Abstract
Oral malodor develops mostly from the metabolic activities of indigenous bacterial populations within the oral cavity, but whether healthy or oral malodor-related patterns of the global bacterial composition exist remains unclear. In this study, the bacterial compositions in the saliva of 240 subjects complaining of oral malodor were divided into groups based on terminal-restriction fragment length polymorphism (T-RFLP) profiles using hierarchical cluster analysis, and the patterns of the microbial community composition of those exhibiting higher and lower malodor were explored. Four types of bacterial community compositions were detected (clusters I, II, III, and IV). Two parameters for measuring oral malodor intensity (the concentration of volatile sulfur compounds in mouth air and the organoleptic score) were noticeably lower in cluster I than in the other clusters. Using multivariate analysis, the differences in the levels of oral malodor were significant after adjustment for potential confounding factors such as total bacterial count, mean periodontal pocket depth, and tongue coating score (P < 0.001). Among the four clusters with different proportions of indigenous members, the T-RFLP profiles of cluster I were implicated as the bacterial populations with higher proportions of Streptococcus, Granulicatella, Rothia, and Treponema species than those of the other clusters. These results clearly correlate the global composition of indigenous bacterial populations with the severity of oral malodor.
Oral malodor is one of the major complaints made by patients visiting the dentist, ranking behind only dental caries and periodontal disease (19). It originates mainly from the mouth itself, and the malodorous substrates most commonly are associated with microbial metabolism (32). Major compounds that contribute to oral malodor are volatile sulfur compounds (VSCs) such as hydrogen sulfide (H2S), methyl mercaptan (CH3SH), and dimethyl sulfide (CH3SCH3) (14, 35). Additionally, short-chain fatty acids, such as propionic acid and butyric acid, cadaverine, indole, and scatole, have been reported to cause oral malodor (8, 16).
The oral surfaces are colonized by large numbers of bacterial species with many members, especially gram-negative anaerobes, which are known to produce malodorous compounds (28). Poor oral hygiene resulting in microbial overgrowth clearly is involved in the development of this condition. Hence, the current major treatment for oral malodor focuses on nonselective antibacterial treatments to reduce the total number of bacteria, with careful attention to anaerobic areas such as the periodontal pockets and tongue dorsum. This approach, however, generally provides short-term benefits, since malodor-causing bacteria quickly recover to their former numbers when treatment is stopped (2).
An alteration in the bacterial population structure would be necessary to completely cure oral malodor. One general and reasonable approach is to specify the causal agent and directly remove it from the oral cavity. A diversity of bacteria, including Porphyromonas gingivalis, Fusobacterium nucleatum, Prevotella intermedia, and Treponema denticola, have been closely associated with oral malodor (19). On the other hand, developing an elimination method that targets various multiple causative agents remains a challenge.
Malodor producers are considered members of the oral microbial ecosystem, which is regulated by numerous interactions among inhabitants. Therefore, adjusting the global composition of indigenous bacterial populations toward a “healthy” pattern may be an alternative approach to effectively prevent oral malodor. Indeed, probiotic treatments have been conducted for the maintenance or manipulation of indigenous bacteria in the gastrointestinal tract (3) and vagina (1), and attempts have been made to apply the same approach to oral malodor (2). However, whether characteristic patterns of healthy and oral malodor-related microbiota or nonspecific bacterial overgrowth result in oral malodor remains unclear. Although the bacterial population structure of tongue microbiota of subjects with and without oral malodor has been revealed comprehensively using a molecular approach (9, 11, 30), a statistical analysis of a larger sample is necessary to correlate the complex microbial community composition with the severity of the condition.
Despite a lack of fully resolved phylogenetic analyses at the species level, terminal-restriction fragment length polymorphism (T-RFLP) analysis is an effective molecular approach for the rapid assessment and comparison of large numbers of complex bacterial communities (18). The relative differences in microbial community structures are reliably reflected in T-RFLP profiles, although some biases may impair the detection of the actual community structure (10). We previously confirmed that major bacterial genera in the oral cavity can be distinguished roughly by this method using a specific primer set and the restriction enzyme HaeIII. Relative bacterial abundances detected in cloning and sequencing methods were fairly consistent with the peak area proportions of the corresponding fragments in the T-RFLP profile (33).
