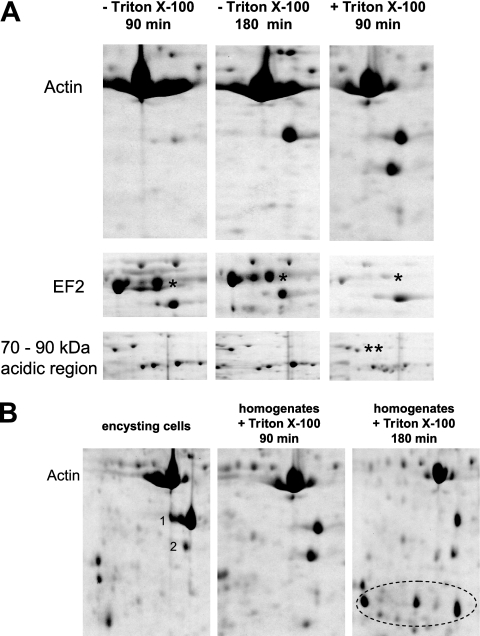
Since the protein synthesis inhibitor cycloheximide did not inhibit the high autoproteolytic activity which is typical for the early stage of encystment, we argued that the proteases required for this activity are already present in the proliferating, noninduced trophozoite rather than having to be synthesized after induction of encystment. To address this hypothesis, we homogenized PAT06 cells in a Dounce homogenizer, incubated the resulting lysates for various periods of time at room temperature (RT), and subsequently analyzed the samples by 2DE. Proteolytic activity proved to be rather low, but after 3 h of incubation, degradation of actin, as observed in encysting cells, could be detected (Fig. 3A). Interestingly, proteolytic activity was greatly enhanced when 0.1% Triton X-100 was added to the lysates. After 90 min of incubation, actin degradation and a decrease of EF2 levels could be observed (Fig. 3A). Thus, the proteolytic activity which leads to the most conspicuous changes in the 2DE protein profile of encysting cells exists in the noninduced trophozoite. It is interesting to note, however, that longer incubation of lysates with Triton X-100 (3 to 5 h) resulted in an extent of proteolysis not observed in encysting cells (Fig. 3B).
Fig. 3.
The protein degradation observed in encysting cells also occurs in homogenates of noninduced trophozoites. (A) Noninduced trophozoites were lysed in a Dounce homogenizer, and the resulting homogenates were incubated at RT under shaking for various periods of time either without any additive or with 0.1% Triton X-100. The asterisks in the acidic, high-molecular-mass region on the gel mark the same proteins as in Fig. 1C. (B) After extended incubation at RT (180 min) with 0.1% Triton X-100, protein fragments that are not observed in encysting cells become apparent (circled).
The proteolytic activity localizes to the large organelle fraction and can be inhibited only by the cysteine protease inhibitor E64.
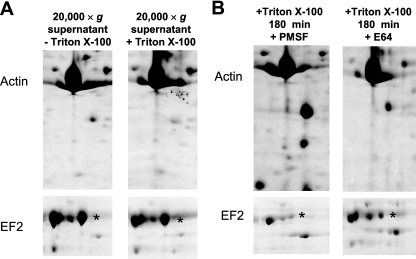
The observation that protein degradation in the homogenates was greatly enhanced by Triton X-100 suggested that the respective proteolytic agent localizes to an intracellular vesicle or another organelle which is permeabilized upon addition of the detergent. This hypothesis was tested by removing the large organelle fraction by centrifugation at 20,000 × g for 20 min after cell lysis, followed by the incubation of the supernatant fraction at RT for 3 h. Such treatment abolished proteolysis even in the presence of Triton X-100 (Fig. 4A), clearly showing that the proteases that mediate the observed proteolytic events reside in a large organelle and not in the cytosol.
Fig. 4.
The proteolytic activity observed in trophozoite homogenates resides in the large organellar fraction and can be inhibited with E64. (A) After homogenization, the large organelle fraction was removed from the trophozoite homogenates by centrifugation, and supernatants were incubated at room temperature. Translation elongation factor 2 is marked by an asterisk in panels A and B. (B) The proteolytic activity in trophozoite homogenates can be inhibited only by E64 (30 μM), not by PMSF (3 mM).
We further investigated the influence of several protease inhibitors on the extent of proteolysis in the cell homogenates. Surprisingly, PMSF had no effect on the extent of protein degradation in the homogenates, even at a concentration of 3 mM (Fig. 4B). Also, another serine protease inhibitor, 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), did not inhibit proteolysis in the samples when applied at a concentration of 1 mM (data not shown). Equally inefficient were the acid protease inhibitor pepstatin A (100 μM), and the metalloprotease inhibitors EDTA (5 mM) and phenanthroline (5 mM). However, the cysteine protease inhibitor E64 (100 μM) efficiently inhibited protein degradation in the homogenates even in the presence of Triton X-100 (Fig. 4B).
