Abstract
Two samples of market oysters, primarily from retail establishments, were collected twice each month in each of nine states during 2007. Samples were shipped refrigerated overnight to five U.S. Food and Drug Administration laboratories on a rotating basis and analyzed by most probable number (MPN) for total and pathogenic Vibrio parahaemolyticus and V. vulnificus numbers and for the presence of toxigenic V. cholerae, Salmonella spp., norovirus (NoV), and hepatitis A virus (HAV). Levels of indicator organisms, including fecal coliforms (MPN), Escherichia coli (MPN), male-specific bacteriophage, and aerobic plate counts, were also determined. V. parahaemolyticus and V. vulnificus levels were distributed seasonally and geographically by harvest region and were similar to levels observed in a previous study conducted in 1998-1999. Levels of pathogenic V. parahaemolyticus were typically several logs lower than total V. parahaemolyticus levels regardless of season or region. Pathogenic V. parahaemolyticus levels in the Gulf and Mid-Atlantic regions were about two logs greater than the levels observed in the Pacific and North Atlantic regions. Pathogens generally associated with fecal pollution were detected sporadically or not at all (toxigenic V. cholerae, 0%; Salmonella, 1.5%; NoV, 3.9%; HAV, 4.4%). While seasonal prevalences of NoV and HAV were generally greater in oysters harvested from December to March, the low detection frequency obscured any apparent seasonal effects. Overall, there was no relationship between the levels of indicator microorganisms and the presence of enteric viruses. These data provide a baseline that can be used to further validate risk assessment predictions, determine the effectiveness of new control measures, and compare the level of protection provided by the U.S. shellfish sanitation system to those in other countries.
The role of molluscan shellfish as a vehicle for transmission of bacterial and viral pathogens to humans is well documented. A century ago, most reported illnesses in the United States were associated with bacterial pathogens from fecal pollution, and the primary causative agent was Salmonella spp. (68, 72). Outbreaks became so prevalent that the National Shellfish Sanitation Program (NSSP) was started in the 1920s. The premise of this program was that shellfish safety was dependent in part on the bacteriological quality of the overlying waters of shellfish growing areas. A water-sampling-based program was developed around indicator bacteria known as coliforms (68). Another important aspect of this program was implementation of sanitary surveys to identify point sources of pollution and to track the distribution of the effluent from these sources. This has been a very successful program, as illnesses from typhoid fever drastically declined within a decade and continue to be very rare in the United States (68).
In the 1970s, new threats that were primarily associated with bacterial pathogens in the genus Vibrio emerged (11, 12, 48). Three Vibrio spp., V. cholerae, V. parahaemolyticus, and V. vulnificus, are the most important vibrios associated with human illness (62). Vibrio spp. are naturally occurring estuarine bacteria, and their distribution and abundance in shellfish are typically unrelated to fecal pollution (35, 49, 65), with the possible exception of toxigenic V. cholerae O1, which is excreted in large numbers in the feces of infected individuals and survives well in sewage (62). Vibrios tend to be most abundant during warmer periods of the year and in growing areas with moderate salinities, depending on the species (62). Vibrios are among the fastest-growing bacteria in nature; they multiply readily in oysters postharvest and likely do so in other molluscan shellfish as well when not promptly refrigerated (26, 44, 57).
Illnesses due to V. vulnificus are the leading cause of mortality associated with seafood consumption in the United States (56). While V. vulnificus has a case fatality rate of approximately 50%, which is the highest for any food-borne pathogen, it causes a relatively rare infection estimated by the CDC to result in 100 food-borne cases per year (56). Nearly all cases are sporadic in nature and linked to consumption of raw oysters harvested on the Gulf Coast from April through November (73). V. vulnificus illnesses frequently present as primary septicemia and occur among individuals with preexisting chronic illnesses, such as liver disease, or who are otherwise immunocompromised (12, 62). Concomitant with the emergence of V. vulnificus, sporadic cases of infection with toxigenic V. cholerae O1 and nontoxigenic V. cholerae non-O1 strains also began to be reported in the 1970s, and these were also frequently associated with consumption of raw oysters harvested from areas along the Gulf of Mexico (11).
V. parahaemolyticus causes moderate to mild gastroenteritis and tends to be distributed more widely than V. cholerae and V. vulnificus, as it occurs in cooler and more saline waters (62). When V. parahaemolyticus first emerged in the United States, outbreaks were typically associated with recontamination of cooked seafood, primarily crustacean shellfish (32). In the 1990s, molluscan shellfish, especially raw oysters, became the primary vehicle of infection, accounting for approximately two-thirds of infections in the United States (32). In the past decade, V. parahaemolyticus has become the leading cause of bacterial gastroenteritis associated with seafood consumption in the United States. Oyster-associated outbreaks have occurred on the Atlantic, Gulf, and Pacific Coasts, including Alaska (an outbreak occurring there in 2004) (55, 56). A pandemic clonal complex that emerged in Asia in the mid-1990s and strains native to North America have been associated with these outbreaks (23, 25, 31, 43, 54). A risk assessment released by the U.S. Food and Drug Administration (FDA) in 2005 attributed most of the risk of V. parahaemolyticus illness to postharvest growth in oysters (3).
Although V. parahaemolyticus and V. vulnificus account for a large proportion of shellfish-associated illnesses since the 1970s, previous illnesses associated with shellfish consumption in the United States were due to unknown etiological agents (68). It has since become apparent that many of these illnesses can be attributed to norovirus (NoV) infections (68, 76). NoVs are the leading cause of nonbacterial illnesses in shellfish consumers, and these diseases occur worldwide (13, 41). NoV illnesses due to shellfish consumption are seasonally related, occurring with higher frequency from late fall through winter (68). This seasonal occurrence of shellfish-associated NoV outbreaks can be attributed to several factors, including increased stability of viruses at low water temperatures, reduced solar inactivation (17), and selective bioaccumulation of these pathogens by the shellfish (16, 40). Typical symptoms of a NoV infection following a mean incubation time of 24 h include diarrhea, vomiting, fever, headache, and muscle aches (15, 22). NoV illnesses are generally self-limiting, with symptoms lasting between 24 to 48 h, and fatalities are rare. Human strains cluster within genogroups I (GI), II (GII), and IV, and these viruses are excreted at high densities in feces (up to 1 × 1012/gram) of infected individuals (4, 67).
NoV and other enteric viruses are more resistant to wastewater treatment and are more environmentally stable than the fecal indicator bacteria (5, 17, 66, 78). Enteric viruses are also concentrated to higher levels and persist longer in shellfish than the indicator bacteria. These differences in viral stability and shellfish interaction relative to indicator bacteria are likely reasons why the indicator microorganisms do not readily index the presence of viruses in marine waters and shellfish and may contribute, in part, to oyster-associated viral outbreaks in the United States (74).
Exposure to V. parahaemolyticus and V. vulnificus among U.S. consumers of raw oysters was investigated in a market study conducted by the FDA in 1998-1999 (29). This study concluded that the highest levels of V. parahaemolyticus and V. vulnificus were observed in oysters harvested from the Gulf Coast, followed by those from the Mid-Atlantic Coast. Generally, the highest levels of both organisms were observed in the summer, and the lowest levels were observed in the winter; intermediate levels were observed in the spring and/or fall, depending on the pathogen. As measured by DNA probe colony hybridization procedures targeting the V. parahaemolyticus thermolabile hemolysin (tlh) and V. vulnificus hemolysin (vvhA) genes (34), levels of these pathogens in market oysters were one to two logs higher than levels reported in previous studies of oysters at harvest and were in close agreement with levels at the point of consumption as predicted by risk assessments (29, 39).
The 1998-1999 market survey measured total V. parahaemolyticus and V. vulnificus levels only, as the DNA probe hybridization methods used lacked the sensitivity and specificity to quantify levels of pathogenic V. parahaemolyticus by the presence of either the thermostable direct hemolysin (tdh) or the thermostability-related hemolysin (trh) gene (29, 34). A number of studies suggest that the proportion of the V. parahaemolyticus population that is pathogenic may vary by season and region and that, consequently, total V. parahaemolyticus levels may not be the best indicator of risk of infection (36, 50, 55). Moreover, real-time PCR methods have been developed and used in a most-probable-number (MPN) format to greatly improve sensitivity and specificity of detection of pathogenic V. parahaemolyticus (64).
Real-time PCR methods for the detection of Salmonella (9), toxigenic V. cholerae (8), NoV (19), and hepatitis A virus (HAV) (42) have also been developed by the FDA. Thus, given the availability of improved methods, a study to determine the levels of important bacterial and viral pathogens associated with oysters was conducted. Additionally, standard culture methods were used to enumerate indicator organisms, including fecal coliforms, Escherichia coli, and male-specific bacteriophage (MSB). This paper describes the results of a year-long nationwide survey of important bacterial and viral pathogens and discusses its comparison to previous surveys in the United States and the use of these data in determining the equivalency of shellfish safety programs in other countries.
MATERIALS AND METHODS
Sample collection.
Two independent samples of market oysters (36 live animals), primarily from retail establishments, were collected twice each month in each of nine states (Washington, Louisiana, Alabama, Florida, South Carolina, Virginia, Rhode Island, Illinois, and Colorado) during 2007. Samples were shipped refrigerated overnight to five FDA laboratories (New York, NY; Atlanta, GA; Bothell, WA; Irvine, CA; and Dauphin Island, AL) on a rotating basis. Data loggers (ACR Systems, Inc., the Data Logger Store, Contoocook, NH) were included in each shipping container to ensure that samples analyzed for pathogens were maintained at temperatures generally between 2 to 10°C to prevent growth of Vibrio spp. and to maintain stability of enteric bacteria and viral populations. The average internal temperature of the oysters upon receipt and analysis was 7.5 ± 2.8°C. Eighty-two percent of the 397 samples were analyzed the day after collection at retail. Due to shipping delays, the balance was analyzed within 48 h after verifying that transport temperatures were in the acceptable range of 0 to 13°C. The number of samples analyzed for each pathogen or indicator varied due to inadequate sample volumes resulting from small or dead oysters and laboratory accidents.
Indicator organisms.
Fecal coliform and E. coli densities were determined using a conventional five-tube, three-dilution MPN procedure with minimal modifications to the FDA Bacteriological Analytical Manual (BAM) and American Public Health Association (APHA) recommended procedures for the examination of shellfish (1, 34). Modifications to this procedure included blending of the shellfish meats and liquors without dilution buffer; this was necessary due to the multiple microbial analyses performed on each shellfish sample. Following homogenization, a 1:10 dilution of homogenate (10 g) was prepared with phosphate-buffered solution (PBS) as previously described (1). Ten milliliters of this dilution, a 1-g equivalent, was transferred to five tubes of 10 ml of double-strength lauryl tryptose broth (LST; Difco Laboratories, Sparks, MD). One-milliliter aliquots (0.1-g equivalent) were also transferred to five tubes of single-strength LST, while five 1-ml aliquots of a 1:100 dilution were also transferred to single-strength LST. Presumptive positive tubes were confirmed for fecal coliforms and E. coli by using EC-MUG (Difco, Sparks, MD) medium (69).
Standard plate count densities were determined using the APHA recommended procedures for the examination of shellfish (1). Necessary dilutions for this test were prepared in PBS by using the same 1:10 shellfish homogenate dilution used for fecal coliforms and E. coli determinations.
MSB densities were determined by using a modified double-agar-overlay method described by Cabelli (21); the E. coli strain HS(pFamp)R (ATCC 700891) was utilized as the suitable bacterial host strain (21, 33, 38).