In the present study, the global composition of bacterial populations found in the saliva of 240 patients complaining of oral malodor, with highly variable levels of severity, was examined using T-RFLP analysis. Although the use of saliva as the most appropriate oral specimen to study the relationship between oral microbiota and health has been questioned, it was considered the sample that would best represent the overall microbial population in the oral cavity. While the bacterial composition in saliva was highly similar to that found in the tongue coating among the different surfaces in the oral cavity (21), bacteria that inhabit other oral surfaces also can be recovered from saliva (4, 15, 20). Microbial community compositions showing a similar pattern were divided into groups using cluster analysis, and the patterns of the microbiota of those exhibiting higher and lower malodor were explored. The significance of malodor severity based on the pattern of microbiota was statistically evaluated using multivariate analysis while controlling for the effect of other oral malodor-associated factors. The aim of this study was to elucidate the relationship between the global composition of indigenous bacterial populations and the severity of oral malodor.
MATERIALS AND METHODS
Subjects and study design.
The study population consisted of 240 patients complaining of oral malodor (110 females and 130 males, aged 12 to 79 years with a mean age of 45 ± 14 years) who visited the Oral Malodor Clinic at Fukuoka Dental College Medical and Dental Hospital in Japan. These 240 patients included both subjects with actual oral malodor and those without oral malodor who were unsure if or believed that they had oral malodor. The Ethics Committee of Fukuoka Dental College and Kyushu University Faculty of Dental Science approved the study design. All participants understood the nature of the study and provided informed consent. Previously to the malodor assessment, each subject was asked to refrain from eating, drinking, chewing, smoking, brushing, or rinsing the mouth for at least 5 h.
Malodor assessment.
The severity of oral malodor in each individual was determined using an organoleptic test (OLT), a method used for the direct sniffing of expelled mouth air, and gas chromatography (model GC14B; Shimadzu Works, Kyoto, Japan). The OLT scores were estimated on a scale of 0 to 5 (0, absence of oral malodor; 1, questionable odor; 2, slight malodor; 3, moderate malodor; 4, strong malodor; 5, severe malodor) (22). In this study, we divided the subjects into four categories: 0 or 1, 2, 3, and 4 or 5. The gas chromatography was used for measuring the concentrations of VSCs (H2S, CH3SH, and CH3SCH3) in mouth air. We divided the subjects into four categories based on the sum of the three VSC concentrations: subjects without oral malodor (≤0.20 ppm, lower than the olfactory threshold for VSC concentrations in mouth air [36]), those with low malodor (0.21 to 0.60 ppm; lower one-third of the total number of subjects with oral malodor), those with moderate malodor (0.61 to 1.10 ppm; middle one-third), and those with high malodor (>1.10 ppm; higher one-third).
Clinical examinations.
For each patient, the periodontal pocket depth and the amount of tongue coating, two major halitosis-inducing factors, were examined. The periodontal pocket depth was measured at six points around each tooth in all subjects. The total area and thickness of the tongue coating was scored based on conventional criteria (26), with a slight modification (1, no tongue coating or thin, with less than one-third covered; 2, thin, with one-third to two-thirds covered or thick, with less than one-third covered; 3, thin with more than two-thirds covered or thick with more than one-third covered).
Saliva sample collection and DNA extraction.
The subjects were asked to bite on paraffin wax for 5.5 min, and stimulated saliva samples produced during the last 5 min were collected in sterile plastic tubes. The samples were stored at −80°C until further analysis. Bacterial DNA extraction was performed as described previously (34).
Quantification of total bacteria by real-time PCR.
