Abstract
Centrioles, cilia, and flagella are ancestral conserved organelles of eukaryotic cells. Among the proteins identified in the proteomics of ciliary proteins in Paramecium, we focus here on a protein, Bug22p, previously detected by cilia and basal-body high-throughput studies but never analyzed per se. Remarkably, this protein is also present in plants, which lack centrioles and cilia. Bug22p sequence alignments revealed consensus positions that distinguish species with centrioles/cilia from plants. In Paramecium, antibody and green fluorescent protein (GFP) fusion labeling localized Bug22p in basal bodies and cilia, and electron microscopy immunolabeling refined the localization to the terminal plate of the basal bodies, the transition zone, and spots along the axoneme, preferentially between the membrane and the microtubules. RNA interference (RNAi) depletion of Bug22p provoked a strong decrease in swimming speed, followed by cell death after a few days. High-speed video microscopy and morphological analysis of Bug22p-depleted cells showed that the protein plays an important role in the efficiency of ciliary movement by participating in the stroke shape and rigidity of cilia. The defects in cell swimming and growth provoked by RNAi can be complemented by expression of human Bug22p. This is the first reported case of complementation by a human gene in a ciliate.
The centriole, a microtubule-based cylindrical structure, is an ancient organelle traced to primitive eukaryotes and present in numerous phyla but lost in some others, such as fungi and higher plants (24). Centriolar function is dual: organization of the centrosome and, as a basal body, nucleation of an axoneme, the backbone of cilia and flagella. The ultrastructure of centrioles and axonemes appears to be highly conserved. In the majority of cases, the centriole is composed of 9 triplets of microtubules, and when the centriole acts as a basal body, the triplets are elongated into 9 doublets of microtubules forming the axoneme, with occurrence of a central pair of microtubules (9-plus-2 pattern) (reviewed by Satir and Christensen [30]). Renewal of interest in cilia in recent years has been prompted by their implication in numerous functions in development and cell physiology through their motile and sensory properties, so that many human genetic diseases, now called ciliopathies, appear to arise from primary defects in ciliary proteins (14). The ultrastructural conservation of centrioles and cilia in evolution is correlated with good conservation of the protein constituents.
High-throughput studies (proteomics, transcriptome analysis, and comparative genomics) have been conducted in several species to identify ciliary and centriolar proteins or proteins involved in their biogenesis. The set of proteins identified comprises both proteins specific to cilia (such as outer- and inner-arm dynein subunits or radial-spoke proteins) or to their biogenesis (specific transcription factors and intraflagellar-transport [IFT] proteins) and ubiquitous proteins (such as tubulins).
In order to identify novel conserved proteins and to specify their roles in ciliary function, we built a new knowledge base, Cildb (1) (http://cildb.cgm.cnrs-gif.fr), which integrates orthology links between the whole proteomes of 18 species and high-throughput studies identifying ciliary proteins, as well as the OMIM database. Cildb includes species for which no ciliary studies are available and species that lack centrioles or cilia in order to distinguish proteins specific to these organelles from ubiquitous proteins present in them.
In the present work, starting from the ciliary proteome analyzed in Paramecium (1), the Bug22p protein, highly conserved through evolution, was selected. Apart from its identification in high-throughput studies of various species (6, 8, 17, 21, 23, 25, 32, 36), no analysis dedicated to Bug22p exists to date for any organism. This protein has amazing sequence conservation, indicating a key role in cellular processes and/or the existence of numerous interacting partners. Intriguingly, the protein is also present in higher plants (e.g., Arabidopsis), a phylum in which centriolar and ciliary structures are absent, which induced us to analyze the relevance of its ciliary function.
We report here the localization of Bug22p in HeLa and Paramecium cells and its functional study in Paramecium. The choice of this organism, beyond the power of functional analysis by RNA interference (RNAi) (13), stems from the fact that we can specifically analyze the motility aspect of ciliary function.
MATERIALS AND METHODS
Strains and culture conditions.
Stock d4-2 of Paramecium tetraurelia, the wild-type reference strain, was used in all RNAi experiments. The mutant nd7-1 (31), which carries a recessive monogenic mutation preventing trichocyst discharge, a dispensable function under laboratory conditions, was used for transformations. Cells were grown at 27°C in wheat grass infusion, BHB (L'arbre de vie, Luçay Le Mâle, France), inoculated with Klebsiella pneumoniae and supplemented with 0.8 μg/ml β-sitosterol according to standard procedures (33).
Expression vectors.
In order to visualize the cellular localization of Bug22p, the Bug22a paramecium gene, as well as the human (Hs) Bug22 cDNA, were cloned into the Paramecium expression vector pPXV-GFP. The human Bug22 cDNA was also cloned into the plasmid pD2EGFP for expression in HeLa cells. In all three constructs, the green fluorescent protein (GFP) sequence was fused at the N terminus of the BUG22 gene.
(i) pPXV-HsBUG22 expression vector for Paramecium.
The human BUG22 sequence was amplified by PCR from a cDNA library kindly provided by François Lacroute using the primers 5′-GGTGCTCGAGATGTTCAAAAACACGTTCCAG-3′ and 5′-GTGGGGTACCTCATTGCTTTGCCTTGTTCTG-3′ and cloned into the XhoI/KpnI sites of the Paramecium expression vector pPXV.
(ii) GFP-PtBUG22 fusion gene to be expressed in Paramecium.
The pPXV-GFP expression vector was used as previously described. The Paramecium (Pt) BUG22c gene was amplified by PCR with primers 5′-GTGGGGTACCGGAGGAGGAATGTTCAAGAACTCATTCCAAAG-3′ and 5′-CCACGGTACCTCATCCTTATTTTTAGATGGGTAAG-3′ and cloned into the KpnI site of pPXV-GFP.
(iii) GFP-HsBUG22 fusion gene to be expressed in Paramecium.
The human sequence of BUG22 was amplified by PCR with primers 5′-GTGGGGTACCGGAGGAGGAATGTTCAAAAACACGTTCCAGAG-3′ and 5′-CCACGGTACCTCATTGCTTTGCCTTGTTCTGAAC-3′ and cloned into the KpnI site of pPXV-GFP.
RNAi vectors.
The sequences of genes to be knocked down were amplified by PCR and cloned into a modified Litmus 28i plasmid (New England Biolabs) in which the EcoRI site had been replaced by an SrfI site, using the method developed for PCRscript cloning by Stratagene (La Jolla, CA), and transferred to the feeding vector L4440 (37) into the XhoI/HindIII sites of the polylinker between two T7 promoters.
(i) Constructs for RNAi of the Paramecium BUG22 genes.
Two constructs were used to knock down the two subfamilies of BUG22 genes. For BUG22a and -b knockdowns, BUG22a was amplified from positions 75 to 391 with the primers 5′-GGATAAGCAAAGTAATTTATGATTGG-3′ and 5′-CTTTATACTAATTACTTAATAATTCGATGC-3′. For BUG22c and -d knockdowns, BUG22d was amplified from positions 75 to 394 with the primers 5′-TCAAAGCCCTTGTAGATTTG-3′ and 5′-GTGTTTTAAGATGTACTTGATAATTAG-3′. A short sequence was also used for RNAi by direct cloning of a pair of complementary oligonucleotides corresponding to the sequence 5′-ATGTTCAAGAACTCATTCCAAAGTGGATTTCTTTCAATTTTATATTCAATAGGATCAAAGCC-3′.
(ii) Constructs for RNAi of human BUG22 transgenes in Paramecium.
