Abstract
The present study compared the performance of commercial PCR-based Salmonella enterica detection methods (BAX System Q7, the iQ-Check Salmonella II kit, and the TaqMan Salmonella enterica detection kit) with culture-based methods (modified semisolid Rappaport-Vassiliadis [MSRV] and NMKL71) in spiked and naturally contaminated samples of feed mill scrapings (FMS), palm kernel meal (PKM), pelleted feed (PF), rape seed meal (RSM), soybean meal (SM), and wheat grain (WG). When results from the various feeds were compared, the number of Salmonella enterica CFU/25 g required to produce a positive were as follows: PKM > FMS = WG > RSM = SM = PF. These data are similar to those developed in earlier studies with culture-based Salmonella detection methods. PCR-based methods were performed similarly to culture-based methods, with respect to sensitivity and specificity. However, many PCR positives could not be confirmed by Salmonella isolation and for that reason the evaluated methods were found to be suitable only when rapid results were paramount. Nevertheless, PCR-based methods cannot presently replace culture-based methods when typing information is required for tracing studies or epidemiological investigations. The observed difference in detection levels is a potential problem when prevalence data are compared as well as when feed ingredients are tested for conformance with microbiological criteria. This paper also presents a statistical model that describes the detection probability when different levels (CFU) of Salmonella contamination are present in feed materials.
The association of Salmonella enterica-contaminated feed with salmonellosis in humans, laboratory animals, poultry, cattle, horses, mink, swine, and dogs has been documented frequently in studies conducted in various countries (11, 16, 19, 22, 25, 27, 36, 42, 45). In addition, a recent review article has highlighted the association of animal feeds with human illness, and a recent opinion from the European Food Safety Authority (EFSA) recommended that microbiological criteria be implemented for Salmonella in feed (6, 14). Consequently, it is essential to reduce the exposure of food-producing animals to Salmonella-contaminated feed. However, the pace of industrial commerce is quite rapid, and there is a great demand for rapid and reliable methods for Salmonella detection.
Traditional culture-based methods for detecting Salmonella in feed require 5 to 7 days and involve the following steps to obtain a positive result: nonselective enrichment, selective enrichment, selective plating, and serological and biochemical confirmation (33). Although culture-based methods detect viable bacterial cells and offer an epidemiological advantage over molecular techniques, such methods are time-consuming, particularly when large numbers of samples are involved (33). Molecular techniques, such as PCR, for Salmonella detection are capable of obtaining positive or negative results in 2 days but still require preenrichment to obtain the 102 to 104 Salmonella CFU/ml required by the method for detection (unpublished studies). While the addition of the preenrichment step increases detection time, it reduces the concern that PCR methods will detect DNA from dead cells since feed is usually contaminated with a very low number of Salmonella cells (15, 40).
Recent studies of spiked and naturally contaminated foods have reported good agreement between culture-based and PCR methods for Salmonella detection (32). The detection of Salmonella in feed samples may be substantially different since the cells present are stressed via low water activities and numerous bacterial species compete for similar survival niches (39). In addition, PCR methods may be more sensitive for some feed ingredients than for others, a finding that may affect the reported prevalence of Salmonella in different feed ingredients (21). Yet several commercial PCR-based methods developed for food are routinely used for detection of Salmonella in feed.
The PCR-based methods evaluated in the present study (i.e., BAX System Q7 [DuPont Qualicon], the iQ-Check Salmonella II kit [Bio-Rad], and the TaqMan Salmonella enterica detection kit [Applied Biosystems]) have all been evaluated previously with food products but not feed ingredients, and, to our knowledge, no comparative studies have been conducted (8, 12, 17, 20, 28, 29, 31, 34, 37, 43).
Therefore, the aim of the present work was to compare the performance of some commercial PCR-based detection methods for Salmonella in feed with culture-based methods under realistic conditions. In addition, we assessed the statistical uncertainty of the experimental results obtained with the different methods and whether or not the PCR-based methods performed according to suppliers' specifications and established detection limits for Salmonella in the different feed ingredients.
MATERIALS AND METHODS
Feed materials.
For spiking experiments, the batches of wheat grain, soybean meal, rape seed meal, palm kernel meal, pellets of pig feed, and scrapings from a feed mill were the same as those used in a previous study by Koyuncu and Haggblom (30). The feed materials were kept at 4°C (water activity, <0.7) until used and analyzed for Salmonella prior to the experiments as previously described (30).
Naturally contaminated samples were scrapings and dust samples from critical control points in feed mills investigated by the national Swedish surveillance program (44) and analyzed immediately after collection on a weekly basis over a 9-week period. A total of 1,350 samples from 16 feed mills were analyzed with the NMKL71 method (2) in parallel with the commercial PCR kits BAX System Q7 (DuPont Qualicon) and iQ-Check Salmonella II (Bio-Rad). Since many samples were analyzed weekly in the laboratories, with a potential risk of contamination between samples, the TaqMan Salmonella enterica detection kit (Applied Biosystems) was not included in the study with the naturally contaminated samples.
Spiking of feed samples with Salmonella.
The Salmonella enterica subsp. enterica serotype Typhimurium ST115506, Salmonella enterica subsp. enterica serotype Cubana ST58403, and Salmonella enterica subsp. enterica serotype Yoruba ST45506 strains that were used in the spiking experiments were obtained from the culture collection of the National Veterinary Institute, Sweden, and all were isolated from animal feed. The procedure for preparing the bacterial cells for the spiking experiments was described in reference 30. The methods (BAX Q7, iQ-Check, and TaqMan) were run in parallel with the modified semisolid Rappaport-Vassiliadis (MSRV) method (3), and an experimental setup similar to that described by Koyuncu and Haggblom (30) was used. Six repetitions were carried out, in which each serotype was used in two repetitions. Twenty-five grams of each feed material was weighed into clean, sanitized plastic jars and spiked with 0.7, 7, 70, or 700 CFU of the respective serotype, except for the palm kernel meal, which was spiked at 10 times higher levels. An inoculum of approximately 370 μl of peptone saline water was used for spiking 25 g of the respective feed sample by dispensing droplets on the surface of the feed sample and immediately mixing thoroughly with a new plastic spoon. The samples were left at room temperature (ca. 20°C) for 4 h before the preenrichment broth was added.