Quantitative real-time PCR was performed using a QuantiFast SYBR green PCR kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The bacterial universal primers 806F (5′-TTA GAT ACC CYG GTA GTG G-3′) and 926R (5′-CCG TCA ATT YCT TTG AGT TT-3′) were used, and the details of the procedure have been described previously (33). The relative amounts of total bacteria were calculated using the comparative CT method, and DNA extracted from Streptococcus mutans Xc was used as a real-time PCR control.
T-RFLP analysis.
From each sample, internal regions of 16S rRNA genes were amplified using the universal forward primer 8F (5′-AGA GTT TGA TYM TGG CTC AG-3′) labeled at the 5′ end with 6-carboxyfluorescein (6-FAM) and the universal reverse primer 806R (5′-GGA CTA CCR GGG TAT CTA A-3′) labeled at the 5′ end with hexachlorofluorescein (HEX). PCR amplification, purification, and digestion by the restriction enzyme HaeIII were performed as previously described (33). The restriction digest products were mixed with 10 μl of deionized formamide and 1 μl of the internal standard, which contained GeneScan-500 ROX standard (Applied Biosystems, Foster City, CA) and six additional DNA fragments (541, 600, 663, 730, 799, and 861 bp) labeled at the 5′ end with ROX. The samples were denatured, electrophoresed, and analyzed with GeneMapper version 4.0 (Applied Biosystems). The unit of fragment size instead of number of bases was estimated based on the molecular weight (MW) of each fragment in the internal standard as previously described (34). The terminal restriction fragments (TRFs) with a peak area of less than 0.5% of the total area were excluded from the analysis.
Cluster classification and characterization.
Cluster analysis was performed using two T-RFLP profiles obtained per subject using two different fluorescent dyes (6-FAM and HEX). The estimated MWs of all TRFs from each subject were aligned, and the TRFs with MWs that differed by 80 or less were considered identical. The aligned T-RFLP profiles composed of the size of the TRFs and the percentage of the peak area in each profile were clustered by hierarchical cluster analysis using Euclidean distance and Ward's algorithm with R, version 2.8.1 (http://www.r-project.org). The aligned T-RFLP profiles for the 240 subjects also were analyzed by principal component analysis (PCA) and displayed as a biplot diagram to visualize the bacterial composition in correlation with the classified clusters. The PCA compressed the information from the T-RFLP profiles to a small number of dimensions, and they were plotted as dots in the two-dimensional display, of which the x and y axes represented the first and second principal components, respectively, and the original variables (each TRF) were indicated by arrows. The direction and length of the arrows indicated how each TRF contributed to the first two components in the biplot. PCA was performed by using the R library ade4 as described previously (33). Candidate bacterial species corresponding to the combination of TRFs (6-FAM-labeled or HEX-labeled TRFs) were selected based on their size from our oral bacterial database using TRFMAW (http://www.trflp.info/trfmaw) as described earlier (33). Based on our previous work (34), the matching window was set to an MW of ±660. The diversity of the bacterial community was evaluated by the number of TRFs and the Shannon diversity index of T-RFLP profiles. The bacterial candidates corresponding to the TRFs with significantly more or fewer subjects in one cluster than in the other clusters were selected using BP-TRFMA (24). The correlation threshold in BP-TRFMA was set at 0.13, which corresponds to a significant correlation coefficient in this sample size.
Statistical analysis.
Tukey's multiple comparison was conducted with R software to look for differences in peak area proportions of TRFs, age, number of total bacteria, mean pocket depth, total number of TRF, Shannon diversity index, and detection frequency of each TRF among the clusters. Fisher's exact test was performed by R to evaluate the statistical differences in gender and tongue coating score. Bivariate and multivariate ordinal logistic regression with the proportional-odds model were used to examine the associations between the intensity of oral malodor and the bacterial population structure in saliva, including other malodor-associated factors that may affect confounding variables. From the ordinal logistic regression model, proportional odds for cumulative probabilities were generated. The statistical analysis was performed using SPSS (version 15.0; SPSS Japan, Tokyo, Japan). The statistical significance was set at P < 0.05 to denote a statistically significant difference.
RESULTS
Differences in malodor productivity associated with T-RFLP profiles.