The HsBUG22 sequence was transferred from the pPXV-HsBUG22 construct by digestion and cloning into the XhoI/HindIII sites of the L4440 vector.
(iii) Constructs for RNAi of the IFT172 genes in Paramecium.
One of the two close paralogous IFT172 genes (GSPATG00033191001) was amplified by PCR using the primers 5′-GCAGTCATTAGATCATCATAGG-3′ and 5′-CAATAACTTCACTTTCAGGAAC-3′ and cloned into the L4440 vector as the BUG22 sequences. Due to high sequence conservation, this single plasmid is supposed to knock down both paralogs.
In all cases, plasmid DNA was prepared using the plasmid midi kit according to the protocol of the manufacturer (Qiagen, Courtaboeuf, France). The sequences of Paramecium genes were retrieved from ParameciumDB (2) (http://paramecium.cgm.cnrs-gif.fr/) and verified by resequencing after cloning.
Transfection. (i) HeLa cells.
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (all from Gibco) at 37°C in 5% CO2. The day prior to transfection, HeLa cells (1 × 105) were plated on a 24-well plate containing presterilized coverslips in DMEM supplemented with 10% FBS. The cells were transfected with 0.5 μg of Hs-Bug22-GFP using Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions.
(ii) Paramecium transformation.
nd7-1 cells were transformed by microinjection into their macronuclei of filtered and concentrated plasmid DNA (5 μg/μl) containing a mixture at a ratio of 10:1 of the plasmids of interest (GFP fusion genes and HsBug22p expression) and a plasmid directing the expression of the ND7 gene (31). Microinjection was done under an inverted Nikon phase-contrast microscope, using a Narishige micromanipulation device and an Eppendorf air pressure microinjector. Successfully transformed cells were screened for the ability to discharge trichocysts on picric acid stimulation and further analyzed.
Antibodies.
The polyclonal GTL3 anti-Bug22p antibody was purchased from Aviva Systems Biology (San Diego, CA). The monoclonal antibody 20H5, raised against Chlamydomonas centrin (29), was used to label centrioles in HeLa cells. The monoclonal antibody ID5 (38), which essentially labels basal bodies and a few microtubule arrays (10), was used to visualize the basal-body pattern. The polyclonal anti-GFP antibody was purchased from Interchim (Montluçon, France). The polyclonal anti-Paramecium ciliary tubulin (9) and anti-ciliary rootlet (34) antibodies were homemade antibodies.
The secondary antibodies used in immunofluorescence were Alexa Fluor 488 or 594 anti-mouse or -rabbit IgG antibodies from Invitrogen, the antibody used in Western blot experiments was an anti-rabbit IgG-alkaline phosphatase (AP) conjugate (Promega), and the antibodies used in immunoelectron microscopy were colloidal-gold-conjugated anti-rabbit immunoglobulins (Gar G5 and Gar G10; Aurion).
Protein extraction, SDS gels, and Western blots.
Paramecium protein extracts were prepared from a pellet of 2 × 105 cells homogenized in a potter with tight clearance in 10 mM Tris HCl, 1 mM EDTA, 250 mM sucrose containing a protease inhibitor cocktail (Set1; Calbiochem). Low-speed pellets and supernatants were separated by centrifugation at 10,000 × g. Total protein extracts of bacteria were obtained by sonication of a bacterial pellet in lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% Na deoxycholate). Gel electrophoresis was performed using Criterion XT gels at neutral pH and 12% Bis-Tris (Bio-Rad) according to the manufacturer's recommendations. The primary antibodies used were anti-GTL3 (1:500) and anti-GFP (1:750). The secondary antibody was anti-rabbit–AP (1:500). Western blots were revealed using the 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (NBT) liquid substrate system from Sigma.
Immunofluorescence microscopy. (i) HeLa cells.
After 24 h of culture, cells were washed with phosphate-buffered saline (PBS), fixed for 6 min at −20°C in 100% methanol, and blocked in 3% bovine serum albumin (BSA) and 0.1% Tween 20 in PBS for 30 min. The cells were incubated at room temperature with either the 20H5 (1:1,000) or the GTL3 (1:200) antibody for 1 h and then washed three times in 1% BSA and 0.1% Tween 20 and incubated for 1 h with the secondary antibody before three additional washes.
(ii) Paramecium.
Immunostaining of cells was carried out as previously described (28) using the anti-ciliary tubulin (1:500), the monoclonal ID5 (1:350), or the anti-ciliary rootlet (1:400) antibody. For both HeLa cells and paramecia, appropriate fluorescent secondary antibodies were applied at 1:500 dilutions, nuclei were stained with Hoechst-33258, and slides were mounted in Cityfluor (London). Preparations were observed under a Zeiss Axioskop 2-plus fluorescence microscope equipped with a Roper Coolsnap-CF intensifying camera with GFP filters. Images were processed with Metamorph software (Universal Imaging).
Electron microscopy.
For morphological observations, cell pellets were processed as previously described (5) using 1% or 2% glutaraldehyde for fixation. For postembedding immunolocalization, cell pellets were processed as described previously (18), using either 2% paraformaldehyde, 0.15% glutaraldehyde, or 3% paraformaldehyde and 0.5% glutaraldehyde for fixation, and embedded in LR White (London Resin). Thin sections were collected on nickel grids and treated with double-distilled water or 0.1 M NH4Cl in 0.1 M PBS and then saturated and processed with 3% BSA in PBS. Primary polyclonal antibodies were diluted 1:50 to 1:75 (anti-GTL3) and 1:10, 1:20, or 1:500 (anti-GFP). After being washed, the sections were incubated with, respectively, 1:40, 1:75, or 1:100 dilutions of 5-nm (GTL3 labeling) or 10-nm (GFP labeling) gold particles. The sections were examined with a Philips CM10 or a Jeol transmission electron microscope.
RNAi by feeding.
RNAi gene knockdown was performed according to the method of Galvani and Sperling (13). HT115, an RNase III-deficient strain of Escherichia coli with an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 polymerase, was transformed by the desired constructs into Litmus or L4440 plasmids. Wild-type paramecia were incubated in double-stranded-RNA (dsRNA)-expressing bacteria and were transferred daily into fresh feeding medium. Control cells were fed with bacteria carrying the complete coding region of the ND7 gene.
Swimming analysis.
Two methods were used for swimming analysis. First, we measured the swimming speed of cells under dark-field microscopy using a Zeiss Axioskop 2-plus microscope with a 10× objective by recording tracks during 650-ms pauses. Second, we used a Zeiss Steni 2000-C dissecting microscope with 1-min time-lapse acquisitions at 7 frames per second with a Roper Coolsnap-CF camera and Metamorph software (Universal Imaging). The speed means and standard errors were calculated by measuring the lengths of the tracks using ImageJ and subtracting a mean length of the paramecia for the first method and using automated analysis of the swimming tracks currently under development for the second method (J. Ferracci, O. Arnaiz, and J. Cohen, unpublished data).
High-speed video microscopy.
All of the observations were made under an inverted microscope (Axiovert 200; Carl Zeiss S.A.S., Le Pecq, France) using a 100× objective on 5 μl of a suspension of paramecia immobilized between a cover glass (diameter, 50 mm; thickness, 0.13 to 0.16 mm) from Menzel-Glaser and a 22- by 22-mm coverslip, with a few hundred calibrated 10-, 15-, or 20-μm polystyrene beads (Fluka), during overall periods shorter than 5 min to avoid changes in ciliary activity. A final dilution of ca. 1/10 of India ink was added to the medium to be visualized as particles that materialized the fluid flow during the acquisitions. Cilium movements were digitally recorded with a camera (PixeLink A741; Ottawa, Canada) at a rate of 355 images per second. A movie was constituted of 1,800 images with a definition of 256 by 192 pixels. The size of a pixel was 0.13 by 0.13 μm2. In each sequence, we selected a cilium that we were able to follow during an entire cycle of beating. For this, the video sequences were played back frame by frame using Streampix software (Norpix Inc., Montreal, Canada). The angle of beating was calculated by trigonometric calculations in the triangle made by the anchoring point and the two extreme positions of the cilium extremities during the power stroke.