To avoid potential sampling errors and the consequential variation between trials, tests were performed as six independent repetitions in which six feed ingredients were spiked at 4 levels, with 5 replicates at each spiking level plus two negative controls. In each trial, samples were preenriched in buffered peptone water (BPW) and analyzed using each of the 4 detection methods (Fig. 1). Each of the three Salmonella enterica strains was used in two repetitions.
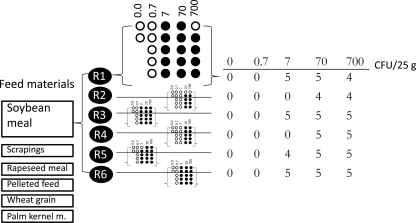
FIG. 1.
Experimental design of the spiking study and example of results from soybean meal (number of positive replicates). Six repetitions (R1 to R6) were used for each feed material, including samples spiked with different levels of Salmonella in five replicates and nonspiked samples in duplicates. Empty circles represent negative replicates, and filled circles (black) represent positive replicates. Each row in the table summarizes the results from one repetition with one detection method. m., meal.
Preenrichment and collection of analytical samples.
Twenty-five grams of each feed sample was preenriched in 225 ml BPW (Oxoid CM 0509, Basingstoke, England) at 37°C ± 1°C for 18 h (16 h for samples analyzed with BAX Q7). Subsamples from the same preenrichment broth were analyzed using each of the compared methods. In the case of samples weighing less than 25 g, e.g., dust samples, one part of feed material and nine parts of BPW were used (the minimum volume of BPW was 90 ml).
After preenrichment, the manufacturer's instructions for each PCR-based method were followed in parallel with the MSRV or NMKL71 method according to Koyuncu and Haggblom (30). In order to reduce the risk for contamination of the samples, DNA extraction, preparation of reaction mixtures, and amplification and detection of PCR products were performed in separate laboratories.
For BAX Q7, after 16-h preenrichment in BPW at 37°C ± 1°C, 10 μl of the sample was transferred to 500 μl brain heart infusion broth (BHI) (Difco; 237500) for a second enrichment at 37°C ± 1°C for 3 h. From the BHI-enriched sample, 5 μl was transferred to the lysis buffer (supplied with the kit) for DNA extraction. From the lysate, 50 μl were transferred to PCR tubes, containing a tablet of all reagents for the PCR. The end-point PCR was performed using the BAX cycler/detector, and the results were analyzed with BAX Q7 software (version 2.4).
For the iQ-Check protocol, after 18-h preenrichment, a 1-ml sample from the BPW was centrifuged and the pellet was resuspended in lysis reagent (supplied with the kit). After heating and a final centrifugation, 5 μl of the supernatant was used for the amplification reaction. In case of inhibition, a 1:10 dilution was performed, and 5 μl of the dilution was used for the amplification reaction. The real-time PCR was performed using the Chromo4 real-time PCR system, and the results were analyzed with Opticon Monitor 3.1 software.
For the TaqMan protocol, after 18-h preenrichment, 1 ml of the BPW was centrifuged and the pellet was resuspended in lysis (PrepMan) reagent. The supernatant was diluted 1:10, and 12 μl was used for the amplification reaction. The results were analyzed with the RapidFinder software.
Confirmation of test results by isolation of Salmonella from naturally contaminated samples.
For the naturally contaminated samples, the BHI tubes were stored at 4 to 8°C (6 to 10 h) after the enrichment. When positive results did not agree among the methods compared, the MSRV method was used for confirming the presence of Salmonella. When the NMKL71 method and both PCR methods (BAX System Q7 and iQ-Check) showed positive results, no confirmation was carried out. In the case of a positive result with at least one PCR method and a negative with the NMKL71 method, the xylose lysine deoxycholate (XLD) agar (XLD supplemented with 1.5% novobiocin; Lab M lab 32, Axel Johnson Lab System Inc., Solna, Sweden) and brilliant green agar (BGA; Oxoid CM 0329) plates were reexamined for suspect colonies. If none was found, a loop of the primary streak of the XLD plate was reinoculated in 5 ml BPW for a second enrichment at 37°C ± 1°C for 16 to 18 h. Reinoculated BPW was analyzed using the MSRV method and serotyped using the Kauffmann-White scheme.
Data analysis and statistical calculations.
The relative accuracy (AC), sensitivity (SE), and specificity (SP) were calculated according to the NordVal validation protocol (5, 32). The agreement between pairs of methods was quantified as Cohen's kappa (κ) (13), as described in NMKL procedure no. 20 (4). The following abbreviations were used: PA, positive agreement; NA, negative agreement; TP, true positive; FN, false negative; and FP, false positive. In the study with spiked samples, FP was defined as a positive result from a nonspiked sample, whereas for the naturally contaminated samples FP was defined as a PCR-positive sample which was positive with neither the NMKL71 method nor the MSRV method following reinoculation.
AC was defined as the compliance between responses by the PCR-based method and the reference method (NMKL71/MSRV) for identical samples: (PA + NA + FP)/(PA + NA + TP + FN + FP). Thus, AC is the proportion of concordant analyses among all samples analyzed, and the confidence interval can be calculated as the shortest confidence interval for a proportion (35), under the assumption that all values are equally probable. An online calculator (http://www.causascientia.org/math_stat/ProportionCI.html; accessed 16 December 2008) was used to calculate a point estimate of AC and to define the boundaries of an interval that, with 95% certainty, contains the true value of the relative AC. For this experimental setup there is no standard method available for the calculation of the confidence interval for relative SE (SErel), as indicated in equation 1. SErel depends on the difference in sensitivity levels between the methods (ΔSE) as well as the average sensitivity of the two methods (SE).