V. parahaemolyticus. (i) Colony hybridization.
Samples were analyzed for total and pathogenic V. parahaemolyticus bacteria by using an MPN format as described in the FDA's BAM (34), with the following modifications. The three-tube MPN dilution series contained 10-g through 10−6-g dilutions. The 10-g and 1-g aliquots were prepared by adding 10 g or 1 g of homogenate to 90 ml or 9 ml of alkaline peptone water (APW; 1% NaCl, 1% peptone, pH 8.5 ± 2), respectively. After overnight incubation (18 to 24 h) at 35°C, a 1-ml aliquot was removed for testing by real-time PCR as described below, and thiosulfate-citrate-bile salts-sucrose (TCBS) agar (Difco Laboratories, Sparks, MD) plates were streaked. After overnight incubation, three typical colonies, or one atypical colony (if no typical colonies were available), were inoculated into individual wells of a 96-well plate containing 100 μl of APW. After 4 to 6 h of incubation at 35 to 37°C, the growth was transferred to triplicate T1N3 (1% tryptone, 3% NaCl, 2% agar, pH 7.6) plates, using a 48-prong replicator (Bokel, Feasterville, PA). Colony lifts and DNA probe hybridization were performed as previously described for tlh (34). Any tube which yielded at least one probe-positive colony was considered positive for that gene target.
(ii) Real-time PCR.
Samples for real-time PCR (1-ml aliquots of overnight enrichments), including V. parahaemolyticus, V. cholerae, and Salmonella, were heated to 100°C for 10 min and then immediately plunged into ice (10). Samples were spun at 12,000 × g for 2 min to pellet debris, and 2 μl of the supernatant was used as the template in the PCR. Reaction setup and cycling parameters for V. parahaemolyticus were as previously described (64) for simultaneous detection of the tlh, tdh, and trh genes on the SmartCycler II system (Cepheid, Sunnyvale, CA). Primer and probe sequences are given in Table 1 .
TABLE 1.
Primer and probe sequences used for detection of bacterial and viral pathogens
| Target organism and gene | Primer | Sequence (5′→3′)a | Reference |
|---|---|---|---|
| V. parahaemolyticus | |||
| tlh | Forward | ACTCAACACAAGAAGAGATCGACAA | Nordstrom et al., 2007 (64) |
| Reverse | GATGAGCGGTTGATGTCCAAA | Nordstrom et al., 2007 (64) | |
| Probe | TxRed-CGCTCGCGTTCACGAAACCGT-BHQ2 | Nordstrom et al., 2007 (64) | |
| tdh | Forward | TCCCTTTTCCTGCCCCC | Nordstrom et al., 2007 (64) |
| Reverse | CGCTGCCATTGTATAGTCTTTATC | Nordstrom et al., 2007 (64) | |
| Probe | 6FAM-TGACATCCTACATGACTGTG-MGBNFQ | Nordstrom et al., 2007 (64) | |
| trh | Forward | TTGCTTTCAGTTTGCTATTGGCT | Nordstrom et al., 2007 (64) |
| Reverse | TGTTTACCGTCATATAGGCGCTT | Nordstrom et al., 2007 (64) | |
| Probe | TET-AGAAATACAACAATCAAAACTGA-MGBNFQ | Nordstrom et al., 2007 (64) | |
| Salmonella spp. (invA) | Forward | CAACGTTTCCTGCGGTACTGT | This study |
| Reverse | CCCGAACGTGGCGATAATT | This study | |
| Probe | TxRed-CTCTTTCGTCTGGCATTATCGATCAGTACCA-BHQ2 | This study | |
| Toxigenic V. cholerae (ctx) | Forward | TTTGTTAGGCACGATGATGGAT | Blackstone et al., 2007 (8) |
| Reverse | ACCAGACAATATAGTTTGACCCACTAAG | Blackstone et al., 2007 (8) | |
| Probe | 6FAM-TGTTTCCACCTCAATTAGTTTGAGAAGTGCCC-BHQ1 | Blackstone et al., 2007 (8) | |
| Norovirus GI | Forward | CGYTGGATGCGNTTYCATGA | Kageyama et al., 2003 (47) |
| Reverse | CTTAGACGCCATCATCATTYAC | Kageyama et al., 2003 (47) | |
| Probe | Cy5-AGATYGCGATCYCCTGTCCA-IBRQ | Kageyama et al., 2003 (47) | |
| Probe 2 | Cy5-AGATCGCGGTCTCCTGTCCA-IBRQ | Kageyama et al., 2003 (47) | |
| Forward | TGGACICGYGGICCYAAYCA | Beuret et al., 2003 (6) | |
| Reverse | GAASCGCATCCARCGGAACAT | Beuret et al., 2003 (6) | |
| Norovirus GII | Forward | CARGARBCNATGTTYAGRTGGATGAG | Kageyama et al., 2003 (47) |
| Reverse | TCGACGCCATCTTCATTCACA | Kageyama et al., 2003 (47) | |
| Probe | Cy3-TGGGAGGGCGATCGCAATCT-IBRQ | Kageyama et al., 2003 (47) | |
| Forward | TGGACICGYGGICCYAAYCA | Beuret, et al., 2003 (6) | |
| Reverse | GAAYCTCATCCAYCTGAACAT | Beuret et al., 2003 (6) | |
| Hepatitis A virus | Forward | ATAGGGTAACAGCGGCGGATAT | Gardner et al., 2003 (42) |
| Reverse | AATGCATCCACTGGATGAG | Gardner et al., 2003 (42) | |
| Probe | Cy5-AGACAAAAACCATTCAACGCCGGAGG-IBRQ | Gardner et al., 2003 (42) | |
| Calicivirus SMSV-17 | Forward | AATCAATTGATATCCTCAGACACTTCAC | Burkhardt et al., 2004 (19) |
| Reverse | CCGCAACCTTGAAAGCCACAC | Burkhardt et al., 2004 (19) | |
| Probe | Cy5-AATTGTTGATTCGGCCTGTGCACCAC-IBRQ | Burkhardt et al., 2004 (19) | |
| Internal control | 46Fb | GACATCGATATGGGTGCCG | Nordstrom et al., 2007 (64) |
| 186Rc | CGAGACGATGCAGCCATTC | Nordstrom et al., 2007 (64) | |
| 185Rd | GAGACGATGCAGCCATTCG | Blackstone et al., 2007 (8) | |
| 194Re | AATATTCGCGAGACGATGCAG | This study | |
| Probef | Cy5-TCTCATGCGTCTCCCTGGTGAATGTG-BHQ2 | Nordstrom et al., 2007 (64) | |
| Probeg | TxRed-TCTCATGCGTCTCCCTGGTGAATGTG-IBRQ | This study |
All primers and probes for this study were purchased from Integrated DNA Technologies (Coralville, IA). TxRed, Texas Red-X N-hydroxysuccinimide ester; BHQ2, black hole quencher-2; 6FAM, 6-carboxyfluorescein; MGBNFQ, TaqMan minor groove binder/nonfluorescent quencher; TET, tetrachlorofluorescein; BHQ1, black hole quencher-1; Cy5, cyanine 5; IBRQ, Iowa black RQ; Cy3, cyanine 3.
IC primer 46F used for amplification of the internal control target DNA or RNA in all assays.
IC primer 186R used for amplification of the internal control target DNA in the V. parahaemolyticus assay.
IC primer 185R used for amplification of the internal control target DNA in the Salmonella and toxigenic V. cholerae assays.
IC primer 194R used for amplification of the internal control target RNA in the norovirus, HAV, and SMSV-17 assays.
Cy5-labeled IC probe used in the V. parahaemolyticus, Salmonella, and toxigenic V. cholerae assays.
TxRed-labeled IC probe used in the norovirus, HAV, and SMSV-17 assays.
V. vulnificus.
Samples were analyzed for V. vulnificus by using the same MPN series as for V. parahaemolyticus. After overnight enrichment of the APW tubes, growth from each turbid tube was streaked to modified cellobiose-polymyxin B-colistin (mCPC) agar (34) plates. The mCPC plates were incubated overnight, and three typical colonies, or one atypical colony (if no typical colonies were available), were picked and transferred into a 96-well plate containing APW. After 4 to 6 h of incubation, growth was replicated on VVA (34). Colony lifts and DNA probe hybridization were performed as previously described for the vvhA gene (34). Any tube which yielded at least one probe-positive colony was considered positive for V. vulnificus.
Toxigenic V. cholerae.
Samples were analyzed for the presence of toxigenic V. cholerae as described in the BAM (34), with the following modifications. A 25-g portion of homogenate was added to 2,475-ml of APW in a 4-liter flask. The mixture was incubated at 42°C overnight (18 to 24 h) in an air incubator and was prepared for the real-time PCR template as described above. Reaction setup and cycling parameters were as previously described (8) for detection of the ctxA gene. Primer and probe sequences are given in Table 1. If a sample was PCR positive for toxigenic V. cholerae, culture isolation was attempted as described in the BAM (34), with a maximum of 50 colonies tested per sample.
Salmonella spp.
Samples were analyzed for the presence of Salmonella as described in the BAM (2), with the following modifications. A 25-g portion of homogenate was added to 225 ml of lactose broth and incubated overnight (24 ± 2 h) at 35°C. A 0.1-ml portion and a 1-ml portion were transferred to 10 ml of Rappaport-Vassiladis (RV) medium (Difco) and 10 ml of tetrathionate (TT) broth (Difco), respectively. The RV and TT broths were incubated at 42°C and 43°C, respectively, for 24 ± 2 h. Real-time PCR templates were prepared from 1-ml aliquots of lactose broth and RV and TT enrichments as described above. Reaction conditions and cycling parameters are given below. Primer and probe sequences are given in Table 1. If Salmonella was detected in any of the enrichment broths, culture isolation was attempted as described in the BAM (2). Real-time PCR was used to confirm suspect isolates as Salmonella, and serology was performed to confirm isolates were different from the laboratory control strains.
The real-time PCR cycling protocol and reaction component concentrations were optimized for detection of the invA gene and included an internal amplification control (IAC). The 25-μl reaction volume contained the following (final concentrations): 1× PCR buffer (Invitrogen, Carlsbad, CA), 5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP; Roche, Indianapolis, IN), 1.25 U Platinum Taq polymerase (Invitrogen), 200 nM each forward and reverse invA and IAC primer, 150 nM of a 5′ nuclease probe for invA, and 200 nM 5′ nuclease probe for IAC. Real-time PCR cycling was run using the SmartCycler II system, utilizing the following cycling parameters: 95°C for 2 min followed by 45 cycles of 94°C for 10 s, 63°C for 15 s, and 72°C for 15 s. Default analysis parameters were used except that the manual threshold fluorescence units setting was adjusted to 15. This real-time PCR assay has been previously shown to have a limit of detection of <10 targets per reaction (9). The specificity for Salmonella was 100% when tested against 86 Salmonella isolates (9) and 10 near neighbors, including Enterobacter aerogenes, E. cloacae, Escherichia coli, Edwardsiella tarda, Citrobacter freundii, Morganella morganii, Proteus mirabilis, Providencia stuartii, Serratia liquefaciens, and Pseudomonas aeruginosa (unpublished data).
Enteric viruses. (i) Virus concentration and RNA extraction.
The extraction method utilized was slightly modified from previous protocols (59, 75). Five to 10 whole oysters were washed and shucked, and the digestive diverticula from 5 to 10 oysters were removed to obtain a total of 25 g of sample. An aliquot of nonhuman calicivirus (San Miguel sea lion virus, serogroup 17 [SMSV-17]) was added as an extraction control prior to homogenization of the digestive diverticula with 7× H2O (7 volumes of 25 g). A total of 105 g of the homogenate was added into a tared 250-ml centrifuge bottle. Conductivity was measured using a 4-ml aliquot of the homogenate (model ARH1; Myron L Company, Carlsbad, CA), and the 105-g homogenate was adjusted to less than 2,000 μS. Viruses were adsorbed onto the particulate by adjusting the pH to 4.8 ± 0.3 and then shaken on an orbital shaker (200 rpm) for 15 min at room temperature. After the absorption step, samples were then centrifuged for 20 min at 2,000 × g at 4°C; following centrifugation, the supernatant was discarded. The pellet was eluted with 105 ml of 0.75 M glycine-0.15 M NaCl, and the pH was adjusted to 7.5 ± 0.2, followed by an additional elution with 52.5 ml of 0.5 M threonine-0.15 M NaCl. The eluates were combined and precipitated with 8% polyethylene glycol (PEG)-0.3 M NaCl and incubated for 3 h or overnight at 4°C. Precipitates were spun, and the pellet was resuspended in 12 ml of tissue culture-grade phosphate-buffered saline (8.0 g NaCl, 0.2 g KCl, 0.12 g KH2PO4, 0.91 g Na2PO4 per liter). Samples were then extracted first with 12 ml of chloroform by vortexing for 1 min and then centrifuged at 1,700 × g for 30 min at 4°C. The upper aqueous phase was transferred to a clean, 50-ml conical tube. The remaining portion was extracted with 6 ml of 0.5 M threonine-0.15 M NaCl and centrifuged as previously described. The two aqueous phases were combined and precipitated with 8% PEG-0.3 M NaCl for 3 h or overnight at 4°C. Following precipitation, samples were centrifuged at 20,800 × g for 15 min at 4°C and pellets were extracted for RNA, utilizing 6 M guanidium isothiocyanate as a lysis solution and the RNeasy minikit (Qiagen, Valencia, CA). Extracted RNA was tested by real-time PCR as described below.