Swimming-behavior assays.
Cells to be assayed were first equilibrated for a few minutes in resting solution (2 mM Na citrate, 1 mM NaH2PO4, 1 mM Na2HPO4, 1.5 mM CaCl2) and then individually transferred to test solutions that consisted of either 30 mM KCl or 5 mM tetraethylammonium, 10 mM NaCl in resting solution (19). Observation of immediate swimming behavior was done during the first minute after transfer under the dissecting microscope.
RESULTS
Conservation of Bug22p in eukaryotes.
Bug22p, a small basic protein of 22 kDa, was one of the abundant proteins identified in a high-throughput proteomics analysis of Paramecium cilia (9 distinct peptides), and its mRNA level in cells regenerating cilia was increased by a 2- to 3-fold induction (reference 1 and unpublished data). A BLAST search for similar proteins in other species for which whole-genome sequence data are available revealed that it is widespread in eukaryotes harboring centrioles/cilia at some stage of their life cycles. Among eukaryotes devoid of centrioles/cilia, plants possess BUG22 genes whereas fungi do not. Interestingly, fungi, such as Batrachochytrium dendrobaditis, which form flagellated gametes also have a BUG22 gene. The protein sequence of Bug22p proteins is highly conserved through evolution (Fig. 1), except for the presence of C-terminal extensions in Trypanosoma and Caenorhabditis. Such lack of sequence variation during evolution suggests a conserved role in different organisms, including but not restricted to ciliary function, since the sequence conservation is also high in plants. Despite high overall amino acid sequence conservation, diagnostic positions in the sequence allow us to distinguish BUG22 genes of acentriolar organisms (higher plants) from the others, as shown in a global alignment (Fig. 1).
Fig. 1.
Sequence conservation of Bug22p in eukaryotes and positions that distinguish ciliary versus nonciliary species. The ClustalW alignment of 58 Bug22p sequences, representative of all the clades of eukaryotic species in which it can be found, was visualized using Boxshade and illustrates the strong conservation of the Bug22p protein through evolution. Careful examination of the divergences at seven specific positions (amino acids 30, 38, 45, 55, 66, 67, and 111 according to the numbering of the Homo sapiens Bug22p sequence) revealed that species having cilia or centrioles in at least some stage of their life cycle have a consensus at these positions (denoted in purple) that dramatically differs from the consensus found in plants (denoted in green), as displayed at the bottom of the figure. Note that both the moss Physcomitrella patens and the fern Selaginella moellendorfii (in the middle of the alignment), which both have several Bug22 genes in their genomes and are plants with occurrence of flagellated cells in their life cycles, display a patchwork of the plant consensus and the ciliary consensus at these positions. In addition, the occurrence of a BUG22 gene with ciliary consensus in the genome of Chlorella vulgaris, harboring a centriole that never develops a cilium (39), suggests that Bug22p has a centriolar function. The species studied and the accession numbers of the proteins are as follows: Aa, Aedes Aegypti XP_001663334.1; At, Arabidopsis thaliana AAG51069.1; Am, Apis mellifera XP_395824.1; Bd, Batrachochytrium dendrobaditis jgi ∣Batde5∣ 32931; Bm, Bombyx mori NP_001040392.1; Brm, Brugia malayi XP_001894987.1; Bt, Bos taurus NP_032213.2; Cca, Capitella capitata jgi ∣Capca1∣77038; Ccl, Citrus clementina EST:FC926856.1, GI:218791863; Ci, Ciona-intestinalis XP_002131975.1; Ce, Caenorhabditis elegans NP_871706.1; Cv, Chlorella vulgaris jgi ∣Chlvu1∣29644∣e; Cr, Chlamydomonas reinhardtii XP_001692489; Cp, Carica papaya EST: EX271746.1, GI:186814042; Cq, Culex quinquefasciatus XP_001869024.1; Dm, Drosophila melanogaster NP_609402.1; Dr, Danio rerio NP_957105.1; Ee, Euphorbia esula EST: DV114861.1; Eg, Elaeis guineensis ACF06559; Gg, Gallus gallus NP_001005833.1; Gh, Gossypium hirsutum EST: EX168390.1 GI:164311590; Gl, Giardia lamblia XP_001708694.1; Gm, Glycine max gb:ACU16157.1; Hm, Hydra magnipapillata XP_002163101.1; Hs, Homo sapiens NP_037374.1; Lb, Leishmania_braziliensis XP_001564754.1; Lj, Lotus japonicus EST: GO028159.1, GI:223439164; Ls, Lactuca serriola EST:BQ986870.1, GI:22404395; Me, Manihot esculenta EST:DV448997; Mg, Mimulus guttatus EST:GR148812.1, GI:238398755; Mm, Mus musculus NP_032213.2; Mp, Micromonas_pusilla jgi ∣MicpuC2∣70275∣AZW; Mt, Medicago trunculata ACJ85502.1; Ng, Naeglaeria gruberi JGI Protein ID: 81674; Ot_1p, Oxytricha trifallax OXADaaa05e09; Ot_2p, Oxytricha trifallax OXAEaaf21g04; Ot_3p, Oxytricha trifallax OXAEaaa06d08; Pf, Plasmodium falciparum XP_001351642.1; Pp_1, Physcomitrella patens XP_001775608.1; Pp_2, Physcomitrella patens XP_001760380.1; Pp_3, Physcomitrella patens XP_001784194.1; Ps, Picea sitchensis ABK26040.1; Pt, Paramecium teraurelia (this article); Ptr, Populus trichocarpa XP_002304809.1; Sb, Sorghum bicolor XP_002460501.1; Sm_1, Selaginella moellendorfii jgi ∣Selmo1∣172529; Sm_2, Selaginella moellendorfii jgi ∣Selmo1∣96514; Sm_3, Selaginella moellendorfii jgi ∣Selmo1∣130480; Sm_4, Selaginella moellendorfii jgi ∣Selmo1∣117290; Ta, Trichophlax adherens jgi ∣Triad1∣ 23937; Tb, Trypanosoma brucei XP_822473.1; Tc, Tribolium castaneum XP_975505.1; Tg, Toxoplasma gondii XP_002364709.1; Tt, Tetrahymena thermophila XP_001007313.2; Tp, Thalassiosira pseudonana XP_002286290.1; Vv_1p, Vitis vinifera XP_002268386.1; Vv_2p, Vitis vinifera XP_002265223.1; Xp, Xenopus tropicalis NP_001004835.1; Zm, Zea mays NP_001148848.1.
The whole Bug22p protein constitutes by itself the Interpro domain DUF667 (for domain of unknown function) that occurs in some proteins other than Bug22p. However, the sequence conservation of the DUF667 domain in these proteins is much lower than within the Bug22p subfamily, suggesting that the functions of the less conserved DUF667 domains may be divergent with respect to the Bug22p domain.
Four Bug22 genes for a single Bug22p protein in Paramecium.