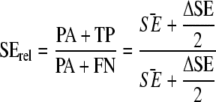 |
(1) |
In this work, we used the confidence interval for ΔSE (equation 2), calculated as described in reference 38, to construct an approximate confidence interval for SErel by substituting true SĒ with (2PA + TP + FN)/2m, where m is the total number of samples.
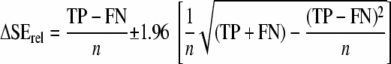 |
(2) |
SP was defined as NA/(NA + FP). Since by definition the spiked samples could be not recorded as FP, a conservative estimate of SP was calculated based on nonspiked samples. For the noncontaminated material, SP is the quotient NA of all noncontaminated samples (n = 30), and, consequently, the confidence interval can be calculated as it was for AC. For the naturally contaminated samples, the denominator NA + FP is dependent on NA and FP and is thus not fixed. However, since FP ≪ NA, we can make the approximation that the number of negatives (N) is fixed as NA + FP. This results in the formula SP = (N − FP)/N, for which the confidence interval can be calculated as for AC. Confidence intervals for Cohen's kappa (κ) were calculated using an online calculator (http://faculty.vassar.edu/lowry/kappa.html). The intervals presented here are calculated using the method described in reference 13.
Interval estimates for detection probability.
In order to quantify the uncertainty and generate interval estimates for the detection probability at different concentrations of Salmonella, we applied Monte Carlo simulation under a hierarchical model. The model assumes that the logarithm of the added number of bacterial cells has a normal variability. We describe the load as P[ln(λ)] ∼ Normal{〈ln(λ)〉,sd[ln(λ)]}, where the parameters are the mean and standard deviation (sd). We estimate the mean of the logarithm by assuming that the nominal level is the median of the load (spiking level, λl0), reflecting our belief that it is equally likely that the stock solution contains a higher or lower concentration than the nominal. Under ideal conditions, a single bacterial cell will multiply in the preenrichment medium and reach a cell density high enough to ensure that any subsample will contain Salmonella DNA and will thus constitute a detectable unit. Under less favorable conditions, not all cells will start to grow and each cell may undergo fewer cell divisions. Thus, a larger initial number of cells are needed in order to reach the same cell density after preenrichment, and, for that reason, a detectable unit would consist of several cells. In our model this is accounted for by introducing a material-specific constant “recovery,” which is defined as 1 divided by the number of cells needed to form a “detectable unit.” The model uses the definitions and abbreviations shown in Table 1 .
TABLE 1.
Symbols and functions used in the detection probability modela
| Definition | Symbol/function |
|---|---|
| Nominal spiking concn at level l | λl0 |
| Standard deviation of logarithmized spiking level [ln(λ)] | sd |
| True spiking level at level l, repetition k | λlk = exp{Normal[ln(λl0)],sd} |
| Recovery in material m | rm |
| Avg no. of detectable units per replicate in repetition k, material m, level l | λmlk = rm × exp{Normal[ln(λl)],sd} |
| No. of detectable units in repetition k, material m, level l, replicate i | λmlki = Poisson(λmlk) |
| Probability for at least one detectable unit in replicate mlki | Pmlki = 1 − Poisson(0|λmlk) = 1 − exp(−λmlk) |
| No. of positive replicates, matrix m, repetition k, level l | posmlk = Binomial{5|[1 − exp(−λmlk)]} |
exp(a), ea; Normal(a,b), random number from the normal distribution, where a is the mean and b is the sd; Poisson(a), random number from normal distribution, where a is the mean; Poisson(b|a), probability that the Poisson distribution with a mean of a returns b; Binomial(a|b), random number from binomial distribution with a trials and probability b.
A point estimate of recovery in matrix m, r̂m, was obtained from data by minimizing Δ2 in equation 3, in which x̄l is the proportion of positive replicates at level l, and λl0 is the nominal spiking level.
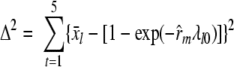 |
(3) |
A global measure of variation (var) in the number of positive replicates (pos) between repetitions at a given level was calculated for all 6 materials (m), with 6 repetitions (k) and 5 levels (l) according to equation 4. The observed value of var was 0.38.
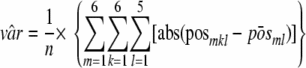 |
(4) |
In order to generate intervals for detection probabilities, approximations of the posterior distributions for sd and rm were generated using Monte Carlo simulations. The hierarchical model was used to simulate 50,000 complete studies with 6 imaginary feed materials and 6 independent repetitions, as illustrated in Fig. 1. For each simulated study, ln(rm) was sampled from Uniform[−7,0] and the sd sampled from Uniform[0,3]. The selection of prior distributions reflects our prior belief that any magnitude of recovery is equally probable, and variation between repetitions can in the worst case be one order of magnitude. For each simulated study, var was calculated for all 6 materials collectively and rm was estimated for each material using equations 3 and 4. The result from each material was recorded as for experimental data. The simulated results from one imaginary material were recorded as a “hit” in relation to material m, if rm from the simulation was within the window.
[r̂m/x···r̂m × x] and the value of var from the simulated study was in the range of 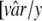 ···
···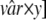 . The frequency distributions of rm and the sd among the scored hits are used as approximations of the posterior probability distributions for these parameters and represent a range of rm and the sd that can explain the experimental data, as illustrated in Fig. 2. Point estimates for detection probability as a function of the concentration were generated by inserting the median value from the frequency distribution of rm in the model (Fig. 3), and 95% intervals were generated from the 2.5th and 95th percentiles from the posterior distribution of rm (Fig. 2 and 3). The width of the window that defines a hit defined by the parameters x and y has no significant effect on the posterior distributions. The results presented are generated with x = 1.05 and y = 1.15.
. The frequency distributions of rm and the sd among the scored hits are used as approximations of the posterior probability distributions for these parameters and represent a range of rm and the sd that can explain the experimental data, as illustrated in Fig. 2. Point estimates for detection probability as a function of the concentration were generated by inserting the median value from the frequency distribution of rm in the model (Fig. 3), and 95% intervals were generated from the 2.5th and 95th percentiles from the posterior distribution of rm (Fig. 2 and 3). The width of the window that defines a hit defined by the parameters x and y has no significant effect on the posterior distributions. The results presented are generated with x = 1.05 and y = 1.15.