(ii) Real-time RT-PCR (norovirus).
Positive controls used for NoV GI and GII were in vitro RNA transcripts of sequences cloned from positive clinical samples previously identified as NoV (20). Primers and probes for NoV GI and GII targeted the most conserved region of the open reading frame 1 (ORF1)-ORF2 junction (47). Real-time reverse transcription (RT)-PCR for detection of NoV GI and NoV GII with an RNA IAC was performed in a 25-μl reaction volume by using a one-step RT-PCR kit (Qiagen). The primer concentrations for the NoV targets were 300 nM each, and the concentrations for the IAC primers (46F and 194R) were 75 nM each. The 5′ nuclease probe concentrations for NoV and the IAC target were 100 and 150 nM each, respectively. The final concentration of MgCl2 in the real-time RT-PCR was 4 mM. Thermal cycling was run using the SmartCycler II system with the following conditions: 50°C for 3,000 s and 95°C for 900 s followed by 50 cycles of 95°C for 10 s, 53°C for 25 s, and 62°C for 70 s. Fluorescence was read at the end of the 62°C elongation step. Default analysis parameters were used, except that the manual threshold fluorescence units were set to 10. Samples positive with the initial primer and probe sets for NoV GI and/or NoV GII (47) were subjected to a secondary detection assay. Amplification of the original RNA extract was performed with primers from the B region (Table 1) by conventional RT-PCR as previously described (7). The primary purpose of this secondary assay was to obtain less-conserved amplicons for sequencing.
(iii) HAV.
The positive control used for HAV was the vaccine strain HM175/18f (subgenotype 1B), propagated in-house by utilizing the FRhK-4 cell line. Real-time RT-PCR for the detection of HAV with an RNA IAC was performed in a 25-μl reaction volume by using a one-step RT-PCR kit (Qiagen). The primer concentrations for HAV and the IAC were 300 nM and 75 nM, respectively, while the 5′ nuclease probe concentrations for HAV and the IAC targets were 200 and 150 mM, respectively. The final concentration of MgCl2 in the RT-PCR was 4 mM. Thermal cycling was run using the SmartCycler II system with the following conditions: 50°C for 3,000 s and 95°C for 900 s followed by 50 cycles of 95°C for 10 s, 53°C for 25 s, and 64°C for 70 s. Fluorescence was read at the end of the 64°C elongation step. Default analysis parameters were used except that the manual threshold fluorescence units were set to 10. Samples showing amplification with the initial primers/probe set were also subjected to a secondary detection assay for the same purpose as described for NoV above by using CDC nested primers and conventional RT-PCR as previously described (74).
(iv) SMSV-17.
The positive control used for San Miguel sea lion virus 17 (SMSV-17) was propagated in-house by utilizing the Vero cell line. Real-time RT-PCR was utilized for the detection of SMSV-17 (the extraction control virus) with an RNA IAC in a 25-μl reaction volume by using a one-step RT-PCR kit (Qiagen). The primer concentrations for SMSV-17 and the IAC were 300 nM and 75 nM, respectively, while the 5′ nuclease probe concentrations for SMSV-17 and the IAC targets were 200 and 150 mM, respectively. The final concentration of MgCl2 in the RT-PCR was 7 mM. Thermal cycling was run using the SmartCycler II system under the following conditions: 50°C for 3,000 s and 95°C for 900 s followed by 45 cycles of 94°C for 10 s, 62°C for 20 s, and 72°C for 40 s. Fluorescence was read at the end of the 72°C elongation step. Default analysis parameters were used except that the manual threshold fluorescence units were set to 10.
Statistical analyses.
Regional (Gulf, Mid-Atlantic [including New Jersey and states to the south], North Atlantic [including New York and states to the north], and Pacific) and seasonal (winter, spring, summer, and fall) estimates of mean levels of Vibrio sp. populations (with tlh, tdh, trh, and vvh) and indicator organisms (fecal coliforms, E. coli, male-specific coliphage [MSC], and aerobic plate counts [APC]) were determined by fitting a Tobit or censored regression for each pathogen endpoint. Seasons were defined as 3-month periods when oyster samples were collected from market establishments, beginning on 1 January (i.e., winter was 1 January through 31 March). Trends in prevalence of viral pathogens were evaluated based on the month of sample collection. The censored regression (45) approach was adopted for quantitative endpoints to appropriately adjust estimates for regional and seasonal differences in the frequency of outcomes above or below specified limits of detection (LOD), corresponding to indeterminate MPN outcomes or observation of a zero plate count. For the Vibrio sp. endpoints, per-sample density estimates were determined using a 3-tube, 1-dilution MPN analysis as previously described (29). Density estimates for indicator organisms were determined using a 5-tube, 3-dilution MPN analysis or plate counts as previously indicated. All observations were log transformed prior to regression analysis and for each regression analysis of the data on each pathogen. A common standard deviation was assumed for the distributions within each region and season category. The significances of differences between geometric means across regional/seasonal categories and their 95% confidence intervals were determined using the large-sample normal approximation, based on the asymptotic variance-covariance matrix corresponding to estimated mean levels. Correlations between the presence of virus (NoV and HAV) and indicator organisms were evaluated using Kendall's tau. Significance of regional differences in the prevalences of viral pathogens was determined by Fisher's exact test. For all bacterial and viral pathogen endpoints, statistical analyses included laboratory as a presumptive categorical factor to test for interlaboratory differences in the determination of abundance or prevalence. A P value of 0.05 was considered the minimum level for significance.
RESULTS
Shell oysters representing 397 lots were collected at retail and wholesale establishments from January through December 2007 in all participating states. Sample collections from Rhode Island (January and first half of February) and South Carolina (February, March, and first half of April) were incomplete, and only on occasion were samples missed from other collecting states. Sixty-seven lots were shipped to the Pacific Northwest Regional Laboratory, 64 lots were shipped to the Pacific Southwest Regional Laboratory, 68 lots were shipped to the New York Regional Laboratory, 65 lots were shipped to the Southeast Regional Laboratory, and 133 lots were shipped to the Gulf Coast Seafood Laboratory for analysis. The internal oyster temperatures of most shellfish lots (85%) were 10°C or below upon receipt and throughout shipment, while the temperatures for 15% of the lots were between 10 and 15°C; only two samples experienced temperatures of >15°C. Temperatures determined by the data loggers (data not shown) indicated that negligible growth of target organisms would likely have occurred in any of the samples during shipment; all 397 samples were analyzed for target organisms as sample volume permitted due to small or dead oysters and/or laboratory accidents.
The market establishments were grouped into three types: (i) restaurants and “raw bars” where shellfish are opened onsite for raw consumption, (ii) seafood markets, including grocery stores, which offer shellstock oysters for retail sale, and (iii) wholesale dealers, who sell oysters in the shell to the above establishments. Oysters were collected at 258 different establishments: 39% were restaurants or raw bars, 59% were seafood markets, and 2% were wholesale dealers. There was no apparent difference between levels of pathogens or indicator organisms for different establishment types (data not shown).
Table 2 summarizes the numbers of oyster lots according to the state of collection and the state where oysters were harvested. The number of lots sampled in each collecting state ranged from 34 to 48. Colorado and Illinois received oysters from 11 and 12 different producing states, respectively, which were more than the corresponding numbers for coastal collecting states. Oysters collected in producing states were usually harvested locally or within the region.
TABLE 2.
Summary by harvest state where retail oyster samples were collected
| Harvest state or country | No. of samples | No. of samples collected in each state |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rhode Island (North Atlantic) | Mid-Atlantic |
Gulf |
Washington (Pacific) | Inland |
||||||
| Virginia | South Carolina | Alabama | Florida | Louisiana | Colorado | Illinois | ||||
| North Atlantic | 53 | 40 | 3 | 7 | 3 | |||||
| Connecticut | 11 | 8 | 2 | 1 | ||||||
| Maine | 4 | 4 | ||||||||
| Massachusetts | 15 | 6 | 1 | 7 | 1 | |||||
| New York | 6 | 6 | ||||||||
| Rhode Island | 17 | 16 | 1 | |||||||
| Mid-Atlantic | 100 | 1 | 35 | 4 | 7 | 2 | 17 | 34 | ||
| Delaware | 14 | 9 | 5 | |||||||
| Maryland | 15 | 8 | 1 | 2 | 4 | |||||
| New Jersey | 19 | 2 | 17 | |||||||
| South Carolina | 3 | 3 | ||||||||
| Virginia | 49 | 1 | 27 | 1 | 7 | 1 | 4 | 8 | ||
| Gulf | 175 | 12 | 29 | 34 | 40 | 48 | 6 | 6 | ||
| Alabama | 4 | 4 | ||||||||
| Florida | 52 | 7 | 19 | 23 | 2 | 1 | ||||
| Louisiana | 86 | 7 | 14 | 8 | 9 | 43 | 3 | 2 | ||
| Texas | 33 | 5 | 4 | 7 | 8 | 5 | 1 | 3 | ||
| Pacific | 60 | 1 | 1 | 43 | 13 | 2 | ||||
| California | 1 | 1 | ||||||||
| Oregon | 1 | 1 | ||||||||
| Washington | 58 | 1 | 43 | 12 | 2 | |||||
| Canada | 6 | 1 | 3 | 2 | ||||||
| Unknown | 3 | 1 | 1 | 1 | ||||||
| Total | 397 | 41 | 47 | 34 | 44 | 47 | 48 | 43 | 46 | 47 |
The temperatures under which oysters were stored at retail were determined by measuring the ambient air temperature in the cooler. Although 29% (105 of 365) of the refrigerator temperatures recorded exceeded 41°F (5°C), the average temperature of the refrigerators in all states met the requirement of the FDA Food Code (<41°F; data not shown). The internal temperatures of the oysters were generally above the refrigerator temperature, probably because temperature checks of shellstock oysters were done after they were removed from the refrigerator. About 15% of the oyster lots exceeded 10°C (minimum growth temperature for V. parahaemolyticus [57]), and 0.3% exceeded 13°C (minimum growth temperature for V. vulnificus [27]); the highest temperature observed was 17°C. Under the temperature ranges observed in these markets at the time of sample collection, only minimal growth of target organisms would be expected in those samples exceeding minimum growth temperatures. The mean storage time (length of time between oyster harvest and sample collection) was 8.9 days. About 11% (41 of 382) of the lots offered for retail sale were at >14 days postharvest.
Tables 3 and 4 show the percentages of lots from each harvest state within eight density ranges of V. vulnificus and V. parahaemolyticus, respectively (MPN counts were determined by DNA colony hybridization of selected isolates). Approximately half of the lots harvested from states in the North Atlantic (43.4%) and Pacific (55%) Coasts had V. vulnificus densities below the detectable level of 0.03 MPN/g, with few (3.8% and 6.7%, respectively) of the lots exceeding 100 MPN/g. V. parahaemolyticus densities were greater than those of V. vulnificus in lots from the same areas; 1.9% and 3.4% of lots from the North Atlantic and Pacific Coasts, respectively, exceeded 1,000 MPN/g. Over all regions and seasons, >15% and >5% of lots exceeded 10,000 MPN/g and 100,000 MPN/g, respectively, for either pathogen. All samples exceeding 10,000 MPN/g for either Vibrio sp., except for a single sample from Washington (V. parahaemolyticus), were harvested in either the Mid-Atlantic (13% for either pathogen) or Gulf Coast (29.5% for V. vulnificus and 25.3% for V. parahaemolyticus) states, with the highest proportion of these being harvested from Louisiana. Approximately 44% and 38% of samples harvested from Louisiana exceeded 10,000 MPN/g for V. vulnificus and V. parahaemolyticus, respectively.