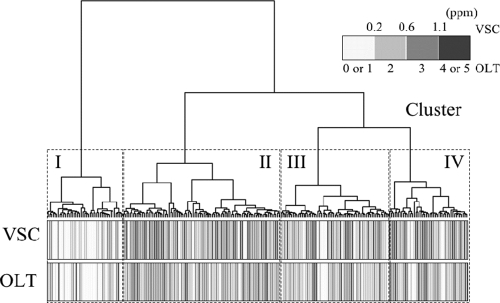
The bacterial composition in the saliva of 240 subjects was assessed using T-RFLP analysis and displayed as peak patterns. The overall profiles contained 164 distinct peaks (TRFs), 99 TRFs in the 6-FAM profiles (F1 to F99) and 65 TRFs in the HEX profiles (R1 to R65). The T-RFLP profiles were compared in a dendrogram generated by hierarchical cluster analysis (Fig. 1). The total concentration of VSCs in mouth air and the organoleptic score of each subject also were displayed. When the bacterial compositions were divided into four groups (clusters I, II, III, and IV), which contained 43, 90, 62, and 45 subjects, respectively, striking differences in malodor productivity were observed among the groups (Fig. 1). Both oral malodor parameters clearly were lower in cluster I, whereas they were higher in clusters III and IV.
FIG. 1.
Cluster analysis of the bacterial population structure in saliva of 240 subjects complaining of oral malodor based on T-RFLP profiles of the 16S rRNA gene. The dendrogram was generated by hierarchical cluster analysis using Euclidean distance and Ward's algorithm. The concentration of total volatile sulfur compounds (VSCs) in mouth air and the organoleptic score (OLT) of each subject were displayed as a gray-scale bar in the lower side of the dendrogram. The bacterial compositions were classified into four clusters: cluster I (n = 43), cluster II (n = 90), cluster III (n = 62), and cluster IV (n = 45).
Statistical comparison of the severity of oral malodor among clusters.
The characteristics of each cluster in terms of age, gender, and other variables possibly associated with oral malodor are shown in Table 1. Significant differences among clusters were observed in mean ages and amount of total bacteria. Tables 2 and 3 show the characteristics of the subjects according to VSC concentrations and organoleptic scores, respectively. To control for the effects of confounding factors, a multivariate analysis was performed that included the above-mentioned variables. After adjusting for potentially confounding variables, subjects belonging to clusters II, III, and IV had significantly higher odds ratios than those for cluster I for both the amount of VSCs in mouth air (Table 2) and organoleptic scores (Table 3). The results suggest that differences in the bacterial colonization pattern were significantly associated with the intensity of oral malodor independently of other variables, including two major halitosis-inducing factors (i.e., an increase in tongue coating and periodontal disease). The adjusted odds ratios for higher levels of VSCs and organoleptic scores were equally high in clusters II (11.0 and 11.4, respectively) and IV (10.8 and 9.8, respectively). Compared to these clusters, the odds ratios were lower for cluster III (4.4 and 3.9, respectively), which exhibited an intermediate pattern in the T-RFLP profile among clusters I, II, and IV.
TABLE 1.
Comparisons of general and clinical parameters among four clustersd
| Variable | Cluster I (n = 43) | Cluster II (n = 90) | Cluster III (n = 62) | Cluster IV (n = 45) | Significant differences between clusters (P < 0.05) |
|---|---|---|---|---|---|
| Agea (yr) | 39 ± 15 | 47 ± 14 | 47 ± 15 | 46 ± 14 | II > I, III > I |
| Amt of total bacteriaa (log deduced CFUb) | 7.6 ± 0.6 | 8.2 ± 0.7 | 7.8 ± 0.6 | 7.8 ± 0.8 | II > I, II > III, II > IV |
| Mean pocket deptha (mm) | 2.9 ± 0.5 | 3.0 ± 0.4 | 3.1 ± 0.6 | 3.2 ± 1.0 | NS |
| Genderc | NS | ||||
| Female | 22 (51.2) | 52 (57.8) | 34 (54.8) | 22 (48.9) | |
| Male | 21 (48.8) | 38 (42.2) | 28 (45.2) | 23 (51.1) | |
| Tongue coating scorec | NS | ||||
| 1 | 23 (53.4) | 28 (31.1) | 20 (32.2) | 17 (37.7) | |
| 2 | 12 (27.9) | 41 (45.5) | 32 (51.6) | 21 (46.6) | |
| 3 | 8 (18.6) | 21 (23.3) | 10 (16.1) | 7 (15.5) |
Significant difference between clusters were evaluated by Tukey's multiple comparison.