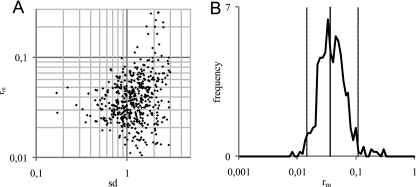
FIG. 2.
Estimating variability in spiking levels between repetitions (sd) and recovery (rm) for Salmonella in wheat grain by Monte Carlo simulation. (A) Distribution of sd and rm from 643 out of 50,000 simulated experiments. Each dot represents a hit, that is, an experiment that resembled the results from the experimental study. (B) Frequency distribution for rm that is used as approximation of posterior distribution of rm in wheat grain. The vertical lines are 2.5th, 50th, and 97.5th percentiles, respectively.
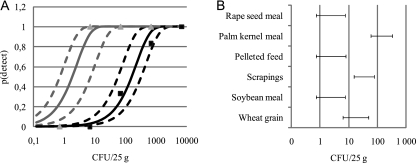
FIG. 3.
Calculated detection probabilities in different feed materials with the MSRV method. (A) Probabilities for Salmonella detection in pelleted feed (gray) and palm kernel meal (black). Data points indicate observed results, and solid lines indicate the predicted detection probability based on the median rm from the Monte Carlo simulation. Broken lines indicate limits of 95% intervals for the detection probability. (B) The 95% intervals for the concentration of Salmonella that results in 50% detection probability.
Estimating probability of detection in naturally contaminated materials.
A rough estimate of the probability of detecting Salmonella, if present in naturally contaminated process control samples, was generated, given the assumptions that a fraction f of all samples was contaminated at the same level, that detection probabilities were equal for all methods, and that detection with the three methods was independent, given that the sample was positive. Under these assumptions, the expected number of positive test results from a contaminated sample is given by Binomial(3,P) where P is P (detection|positive). Point estimates of f and p were generated by minimizing
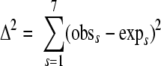 |
where obss is the observed frequency of positives in sector s of Fig. 4 and exps is the frequency predicted by the binomial distribution.
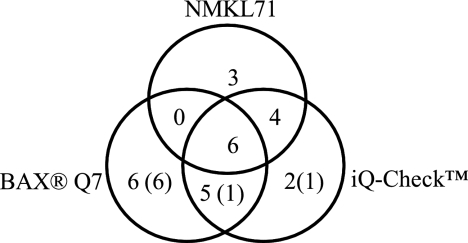
FIG. 4.
Detection of Salmonella in process control samples from feed mills. The figures in the different sectors indicate the number of samples that were positive with that combination of methods when applied in parallel on the same preenrichment. Positive results not confirmed by isolation are shown within parentheses.
RESULTS
Performance of the PCR-based methods.
Sample preparation and cell lysis procedures were quite similar for the iQ-Check and TaqMan assays, except that the DNA was routinely diluted 1:10 in the TaqMan assay. However, the BAX Q7 procedures were less laborious than other methods due to prepackaged reagents and PCR tablets. When analyzing spiked scraping samples, the iQ-Check PCR was, in most cases, inhibited and could not detect Salmonella unless diluted 1:10. Such inhibition was not observed with the TaqMan and BAX Q7 assays for any samples.
No differences in detection limits were observed, with respect to serotype (data not shown). Therefore, data from the various serotypes were pooled. In an earlier study using culture-based detection methods, we observed that even though feed ingredients were spiked with identical Salmonella cell numbers, detection limits varied with ingredient type (30). Similar results were obtained when PCR-based methods were employed in this study. Palm kernel meal had the highest detection limit, and rape seed meal had the lowest detection limit (Fig. 3).
Detection limits for Salmonella in feed materials.
Spiking experiments were performed in order to establish the detection limits for Salmonella in the different feed ingredients and to study the agreement of the three PCR-based methods with the MSRV method. A total of 720 samples were spiked according to the outline shown in Fig. 1.
The first aim was to generate interval predictions for the probability of detection at different levels of contamination, and, for this purpose, the results from each detection method were analyzed separately.
Due to the limited number of data points and spiking levels, it was necessary to select a model with few parameters. Attempts to model the data with classical logistic regression showed that the results were extremely sensitive to single data points (results not shown). For this reason, the data were modeled using a simple hierarchical model, in which the shape of the response curve is given by the Poisson distribution and the detection level was determined by a single material-specific constant (recovery, rm). A characteristic feature of this model is that it predicts that the concentrations that give 5% or 95% detection probability differ by approximately two orders of magnitude, which is similar to what has been observed for purified DNA in water (7) and agrees well with our earlier observations of Salmonella in feed (30).
Replicates typically showed the same test results; however, close to the detection limit, the result often varied between repetitions (Fig. 1), indicating a variability in true spiking levels. Generating confidence intervals for the detection limit in feed materials is not trivial, since it is necessary to take into account the uncertainty about the true spiking level in each repetition as well as the limited number of replicates, and, for this reason, a Bayesian approach was used. As a first step, we obtained a point estimate of recovery in each material by fitting the model to the experimental data using the least-squares method. As a second step, interval predictions for the recovery in each material were obtained by Monte Carlo simulation (Fig. 3A and B).
As shown in Fig. 3A, an average contamination level of 1 CFU/25 g in pelleted feed will give a detection probability of 10 to 60%, whereas in palm kernel meal the detection probability at the same level of contamination is close to zero (<1.25%) when the MSRV method is used. At a contamination level of 100 CFU/25 g, the detection probability in pelleted feed is close to 100% (>99%), whereas in palm kernel meal the probability is in the range of 20 to 80%. At a contamination level of 10 CFU Salmonella/25 g, Salmonella will be detected in >80% of samples from rape seed meal, soybean meal, and pelleted feed (Fig. 3A and B), in approximately 50% of samples from wheat grain, and in <10% of samples from palm kernel meal (Fig. 3A). Figure 3B shows 95% intervals for the detection limit in six different feed materials, defined as the contamination level at which the probability of detection is 50%.