Four paralogous BUG22 genes encoding four 100% identical proteins are present in the Paramecium genome. These paralogous genes arose from the last two whole-genome duplications (WGDs) that occurred in this organism (3). The four genes, with the identifiers (IDs) GSPATG00007012001, GSPATG00016658001, GSPATG00012810001, and GSPATG00013199001, were, respectively, called BUG22a, -b, -c, and -d. The DNA sequences are almost identical (96.1% identity between BUG22a and -b and 96.9% between BUG22c and -d [both pairs are paralogs from the most recent WGD] and 83.2% to 83.8% between the pair BUG22a plus -b and the pair BUG22c plus -d). Most of the divergence occurs in intron sequences, as visualized by multiple alignments (see Fig. S1 in the supplemental material). The few differences in exons are all silent.
Localization of Bug22p in HeLa cells and Paramecium.
HeLa cells transfected with the GFP-HsBUG22 cDNA displayed labeling of two dots close to the nucleus, which colocalized with centrin, a centriolar marker (11), as revealed by the 20H5 monoclonal antibody (see Fig. S2A in the supplemental material), indicating that HsBug22p is present at the centrosome. This observation is similar to images published by Keller et al. (16), who transfected U2OS cells with a C-terminal GFP-Bug22p fusion protein in a systematic study of centriolar proteins. Our results thus confirm the centriolar localization of Bug22p.
Both the HsBUG22 and the PtBUG22 GFP fusion genes were used to transform Paramecium by DNA microinjection into the macronucleus, and the fluorescence was observed in the clonal descendants of the injected cells. Labeling of basal bodies and the proximal two-thirds of cilia was observed with both GFP fusions (Fig. 2), showing that, in agreement with the proteomics of cilia in several species, Bug22p localizes to basal bodies and cilia and that the human ortholog HsBug22p is targeted to these organelles, as well.
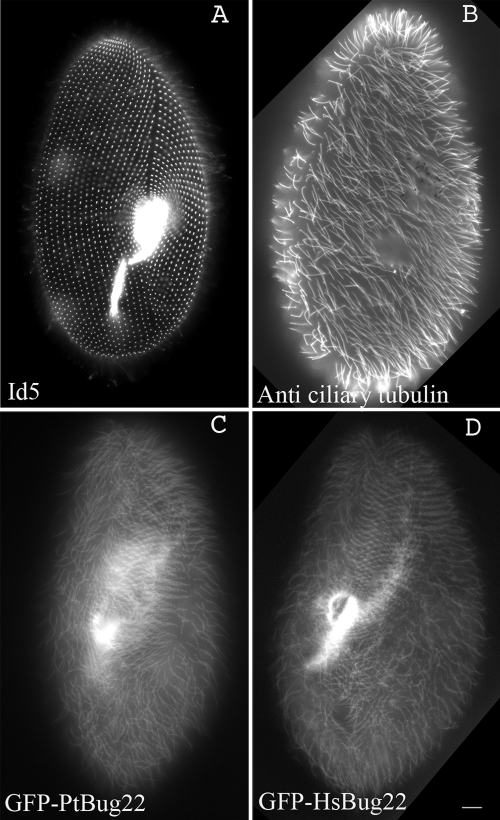
Fig. 2.
Ciliary and basal-body localization of Paramecium and human Bug22p GFP fusions in Paramecium. (A and B) Wild-type paramecia immunofluorescently labeled with the ID5 monoclonal antibody to visualize basal bodies (A) and with the anti-ciliary tubulin to show cilia (B). (C) Paramecium expressing GFP-PtBug22p showing clear labeling of basal bodies and cilia. (D) Same as panel C for a Paramecium expressing GFP-HsBug22p. Bar = 10 μm.
GTL3 is a commercial antibody raised against the C-terminal portion of the human Bug22p protein. As the primary sequence is highly conserved between Paramecium and humans (89% identity in the amino acid sequences) (Fig. 1), we tested the GTL3 antibody by Western blotting (Fig. 3) and found that it specifically recognized a 22-kDa peptide in Paramecium extracts (Fig. 3A), as well as the GFP-HsBug22p fusion expressed in E. coli (Fig. 3B and C). In HeLa cells, the GTL3 labeling coincided with the localization of centrin (see Fig. S2B in the supplemental material) and GFP-HsBug22p (see Fig. S2C in the supplemental material), confirming its Bug22p specificity.
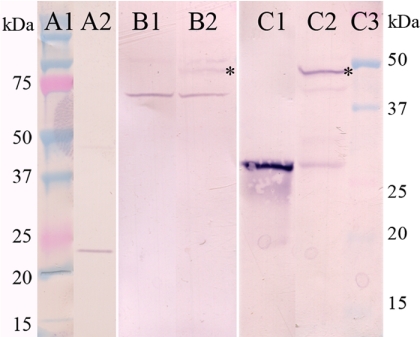
Fig. 3.
Western blot evidence that Bug22p is the antigen of a commercial anti-GTL3 antibody. Lanes: A1, molecular mass markers for lane A2; A2, anti-GTL3 labeling of a 10,000 × g supernatant of whole homogenized paramecia showing a positive band between 20 and 25 kDa; B1 and C1, total extracts of E. coli expressing GFP in the pPXV plasmid normally used for expression in Paramecium but which allows low expression in the bacteria; B2 and C2, E. coli expressing the GFP-Bug22p fusion; C3, molecular mass markers for lanes B1 to C2. The B series lanes were revealed with the anti-GTL3 antibody and the C series with the anti-GFP antibody. The asterisk indicates the level of the fusion protein that is recognized by both GTL3 and GFP antibodies.
In immunofluorescence, no labeling of Paramecium by GTL3 was observed. This negative result may be due to the solubility of Bug22p molecules in the medium after detergent permeabilization or to lack of accessibility to the antigen under the fixation and labeling procedures used. The protein localization was then examined by postembedding immunoelectron microscopy, both with the GTL3 antibody on wild-type cells and with an anti-GFP antibody on GFP-Bug22p-expressing cells (Fig. 4). Examination of 142 sections of cilia and basal bodies using the GTL3 antibody and 115 sections with the anti-GFP antibody indicated that both antibodies gave the same labeling in the basal bodies and cilia: gold particles occurred preferentially in the terminal plate of the basal body and the transition zone to the cilium, as well as in the cilium, in the vicinity of the outer doublets of the axoneme, and often between the axoneme and the membrane. In GFP-expressing cells, additional labeling was often found within the basal body, close to its proximal part (Fig. 4G). In GFP-expressing cells, 34 longitudinal sections of cilia revealed an apparent periodicity of the labeling as a regular disposition of the gold particles at 500- to 550-nm intervals (Fig. 4J and K) and, less often, at 250- and 800-nm intervals.
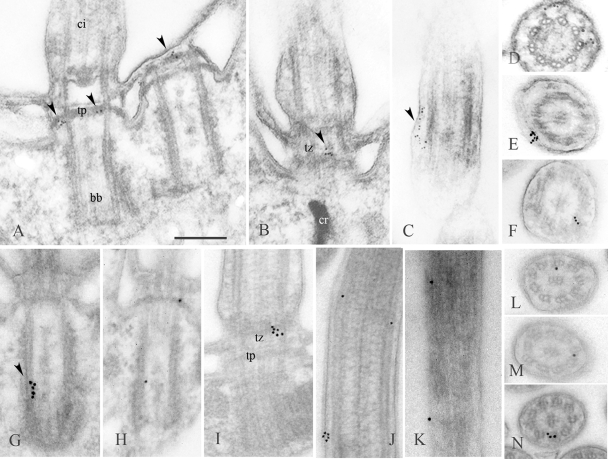
Fig. 4.