The copy number of the total bacterial 16S rRNA gene was divided by that of Streptococcus mutans UA159, corresponding to 1 CFU per 500 μl.
Significant difference was evaluated by Fisher's exact test.
Data are given either as means ± standard deviations or as the number of data with the percentage in parentheses. NS indicates lack of significance.
TABLE 2.
Effects of each variable on the concentration of VSCsb
| Variable | Total (n = 240) | Concn of VSCs in mouth air (ppm) |
Crude odds ratio (95% CI) | Multivariate odds ratio (95% CI) | |||
|---|---|---|---|---|---|---|---|
| ≤0.2 (n = 41) | >0.2 (n = 64) | >0.6 (n = 71) | >1.1 (n = 64) | ||||
| Age (yr) | 45 ± 14 | 39 ± 14 | 45 ± 15 | 45 ± 12 | 50 ± 15 | 1.0 (1.0-1.0)*** | 1.0 (1.0-1.0) |
| Amt of total bacteria (log deduced CFUa) | 8.0 ± 0.7 | 7.8 ± 0.7 | 7.9 ± 0.7 | 8.0 ± 0.7 | 8.1 ± 0.7 | 1.5 (1.1-2.0)* | 1.0 (0.7-1.4) |
| Mean pocket depth (mm) | 3.1 ± 0.7 | 2.8 ± 0.4 | 3.1 ± 0.8 | 3.2 ± 0.6 | 3.3 ± 0.7 | 1.9 (1.3-2.8)** | 1.8 (1.2-2.8)*** |
| Gender | |||||||
| Female | 110 (45.8) | 18 (43.9) | 30 (46.9) | 27 (38.0) | 35 (54.7) | 1 | 1 |
| Male | 130 (54.2) | 23 (56.1) | 34 (53.1) | 44 (62.0) | 29 (45.3) | 0.8 (0.5-1.3) | 0.9 (0.5-1.4) |
| Tongue coating score | |||||||
| 1 | 88 (36.7) | 26 (63.4) | 28 (43.8) | 15 (21.1) | 19 (29.7) | 1 | 1 |
| 2 | 106 (44.1) | 12 (29.3) | 29 (45.3) | 38 (53.5) | 27 (42.2) | 2.3 (1.4-3.8)** | 1.9 (1.1-3.3)* |
| 3 | 46 (19.2) | 3 (7.3) | 7 (10.9) | 18 (25.4) | 18 (28.1) | 4.3 (2.2-8.3)*** | 4.0 (2.0-8.3)*** |
| Cluster of T-RFLP profile | |||||||
| I | 43 (17.9) | 21 (51.2) | 13 (20.3) | 9 (12.7) | 0 (0) | 1 | 1 |
| II | 90 (37.5) | 6 (14.6) | 19 (29.7) | 33 (46.5) | 32 (50.0) | 11.1 (5.4-23.2)*** | 11.0 (5.0-24.2)*** |
| III | 62 (25.8) | 10 (24.4) | 22 (34.4) | 14 (19.7) | 16 (25.0) | 5.0 (2.4-10.5)*** | 4.4 (2.0-9.7)*** |
| IV | 45 (18.8) | 4 (9.8) | 10 (15.6) | 15 (21.1) | 16 (25.0) | 10.2 (4.5-23.3)*** | 10.8 (4.5-25.5)*** |
Copy number of the total bacterial 16S rRNA gene was divided by that of Streptococcus mutans UA159, corresponding to 1 CFU per 500 μl.