The results show that the difference in detection limits between feed materials is statistically significant and spans over approximately two orders of magnitude. High detection limits were observed with palm kernel meal, wheat grain, and scrapings, while low detection limits were seen with rape seed meal, pelleted feed, and soybean meal. The graphs shown in Fig. 2 and 3 were generated using data from the MSRV method, but similar results were obtained with all four methods (results not shown).
Agreement between PCR- and culture-based methods for Salmonella-spiked feed materials.
In order to study the agreement between the PCR-based methods and the MSRV method, as well as possible differences in sensitivity and accuracy, statistical parameters were calculated using paired samples and following the guidelines of NordVal (5, 32). All PCR-based methods showed good agreement with the MSRV method (Table 2). The AC for all PCR-based methods was >98% at a 95% confidence level, and the SE was close to 100% (Table 2). The agreement between the PCR-based methods and the MSRV method was quantified using Cohen's kappa (κ) (4). For all three methods, κ was >0.95 (at a 95% confidence level). NMKL71 defines almost perfect agreement between the methods as a κ value of >0.81 (4).
TABLE 2.
Agreement between PCR-based methods and culture-based methods with spiked feed materials and naturally contaminated process control samples
| Method | PAa | NA | FN | TP | FP | Total | AC (%) | SE (%) | SP (%) | κ |
|---|---|---|---|---|---|---|---|---|---|---|
| Spiked | ||||||||||
| iQ-Check | 443 | 340 | 0 | 9 | 0 | 792 | 98.9 (98.2-99.6) | 102.0 (100.7-103.4) | 100 (0.907-1) | 0.98 (0.96-0.99) |
| BAX Q7 | 439 | 345 | 4 | 4 | 0 | 792 | 99.0 (98.1-99.6) | 100.0 (98.7-101.3) | 100 (0.907-1) | 0.98 (0.96-0.99) |
| TaqMan | 443 | 338 | 0 | 8 | 3 | 792 | 99.0 (98.1-99.6) | 101.8 (100.6-103.1) | 90 (0.761-0.974) | 0.971 (0.95-0.99) |
| Naturally contaminated | ||||||||||
| iQ-Check | 10 | 1,330 | 3 | 5 | 2 | 1,350 | 99.4 (98.8-99-7) | 115.4 (77.5-174) | 99.8 (99.4-99.9) | 0.66 (0.45-0.87) |
| BAX Q7 | 6 | 1,326 | 7 | 4 | 7 | 1,350 | 99.2 (98.5-99.5) | 76.9 (41.5-136) | 99.5 (98.9-99.7) | 0.39 (0.11-0.67) |
PA, positive agreement; NA, negative agreement; FN, false negative; TP, true positive; FP, false positive; AC, accuracy (PA + NA + FP)/(PA + NA + TP + FN + FP); SE, sensitivity (PA + TP)/(PA + FN); SP, specificity (total − FP)/total; κ, Cohen's kappa. Values in parentheses indicate a 95% confidence interval for the respective parameter.
From the spiked materials, three FP samples were obtained with TaqMan (SP = 90%), whereas no FP samples were obtained with iQ-Check, BAX Q7, and NMKL71 (SP = 100%). The difference in specificity levels between methods was, however, not significant (McNemar's test, P = 0.25) (38). Both iQ-Check and TaqMan showed slightly higher sensitivities for spiked material than the MSRV method. McNemar's test was significant (at a 95% confidence level) for iQ-Check and TaqMan (P = 0.023 and P = 0.04, respectively) after a Bonferroni correction (three degrees of freedom).
Agreement between detection methods on naturally infected samples.
A total of 1,350 samples (scrapings) were obtained from feed mills and were analyzed using the BAX Q7, iQ-Check, and NMKL71 methods. Salmonella was detected by at least one method in a total of 26 samples (Fig. 4). The agreement between the results from each method is illustrated, and with six samples, Salmonella was detected with all three methods. The NMKL71 method detected 13 samples as positive (defined as positive when Salmonella was isolated from the first preenrichment), and both PCR-based methods detected 17 samples as positive (defined as positive when PCR-positive results were obtained). Out of 26 positive samples, three were obtained exclusively with the NMKL71 method, and 13 with the PCR-based methods. Five samples were positive with both PCR-based methods but negative with the NMKL71 method.
In order to confirm the presence of Salmonella in samples that were PCR positive but NMKL71 (culture) negative, the following two procedures were applied: (i) for iQ-Check and BAX Q7, samples were reinoculated using a loop from the primary streak on XLD plates into BPW; and (ii) if the BAX Q7 method was concerned, an additional sample from the BHI broth was transferred to an MSRV plate. Confirmation was accomplished using the MSRV method. For samples that were negative with NMKL71, the values in parentheses in Fig. 4 indicate the number of samples (six samples with BAX Q7, one sample with iQ-Check, and one sample with both PCR-based methods) that were not confirmed by isolation.
A total of six different serotypes were isolated from the naturally contaminated environmental samples from feed mills. The most commonly isolated serotypes were S. Typhimurium and S. Cubana, in eight and six samples, respectively. Other serotypes detected included Salmonella enterica subsp. enterica serotype Reading, Salmonella enterica subsp. enterica serotype Oranienburg, Salmonella enterica subsp. enterica serotype Düsseldorf, Salmonella enterica subsp. enterica serotype Anatum, and a nontypeable Salmonella strain. Positive samples were obtained from all control points monitored in the feed mills sampled.
The data in Table 2 indicate that both PCR-based methods showed a good relative accuracy and specificity for naturally contaminated samples. In a comparison of spiked samples, confidence intervals for κ were large and significantly smaller agreement was observed. Using the scale from NMKL (4), the agreement between iQ-Check and NMKL71 was in the range of “fair” to “good,” whereas for BAX Q7, the agreement was “slight” or “substantial.” However, neither BAX Q7 nor iQ-Check showed a sensitivity that was significantly different from that of the NMKL71 method.