DISCUSSION
In the present study, we analyzed the early and advanced stages of the encystment process in A. castellanii (strain PAT06) by using 2DE. We showed that most of the changes in the protein profile occur early during encystment (Fig. 1A) and result from proteolytic activity in the encysting cell. This proteolytic activity is totally inhibited by the serine protease inhibitor PMSF (1 mM) and widely inhibited by the cysteine protease inhibitor E64d (100 μM) (Fig. 2A and B). The effectiveness of the latter was surprising because cysteine proteases have been suggested to be dispensable for encystment due to the inability of E64 to inhibit encystment (20). However, E64d permeates membranes much more efficiently than E64 and is therefore arguably present in far higher concentrations within the cells.
The prominent role of proteolytic activity during encystment, as described in this study, is in accordance with earlier reports on autophagic processes in encysting A. castellanii (2). This study also provides further support for a causal connection between protease activity and encystment capacity of A. castellanii, a connection which has been indicated by our previously published observation that both characteristics are less marked in aged and axenically grown cultures (11).
We further show here that at least most of the protease activity required in the early phase of encystment is already present in the proliferating trophozoite. This is indicated by the inability of cycloheximide, a potent inhibitor of protein synthesis in eukaryotic cells, to prevent Acanthamoeba from starting the encystment process (Fig. 2B). In addition, 2D gels of trophozoite homogenates displayed significant similarities to 2D gels of encysting cells after incubation at RT for several hours (Fig. 2 and 3). Interestingly, however, this proteolytic activity in homogenates was inhibited only by the cysteine protease inhibitor E64 (Fig. 4B). The serine protease inhibitor PMSF had no effect in trophozoite homogenates (Fig. 4B), which is in stark contrast to our observation that PMSF effectively inhibits protein degradation in encysting cells (Fig. 2). Also, another serine protease inhibitor, AEBSF, was ineffective. It is, therefore, likely that serine proteases do not degrade cellular protein during encystment on a large scale but that they are necessary to mediate certain discrete processes in the encysting cell. Such processes of a more subtle nature would be hard or even impossible to monitor via 2DE, which mainly visualizes intermediately and highly expressed proteins. For example, one possible role of serine proteases could be mediating the release of cysteine proteases from their intracellular confinement (Fig. 4A) by promoting the maturation of autophagosomes. There are certain indications for this assumption, because the autoproteolytic activity is not cytosolic but resides in the large organelle fraction (Fig. 4A). Moreover, AhCP, a cysteine protease in Acanthamoeba healyi, was found to localize to lysosomes (18). Nevertheless, other physiological roles of serine proteases during encystment are also conceivable, and currently, there are no data available that further substantiate the above-stated hypothesis. Indisputably, however, serine proteases are necessary for encystment, as repeatedly demonstrated in gene silencing experiments with small interfering RNA (siRNA) (5, 20).
Unfortunately, protein identification by mass spectrometry was often unsuccessful due to the deficiency of reliable genomic data, low sequence similarity to well-described, related organisms, and the fact that many proteins on the gels of encysting cells and cysts were partially degraded isoforms of their respective counterparts in the trophozoite. In fact, the identification of partially degraded proteins by peptide mass fingerprinting (PMF) is greatly complicated by aberrant tryptic peptide lengths, a problem that was certainly exacerbated by the use of a genotype T4 Acanthamoeba isolate (i.e., PAT06) other than NEFF, the genome strain. Unfortunately, it was not possible to avoid the selection of partially degraded proteins for identification, because these were mistaken for de novo-synthesized proteins. Indeed, many of the partially degraded proteins were stable and persisted throughout the time interval studied, i.e., at least 36 h after induction of encystment (Fig. 1B and C). Taken together, these issues made highly confident (in