Localization of Bug22p in Paramecium by postembedding immunoelectron microscopy. (A to F) Labeling of wild-type cells with the GTL3 antibody using 5-nm gold particles. (G to N) Labeling of GFP-Bug22p-expressing cells with an anti-GFP antibody using 10-nm gold particles. The arrowheads point to gold particles. (A and H) Labeling of the terminal plates of basal bodies. (B, D, and I) Labeling of the transition zone to the cilium. (F and J to N) Labeling in the vicinity of the outer doublets of the axoneme. (C, E, and J) Labeling between the axoneme and the membrane. (G) Labeling within the basal body close to its proximal part. (J and K) Examples of regular disposition of the gold particles along the axoneme. ci, cilium; bb, basal body; tp, terminal plate; tz, transition zone. Bar = 250 nm.
Growth and swimming defects provoked by Bug22p depletion in Paramecium can be complemented by the human ortholog.
The function of Bug22p was investigated in Paramecium using the RNAi feeding method. Since the four Paramecium BUG22 genes encode the same protein, we would expect not qualitative but quantitative differences by inactivating each paralog of the gene family. We used BUG22a and BUG22c sequences for double-stranded-RNA production. The two dsRNAs were expected to silence all four paralogs, based on sequence conservation (see Fig. S1 in the supplemental material). Double silencing using the two sequences was needed to improve the knockdown efficiency on all four genes.
We first evaluated the efficiency of the RNAi procedure on fluorescent Paramecium cells from a clone transformed with the GFP-PtBUG22a fusion gene: the fluorescence was completely abolished by the double BUG22a-BUG22c knockdown, showing that the RNAi efficiency depleted Bug22p (data not shown). Phenotypic analyses obtained in further RNAi experiments were performed on nontransformed wild-type cells.
Knocking down each pair of BUG22a-BUG22b or BUG22c-BUG22d genes resulted in a growth rate of 1 or 2 fissions per day compared to 4 or 5 in control cells and led to lethality in 2 to 3 days. When all 4 genes products were knocked down, cells divided 1 or 2 times on the first day and 0 or 1 time on the second day and died within 3 days. Lethality might be the result of starvation owing to impaired ciliary beating (see below) that prevented food capture in the oral apparatus, as observed by defective take-up of India ink (not shown).
Swimming speed was measured by recording swimming tracks under dark-field microscopy, which showed that, after 1 day of silencing, knocking down each pair of gene products decreased the rate of motility by 75%, while knocking down all four gene products decreased the rate of motility by 90% (Fig. 5).
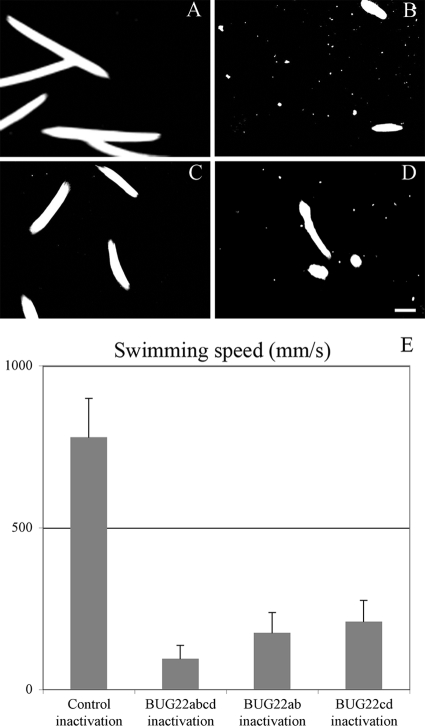
Fig. 5.
Swimming-speed diminution in Bug22p-depleted cells. Shown is a track recording of paramecia swimming under a dark-field microscope equipped with a 10× objective during a 650-ms pause. (A) Control Nd7p-depleted cells. (B) Double RNAi targeting all four BUG22 genes. (C and D) Simple RNAi targeting BUG22a and BUG22b, and BUG22c and BUG22d, respectively. Bar = 100 μm. (E) Histograms of measured speeds under all conditions showing quantization of the cumulative effects of double RNAi compared to simple RNAi. The error bars indicate standard errors.
To test whether a human Bug22 protein could complement RNAi depletion of Paramecium orthologs, we transformed Paramecium for heterolog expression of the human BUG22 cDNA and submitted the cells to RNAi against the BUG22a and BUG22c Paramecium sequences, which do not share nucleotide identity stretches sufficient to induce silencing of the human gene, and reciprocally to RNAi against the human BUG22 cDNA, which did not cross-react with Paramecium sequences (see Fig. S1 in the supplemental material). A control with double human and Paramecium BUG22 RNAi was also performed. We observed that neither HsBUG22 nor PtBug22 RNAi alone could alter the growth rate or the swimming speed, whereas the double Hs/Pt RNAi could (Fig. 6 and its legend provide details). This demonstrates the ability of the human protein to rescue depletion of Paramecium Bug22p.
Fig. 6.
Complementation of the effects of Bug22p depletion by expression of the human gene in Paramecium. To test possible rescue of Bug22p-depleted paramecia by expression of human Bug22p, cells were first transformed by a plasmid containing the human BUG22 cDNA under Paramecium regulatory sequences and then subjected to RNAi against the BUG22 endogene (PtBUG22), the human BUG22 transgene (HsBUG22), or both (HsBUG22 and PtBUG22) to follow the phenotypes. Transformation of Paramecium nd7-1 cells (unable to undergo trichocyst exocytosis) was performed by microinjection of a mixture of two plasmids, one driving the expression of the wild-type ND7 gene to monitor the efficiency of the transformation and the other for expression of the human BUG22 cDNA. The clones derived from transformed cells were cultured and submitted to RNAi, with the ND7 gene used as a target for control RNAi. The phenotypic analysis consisted of monitoring the growth rate by cell counting and the swimming speed by automated analysis of recorded tracks. Each measurement was performed on accumulations of tracks of ca. 20 cells during 1-min acquisitions and was repeated 3 times on independent samples. Shown are the swimming speed (histograms and standard errors) and growth rate (values below the histograms) of an HsBug22 transformant (B) compared to a control clone (A). This transformant appeared to be resistant to RNAi targeting the endogenous Paramecium BUG22 genes (PtBUG22 RNAi) compared to the control clone under the same conditions, as shown by the heights of the bars labeled PtBUG22 RNAi in both histograms, and to be resistant to the human BUG22 transgene (HsBUG22 RNAi), as was the control clone, but not to double RNAi (Hs/PtBUG22 RNAi). This indicates that expression of the human cDNA is able to replace the depleted Paramecium protein, thus showing complementation ability, but does not represent a nonspecific effect, since the double RNAi inactivates any BUG22 function in these cells. Altogether, six independently transformed clones have been extensively analyzed with similar results. Some variation was observed in the levels of the complementation: PtBUG22- depleted cells swam at 10% of the normal speed, while cells rescued by transformation swam at 35% to 90% of the normal speed. Such variation is routinely observed in transformation of Paramecium, depending on the amount of transforming DNA introduced via microinjection. In three cases where the amount of injected DNA was optimal, the rescue was abolished when both Paramecium and human sequences were targeted together by RNAi.
Bug22p depletion affects swimming behavior.