DISCUSSION
In order to prevent transmission of Salmonella from feed to animals and further in the food chain, rapid and reliable detection methods to be used in the feed sector are needed. It has been known for many years that young chickens may become infected by only a few cells of Salmonella in the feed (41). For other animal species, such as pigs, the knowledge about infectious doses is limited, particularly when animals are exposed to low levels of Salmonella in the feed for long periods of time. Efficient isolation methods and sampling plans are important tools in tracing scenarios, control programs, and decision-making processes by the industry or authorities. The low levels of Salmonella in compound feed and feed raw ingredients, the dehydration of the Salmonella present, the uneven distribution of contamination in feed commodities, and the high levels of competing aerobic bacteria combine to put great demands on the isolation methods (15, 23, 39).
In 2008, the European Food Safety Authority (EFSA) reported that rape seed meal and soybean meal had the highest prevalence of Salmonella (1). While the EFSA reported that the prevalence of Salmonella in other feed raw materials, e.g., grain, was significantly lower (6), Koyuncu and Haggblom observed that the sensitivity of cultural detection methods varied with the type of ingredient tested (30). Data collected here with PCR-based methods confirm the observations of Koyuncu and Haggblom (Fig. 3A and B). The wide intervals for the predicted detection probabilities are likely due to the variation between repetitions.
The data in Fig. 3B show that, in terms of the number of CFU of Salmonella/25 g required to produce a 50% chance of a positive test result, palm kernel meal required over 100, feed mill scrapings and wheat grain required 10 to 100, and the other materials tested (i.e., rape seed meal, soybean meal, and pelleted feed, which were equal) required 1 to 10. The reason that palm kernel meal requires more CFU compared to other feed materials is presently unknown. However, Becker and Galletti observed that palm kernel meal effectively adhered to Salmonella, possibly because of high levels of mannans (9). Consequently, these results suggest that regardless of the analytical procedure employed, Salmonella contamination rates for palm kernel meal are likely underestimated.
The data obtained from spiked samples (Table 2) show that, even though the TaqMan and iQ-Check assays had a slightly higher sensitivity than the other method tested, the PCR-based methods tested were in almost perfect agreement with the MSRV method. Slight discrepancies between methods were most likely due to the low concentration of cells after preenrichment and subsampling errors. Based on this hypothesis, the slightly higher sensitivity values observed for the TaqMan and iQ-Check procedures were not unexpected since both these methods use larger subsamples from preenrichment broth (1 ml) than the NMKL71 and BAX Q7 (100 μl and 10 μl, respectively) assays.
In the naturally contaminated samples, the most common serotype was S. Typhimurium (data not shown). The serotype was isolated over several weeks in the environment of one feed mill, indicating a persistent contamination, and then later resulted in infected pig farms.
In contrast to the spiked samples, the agreement between methods with naturally contaminated process control samples (scrapings) was relatively low (Table 2; Fig. 4). However, this observation does not seem to be the result of differences in performance between methods, because the positive results seem to be randomly distributed (Fig. 4). Another explanation is that the concentration of Salmonella in the broth after preenrichment was low, and some subsamples did not contain Salmonella. If the detection probabilities for all positive samples were the same and there were no differences in sensitivity between methods, we could estimate that the probability of detecting Salmonella with either of the methods was approximately 50 to 60% in the samples in which Salmonella was present in the preenrichment broth. For that reason, we can predict that 2 to 4 of the samples that were negative with all three methods still contained Salmonella at high levels equal to those of the positive samples. Using the definitions from NMKL procedure 20 (4), the BAX Q7 and iQ-Check assays showed a relative sensitivity of 77% and 115%, respectively (Table 2). As illustrated in Fig. 4, random effects seem to play a major role in the detection and isolation of Salmonella. Thus, it is likely that some of the samples recorded as false positives with the PCR-based methods are the result of false negatives from NMKL71 that coincided with the failure to isolate Salmonella from the preenrichment medium (BPW) or BHI. A fact which is sometimes overlooked is that the quality parameters (AC, SE, SP, and κ) are sensitive to random variation when the sample size is small, and this uncertainty must be taken into account before conclusions are drawn about the performance of a new method. In this paper we introduce methods for estimating these intervals and, when possible, provide links to tools that can facilitate the calculations.
In this study a model based on the Poisson distribution was used to estimate the probability of detecting Salmonella at different levels of concentration. The difference from the system studied by Evers et al. (18) is that Salmonella detection involves a preenrichment step. In our model this is accounted for by the introduction of a material-specific constant (rm). The constant rm depends on the average number of cell divisions (n) a cell undergoes during preenrichment and thus on the length of the lag phase as well as competition from the background flora. However, other factors, such as aggregation of cells and adhesion of cells to the material, may also influence rm. For low values of rm, we can expect that the constant is approximately proportional to 2n. However, the number of bacterial cells in a 25-g sample is given by Poisson(λlk), and in situations with a high n and low λlk, this sampling event will dominate. Consequently, rm will asymptotically approach 1 when n is high.
The intervals for the predicted detection probabilities are dependent on the chosen prior distributions (26) and should thus be interpreted as (Bayesian) credibility intervals. In this study, we used a noninformative prior for rm, with a uniform distribution on the logarithmic scale. A natural alternative is to use a linear prior distribution for rm on a scale from [0 . . 1]. As expected, using a linear prior results in intervals with wide “left-side” tails arising from hits, with a combination of high rm and very high sd values (results not shown).
The evaluated PCR-based methods showed sensitivities equal to or higher than the culture-based methods, and, in addition, the specificity was very high, which agrees with studies of food materials (10, 12, 24, 32).
PCR-based methods clearly have a function in process control, primarily because of rapid indications of the presence or absence of Salmonella. An important problem is that isolation of Salmonella often fails. Particularly in epidemiological investigations, there is a need for typing, and the investigated PCR-based methods need to be accompanied by suitable isolation methods that can find Salmonella also when the enrichment broth contains low levels.