TABLE 3.
Abundance of Vibrio vulnificus in oysters at retaila
| Harvest state or country | No. of samples | % of samples with V. vulnificus densities with the indicated MPN/g |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| None detected | ≤1 | >1-10 | >10-102 | >102-103 | >103-104 | >104-105 | >105 | ||
| North Atlantic | 53 | 43.4 | 22.6 | 9.4 | 20.8 | 3.8 | |||
| Connecticut | 11 | 36.4 | 9.1 | 9.1 | 36.4 | 9.1 | |||
| Maine | 4 | 75.0 | 25.0 | ||||||
| Massachusetts | 15 | 60.0 | 13.3 | 13.3 | 13.3 | ||||
| New York | 6 | 66.7 | 33.3 | ||||||
| Rhode Island | 17 | 41.2 | 29.4 | 11.8 | 11.8 | 5.9 | |||
| Mid-Atlantic | 100 | 22.0 | 15.0 | 11.0 | 11.0 | 13.0 | 15.0 | 11.0 | 2.0 |
| Delaware | 14 | 21.4 | 7.1 | 14.3 | 21.4 | 7.1 | 21.4 | 7.1 | |
| Maryland | 15 | 20.0 | 46.7 | 20.0 | 6.7 | 6.7 | |||
| New Jersey | 19 | 10.5 | 15.8 | 5.3 | 10.5 | 5.3 | 31.6 | 15.8 | 5.3 |
| South Carolina | 3 | 66.7 | 33.3 | ||||||
| Virginia | 49 | 24.5 | 8.2 | 14.3 | 10.2 | 16.3 | 16.3 | 10.2 | |
| Gulf | 173 | 12.1 | 9.3 | 8.7 | 12.1 | 13.3 | 15.0 | 19.1 | 10.4 |
| Alabama | 4 | 25.0 | 50.0 | 25.0 | |||||
| Florida | 52 | 11.5 | 13.5 | 15.4 | 19.2 | 7.7 | 21.2 | 9.6 | 1.9 |
| Louisiana | 84 | 7.1 | 6.0 | 4.8 | 8.3 | 16.7 | 13.1 | 26.2 | 17.9 |
| Texas | 33 | 27.3 | 12.1 | 9.1 | 9.1 | 9.1 | 9.1 | 18.2 | 6.1 |
| Pacific | 60 | 55.0 | 25.0 | 6.7 | 6.7 | 6.7 | |||
| California | 1 | 100.0 | |||||||
| Oregon | 1 | 100.0 | |||||||
| Washington | 58 | 53.5 | 25.9 | 6.9 | 6.9 | 6.9 | |||
| Canada | 6 | 50.0 | 16.7 | 33.3 | |||||
| Unknown | 2 | 50.0 | 50.0 | ||||||
| Total | 394 | 26.1 | 14.7 | 9.1 | 12.2 | 10.7 | 10.4 | 11.7 | 5.1 |
Data are shown by harvest state and region and are based on DNA probe colony hybridization results.
TABLE 4.
Abundance of Vibrio parahaemolyticus in oysters at retaila
| Harvest state or country | No. of samples | % of samples with V. parahaemolyticus densities with the indicated MPN/g |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| None detected | ≤1 | >1-10 | >10-102 | >102-103 | >103-104 | >104-105 | >105 | ||
| North Atlantic | 53 | 50.9 | 5.7 | 15.1 | 18.9 | 7.6 | 1.9 | ||
| Connecticut | 11 | 36.4 | 18.2 | 36.4 | 9.1 | ||||
| Maine | 4 | 50.0 | 25.0 | 25.0 | |||||
| Massachusetts | 15 | 60.0 | 6.7 | 20.0 | 13.3 | ||||
| New York | 6 | 33.3 | 16.7 | 16.7 | 33.3 | ||||
| Rhode Island | 17 | 58.8 | 5.9 | 11.8 | 11.8 | 5.9 | 5.9 | ||
| Mid-Atlantic | 100 | 21.0 | 14.0 | 11.0 | 9.0 | 15.0 | 17.0 | 6.0 | 7.0 |
| Delaware | 14 | 21.4 | 14.3 | 7.1 | 14.3 | 21.4 | 14.3 | 7.1 | |
| Maryland | 15 | 6.7 | 53.3 | 13.3 | 13.3 | 6.7 | 6.7 | ||
| New Jersey | 19 | 21.1 | 10.5 | 5.3 | 10.5 | 31.6 | 15.8 | 5.3 | |
| South Carolina | 3 | 33.3 | 33.3 | 33.3 | |||||
| Virginia | 49 | 26.5 | 8.2 | 12.2 | 8.2 | 18.4 | 14.3 | 4.1 | 8.2 |
| Gulf | 174 | 5.8 | 9.8 | 9.2 | 12.1 | 19.0 | 19.0 | 15.5 | 9.8 |
| Alabama | 4 | 25.0 | 50.0 | 25.0 | |||||
| Florida | 52 | 5.8 | 17.3 | 7.7 | 17.3 | 26.9 | 11.5 | 7.7 | 5.8 |
| Louisiana | 85 | 4.7 | 5.9 | 9.4 | 3.5 | 14.1 | 24.7 | 21.2 | 16.5 |
| Texas | 33 | 9.1 | 9.1 | 12.1 | 24.2 | 21.2 | 12.1 | 12.1 | |
| Pacific | 60 | 50.0 | 15.0 | 11.7 | 3.3 | 16.7 | 1.7 | 1.7 | |
| California | 1 | 100.0 | |||||||
| Oregon | 1 | 100.0 | |||||||
| Washington | 58 | 48.3 | 15.5 | 12.1 | 3.5 | 17.2 | 1.7 | 1.7 | |
| Canada | 6 | 33.3 | 16.7 | 16.7 | 33.3 | ||||
| Unknown | 3 | 33.3 | 33.3 | 33.3 | |||||
| Total | 396 | 23.0 | 11.1 | 10.9 | 10.6 | 15.7 | 13.6 | 8.8 | 6.3 |
Data are shown by harvest state and regions and is based on DNA probe colony hybridization results.
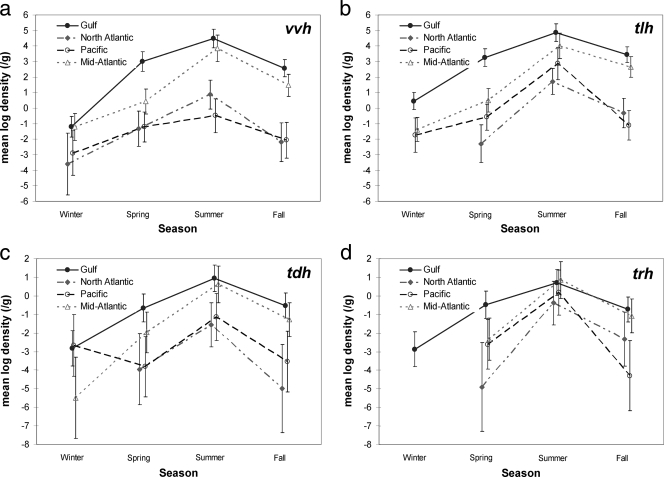
Figure 1a illustrates estimated geometric means and 95% confidence intervals of V. vulnificus levels (DNA colony hybridization results) in oysters collected for each season and region. While Gulf oysters had significantly higher V. vulnificus levels than either the North Atlantic or Pacific region during each season, differences between Gulf and Mid-Atlantic (New Jersey, Delaware, Maryland, Virginia, and South Carolina) levels were statistically significant only during the spring and fall (Table 5) . Geometric mean V. vulnificus levels in the Pacific and North Atlantic regions were generally <1 MPN/g, except during the summer in the North Atlantic region; no significant differences were observed between these regions for any season.
FIG. 1.
Seasonal trends for V. vulnificus and V. parahaemolyticus densities in market oysters harvested from various regions of the United States; V. vulnificus densities were determined by use of MPN and DNA probe for vvh (a), and total and pathogenic V. parahaemolyticus densities were determined by MPN-PCR for tlh (b), tdh (c), and trh (d).
TABLE 5.
Regional and seasonal estimates of mean log Vibrio densities in retail oysters harvested from different coastal areas
| Target/method | Season | Mean log Vibrio density (MPN/g)a |
|||
|---|---|---|---|---|---|
| Gulf | Mid-Atlantic | North Atlantic | Pacific | ||
| vvh/DNA probe | Winter | −1.25† | −1.21†‡ | −3.60 A§ | −2.89 A‡§ |
| Spring | 3.00 A | 0.44 A | −1.32 A† | −1.22 AB† | |
| Summer | 4.49† | 3.85† | 0.87‡ | −0.48 BC‡ | |
| Fall | 2.56 A | 1.47 A | −2.19 A† | −2.06 AC† | |
| tlh/DNA probe | Winter | 0.06 | −1.53 | −3.43 | |
| Spring | 2.57 A | −0.12 | −4.11 | −1.52 A | |
| Summer | 4.36† | 3.45 A†‡ | 1.77§ | 2.45‡§ | |
| Fall | 2.85 A† | 2.91 A† | −0.69‡ | −1.50 A‡ | |
| tlh/PCR | Winter | 0.46 | −1.38† | −1.74 A† | |
| Spring | 3.25 A | 0.51† | −2.29 | −0.57 A† | |
| Summer | 4.86† | 4.01†‡ | 1.71§ | 2.86‡§ | |
| Fall | 3.44 A† | 2.65† | −0.32‡ | −1.10 A‡ | |
| tdh/PCR | Winter | −2.84† | −5.50 | −2.67 AB† | |
| Spring | −0.66 A† | −1.96 A†‡ | −3.95 A‡ | −3.81 A‡ | |
| Summer | 0.94† | 0.62† | −1.57‡ | −1.14 B‡ | |
| Fall | −0.53 A† | −1.28 A† | −4.99 A‡ | −3.54 A‡ | |
| trh/PCR | Winter | −2.88 | |||
| Spring | −0.48 A | −2.32 A† | −4.90 A† | −2.61 A† | |
| Summer | 0.72† | 0.86† | −0.39† | 0.18† | |
| Fall | −0.73 A† | −1.07 A† | −2.33 A†‡ | −4.29 A‡ | |
Region/season estimates for each target/method followed by the same letter (A, B, or C) within a column are not significantly different (P > 0.05). Region/season estimates for each target/method followed by the same symbol (†, ‡, or §) within a row are not significantly different (P > 0.05).
Geometric means and 95% confidence intervals of total V. parahaemolyticus levels are shown in Fig. 1b. V. parahaemolyticus levels in the Gulf and Mid-Atlantic regions followed a similar seasonal and regional trend as V. vulnificus levels. However, V. parahaemolyticus levels in the Pacific and North Atlantic regions were typically much higher than V. vulnificus levels for all seasons, with >3-log higher levels in the Pacific region during the summer (Fig. 1a and b and Table 5). V. parahaemolyticus levels in Gulf oysters were generally at least 2 logs greater than those observed in those from the Pacific and North Atlantic regions during all seasons. Total V. parahaemolyticus levels were determined by DNA colony hybridization and real-time PCR (Table 5). While higher MPN estimates were nearly always obtained using the real-time PCR data, differences in estimates were generally within one log. Much larger differences were observed between real-time PCR and colony hybridization for estimating levels of pathogenic V. parahaemolyticus, and only the PCR data are presented in Fig. 1c and d and Table 5.