When carefully observed under the dissecting microscope, live Bug22p-depleted cells seem to undergo less frequent backward swimming than control cells. Since these events are rare under control conditions, we tested swimming behavior in ion solutions, as described for the identification of Mendelian mutants (19). Control cells immersed in 30 mM KCl or in 5 mM tetraethylammonium, 10 mM NaCl (see Materials and Methods) immediately underwent continuous backward swimming for more than 20 s and then whirled for a few seconds and resumed normal forward swimming. In contrast, Bug22p-depleted cells showed a very much weakened reaction, with very short or no backward swimming and whirling for less than 5 s. This suggests abnormal ciliary-channel activity or abnormal response of the axonemal motors to ion currents, induced by RNAi knockdown.
Bug22p depletion disturbs basal-body organization in the cortex of Paramecium.
Since the Bug22p protein is present in basal bodies, in addition to cilia, we wondered whether its depletion causes basal body defects. We therefore examined Bug22p-depleted cells using antibody labeling that revealed, not only the basal bodies themselves (antibody ID5), but also the ciliary rootlets (anti-KD), which provide good markers of the orientation of individual basal bodies in the global pattern of the polarized cell cortex. By the second day under BUG22 RNAi conditions, a sporadic disorganization started to appear, with ciliary rootlets showing diverse erratic orientations (see Fig. S3 in the supplemental material). This effect on basal-body organization and polarity could be a direct consequence of Bug22p depletion or an indirect effect secondary to alteration of ciliary activity.
Bug22p depletion alters ciliary beating efficiency and stroke shape in Paramecium.
To investigate the cause of the decrease in swimming speed induced by RNAi depletion of Bug22p, we first checked the presence of cilia by immunofluorescence using an anti-tubulin antibody. Cilia appeared unaffected in number, density, shape, or length, in contrast to what is obtained by silencing IFT172, a component of the intraflagellar-transport machinery involved in ciliary growth and maintenance (26), used as a control (see Fig. S4 in the supplemental material). In living cells observed under phase-contrast microscopy at high magnification, the cilia appeared to beat vigorously, in apparent contradiction of the strong decrease in the cell swimming speed. This indicates that other factors than beating frequency can affect swimming speed. We therefore studied the ciliary beating of Bug22p-depleted cells by high-speed video microscopy (Fig. 7; see Videos S1 and S2 in the supplemental material). The recorded images revealed that although the cilia beat at an even higher frequency than normal (42 Hz versus 14 Hz), they beat with reduced efficiency, since (i) the angle of beating was narrower (55° versus 97°) and (ii) the apparent distinction between power and recovery strokes, as well as the coordination of beating over the cell surface, was lost. In addition, cilia appeared “floppy” when they moved, as though they were less rigid, which could explain their altered beating. The global impairment in the beating efficiency of fluid movement could be visualized by particles in the medium moving in a disorganized manner, more or less randomly, compared to the control, where particles were rapidly swept in a dominant direction.
Fig. 7.
Effects of Bug22p depletion on ciliary beating viewed by high-speed video microscopy. (A and B) Images of control Nd7p- and Bug22p-depleted cells, respectively, captured from high-speed video microscopy movies (see Videos S1 and S2 in the supplemental material) showing the ciliary beating, on which are superimposed the representation of a single cilium (in red) plus the representation of the same cilium in the 8 adjacent frames (blue), representing one power stroke in the control (25 ms) and erratic beating in the Bug22p-depleted cell. (C to F) Quantization of four parameters manually extracted from the movies on 14 beating cilia for BUG22 RNAi and 12 cilia for the control, with standard deviations. (C) Frequency of beating. (D) Angle between extreme positions taken by beating cilia. (E) Length of the trajectory of the tips of beating cilia. (F) Triangular surface swept by beating cilia. Bug22p depletion increased the frequency but decreased all the other parameters.
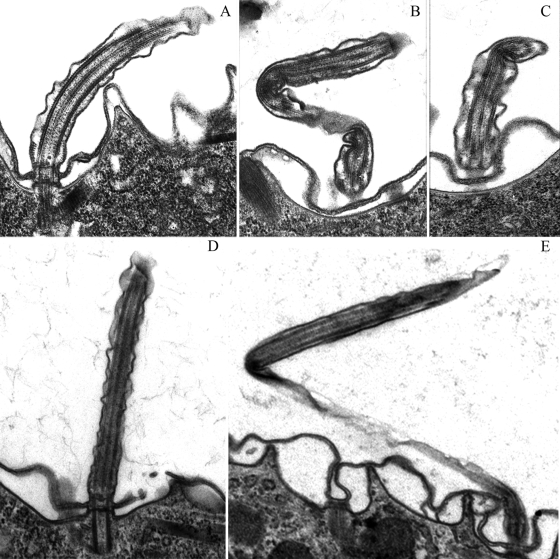
Examination of Bug22p-depleted cells under transmission electron microscopy did not reveal particular defects in the general organization of the basal body or cilium (9-fold symmetry, a central pair of microtubules, and radial spokes in the axoneme). However, the overall shape of the cilia was strongly altered in 30% of the longitudinal sections observed, with sharp bending that was never observed in controls, accompanied by shrinking of the axoneme diameter in the region of the bend (Fig. 8). This indicated that the natural rigidity of cilia was greatly impaired by the depletion of Bug22p, in good agreement with the observation of the shape of beating cilia in live cells.
Fig. 8.
Ciliary defects in Bug22p-depleted paramecia observed in electron microscopy. (A to C) Observation of RNAi after 24 h. Magnification, ×22,000. (D and E) Observations of RNAi after 48 h. Magnification, ×16,000. (A and D) Control ND7 RNAi. (B, C, and D) BUG22 RNAi. Note the unusual bending of cilia in Bug22p-depleted cells, as viewed in the longitudinal axis of cilia in panels B and E and with contiguous longitudinal and cross sections of the same cilium in panel C. Images of exaggerated ciliary bending were never observed earlier and were detected for the first time in Bug22p-depleted cells. Among 140 longitudinal sections of cilia from 15 different cells, 43 cilia displayed such exaggerated bending, whereas none were observed in a similar number of sections in the control.
DISCUSSION
Bug22p, a novel ciliary protein.
The present study of Bug22p was stimulated by finding the protein in proteomic analysis of Paramecium cilia (1) and by the fact that we could extract it by mining the results of high-throughput studies of various species. In organisms with centrioles and cilia, high-throughput studies provided strong evidence that Bug22p has a localization/function related to these organelles. In Chlamydomonas, Bug22p has been detected in proteomic analysis of flagella (25) and basal bodies (17), and BUG22 mRNA is upregulated during flagellar biogenesis (17, 36). The protein has also been found in Trypanosoma flagella (8), in Tetrahymena cilia (32), in human cilia (23), and in mouse photosensory receptors (21). The promoter of the BUG22 gene in Caenorhabditis displays an X box (6), characteristic of genes encoding proteins related to ciliary biogenesis and regulated by the daf19 transcription factor. The mouse ortholog of BUG22, GTL3, has been identified in a gene trap screen (transcript T10-2A2) as expressed in many tissues, but preferentially in the testis (27), an organ where flagellated sperm cells develop. A large-scale cDNA screen by in situ hybridization of zebra fish embryos (35; http://zfin.org/) localized major expression of BUG22 in tissues where cilia are known to play an important role in embryonic development—Kupffers vesicle (equivalent to the embryonic node of mammals), neural tube, spinal cord, and pronephric duct—which is compatible with preferential expression in ciliated tissues. In situ hybridization in Drosophila (Berkeley Drosophila Genome Project [http://www.fruitfly.org/]) revealed preferential expression of BUG22 in sensory complexes during embryogenesis, again compatible with expression in ciliary tissues. The high sequence conservation and apparently ubiquitous presence of Bug22p in centrosomes, cilia, and flagella stimulated its further study.