Acknowledgments
This study was funded by the Swedish Board of Agriculture and by the European Union-funded Integrated Project Biotracer (contract 036272) under the 6th RTD Framework.
We thank Eva Jonsson for her technical assistance and Judith Straver and Gary Barker for valuable discussions on the statistical model. Frank T. Jones is acknowledged for excellent review of the manuscript.
Footnotes
Published ahead of print on 12 March 2010.
REFERENCES
- 1.Anonymous. 2009. Community summary report on food-borne outbreaks in the European Union in 2007. EFSA J. 271:1-128. [Google Scholar]
- 2.Anonymous. 1999. Salmonella. Detection in foods, 5th ed. Method no. 71. Nordic Committee on Food Analysis (NKML), Oslo, Norway.
- 3.Anonymous. 2002. Microbiology of food and animal feeding stuffs—horizontal method for the detection of Salmonella spp., ISO 6579:2002 E standard, 4th ed. ISO Central Secretariat 1, Geneva, Switzerland.
- 4.Anonymous. 2007. NMKL procedure no. 20. Evaluation of results from qualitative methods. Nordic Committee on Food Analysis (NMKL), Oslo, Norway.
- 5.Anonymous. 2002. Protocol for the validation of alternative microbiological methods. NV-DOC.D-20021022. NordVal, Søborg, Denmark.
- 6.Anonymous. 2008. Scientific opinion of the panel on biological hazards on a request from the Health and Consumer Protection, Directorate General, European Commission on Microbiological Risk Assessment in feedingstuffs for food producing animals. EFSA J. 720:1-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artin, I., P. Bjorkman, J. Cronqvist, P. Radstrom, and E. Holst. 2007. First case of type E wound botulism diagnosed using real-time PCR. J. Clin. Microbiol. 45:3589-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey, J. S., and D. E. Cosby. 2003. Detection of Salmonella from chicken rinses and chicken hot dogs with the automated BAX PCR system. J. Food Prot. 66:2138-2140. [DOI] [PubMed] [Google Scholar]
- 9.Becker, P. M., and S. Galletti. 2008. Food and feed components for gut health-promoting adhesion of E. coli and Salmonella enterica. J. Sci. Food Agric. 88:2026-2035. [Google Scholar]
- 10.Bennett, A. R., D. Greenwood, C. Tennant, J. G. Banks, and R. P. Betts. 1998. Rapid and definitive detection of Salmonella in foods by PCR. Lett. Appl. Microbiol. 26:437-441. [DOI] [PubMed] [Google Scholar]
- 11.Boyer, C. I., S. Narotsky, D. W. Bruner, and J. A. Brown. 1962. Salmonellosis in turkeys and chickens associated with contaminated feed. Avian Dis. 6:43-50. [Google Scholar]
- 12.Cheung, P. Y., K. K. Kwok, and K. M. Kam. 2007. Application of BAX system, Tecra Unique Salmonella test, and a conventional culture method for the detection of Salmonella in ready-to-eat and raw foods. J. Appl. Microbiol. 103:219-227. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, J. 1960. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 20:213-220. [Google Scholar]
- 14.Crump, J. A., P. M. Griffin, and F. J. Angulo. 2002. Bacterial contamination of animal feed and its relationship to human foodborne illness. Clin. Infect. Dis. 35:859-865. [DOI] [PubMed] [Google Scholar]
- 15.D'Aoust, J. Y., and A. M. Sewell. 1986. Slow rehydration for detection of Salmonella spp. in feeds and feed ingredients. Appl. Environ. Microbiol. 51:1220-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietz, H. H., M. Chriel, T. H. Andersen, J. C. Jorgensen, M. Torpdahl, H. Pedersen, and K. Pedersen. 2006. Outbreak of Salmonella Dublin-associated abortion in Danish fur farms. Can. Vet. J. 47:1201-1205. [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson, E., and A. Aspan. 2007. Comparison of culture, ELISA and PCR techniques for salmonella detection in faecal samples for cattle, pig and poultry. BMC Vet. Res. 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evers, E. G., J. Post, F. F. Putirulan, and F. J. van der Wal. 2009. Detection probability of Campylobacter. Food Control 21:247-252. [Google Scholar]
- 19.Ferraro, A., M. Deasy, V. Dato, C. Sandt, J. Tait, B. Perry, L. Lind, N. Rea, R. Rickert, C. Marriott, C. Teacher, P. Fox, K. Bluhm, V. Urdaneta, S. Ostroff, E. Villamil, and P. Smith. 2007. Management of transmissible spongiform encephalopathies in livestock feeds and feeding. In Capacity building for surveillance and prevention of BSE and other zoonotic diseases: course manual. Food and Agriculture Organization of the United Nations, Rome, Italy.
- 20.Fratamico, P. M. 2003. Comparison of culture, polymerase chain reaction (PCR), TaqMan Salmonella, and Transia Card Salmonella assays for detection of Salmonella spp. in naturally-contaminated ground chicken, ground turkey, and ground beef. Mol. Cell. Probes 17:215-221. [DOI] [PubMed] [Google Scholar]
- 21.Gardner, I. A. 2004. An epidemiologic critique of current microbial risk assessment practices: the importance of prevalence and test accuracy data. J. Food Prot. 67:2000-2007. [DOI] [PubMed] [Google Scholar]
- 22.Griffin, C. A. 1952. A study of prepared feeds in relation to Salmonella infection in laboratory animals. J. Am. Vet. Med. Assoc. 121:197-200. [PubMed] [Google Scholar]
- 23.Gunnert, K., and B. Brest. 1969. Salmonella types isolated from the Gulf of Aarhus compared with types from infected human beings, animals and feed products in Denmark. Appl. Microbiol. 18:985-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hein, I., G. Flekna, M. Krassnig, and M. Wagner. 2006. Real-time PCR for the detection of Salmonella spp. in food: an alternative approach to a conventional PCR system suggested by the FOOD-PCR project. J. Microbiol. Methods 66:538-547. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch, W., and R. Sapiro-Hirsch. 1958. The role of certain animal feeding stuffs, especially bone meal, in the epidemiology of salmonellosis. Bull. Hyg. 33:647. [PubMed] [Google Scholar]
- 26.Jaynes, E. T. 1976. Confidence intervals vs. Bayesian intervals, p. 175. In W. L. Harper and C. A. Hooker (ed.), Foundations of probability theory, statistical inference, and statistical theories of science. D. Reidel, Dordrecht, Netherlands.