Geometric means and 95% confidence intervals of pathogenic V. parahaemolyticus (real-time PCR results) are shown in Fig. 1c (tdh) and d (trh). The highest levels of pathogenic V. parahaemolyticus based on detection of either gene were observed in the summer for all regions. Pathogenic V. parahaemolyticus levels were generally several logs lower than total V. parahaemolyticus levels and were considerably more variable. Levels based on MPN-PCR detection of either tdh or trh were similar and generally <1 MPN/g, except during the summer in the Gulf and Mid-Atlantic regions; trh levels were also >1 MPN/g in the Pacific region during the summer. During the summer, the Gulf and Mid-Atlantic samples had significantly higher pathogenic V. parahaemolyticus levels than those from the North Atlantic or Pacific based on tdh results (Table 5); similar levels were observed with trh results, but fewer significant differences were observed (Table 5). Two samples from the Gulf region and three from the Mid-Atlantic region had pathogenic V. parahaemolyticus levels of >1,000 MPN/g based on detection of one or both virulence genes.
Table 6 provides mean levels of various microbial indicator organisms in market oysters by region and season. Fecal coliform, E. coli, and APC levels were generally higher in Gulf oysters than those from other regions during all seasons. The highest geometric mean levels of fecal coliforms and E. coli were observed in the Gulf region oysters during the summer and were >2,000 MPN/100 g and ∼200 MPN/100 g, respectively. While Gulf oysters showed a seasonal trend with higher levels of fecal coliforms and E. coli in the warmer months, this was less apparent in the Mid-Atlantic region, and no significant differences were observed between seasons in the North Atlantic or Pacific region. With regard to MSB levels, there were few significant differences between seasons or regions except for the Gulf, where the highest levels were observed in the summer and the lowest levels were observed in the winter. Aerobic plate counts were generally higher for market oysters from the Gulf and Mid-Atlantic regions than for those from the North Atlantic and Pacific regions. No significant seasonal trends in aerobic plate counts were observed. Overall, there was a strong, significant relationship between fecal coliform levels and the levels of E. coli (P < 0.0001), MSB (P = 0.0006), and APC (P < 0.0001) microorganisms (Table 7) .
TABLE 6.
Regional and seasonal estimates of mean log densities (per 100 g) of microbial indicator organisms in retail oysters harvested from different coastal areas
| Indicator microbe | Season | Mean log density/100 ga |
|||
|---|---|---|---|---|---|
| Gulf | Mid-Atlantic | North Atlantic | Pacific | ||
| Fecal coliform | Winter | 1.61 A†‡ | 0.61 A§ | 0.59 A†§ | 1.03 A‡§ |
| Spring | 1.97 A | 1.19 AB† | 0.66 A† | 0.95 A† | |
| Summer | 3.34 | 1.51 B† | 1.44 A† | 1.17 A† | |
| Fall | 2.67 | 1.46 B† | 1.56 A† | 1.83 A† | |
| E. coli | Winter | 1.33 A†‡ | 0.62 A§ | 0.56 A†§ | 0.94 A‡§ |
| Spring | 1.52 A† | 1.10 AB†‡ | 0.35 A‡ | 0.61 A‡ | |
| Summer | 2.30 | 1.48 B† | 0.94 A† | 1.11 A† | |
| Fall | 1.45 A† | 0.69 A‡ | 0.59 A‡ | 0.85 A†‡ | |
| Male-specific coliphage | Winter | −1.53† | 0.94‡ | 0.11 A‡§ | −0.63 A†§ |
| Spring | −0.32 A† | −0.25 A†‡ | 0.09 A‡ | −1.95 A†§ | |
| Summer | 0.92† | −1.90 A‡ | −0.14 A†‡ | −0.68 A‡ | |
| Fall | 0.02 A† | −0.61 A† | 0.27 A† | −1.09 A† | |
| Aerobic plate count | Winter | 6.06 A | 6.74 A | 4.17† | 5.14 AB† |
| Spring | 6.76 B† | 6.56 A† | 5.78 A | 4.65 A | |
| Summer | 7.01 B† | 6.66 A† | 5.88 A‡ | 5.15 AC‡ | |
| Fall | 6.28 A†‡ | 5.81†§ | 5.26 A§ | 5.72 BC‡§ | |
Region/season estimates for each pathogen followed by the same letter (A, B, or C) within a column are not significantly different (P > 0.05). Region/season estimates for each pathogen followed by the same symbol (†, ‡, or §) within a row are not significantly different (P > 0.05).
TABLE 7.
Correlations between presence of virus and indicator microbes based on Kendall's tau
| Microbe | Correlation (associated P value) |
|||||
|---|---|---|---|---|---|---|
| NoV | HAV | NoV or HAV | Fecal coliforms | E. coli | MSC | |
| HAV | −0.043 (0.40) | |||||
| NoV or HAV | 0.67 (<0.0001) | 0.71 (<0.0001) | ||||
| Fecal coliforms | −0.030 (0.49) | 0.033 (0.45) | 0.0033 (0.94) | |||
| E. coli | 0.004 (0.93) | 0.0053 (0.91) | 0.0064 (0.89) | 0.67 (<0.0001) | ||
| MSC | 0.045 (0.30) | 0.0073 (0.87) | 0.037 (0.39) | 0.13 (0.0006) | 0.052 (0.18) | |
| APC | −0.023 (0.59) | −0.036 (0.39) | −0.042 (0.31) | 0.29 (<0.0001) | 0.25 (<0.0001) | 0.067 (0.057) |
The detection frequencies of bacterial and viral pathogens are shown in Table 8. Salmonella was detected in 8.6% of market oyster samples by real-time PCR of the invA gene in at least one of the three enrichment broths (lactose broth, RV broth, or TT broth). However, only 6 of 34 presumptive positive samples by real-time PCR yielded Salmonella isolates when culture confirmation was attempted, for an overall detection rate of 1.5%. Salmonella isolation occurred only in samples with threshold cycle (CT) values of <30. Three of the six culture-confirmed samples were from a single estuary in Florida.
TABLE 8.
Detection frequencies of bacterial and viral pathogens in market oysters
| Pathogen(s) | No. of pathogens detected/total (%) |
|
|---|---|---|
| PCR positive | Culture confirmed or secondary detectiona | |
| Salmonella spp. | 34/395 (8.6)b | 6/395 (1.5)c |
| Toxigenic V. cholerae | 4/397 (1.0)d | 0/397 |
| Norovirus | 15/388 (3.9) | 5/15 (33) |
| Genogroup I | 4/388 (1.0) | 0/4 |
| Genogroup II | 11/388 (2.8) | 5/11 (45) |
| Hepatitis A virus | 17/388 (4.4)e | 0/17 |
Culture confirmation for Salmonella spp. and toxigenic V. cholerae was as described in Materials and Methods. Secondary detection of norovirus and HAV was done by amplification using an additional primer set as described in Materials and Methods.
Only 395 samples were analyzed for the presence of Salmonella due to insufficient weight of oysters.
50% detected in oysters from Apalachicola Bay, FL.
75% detected in oysters harvested from the Gulf of Mexico.
Sequencing of 89-bp products identified HAV.
The ctx gene was detected in APW enrichments (CT > 30) of 4 samples (1% detection frequency), but no toxigenic strains of V. cholerae were isolated from these samples in spite of intensive efforts (examination of ∼50 suspect colonies from TCBS agar). Three of these samples were harvested from the Gulf Coast, and the other was from Washington State.
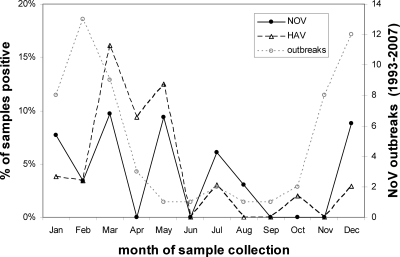
NoV was detected in 15 of 388 samples (3.9% detection frequency); 4 were genotype I and 11 were genotype II (Table 8). Only 5 of the 15 samples that were detected initially by the NoV real-time RT-PCR assay amplified with the conventional RT-PCR assay, which targeted a different genomic region; all five were identified as belonging to genotype II. A majority (9 of 15 samples; 60%) of NoV-positive samples were collected between December and March, when shellfish-associated NoV outbreaks are most prevalent (Fig. 2). While NoV was most frequently detected in oysters from the North Atlantic region (7.5%) and least frequently detected in oysters from the Gulf (2.9%), no significant differences (P < 0.05) were observed (Table 9 ).
FIG. 2.
Monthly distribution of detection frequencies of NoV and HAV in raw market oysters and incidence of oyster-associated NoV outbreaks.
TABLE 9.
Prevalence of virus by regiona
| Region | No. of samples positive/no. of samples tested (%) |
||
|---|---|---|---|
| NoV | HAV | NoV or HAV | |
| Gulf | 5/171 (2.9) | 11/171 (6.4) | 16/172 (9.3) |
| Mid-Atlantic | 3/97 (3.1) | 2/97 (2.1) | 5/97 (5.2) |
| North Atlantic | 4/53 (7.5) | 2/53 (3.8) | 6/53 (11.3) |
| Pacific | 3/59 (5.1) | 2/59 (3.4) | 5/59 (8.5) |
Data for which the region is Canada or unknown are excluded. No significant differences were observed (by Fisher's exact test [two sided]).
HAV was detected in 17 of 388 market oyster samples (4.4%), but none were determined to be positive by conventional RT-PCR (Table 8). While the prevalence of HAV in oysters was highest in oysters collected between March and May (70.6%), HAV was detected during most months (Fig. 2). HAV was detected most frequently in oysters harvested from the Gulf region, at a frequency of 6.4%, but there was no significant difference (P < 0.05) between regions (Table 9). NoV and HAV were not detected simultaneously in any one oyster sample, and there was no significant relationship between the occurrences of either of these enteric pathogens and the levels of fecal coliforms, E. coli, MSB, or APC microorganisms (Table 7).
DISCUSSION
To our knowledge, this market survey of bacterial and viral pathogens in raw oysters is the most comprehensive and systematic national survey of a commodity in the food supply that has been conducted to date. Major U.S. markets selling oysters harvested from the Pacific, Gulf, and Atlantic Coasts were sampled for a 1-year period to investigate both seasonal and geographical trends in the abundance of total and pathogenic V. parahaemolyticus and V. vulnificus and the incidence of toxigenic V. cholerae, Salmonella, norovirus, and HAV in oysters at the point of sale immediately prior to consumption. Additionally, indicator organisms, including fecal coliforms, E. coli, MSB, and APCs, were also determined. These data near the point of consumption provide considerable new insight on the level of protection provided by the current U.S. system of shellfish safety controls. While the level and risk of enteric viruses in raw oysters are reasonably assumed to be constant from harvest to consumption, bacterial levels, especially of vibrios, can increase or decrease substantially depending on handling practices. The performance of the overall system of controls for bacterial pathogens can be most appropriately evaluated at the point of consumption. However, this approach does not identify which aspects of the system (e.g., harvest, transportation, and retail) account for bacterial growth or die-off. Caution should also be used to avoid overinterpreting the results from individual samples. Emphasis is more appropriately placed on average levels and how these vary across different regions and seasons. The data for total V. parahaemolyticus and V. vulnificus levels can be compared directly to levels found in a previous study in 1998-1999 (29) that used a similar sampling plan and enumeration methods.
Due to the broad scope and intensive analytical protocols used in this study, it was necessary to enlist the support of numerous analysts from five laboratories. The FDA has an extensive laboratory network of well-trained and experienced microbiologists. Nearly all had been trained in real-time PCR, but minimal virus detection capability existed at the Office of Regulatory Affairs (ORA) laboratories prior to this study. The following steps were taken to minimize any potential laboratory bias. Representatives from each participating laboratory attended a 1-week training course specific to testing oysters for HAV and NoV. After this training, all laboratories received several sets of samples with increasing challenge for detection of all target organisms. In the study, no statistical differences between laboratories were noted for detection frequencies or levels of any of the indicator organisms or bacterial pathogens. However, there were some significant differences for detection frequencies of the viral pathogens, but there was no pattern, and these differences did not appear to be related to laboratory proficiency (data not shown). For example, the Gulf Coast Seafood Laboratory was the lead laboratory, with the most experience in virology, but had one of the lowest detection frequencies for NoV and the highest for HAV. All virus-positive amplicons were sequenced, and there was no evidence of laboratory contamination. Finally, collecting states systematically rotated shipments to each of the analytical laboratories to minimize any seasonal or regional bias that could have resulted from differences in proficiency of analysts from the various laboratories.