terms of statistical scoring parameters) protein identification with the methodology applied extremely difficult. However, we succeeded in identifying several proteins that are affected during the encystment process, including actin and EF2 (Table 1). Actin is partially degraded in the early stage of encystment (28), resulting in the appearance of persistent actin degradation products in the encysting cell (this study). During early encystment, EF2 was found to be present in sharply decreased amounts, i.e., less than 15% of the original level (Fig. 1B). This is in line with a recent study (1) in which a proteolytic fragment of EF2 was found in 2D gels of encysting cells of another A. castellanii strain. This suggests that the rate of protein synthesis must be very low during early encystment, which is corroborated by previous studies that reported that rRNA synthesis shut off early in encystment (25) and reported a strongly increased proportion of monosomal ribosomes (7). Later in the encystment process, however, the synthesis of encystment-specific proteins is required, including Csp21, serine proteases, glycogen phosphorylase, and cellulose synthase (5, 6, 14, 20, 21). It has to be emphasized, though, that defining “later in the encystment process” or “more advanced stage of encystment” in a strictly temporal sense can be misleading because the Acanthamoeba strains used by different research groups do not encyst equally fast (5, 6, 14, 20–22), probably due to physiological adaptations to prolonged axenic culture (10, 11). Thus, the time points given in this study apply only to strain PAT06 after approximately 2 years of axenic culture. Still, it seems safe to state that one of the hallmarks of advanced encystment must be the partial recovery of EF2 levels, as observed on our 2D gels of cells 24 h after induction of encystment (Fig. 1B). This was also accompanied by the reappearance of proteins that had been present in the trophozoite but were absent 8 h after induction (Fig. 1C), as well as of novel encystment-specific proteins, e.g., spot 5 (Fig. 1A). The partial restoration of the trophozoite proteome suggests that encystment is not a gradual process but a bipartite process, i.e., comprising an early phase of autolysis and protein degradation as well as an advanced phase of partial restoration and expression of encystment-specific genes. This finding reveals yet another layer of complexity of the Acanthamoeba encystment process.
ACKNOWLEDGMENT
This work was supported by grant P19044 of the Austrian Science Fund (FWF).
Footnotes
Published ahead of print on 26 February 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Bouyer S., Rodier M. H., Guillot A., Héchard Y. 2009. Acanthamoeba castellanii: proteins involved in actin dynamics, glycolysis, and proteolysis are regulated during encystation. Exp. Parasitol. 123:90–94 [DOI] [PubMed] [Google Scholar]
- 2. Bowers B., Korn E. D. 1969. The fine structure of Acanthamoeba castellanii (Neff strain). II. Encystment. J. Cell Biol. 41:786–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen L., Orfeo T., Gilmartin G., Bateman E. 2004. Mechanism of cyst specific protein 21 mRNA induction during Acanthamoeba differentiation. Biochim. Biophys. Acta 1691:23–31 [DOI] [PubMed] [Google Scholar]
- 4. De Jonckheere J. F. 1991. Ecology of Acanthamoeba. Rev. Infect. Dis. 13(Suppl. 5):S385–S387 [DOI] [PubMed] [Google Scholar]
- 5. Dudley R., Alsam S., Khan N. A. 2008. The role of proteases in the differentiation of Acanthamoeba castellanii. FEMS Microbiol. Lett. 286:9–15 [DOI] [PubMed] [Google Scholar]
- 6. Hirukawa Y., Nakato H., Izumi S., Tsuruhara T., Tomino S. 1998. Structure and expression of a cyst specific protein of Acanthamoeba castellanii. Biochim. Biophys. Acta 1398:47–56 [DOI] [PubMed] [Google Scholar]
- 7. Jantzen H. 1981. Ribosomal phosphoproteins in Acanthamoeba castellanii. Eur. J. Biochem. 119:347–352 [DOI] [PubMed] [Google Scholar]
- 8. Keller B. O., Sui J., Young A. B., Whittal R. M. 2008. Interferences and contaminants encountered in modern mass spectrometry. Anal. Chim. Acta 627:71–81 [DOI] [PubMed] [Google Scholar]
- 9. Khunkitti W., Hann A. C., Lloyd D., Furr J. R., Russell A. D. 1998. Biguanide-induced changes in Acanthamoeba castellanii: an electron microscopic study. J. Appl. Microbiol. 84:53–62 [DOI] [PubMed] [Google Scholar]