Data are given either as means ± standard deviations or as the number of data, with the percentage in parentheses. CI, confidence interval. Each odds ratio and P values were calculated by ordinal logistic regression analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
TABLE 3.
Effects of each variable on the organoleptic scoreb
| Variable | Total (n = 240) | Organoleptic score |
Crude odds ratio (95% CI) | Multivariate odds ratio (95% CI) | |||
|---|---|---|---|---|---|---|---|
| 0 or 1 (n = 44) | 2 (n = 79) | 3 (n = 89) | 4 or 5 (n = 28) | ||||
| Age (yr) | 45 ± 14 | 38 ± 14 | 44 ± 15 | 48 ± 14 | 53 ± 15 | 1.0 (1.0-1.1)*** | 1.0 (1.0-1.0) |
| Amt of total bacteria (log deduced CFUa) | 8.0 ± 0.7 | 7.7 ± 0.7 | 7.9 ± 0.7 | 8.1 ± 0.8 | 8.2 ± 0.7 | 1.6 (1.2-2.3)** | 0.9 (0.6-1.3) |
| Mean pocket depth (mm) | 3.1 ± 0.7 | 2.8 ± 0.4 | 3.0 ± 0.5 | 3.3 ± 0.7 | 3.6 ± 0.9 | 3.0 (2.0-4.5)** | 1.8 (1.8-4.5)*** |
| Gender | |||||||
| Female | 110 (45.8) | 18 (40.9) | 29 (36.7) | 42 (47.2) | 21 (75.0) | 1 | 1 |
| Male | 130 (54.2) | 26 (59.1) | 50 (63.3) | 47 (52.8) | 7 (25.0) | 0.5 (0.3-0.8)** | 0.5 (0.3-0.9)* |
| Tongue coating score | |||||||
| 1 | 88 (36.7) | 28 (63.6) | 34 (43.0) | 26 (29.2) | 0 (0.0) | 1 | 1 |
| 2 | 106 (44.1) | 14 (31.8) | 38 (48.1) | 41 (46.1) | 13 (46.4) | 3.0 (1.8-5.2)** | 2.9 (1.6-5.3)*** |
| 3 | 46 (19.2) | 2 (4.5) | 7 (8.8) | 22 (24.7) | 15 (53.6) | 12.0 (5.8-25.2)*** | 13.1 (5.9-29.0)*** |
| Cluster of T-RFLP profile | |||||||
| I | 43 (17.9) | 23 (52.3) | 10 (12.7) | 7 (7.9) | 3 (10.7) | 1 | 1 |
| II | 90 (37.5) | 5 (11.4) | 27 (34.2) | 47 (52.8) | 11 (39.3) | 9.3 (4.5-19.4)*** | 11.4 (5.0-26.0)*** |
| III | 62 (25.8) | 11 (25.0) | 27 (34.2) | 18 (20.2) | 6 (21.4) | 3.9 (1.8-8.2)*** | 3.9 (1.7-8.8)** |
| IV | 45 (18.8) | 5 (11.4) | 15 (19.0) | 17 (19.1) | 8 (28.6) | 8.3 (3.7-19.0)*** | 9.8 (4.0-24.2)*** |
Copy number of total bacterial 16S rRNA gene was divided by that of Streptococcus mutans UA159 corresponding to 1 CFU per 500 μl.
Data are given either as means ± standard deviations or as the number of data with the percentage in parentheses. Each odds ratio and P values were calculated by ordinal logistic regression analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Mean periodontal pocket depth and tongue coating score also were associated with significantly higher odds ratios for higher levels of VSCs (Table 2) and organoleptic scores (Table 3), as expected. In addition, gender was significantly associated with the organoleptic score (Table 3). Although the total bacterial count in saliva had a significantly increased odds ratio for both a higher VSC level (Table 2) and higher organoleptic score (Table 3) in the univariate analysis, the relationship dissipated after multivariate adjustment.