- 27.Jones, P. W., P. Collins, G. T. Brown, and M. Aitken. 1982. Transmission of Salmonella mbandaka to cattle from contaminated feed. J. Hyg. (Lond.) 88:255-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawasaki, S., B. Kimura, and T. Fujii. 2001. Comparison of TaqMan Salmonella amplification/detection kit with standard culture procedure for detection of Salmonella in meat samples. Shokuhin Eiseigaku Zasshi 42:33-39. [DOI] [PubMed] [Google Scholar]
- 29.Kimura, B., S. Kawasaki, T. Fujii, J. Kusunoki, T. Itoh, and S. J. Flood. 1999. Evaluation of TaqMan PCR assay for detecting Salmonella in raw meat and shrimp. J. Food Prot. 62:329-335. [DOI] [PubMed] [Google Scholar]
- 30.Koyuncu, S., and P. Haggblom. 2009. A comparative study of cultural methods for the detection of Salmonella in feed and feed ingredients. BMC Vet. Res. 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liming, S. H., and A. A. Bhagwat. 2004. Application of a molecular beacon-real-time PCR technology to detect Salmonella species contaminating fruits and vegetables. Int. J. Food Microbiol. 95:177-187. [DOI] [PubMed] [Google Scholar]
- 32.Löfström, C., C. E. Axelsson, and P. Rådström. 2008. Validation of a diagnostic PCR method for routine analysis of Salmonella spp. in animal feed samples. Food Anal. Methods 1:23-27. [Google Scholar]
- 33.Maciorowski, K. G., P. Herrera, F. T. Jones, S. D. Pillai, and S. C. Ricke. 2006. Cultural and immunological detection methods for Salmonella spp. in animal feeds—a review. Vet. Res. Commun. 30:127-137. [DOI] [PubMed] [Google Scholar]
- 34.Nde, C. W., M. K. Fakhr, C. Doetkott, and C. M. Logue. 2008. An evaluation of conventional culture, invA PCR, and the real-time PCR iQ-Check kit as detection tools for Salmonella in naturally contaminated premarket and retail turkey. J. Food Prot. 71:386-391. [DOI] [PubMed] [Google Scholar]
- 35.Nicholson, B. J. 1985. On the F-distribution for calculating Bayes credible intervals for fraction nonconforming. IEEE Trans. Reliab. 34:227-228. [Google Scholar]
- 36.Osterberg, J., I. Vagsholm, S. Boqvist, and S. S. Lewerin. 2006. Feed-borne outbreak of Salmonella Cubana in Swedish pig farms: risk factors and factors affecting the restriction period in infected farms. Acta Vet. Scand. 47:13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel, J. R., A. A. Bhagwat, G. C. Sanglay, and M. B. Solomon. 2006. Rapid detection of Salmonella from hydrodynamic pressure-treated poultry using molecular beacon real-time PCR. Food Microbiol. 23:39-46. [DOI] [PubMed] [Google Scholar]
- 38.Petrie, A., and P. Watson. 1999. Statistics for veterinary and animal sciences, 1st ed. Blackwell Science Ltd., Oxford, United Kingdom.
- 39.Salomonsson, A. C., A. Aspán, S. Johansson, A. Heino, and P. Häggblom. 2005. Salmonella detection by polymerase chain reaction after pre-enrichment of feed samples. J. Rapid Methods Autom. Microbiol. 13:96-110. [Google Scholar]
- 40.Sauli, I., J. Danuser, A. H. Geeraerd, J. F. Van Impe, J. Rufenacht, B. Bissig-Choisat, C. Wenk, and K. D. Stark. 2005. Estimating the probability and level of contamination with Salmonella of feed for finishing pigs produced in Switzerland—the impact of the production pathway. Int. J. Food Microbiol. 100:289-310. [DOI] [PubMed] [Google Scholar]
- 41.Schleifer, J. H., B. J. Juven, C. W. Beard, and N. A. Cox. 1984. The susceptibility of chicks to Salmonella montevideo in artificially contaminated poultry feed. Avian Dis. 28:497-503. [PubMed] [Google Scholar]
- 42.Schotte, U., D. Borchers, C. Wulff, and L. Geue. 2007. Salmonella Montevideo outbreak in military kennel dogs caused by contaminated commercial feed, which was only recognized through monitoring. Vet. Microbiol. 119:316-323. [DOI] [PubMed] [Google Scholar]
- 43.Silbernagel, K., R. Jechorek, C. Carver, W. M. Barbour, and P. Mrozinski. 2003. Evaluation of the BAX system for detection of Salmonella in selected foods: collaborative study. J. AOAC Int. 86:1149-1159. [PubMed] [Google Scholar]
- 44.Sternberg Lewerin, S., B. Boquist, P. Engström, and P. Häggblom. 2005. The effective control of Salmonella in Swedish poultry, p. 195-215. In G. C. Mead (ed.), Food safety control in the poultry industry. Woodhouse Publishing in Food Science and Technology, CRC Press, Cambridge, England.
- 45.Walker, R. L., T. L. de Peralta, M. R. Villanueva, K. P. Snipes, J. E. Madigan, D. W. Hird, and R. W. Kasten. 1995. Genotypic and phenotypic analysis of Salmonella strains associated with an outbreak of equine neonatal salmonellosis. Vet. Microbiol. 43:143-150. [DOI] [PubMed] [Google Scholar]