Potential roles of Bug22p in motile cilia.
Cilia and flagella are organelles composed of hundreds of proteins whose assembly and function, apart from main components of the backbone of the axoneme, are not well understood. Even the most abundant proteins found in high-throughput proteomics studies of cilia remain to be studied, and we focused on one of them, Bug22p.
In Paramecium, four genes issuing from two successive whole-genome duplications encode four identical proteins, so that only one Bug22p protein is synthesized in the cell although the nucleotide sequences had time to diverge at silent positions. An explanation is that the function of Bug22p is so constrained that there is no latitude for mutation without function impairment. Thus, Bug22p is likely to have an essential role in the cell, interacting with many different partners.
The localization experiments described here using a GFP-PtBug22p fusion in vivo, or using anti-Bug22p or anti-GFP antibodies on wild-type or GFP-PtBug22p-expressing cells in immunoelectron microscopy, confirmed the basal-body and ciliary localization of the protein in Paramecium. More precisely, the labeling showed a preferential concentration of the gold particles close to the triplet (basal bodies) or doublet (cilia) microtubules, with frequent labeling between the axoneme and the ciliary membrane. Bug22p could therefore be a protein more or less stably associated with microtubules. Its basic isoelectric point (∼10) is compatible with possible interaction with tubulin, although this remains to be demonstrated biochemically.
The localization of Bug22p in the transition zone between the basal body and cilium, as well as in foci along the cilia, suggests a role in IFT. Such a view could be supported by the facts that high-throughput 2-hybrid screens revealed CLUAP1/qilin/dyf-3 as a potential interactant of Bug22p in Drosophila melanogaster (15) and that dyf-3 has been shown to belong to the IFT in Caenorhabditis elegans (12, 22). More precisely, dyf-3 is a component of the IFTB particle that is involved in the anterograde movements in cilia that control ciliary growth and turnover. However, knocking down Bug22p in Paramecium does not yield lack-of-cilia phenotypes found in IFTB knockdowns in other species, and in Paramecium, as verified by IFT172 RNAi experiments (see Fig. S4 in the supplemental material). The question of an interaction of Bug22p with dyf-3 in Paramecium, or its potential role in the IFT itself, or as IFT cargo, is still open.
The most obvious effect of Bug22p depletion is a reduction in cell swimming speed resulting from inefficient ciliary beating (Fig. 7; see Videos S1 and S2 in the supplemental material) and apparent loss of ciliary rigidity (Fig. 8). Little is known about the factors that rigidify membrane-bound microtubular complexes. Ciliary bending results from multiple interactions within the axoneme, where the nine microtubular doublets slide over neighboring doublets through dynein-tubulin interactions coordinated by the central pair and radial spokes (30). Interestingly, it has recently been shown that hydin, a central-pair-associated protein, is critical for regulation of ciliary bending during beating strokes and of beat frequency in a manner opposite to our observations for Bug22p. Indeed, hydin depletion in mutants produces excessive ciliary rigidity and reduces beat frequency (20), in contrast to Bug22p-depleted cilia, which display excessive flexibility and beating frequency.
Functional conservation of Bug22p between Paramecium and humans.
Bug22p sequence conservation from Paramecium to humans suggests conservation of its biological function, tested here by functional complementation. The expression of the human HsBug22p protein can rescue the depletion of the PtBug22p protein under RNAi conditions. Moreover, a balance can be found in which the expression level of the HsBug22p protein is in a physiological range, rescuing the PtBUG22 RNAi phenotype, while being susceptible to RNAi silencing itself. This is the first example of functional complementation by a human gene in a ciliate, which opens the use of Paramecium as a model to study human proteins by comparing the rescue properties of wild-type and mutated versions associated with diseases.
Different statuses for BUG22 genes through evolution and potential roles of the proteins.
The Bug22p protein is a highly conserved eukaryotic protein; however, it is not found in all eukaryotes. All species that display centrioles and cilia, be they motile or not, at some stages of their life cycle have one or several BUG22 genes. With a few exceptions, plants and fungi do not have centrioles and cilia. Fungi do possess a centrosome-like structure, the spindle pole body, while plants have MTOC structures to organize the microtubules that govern chromosome separation at mitosis. Interestingly, only plants, not fungi, have BUG22 genes in their genomes. Examination of the aligned sequences reveals a consensus at critical positions in Bug22p proteins of species with centriolar/ciliary function (Fig. 1). Plant Bug22p proteins display another consensus at the same positions and not only a lack of the ciliary consensus, suggesting that there is selective pressure to prevent divergence from this consensus. Plants that do possess centrioles and cilia/flagella, such as Physcomitrella and Selaginella, have several Bug22p proteins encoded in their genomes that display different combinations of ciliary and plant consensus residues at the critical positions, indicating that both putative centriolar/ciliary and plant functions of Bug22p are needed in these plants.
In the present study of the motile cilia of Paramecium, we have found a striking role of Bug22p in ciliary rigidity and beating efficiency, as well as a possible role in the cortical organization of basal bodies. Bug22p proteins seem to exist in other types of cilia, sensory and primary cilia. Indeed, a BUG22 gene with a cilium-specific X box in its promoter is found in C. elegans (6), which possesses only sensory cilia. Bug22p peptides have also been detected in the primary cilium-derived photosensory rod outer segments of mouse retina (21). Although the role of Bug22p in sensory and primary cilia is unknown, we can infer from its localization in the motile cilia of Paramecium that Bug22p plays a role associated with axonemal doublets and the ciliary membrane, potentially important for sensory and signaling functions.
The presence of BUG22 genes in higher plants devoid of centrioles and cilia, with such sequence conservation, suggests that functions other than those associated with centrioles and cilia also exist for Bug22p. However, although higher plants have lost centrioles, they have kept microtubule-organizing centers, as well as proteins with conserved domains characteristic of centrosomes, such as TOF, LisH, and PLL motif-containing proteins (TONNEAU1 in plants, FOP and OFD in animals) and centrin 2 (4). The prominent interactions of microtubules with the plasma membrane in these cells could be a clue to the function of Bug22p in plants, by extrapolation of what we have found in motile cilia.
Interestingly, coevolution of the centrin 2 and Bug22p proteins seems to have occurred in eukaryotes. Among the different centrin genes that exist in eukaryotes, the centrin 2 form is found, not only in species with centrioles and cilia, in functional association with these organelles, but also, as a derived form called centrin 2′, in higher plants lacking centrioles and cilia (7). No Bug22p or centrin 2 sequences are present in fungi devoid of centrioles and cilia, which rely on centrin 3 isoforms only for centrosomal-like function of the spindle pole body. If we call BUG22 the form found with the ciliary consensus and BUG22′ the form with the plant consensus, a good parallel thus appears between the evolution of the centrin 2 and BUG22 families. A functional relationship between Bug22 and centrin 2 proteins may exist and should now be investigated to understand the essential role of Bug22p.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joëlle Marie for hosting our HeLa cell cultures; François Lacroute for sharing his human cDNA libraries; J. Salisbury for the generous gift of the 20H5 antibody; Serge Amselem, Estelle Escudier, and Denise Escalier for helpful advice on ciliary-beating measurements; and Jean-Pierre Denizot and the Imagif platform for providing electron microscopy facilities. We are grateful to Janine Beisson and Linda Sperling for critical reading of the manuscript.