Overall, the V. vulnificus and V. parahaemolyticus densities in the oysters were correlated. This may be expected, since the abundances of both species in shellfish at the time of harvest are correlated with water temperature, and the two vibrios multiply in shellfish after harvest at similar rates (3, 39). Consequently, the overall correlation is attributable, in part, to the seasonal shift in the mean densities with water temperature. The V. vulnificus and V. parahaemolyticus densities were dependent on the season and harvest region of the oysters. Samples from the Gulf Coast consistently had the highest numbers of both V. vulnificus and V. parahaemolyticus year-round. V. vulnificus and V. parahaemolyticus densities in Gulf Coast oysters at retail were generally 10- to 100-fold greater than those observed at harvest (28, 58).
V. parahaemolyticus.
The original aim of this study was to gather data on levels of pathogenic V. parahaemolyticus bacteria in market oysters, as the methods used in the 1998-1999 survey permitted reliable enumeration of only total V. parahaemolyticus bacteria. In oysters harvested from the Gulf Coast, which had the highest total V. parahaemolyticus levels, similar seasonal levels were observed relative to the previous study based on the DNA probe data (29). Total V. parahaemolyticus levels in the Pacific Northwest oysters were also in relatively good agreement with the 1998-1999 study. While a majority of oyster-associated V. parahaemolyticus cases are reported from the Pacific Northwest, pathogenic V. parahaemolyticus levels in this region were found to be approximately 2 logs lower than the Gulf Coast level during the summer season, when most Pacific Northwest cases are reported. These findings suggests that tdh+ V. parahaemolyticus strains from the Pacific Northwest may be more virulent or infect at a lower dose than those from the Gulf. This hypothesis is also supported by the high attack rates observed during the 2004 V. parahaemolyticus outbreak in Alaska among cruise ship passengers that were exposed to relatively low levels of this pathogen (55). The possibility of reporting biases may also contribute to regional differences in oyster-associated illness. Sequencing representative V. parahaemolyticus genomes may identify geographically unique DNA sequences that are potential virulence determinants or markers.
V. vulnificus.
V. vulnificus infections, while rare and limited to individuals with chronic underlying illnesses, are the most serious threat to raw oyster consumers, as they account for nearly all fatalities associated with seafood consumption in the United States. The highest V. vulnificus levels were observed in oysters harvested from the Gulf Coast, which is the harvest region epidemiologically linked to nearly all such illnesses. V. vulnificus levels observed in Gulf oysters in 2007 followed a seasonal trend similar to what was reported in the 1998-1999 survey, except these levels were lower in the fall of 2007 than in 1998 (29). Although a plan was adopted by the Interstate Shellfish Sanitation Conference (ISSC) in 2000 to reduce V. vulnificus illnesses by 60%, the efforts thus far have focused on educating high-risk consumers, and time-temperature controls had not changed since the prior survey. Thus, it is not surprising that V. vulnificus levels in these two surveys are similar. Additionally, the number of oyster-associated V. vulnificus illnesses reported nationally by the CDC (Marc Glatzer, FDA, personal communication) in 2007 was 33, compared to an average of 31 cases/year from 1995 to 1999. The data from the current study and the 1998-1999 survey could be compared against future study data to measure the effectiveness of new controls aimed at reducing exposure.
V. vulnificus levels in oysters from the Mid-Atlantic region during the summer season were nearly as high as those from the Gulf region. Few V. vulnificus illnesses have been linked to Atlantic oysters, but summertime oyster harvest landings are much more infrequent there than in the Gulf region. Recent oyster restoration efforts along the Atlantic Coast have been increasingly successful and increase the likelihood of V. vulnificus illnesses occurring in the future. V. vulnificus was also detected frequently in the Pacific Northwest but at levels several logs lower than in the Gulf region. These levels were usually below 30 MPN per gram, which is the level required for labeling “postharvest processed to reduce V. vulnificus to nondetectable levels.” Additionally, oysters from the Pacific Northwest have never been incriminated in a V. vulnificus case.
A V. vulnificus risk assessment conducted by the Food and Agricultural Organization and the World Health Organization assumed that all V. vulnificus strains were equally virulent (39), but an accumulating body of evidence from recent studies indicates that the strains causing most human septicemia illnesses are a relatively small fraction of those found in oysters (61). Markers, including rRNA sequence type (61), have been used to differentiate these strains, but little information on the regional and seasonal distribution of the more virulent types is available. We are currently conducting studies with representative isolates from this study to determine the distribution of the various rRNA sequence types in U.S. market oysters.
Toxigenic V. cholerae.
The failure to isolate toxigenic V. cholerae in the current study is not unexpected, as it has rarely been isolated from U.S. oysters, with an exception being during the Latin American cholera epidemic in 1991-1992. Three of the four samples that were positive when screened for ctx by using real-time PCR were harvested from Gulf states in August and September, which is coincidentally the peak period for reported cholera cases in the United States. A fourth real-time-PCR-positive case occurred in an oyster sample harvested from Washington, where a few non-O1 V. cholerae-related cases have been previously reported.
Salmonella spp.
The frequency of Salmonella detection in the current study (1.5%) was much lower than that reported in a previous survey of oysters from growing areas of the United States (14), which reported a detection rate of 6%. While the current study was more systematic in seasonal and geographical representation, both studies reported a higher prevalence of Salmonella detection in oysters harvested from the panhandle of Florida than in those from other states. Half of the culture-confirmed (3/6) Salmonella samples in the current study originated from Apalachicola Bay, even though these accounted for only 12% of the samples analyzed. However, levels of fecal coliforms in these oysters were at levels consistent with shellfish harvested from an approved growing area. Nearly 10% of samples yielded positive real-time PCR results for the invA gene of Salmonella, but similar to the case for detection of the V. cholerae ctx gene, none were culture positive at CT values greater than 30. The significance of positive real-time PCR results for Salmonella in the absence of an isolate was likely the result of overgrowth by competing flora in the enrichments or the isolation media.
Indicator microbes.
Levels of the bacterial indicators, fecal coliforms and E. coli, followed both a seasonal and geographical trend, with greater abundance associated with warmer ambient conditions (summer and fall; Gulf Coast), and this trend was more evident with fecal coliforms (Table 6). Some bacterial species that comprise the fecal coliform group, such as Klebsiella spp., multiply readily in oyster tissues when held at warm temperatures, and to a lesser extent, this has also been observed with E. coli (46). Except in the Gulf Coast, seasonal and regional trends in MSB abundance in U.S. market oysters were not observed. These observations suggest that elevated fecal coliform and E. coli levels were usually the result of postharvest multiplication rather than growing area water quality. Thus, it would not be appropriate to apply the standards intended for levels in shellfish at the time of harvest. Overall, MSB levels in 74% of the shellfish samples were below the level of assay sensitivity (10 PFU/100 g), while only 10% of the market samples harbored MSB levels exceeding a level of 50 PFU/100 g. Two predominant bacterial host strains are used for the enumeration of MSB, E. coli HS(pFamp)R and Salmonella enterica serovar Typhimurium WG49. The E. coli strain was selected for this study instead of the Salmonella strain for several reasons, including similar recovery and enumeration of MSB (18, 71), lack of pathogenicity, and stability of the strain to retain the plasmid encoding pilus production (60).
Norovirus and HAV.
Interpreting the public health significance of detecting either HAV or NoV in ∼4% of U.S. market oysters is complicated by the inability to determine infectivity, because these viruses cannot be cultured using conventional tissue culture techniques. Human feeding studies with isolates from fresh or well-preserved stool samples containing NoV have demonstrated attack rates of >50% with relatively low doses (1 to 10 viral particles) (4, 79). Only one oyster-associated HAV outbreak has been reported in the United States since 1989 (74). Given the potential severity of HAV infections, it is unlikely that underreporting of these infections is as great as that of NoV infections. HAV detection in the absence of reported outbreaks may be attributed to HAV RNA persistence in marine environments well after infectivity is lost, providing an opportunity for lengthy transport from pollution sources.
Unpublished CDC data (24) list 59 potential oyster-associated NoV outbreaks between 1993 and 2007, and this information provides a basis to evaluate the significance of NoV detection in the current study. The relatively high NoV prevalence in oysters collected between December and March (9 of 15 total positive samples during 2007) corresponds well with reported outbreaks (42 of 59 total outbreaks in 2007) for the same period as shown in Fig. 2. NoV was detected in only three samples collected between June and October, and only seven NoV outbreaks were reported to the CDC during the corresponding period. The lack of a statistical relationship with seasonal occurrence (or other factors, such as region and indicator levels) is in part due to the low frequency of NoV detection in the current study. A more limited survey of U.S. market oysters (45 samples) reported a higher prevalence of NoV in the summer than in the winter, but differences were not significant (30). Thus, the association between NoV detection in oysters and risk is tenuous at best, because the ratio of infectious NoV to noninfectious NoV cannot be established and may vary by season, environmental conditions, and efficacy of wastewater treatment. It is generally assumed that NoV behaves similarly to related culturable surrogates that lose infectivity with exposure to sunlight and warm marine waters (17); this may in part explain the low frequency of NoV outbreaks in summer relative to detection frequency.
The real-time PCR virus detection approach used in the current study incorporated an internal amplification control to determine matrix inhibition and an extraction control (SMSV-17) to determine extraction efficiency. Normalization for matrix inhibition and extraction efficiency permits semiquantitative estimation of viral contamination. In general, most of the positive virus samples had CT values of >45, which is near the limit of detection of these real-time PCR assays. Our laboratory has used the methods described in the current study to test oysters implicated in several U.S. outbreaks in recent years, and in each case, the CT values were in the high 30s or low 40s (unpublished data). None of the oyster lots collected in this market survey was linked to a NoV outbreak.
NoV detection frequency is on the lower end of what had been reported in most previous studies in France (6, 51), Greece (41), India (77), Japan (63), The Netherlands (13), Norway (60), Spain (41), Sweden (41), the United Kingdom (53), and the United States (30), which reported NoV occurrences in shellfish of 9 and 23, <1, 2, 5, 4.8, 5.6, 12 to 14, 17 to 24, 52, and 18%, respectively. NoV genogroup II was detected ∼4-fold more frequently than genogroup I, consistent with previous reports (30, 41, 60). This wide difference in reporting can be attributed to multiple factors, including experimental design, virus stability, season, location, virus extraction protocol, or perhaps molecular test protocol employed (conventional RT-PCR or real-time RT-PCR). The current study data illustrate the last point. Only 5 of 15 NoV-positive extracts and none of the 17 HAV-positive extracts were detected by conventional RT-PCR assays directed at different genome regions. While real-time RT-PCR is generally considered to be more sensitive than conventional RT-PCR, other factors, such as amplicon size and sequence conservation, also affect assay sensitivity. The secondary NoV and HAV targets were more variable, and the amplicon sizes were larger than the primary targets of the real-time RT-PCR assay. The 89-bp fragments of the positive HAV isolates were sequenced and found to be genetically different from the HM175/18f vaccine strain used as a positive control.
Detection of either viral pathogen in oysters was not significantly correlated to fecal coliform, E. coli, or MSB levels. A lack of correlation between indicator bacteria and viral prevalence has been reported previously (40, 52, 70), but others have observed a relationship between MSB levels and NoV occurrence (37, 53, 60). Our study's inability to establish a relationship could be attributed to the low frequency (10%) of MSB levels exceeding 50 PFU/100 g and the relatively low frequency of NoV detection. In contrast, a United Kingdom study reported that 31% of samples with MSB levels of <100/100 g were NoV positive, while the frequency of NoV-positive samples increased to 67 to 70% when MSB levels exceeded 100/100 g (53).