- 10. Köhsler M., Leitsch D., Fürnkranz U., Duchêne M., Aspöck H., Walochnik J. 2008. Acanthamoeba strains lose their abilities to encyst synchronously upon prolonged axenic culture. Parasitol. Res. 102:1069–1072 [DOI] [PubMed] [Google Scholar]
- 11. Köhsler M., Leitsch D., Duchêne M., Nagl M., Walochnik J. 2009. Acanthamoeba castellanii: growth on human cell layers reactivates attenuated properties after prolonged axenic culture. FEMS Microbiol. Lett. 299:121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leitsch D., Radauer C., Paschinger K., Wilson I. B. H., Breiteneder H., Scheiner O., Duchêne M. 2005. Entamoeba histolytica: analysis of the trophozoite proteome by two-dimensional polyacrylamide gel electrophoresis. Exp. Parasitol. 110:191–195 [DOI] [PubMed] [Google Scholar]
- 13. Lloyd D., Turner N. A., Khunkitti W., Hann A. C., Furr J. R., Russell A. D. 2001. Encystation in Acanthamoeba castellanii: development of biocide resistance. J. Eukaryot. Microbiol. 48:11–16 [DOI] [PubMed] [Google Scholar]
- 14. Lorenzo-Morales J., Kliescikova J., Martinez-Carretero E., De Pablos L. M., Profotova B., Nohynkova E., Osuna A., Valladares B. 2008. Glycogen phosphorylase in Acanthamoeba spp.: determining the role of the enzyme during the encystment process using RNA interference. Eukaryot. Cell 7:509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marchetti-Deschmann M., Kemptner L., Reichel C., Allmaier G. 2009. Comparing standard microwave-assisted staining protocols for SDS-PAGE of glycoproteins followed by subsequent PMF with MALDI MS. J. Proteomics 72:628–639 [DOI] [PubMed] [Google Scholar]
- 16. Marciano-Cabral F., Cabral G. 2003. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 16:273–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mergeryan H. 1991. The prevalence of Acanthamoeba in the human environment. Rev. Infect. Dis. 13:S390–S391 [DOI] [PubMed] [Google Scholar]
- 18. Moon E. K., Lee S. T., Chung D. I., Kong H. H. 2006. Intracellular localization and trafficking of serine proteinase AhSub and cysteine proteinase AhCP of Acanthamoeba healyi. Eukaryot. Cell 5:125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moon E. K., Chung D. I., Hong C. C., Kong H. H. 2007. Differentially expressed genes of Acanthamoeba castellanii during encystations. Korean J. Parasitol. 45:283–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moon E. K., Chung D. I., Hong Y. C., Kong H. H. 2008. Characterization of a serine proteinase mediating encystations of Acanthamoeba. Eukaryot. Cell 7:1513–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moon E. K., Chung D. I., Hong Y. C., Ahn T. I., Kong H. H. 2008. Acanthamoeba castellanii: gene profile of encystations by ESTs analysis and KOG assignment. Exp. Parasitol. 119:111–116 [DOI] [PubMed] [Google Scholar]
- 22. Moon E. K., Chung D. I., Hong Y. C., Kong H. H. 2009. Autophagy protein 8 mediating autophagosome in encysting Acanthamoeba. Mol. Biochem. Parasitol. 168:43–48 [DOI] [PubMed] [Google Scholar]
- 23. Moore M. B., McCulley J. P., Newton C., Cobo L. M., Foulks G. N., O'Day D. M., Johns K. J., Driebe W. T., Wilson L. A., Epstein R. J. 1987. Acanthamoeba keratitis. A growing problem in soft and hard contact lens wearers. Ophthalmology 94:1654–1661 [PubMed] [Google Scholar]
- 24. Park J. T., Jeong Y. E., Ahn T. I. 2002. Changes in profiles of major proteins in encysting Acanthamoeba castellanii. Korean J. Biol. Sci. 6:341–347 [Google Scholar]
- 25. Paule M. R., Iida C. T., Perna P. J., Harris G. H., Knoll D. A., D'Alessio J. M. 1984. In vitro evidence that eukaryotic ribosomal RNA transcription is regulated by modification of RNA polymerase I. Nucleic Acids Res. 12:8161–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567 [DOI] [PubMed] [Google Scholar]
- 27. Rodriguez-Zaragoza S. 1994. Ecology of free-living amoebae. Crit. Rev. Microbiol. 20:225–241 [DOI] [PubMed] [Google Scholar]
- 28. Rubin R. W., Maher M. 1976. Actin turnover during encystation in Acanthamoeba. Exp. Cell Res. 103:159–168 [DOI] [PubMed] [Google Scholar]
- 29. Turner N. A., Russell A. D., Furr J. R., Lloyd D. 2000. Emergence of resistance to biocides during differentiation of Acanthamoeba castellanii. J. Antimicrob. Chemother. 46:27–34 [DOI] [PubMed] [Google Scholar]
- 30. Weisman R. A., Moore M. O. 1969. Bead uptake as a tool for studying differentiation in Acanthamoeba. Exp. Cell Res. 54:17–22 [DOI] [PubMed] [Google Scholar]
- 31. Weisman R. A. 1976. Differentiation in Acanthamoeba castellanii. Annu. Rev. Microbiol. 30:189–219 [DOI] [PubMed] [Google Scholar]