A fellowship to C.L. from the Ministère de la Recherche and grant NT05_1522 from the Agence Nationale de la Recherche are gratefully acknowledged.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Arnaiz O., Malinowska A., Klotz C., Sperling L., Dadlez M., Koll F., Cohen J. 2009. Cildb: a knowledgebase for centrosomes and cilia. Database doi:10.1093/database/bap022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaiz O., Cain S., Cohen J., Sperling L. 2007. ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 3:D439–D444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aury J. M., Jaillon O., Duret L., Noël B., Jubin C., Porcel B. M., Ségurens B., et al. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444:171–178 [DOI] [PubMed] [Google Scholar]
- 4.Azimzadeh J., Nacry P., Christodoulidou A., Drevensek S., Camilleri C., Amiour N., Parcy F., Pastuglia M., Bouchez D. 2008. Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20:2146–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beisson J., Clérot J. C., Fleury-Aubusson A., Garreau de Loubresse N., Ruiz F., Klotz C. 2001. Basal body-associated nucleation center for the centrin-based cortical cytoskeletal network in Paramecium. Protist 152:339–354 [DOI] [PubMed] [Google Scholar]
- 6.Blacque O. E., Perens E. A., Boroevich K. A., Inglis P. N., Li C., Warner A., Khattra J., et al. 2005. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 15:935–941 [DOI] [PubMed] [Google Scholar]
- 7.Bornens M., Azimzadeh J. 2007. Origin and evolution of the centrosome. Adv. Exp. Med. Biol. 607:119–129 [DOI] [PubMed] [Google Scholar]
- 8.Broadhead R., Dawe H. R., Farr H., Griffiths S., Hart S. R., Portman N., Shaw M. K., et al. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440:224–227 [DOI] [PubMed] [Google Scholar]
- 9.Cohen J., Adoutte A., Grandchamp S., Houdebine L. M., Beisson J. 1982. Immunocytochemical study of microtubular structures throughout the cell cycle of Paramecium. Biol. Cell 44:35–44 [Google Scholar]
- 10.Dupuis-Williams P., Klotz C., Mazarguil H., Beisson J. 1996. The tubulin gene family of Paramecium: characterization and expression of the alpha PT1 and alpha PT2 genes which code for alpha-tubulins with unusual C-terminal amino acids, GLY and ALA. Biol. Cell 87:83–93 [DOI] [PubMed] [Google Scholar]
- 11.Errabolu R., Sanders M. A., Salisbury J. L. 1994. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J. Cell Sci. 107:9–16 [DOI] [PubMed] [Google Scholar]
- 12.Follit J. A., Xu F., Keady B. T., Pazour G. J. 2009. Characterization of mouse IFT complex B. Cell Motil. Cytoskeleton 66:457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galvani A., Sperling L. 2002. RNA interference by feeding in Paramecium. Trends Genet. 18:11–12 [DOI] [PubMed] [Google Scholar]
- 14.Gerdes J. M., Davis E. E., Katsanis N. 2009. The vertebrate primary cilium in development, homeostasis, and disease. Cell 137:32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., et al. 2003. A protein interaction map of Drosophila melanogaster. Science 302:1727–1736 [DOI] [PubMed] [Google Scholar]
- 16.Keller L. C., Geimer S., Romijn E., Yates J., Zamora I., Marshall W. F. 2009. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell 20:1150–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller L. C., Romijn E. P., Zamora I., Yates J. R., Marshall W. F. 2005. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15:1090–1098 [DOI] [PubMed] [Google Scholar]
- 18.Klotz C., Garreau de Loubresse N., Ruiz F., Beisson J. 1997. Genetic evidence for a role of centrin-associated proteins in the organization and dynamics of the infraciliary lattice in Paramecium. Cell Motil. Cytoskeleton 38:172–186 [DOI] [PubMed] [Google Scholar]
- 19.Kung C., Chang S. Y., Satow Y., van Houten J., Hansma H. 1975. Genetic dissection of behavior in Paramecium. Science 188:898–904 [PubMed] [Google Scholar]
- 20.Lechtreck K. F., Delmotte P., Robinson M. L., Sanderson M. J., Witman G. B. 2008. Mutations in Hydin impair ciliary motility in mice. J. Cell Biol. 180:633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q., Tan G., Levenkova N., Li T., Pugh E. N., Rux J. J., Speicher D. W., Pierce E. A. 2007. The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics 6:1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murayama T., Toh Y., Ohshima Y., Koga M. 2005. The dyf-3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. J. Mol. Biol. 346:677–687 [DOI] [PubMed] [Google Scholar]
- 23.Ostrowski L. E., Blackburn K., Radde K. M., Moyer M. B., Schlatzer D. M., Moseley A., Boucher R. C. 2002. A proteomic analysis of human cilia: identification of novel components. Mol. Cell. Proteomics 6:451–465 [DOI] [PubMed] [Google Scholar]
- 24.Pazour G. J. 2004. Comparative genomics: prediction of the ciliary and basal body proteome. Curr. Biol. 4:R575–R577 [DOI] [PubMed] [Google Scholar]
- 25.Pazour G. J., Agrin N., Leszyk J., Witman G. B. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen L. B., Miller M. S., Geimer S., Leitch J. M., Rosenbaum J. L., Cole D. G. 2005. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr. Biol. 15:262–266 [DOI] [PubMed] [Google Scholar]
- 27.Rijkers T., Rüther U. 1996. Sequence and expression pattern of an evolutionarily conserved transcript identified by gene trapping. Biochim. Biophys. Acta 1307:294–300 [DOI] [PubMed] [Google Scholar]
- 28.Ruiz F., Krzywicka A., Klotz C., Keller A., Cohen J., Koll F., Balavoine G., Beisson J. 2000. The SM19 gene, required for duplication of basal bodies in Paramecium, encodes a novel tubulin, eta-tubulin. Curr. Biol. 10:1451–1454 [DOI] [PubMed] [Google Scholar]
- 29.Sanders M. A., Salisbury J. L. 1994. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 124:795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satir P., Christensen S. T. 2008. Structure and function of mammalian cilia. Histochem. Cell Biol. 129:687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skouri F., Cohen J. 1997. Genetic approach to regulated exocytosis using functional complementation in Paramecium: identification of the ND7 gene required for membrane fusion. Mol. Biol. Cell 8:1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith J. C., Northey J. G. B., Garg J., Pearlman R. E., Siu K. W. M. 2005. Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J. Proteome Res. 4:909–919 [DOI] [PubMed] [Google Scholar]
- 33.Sonneborn T. M. 1970. Methods in Paramecium research. Methods Cell Physiol. 4:241–339 [Google Scholar]
- 34.Sperling L., Keryer G., Ruiz F., Beisson J. 1991. Cortical morphogenesis in Paramecium: a transcellular wave of protein phosphorylation involved in ciliary rootlet disassembly. Dev. Biol. 148:205–218 [DOI] [PubMed] [Google Scholar]
- 35.Sprague J., Bayraktaroglu L., Clements D., Conlin T., Fashena D., Frazer K., Haendel M., et al. 2006. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 34:D581–D585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stolc V., Samanta M. P., Tongprasit W., Marshall W. F. 2005. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. U. S. A. 102:3703–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timmons L., Court D. L., Fire A. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263:103–112 [DOI] [PubMed] [Google Scholar]
- 38.Wehland J., Weber K. 1987. Turnover of the carboxy-terminal tyrosine of alpha-tubulin and means of reaching elevated levels of detyrosination in living cells. J. Cell Sci. 88:185–203 [DOI] [PubMed] [Google Scholar]
- 39.Woodland H. R., Fry A. M. 2008. Pix proteins and the evolution of centrioles. PLoS One 3:e3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.