For the purposes of this study, the FDA established a set of triggers to notify states when levels of concern were encountered. In addition to detection of norovirus or HAV, corresponding MSB levels of >50 PFU/100 g were established as a criterion for notifying a state that it had a potential public health issue. The rationale was that elevated levels of viable MSB levels were indicative of possible recent fecal contamination. For NoV, these two triggers occurred only once in the entire study, with a single sample that was collected in Rhode Island. During the follow-up investigation, it was determined that the source oysters were harvested in an area that was unclassified and potentially subject to human pollution. Three additional oyster samples were collected from the implicated area, and two of these were NoV positive and also had MSB levels of >50 PFU/100 g. These findings suggest that a dual criterion of NoV detection and elevated MSB has some merit and should be explored further.
While the real-time PCR methods used in this study for bacterial and viral pathogens have not been validated, we believe that they provide a more accurate and robust measure of risk than standard culture methods. The use of real-time PCR to screen/test oyster enrichments greatly increased detection frequency and efficiency of sample analysis, especially for pathogenic V. parahaemolyticus, toxigenic V. cholerae, and Salmonella, but relatively few of these PCR-positive samples were culture confirmed. The culture methods currently available in the FDA BAM require resource-intensive procedures for screening and identification of suspect pathogen isolates and are not effective when few of the typical colonies on the isolation medium are the target pathogen or the target pathogen does not reach high levels in the enrichment. Culture methods are currently unavailable for NoV and difficult for wild-type HAV, and therefore, RT-PCR-based methods are the only viable means of detecting low levels of virus in food.
We believe that the results of this study provide a baseline for the level of protection provided by the U.S. molluscan shellfish safety program in 2007 with regard to the key human pathogens. The availability of these baseline data provide an appropriate basis for the evaluation of any future controls aimed at reducing exposure and risk to any of these pathogens. Additionally, changes in exposure after a control is implemented would provide insight into compliance (i.e., predicted reductions not obtained) and validate accuracy of risk assessment models with regard to exposure and the association between exposure and risk.
Acknowledgments
We thank the ISSC for financial support for the purchase of supplies used in this study.
This study would not have been possible without the support from personnel in the states where market samples were collected. We greatly appreciate the support and efforts of Alabama Department of Public Health, Seafood Branch, of the Colorado Department of Public Health and Environment, Consumer Protection Division, of the U.S. Food and Drug Administration, Miami Resident Post, Domestic Operations, of the Illinois Department of Public Health, Division of Food, Drugs and Dairies, of the Louisiana Department of Health and Hospitals, of William Long (for conducting the sampling in Rhode Island), of the South Carolina Department of Health and Environmental Control, State Shellfish Program, and of the Virginia Department of Health and Washington Department of Health. We thank the following for analytical support: the FDA Northeast Regional Laboratory (William Baroletti, Virginia Chung, Eufemia Gonzalez, Thomas Herbst, Kent Hermann, Lawrence James, Patty Kaewussdangkul, Angela Lara, Allan Littell, Jose Obando, Gloria Parra, Morris Paul, Haydee Romero, Juanita Versace, and Joyce Williams), the FDA Southeast Regional Laboratory (Lacresha Chatman, Stacy Eliasberg, Jeffrey Hunsucker, Kathy Noe, LaKenya Patton, Angela Swinford, and Jacquelyn Welch), the FDA Pacific Regional Laboratory—Northwest (Carlos Abeyta, Michael Brown, Moises O'Neill, Cheryl Eklund, Janelle Johnson, Doan Nguyan, and June Wetherington), and the FDA Pacific Regional Laboratory—Southwest (Carlota Agpaoa, Michael Kawalek, Lieuchi Phan, and Nelly Tran). We also acknowledge George Blackstone for his assistance with development of the Salmonella real-time PCR assay.
Footnotes
Published ahead of print on 15 March 2010.
REFERENCES
- 1.American Public Health Association. 1970. Recommended procedures for the examination of seawater and shellfish. American Public Health Association, Washington, DC.
- 2.Andrews, W. H., and T. Hammack. 2004. Salmonella, chapter 5. In U.S. Food and Drug Administration (ed.), Bacteriological analytical manual. U.S. Food and Drug Administration, Washington, DC.
- 3.Anonymous. 2005. Quantitative risk assessment on the public health impact of pathogenic Vibrio parahaemolyticus in raw oysters. U.S. Food and Drug Administration, Washington, DC.
- 4.Atmar, R. L., A. R. Opekun, M. A. Gilger, M. K. Estes, S. E. Crawford, F. H. Neill, and D. Y. Graham. 2008. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 14:1553-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baggi, F., A. Demarta, and R. Peduzzi. 2001. Persistence of viral pathogens and bacteriophages during sewage treatment: lack of correlation with indicator bacteria. Res. Microbiol. 152:743-751. [DOI] [PubMed] [Google Scholar]
- 6.Beuret, C., A. Baumgartner, and J. Schluep. 2003. Virus-contaminated oysters: a three-month monitoring of oysters imported to Switzerland. Appl. Environ. Microbiol. 69:2292-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beuret, C., D. Kohler, A. Baumgartner, and T. M. Luthi. 2002. Norwalk-like virus sequences in mineral waters: one year monitoring of three brands. J. Food Prot. 68:1925-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackstone, G. M., J. L. Nordstrom, M. D. Bowen, R. F. Meyer, P. Imbro, and A. DePaola. 2007. Use of a real time PCR assay for detection of the ctxA gene of Vibrio cholerae in an environmental survey of Mobile Bay. J. Microbiol. Methods 68:254-259. [DOI] [PubMed] [Google Scholar]
- 9.Blackstone, G. M., J. L. Nordstrom, and A. DePaola. 2005. A same day detection method for Salmonella species in shrimp using real time PCR, p. 277. In Abstr. 105th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 10.Blackstone, G. M., J. L. Nordstrom, M. C. L. Vickery, M. D. Bowen, R. F. Meyer, and A. DePaola. 2003. Detection of pathogenic Vibrio parahaemolyticus in oyster enrichments by real time PCR. J. Microbiol. Methods 53:149-155. [DOI] [PubMed] [Google Scholar]
- 11.Blake, P. A., D. T. Allegra, J. D. Snyder, T. J. Barrett, L. McFarland, C. T. Caraway, J. C. Feeley, J. P. Craig, J. V. Lee, N. D. Puhr, and R. A. Feldman. 1980. Cholera—a possible endemic focus in the United States. New Engl. J. Med. 302:305-309. [DOI] [PubMed] [Google Scholar]
- 12.Blake, P. A., M. H. Merson, R. E. Weaver, D. G. Hollis, and P. C. Heublein. 1979. Disease caused by a marine vibrio: clinical characteristics and epidemiology. N. Engl. J. Med. 300:1-5. [DOI] [PubMed] [Google Scholar]
- 13.Boxman, I. L. A., J. J. H. C. Tilburg, N. A. J. M. te Loeke, H. Vennema, K. Jonker, E. de Boer, and M. Koopmans. 2006. Detection of noroviruses in shellfish in the Netherlands. J. Food Microbiol. 108:391-396. [DOI] [PubMed] [Google Scholar]
- 14.Brands, D. A., A. E. Inman, C. P. Gerba, C. J. Mare, S. J. Billington, L. J. Saif, J. F. Levine, and L. A. Joens. 2005. Prevalence of Salmonella spp. in oysters in the United States. Appl. Environ. Microbiol. 71:893-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bresee, J. S., M.-A. Widdowson, S. S. Monroe, and R. I. Glass. 2002. Foodborne viral gastroenteritis: challenges and opportunities. Clin. Infect. Dis. 35:748-753. [DOI] [PubMed] [Google Scholar]
- 16.Burkhardt, W., III., and K. R. Calci. 2000. Selective accumulation may account for shellfish-associated viral illness. Appl. Environ. Microbiol. 66:1375-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burkhardt, W., III., K. R. Calci, W. D. Watkins, S. R. Rippey, and S. J. Chirtel. 2000. Inactivation of indicator microorganisms in estuarine waters. Water Res. 34:2207-2214. [Google Scholar]
- 18.Burkhardt, W., III., and W. D. Watkins. 1993. Evaluation of host bacterial strains for the enumeration of male-specific bacteriophage from wastewater. Abstr. 1993 Am. Soc. Microbiol. Conference on Water Quality in the Western Hemisphere, San Juan, PR.
- 19.Burkhardt, W., III., J. Woods, and M. C. L. Vickery. 2004. Development of a quantitative multiplex RT-PCR assay for the detection of noro- and enteroviruses, abstr. Q.-308. Abstr. 104th Gen. Meet. Am. Soc. Microbiol. American Society Microbiology, Washington, DC.
- 20.Burkhardt, W., III., J. W. Woods, J. Nordstrom, and G. Hartman. 2006. A real-time RT-PCR protocol for the simultaneous detection of norovirus and enteroviruses. U.S. Food and Drug Administration, Washington, DC.
- 21.Cabelli, V. J. 1988. Microbial indicator levels in shellfish, water and sediments from the upper Narragansett Bay conditional shellfish-growing area. Report to the Narragansett Bay Project. Narragansett Bay Project, Providence, RI.
- 22.Centers for Disease Control and Prevention. 2006. Norovirus: technical fact sheet. Centers for Disease Control and Prevention, Atlanta, GA.
- 23.Centers for Disease Control and Prevention. 2006. Vibrio parahaemolyticus infections associated with consumption of raw shellfish—three states, 2006. MMWR Morb. Mortal. Wkly. Rep. 55:854-856. [PubMed] [Google Scholar]
- 24.Centers for Disease Control and Prevention. 2009. Outbreak surveillance data. Centers for Disease Control and Prevention, Atlanta, GA.
- 25.Centers for Disease Control and Prevention. 1998. Outbreak of Vibrio parahaemolyticus infections associated with eating raw oysters—Pacific Northwest, 1997. MMWR Morb. Mortal. Wkly. Rep. 47:457-462. [PubMed] [Google Scholar]
- 26.Cook, D. W. 1994. Effect of time and temperature on multiplication of Vibrio vulnificus in postharvest Gulf coast shellstock oysters. Appl. Environ. Microbiol. 60:3483-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook, D. W. 1997. Refrigeration of oyster shellstock: conditions which minimize the outgrowth of Vibrio vulnificus. J. Food Prot. 60:349-352. [DOI] [PubMed] [Google Scholar]
- 28.Cook, D. W., J. C. Bowers, and A. DePaola. 2002. Density of total and pathogenic (tdh+) Vibrio parahaemolyticus in Atlantic and Gulf Coast molluscan shellfish at harvest. J. Food Prot. 65:1873-1880. [DOI] [PubMed] [Google Scholar]
- 29.Cook, D. W., P. O'Leary, J. C. Hunsucker, E. M. Sloan, J. C. Bowers, R. J. Blodgett, and A. DePaola. 2002. Vibrio vulnificus and Vibrio parahaemolyticus in U.S. retail shell oysters: a national survey June 1998 to July 1999. J. Food Prot. 65:79-87. [DOI] [PubMed] [Google Scholar]
- 30.Costantini, V., F. Loisy, L. Joens, F. S. LeGuyader, and L. J. Saif. 2006. Human and animal enteric caliciviruses in oysters from different coastal regions of the United States. Appl. Environ. Microbiol. 72:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daniels, N. A., B. Ray, A. Easton, N. Marano, E. Kahn, A. L. McShan, L. Del Rosario, T. Baldwin, M. A. Kingsley, N. D. Puhr, J. G. Wells, and F. J. Angulo. 2000. Emergence of a new Vibrio parahaemolyticus serotype in raw oysters. JAMA 284:1541-1545. [DOI] [PubMed] [Google Scholar]
- 32.Daniels, N., L. MacKinnon, R. Bishop, S. Altekruse, B. Ray, R. Hammond, S. Thompson, S. Wilson, N. Bean, P. Griffin, and L. Slutsker. 2000. Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181:1661-1666. [DOI] [PubMed] [Google Scholar]
- 33.Debartolomeis, J., and V. J. Cabelli. 1991. Evaluation of an Escherichia coli host strain for enumeration of F male-specific bacteriophages. Appl. Environ. Microbiol. 57:1301-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DePaola, A., Jr., and C. A. Kaysner. 2004. Vibrio, chapter 9. In U.S. Food and Drug Administration (ed.), Bacteriological analytical manual online. U.S. Food and Drug Administration, Washington, DC.
- 35.DePaola, A., L. H. Hopkins, J. T. Peeler, B. Wentz, and R. M. McPhearson. 1990. Incidence of Vibrio parahaemolyticus in U.S. coastal waters and oysters. Appl. Environ. Microbiol. 56:2299-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DePaola, A., J. L. Nordstrom, J. C. Bowers, J. G. Wells, and D. W. Cook. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69:1521-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doré, W. J., K. Henshilwood, and D. N. Lees. 2000. Evaluation of F-specific RNA bacteriophage as a candidate human enteric virus indicator for bivalve molluscan shellfish. Appl. Environ. Microbiol. 66:1280-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Environmental Protection Agency. 2001. Male-specific (F+) and somatic coliphage in water by two-step enrichment procedure. Environmental Protection Agency, Washington, DC.
- 39.FAO and WHO. 2005. Risk assessment of Vibrio vulnificus in raw oysters: interpretative summary and technical report. FAO, Rome, Italy.
- 40.Formiga-Cruz, M., A. K. Allard, A. C. Conden-Hansson, K. Henshilwood, B. E. Hernroth, J. Jofre, D. N. Lees, F. Lucena, M. Papapetropoulou, R. E. Rangdale, A. Tsibouxi, A. Vantarakis, and R. Girones. 2003. Evaluation of potential indicators of viral contamination in shellfish and their applicability to diverse geographical areas. Appl. Environ. Microbiol. 69:1556-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Formiga-Cruz, M., G. Tofino-Quesada, S. Bofill-Mas, D. N. Lees, K. Henshilwood, A. K. Allard, A. C. Conden-Hansson, B. E. Hernroth, A. Vantarakis, A. Tsibouxi, M. Papapetropoulou, M. D. Furones, and R. Girones. 2002. Distribution of human virus contamination in shellfish from different growing areas in Greece, Spain, Sweden, and the United Kingdom. Appl. Environ. Microbiol. 68:5990-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardner, S. N., T. A. Kuczmarski, E. A. Vitalis, and T. R. Slezak. 2003. Limitations of Taqman PCR for detecting divergent viral pathogens illustrated by hepatitis A, B, C, E viruses and human immunodeficiency virus. J. Clin. Microbiol. 41:2417-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.González-Escalona, N., J. Martinez-Urtaza, J. Romero, R. T. Espejo, L. A. Jaykus, and A. DePaola. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190:2831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gooch, J. A., A. DePaola, J. C. Bowers, and D. L. Marshall. 2002. Growth and survival of Vibrio parahaemolyticus in postharvest American oysters. J. Food Prot. 65:970-974. [DOI] [PubMed] [Google Scholar]
- 45.Helsel, D. 2005. Nondetects and data analysis: statistics for censored environmental data. John Wiley, New York, NY.
- 46.Hood, M. A., G. E. Ness, and N. J. Blake. 1983. Relationship among fecal coliforms, Escherichia coli, and Salmonella spp. in shellfish. Appl. Environ. Microbiol. 45:122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaneko, T., and R. R. Colwell. 1975. Incidence of Vibrio parahaemolyticus in Chesapeake Bay. Appl. Microbiol. 30:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaper, J., H. Lockman, R. R. Colwell, and S. W. Joseph. 1979. Ecology, serology, and enterotoxin production of Vibrio cholerae in Chesapeake Bay. Appl. Environ. Microbiol. 37:91-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaysner, C. A., C. Abeyta, Jr., R. F. Stott, J. L. Lilja, and M. M. Wekell. 1990. Incidence of urea-hydrolyzing Vibrio parahaemolyticus in Willapa Bay, Washington. Appl. Environ. Microbiol. 56:904-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Guyader, F., L. Haugarreau, L. Miossec, E. Dubois, and M. Pommepuy. 2000. Three-year study to assess human enteric viruses in shellfish. Appl. Environ. Microbiol. 66:3241-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Guyader, F., L. Miossec, L. Haugarreau, E. Dubois, H. Kopecka, and M. Pommepuy. 1998. RT-PCR evaluation of viral contamination in five shellfish beds over a 21-month period. Water Sci. Technol. 38:45-50. [Google Scholar]
- 53.Lowther, J. A., K. Henshilwood, and D. N. Lees. 2008. Determination of norovirus contamination in oysters from two commercial harvesting areas over an extended period, using semiquantitative real-time reverse transcription PCR. J. Food Prot. 71:1427-1433. [DOI] [PubMed] [Google Scholar]
- 54.Matsumoto, C., J. Okuda, M. Ishibashi, M. Iwanaga, P. Garg, T. Rammamurthy, H. Wong, A. DePaola, Y. B. Kim, M. J. Albert, and M. Nishibuchi. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analysis. J. Clin. Microbiol. 38:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLaughlin, J. B., A. DePaola, C. A. Bopp, K. A. Martinek, N. P. Napolilli, C. G. Allison, S. L. Murray, E. C. Thompson, M. M. Bird, and J. P. Middaugh. 2005. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 353:1463-1470. [DOI] [PubMed] [Google Scholar]
- 56.Mead, P. S., L. Slutsker, V. Dietz, L. F. McGaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miles, D. W., T. Ross, J. Olley, and T. A. McMeekin. 1997. Development and evaluation of a predictive model for the effect of temperature and water activity on the growth rate of Vibrio parahaemolyticus. Int. J. Food Microbiol. 38:133-142. [DOI] [PubMed] [Google Scholar]
- 58.Motes, M. L., A. DePaola, D. W. Cook, J. E. Veazey, J. C. Hunsucker, W. E. Garthright, R. J. Blodgett, and S. J. Chirtel. 1998. Influence of water temperature and salinity on Vibrio vulnificus in northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64:1459-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mullendore, J. L., M. D. Sobsey, and Y. C. Shieh. 2001. Improved method for the recovery of hepatitis A virus from oysters. J. Virol. Methods 94:25-35. [DOI] [PubMed] [Google Scholar]
- 60.Myrmel, M., E. M. M. Berg, E. Rimstad, and B. Grinde. 2004. Detection of enteric viruses in shellfish from the Norwegian coast. Appl. Environ. Microbiol. 70:2678-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nilsson, W. B., R. N. Paranjpye, A. DePaola, and M. S. Strom. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 41:442-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishibuchi, M., and A. DePaola. 2005. Vibrio species, p. 251-271. In P. M. Fratamico, A. K. Bhunia, and J. L. Smith (ed.), Foodborne pathogens: microbiology and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 63.Nishida, T., O. Nishio, M. Kato, T. Chuma, H. Kato, H. Iwata, and H. Kimura. 2007. Genotyping and quantitation of noroviruses in oysters from two distinct sea areas in Japan. Microbiol. Immunol. 51:177-184. [DOI] [PubMed] [Google Scholar]
- 64.Nordstrom, J. L., M. C. L. Vickery, G. M. Blackstone, S. L. Murray, and A. DePaola. 2007. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl. Environ. Microbiol. 73:5840-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oliver, J. D., R. A. Warner, and D. R. Cleland. 1982. Distribution and ecology of Vibrio vulnificus and other lactose-fermenting marine vibrios in coastal waters of the southeastern United States. Appl. Environ. Microbiol. 44:1404-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ottoson, J., A. Hansen, T. Westrell, K. Johansen, H. Norder, and T. A. Stenstrom. 2006. Removal of noro- and enteroviruses, Giardia cysts, Cryptosporidium oocysts, and fecal indicators at four secondary wastewater treatment plants in Sweden. Water Environ. Res. 78:828-834. [DOI] [PubMed] [Google Scholar]
- 67.Patel, M. M., A. J. Hall, J. Vinge, and U. D. Parashar. 2009. Norovirus: a comprehensive review. J. Clin. Virol. 44:1-8. [DOI] [PubMed] [Google Scholar]
- 68.Rippey, S. R. 1994. Infectious diseases associated with molluscan shellfish consumption. Clin. Microbiol. Rev. 7:419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rippey, S. R., L. A. Chandler, and W. D. Watkins. 1987. Fluorometric method for enumeration of Escherichia coli in molluscan shellfish. J. Food Prot. 50:685-690. [DOI] [PubMed] [Google Scholar]
- 70.Romalde, J. L., E. Area, G. Sanchez, C. Ribao, I. Torrado, X. Abad, R. M. Pinto, J. L. Barja, and A. Bosch. 2002. Prevalence of enterovirus and hepatitis A virus in bivalve molluscs from Galicia (NW Spain): inadequacy of the EU standards of microbiological quality. Int. J. Food Microbiol. 74:119-130. [DOI] [PubMed] [Google Scholar]
- 71.Schaper, M., A. E. Duran, and J. Jofre. 2002. Comparative resistance of phage isolates of four genotypes of F-specific RNA bacteriophages to various inactivation processes. Appl. Environ. Microbiol. 68:3702-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sedgwick, W. T., and S. Macnutt. 1908. An examination of the theorem of Allen Hazen that for every death from typhoid fever avoided by the purification of public water supplies two of three deaths are avoided from other causes. Science 14:216. [DOI] [PubMed] [Google Scholar]
- 73.Shapiro, R. L., S. Altekruse, S. Hutwagner, R. Bishop, R. Hammond, S. Wilson, B. Ray, S. Thompson, R. V. Tauxe, P. M. Griffin, and Vibrio Working Group. 1998. The role of Gulf Coast oysters harvested in warmer months in Vibrio vulnificus infections in the United States, 1988-1996. J. Infect. Dis. 178:752-759. [DOI] [PubMed] [Google Scholar]
- 74.Shieh, Y. C., Y. E. Khudyakov, L. M. Ganova-Raeva, F. M. Khambaty, J. W. Woods, J. E. Veazey, M. L. Motes, M. Glatzer, S. R. Bialek, and A. E. Fiore. 2007. Molecular confirmation of oysters as the vector for hepatitis A in a 2005 multistate outbreak. J. Food Prot. 70:145-150. [DOI] [PubMed] [Google Scholar]
- 75.Shieh, Y. C., J. W. Woods, and K. R. Calci. 2003. Molecular surveillance of enterovirus and Norwalk-like virus in oysters relocated to a municipal sewage-impacted gulf estuary. Appl. Environ. Microbiol. 69:7130-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shieh, Y. S. C., S. S. Monroe, R. L. Frankhauser, G. W. Langois, W. Burkhardt III, and S. Baric. 2000. Detection of Norwalk-like virus in shellfish implicated in illness. J. Infect. Dis. 181:S360-S366. [DOI] [PubMed] [Google Scholar]
- 77.Umesha, K. R., N. C. Bhavani, M. N. Venugopal, I. Karunasagar, G. Krohne, and I. Karunasagar. 2008. Prevalence of human pathogenic enteric viruses in bivalve molluscan shellfish and cultured shrimp in south west coast of India. Int. J. Food Microbiol. 122:279-286. [DOI] [PubMed] [Google Scholar]
- 78.Woods, J. W., K. R. Calci, and W. Burkhardt III. 2005. Evaluation of wastewater treatment plant efficiency to reduce bacterial and viral loading using real time RT-PCR. Mississippi-Alabama Sea Grant Consortium, Ocean Springs, MS.
- 79.Wyatt, R. G., R. Dolin, N. R. Blacklow, H. L. Dupont, R. F. Buscho, T. S. Thornhill, A. Z. Kapikian, and R. M. Chanock. 1974. Comparison of three agents of acute infectious nonbacterial gastroenteritis by cross-challenge in volunteers. J. Infect. Dis. 129:709-714. [DOI] [PubMed] [Google Scholar]