Bacterial colonization patterns of each predicted cluster based on T-RFLP profiles.
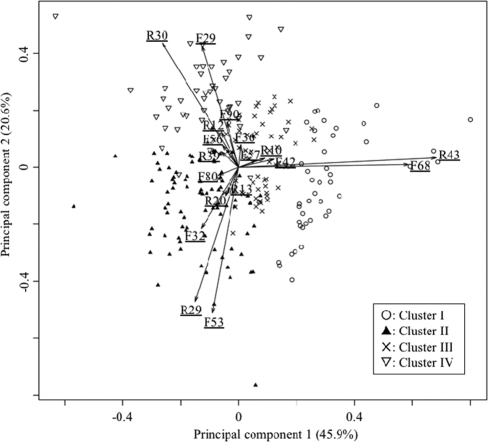
To visualize the differences in the T-RFLP profiles among the four clusters, they were analyzed by PCA and represented as four types of dots on a PCA biplot diagram of the first principal component (PC1) and the second principle component (PC2) (Fig. 2). These two components explain 66.5% of the total variance. The four clusters were localized in the right (cluster I), lower-left (cluster II), upper-left (cluster IV), and center (cluster III) areas of the diagram. Of a total of 164 TRFs, 18 were selected that had large loadings (>0.05 in absolute value on the PC1 and PC2) (Table 4) and are represented by arrows (Fig. 2). All of the TRFs were predominately and commonly detected in the T-RFLP profiles of most of the subjects (Table 4).
FIG. 2.
Principal component analysis (PCA) biplot diagram showing the relationship between variables (each TRF; arrows) and 240 T-RFLP profiles classified into four clusters by hierarchical cluster analysis (four types of dots). Only 18 TRFs with large loadings (>0.05 in absolute value on the first and second principal components) of 164 TRFs were selected and are represented by arrows. These two components explain 66.5% of the point variability.
TABLE 4.
Comparisons of peak area proportions of 18 TRFs with large loadings on the first two components in PCA in each cluster
| TRF | Loading of PC |
No. (%) of subjects detected in the TRF | Peak area proportions (%, means ± SD) for clusterb: |
Significant differences between clustersa | ||||
|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | I | II | III | IV | |||
| 6-FAM profile | ||||||||
| F29 | −0.13 | 0.43 | 237 (98.8) | 4.7 ± 3.1 | 6.8 ± 3.4 | 9.3 ± 5.7 | 15.3 ± 5.7 | IV > III, IV > II, IV > I, III > II, III > I, II > I |
| F32 | −0.13 | −0.22 | 240 (100) | 6.6 ± 4.0 | 10.9 ± 6.2 | 6.5 ± 3.3 | 5.1 ± 3.3 | II > I, II > III, II > IV |
| F36 | 0.01 | 0.08 | 151 (62.9) | 1.4 ± 3.1 | 0.9 ± 1.4 | 1.0 ± 6.3 | 3.8 ± 6.3 | IV > I, IV > II, IV > III |
| F42 | 0.12 | 0.03 | 240 (100) | 9.8 ± 4.7 | 5.1 ± 2.2 | 8.0 ± 3.1 | 6.3 ± 3.1 | I > II, I > III, I > IV, III > II, III > IV |
| F53 | −0.09 | 0.51 | 236 (98.3) | 9.1 ± 5.4 | 13.2 ± 5.5 | 8.4 ± 3.7 | 5.4 ± 3.7 | II > I, II > III, II > IV, I > IV, III > IV |
| F56 | −0.06 | 0.08 | 217 (90.4) | 1.0 ± 1.1 | 2.5 ± 1.6 | 2.3 ± 2.1 | 3.7 ± 2.1 | IV > I, IV > II, IV > III, II > I, III > I, |
| F57 | 0.06 | 0.02 | 239 (99.6) | 4.8 ± 1.9 | 2.8 ± 1.1 | 4.2 ± 1.3 | 3.2 ± 1.3 | I > II, I > IV, III > II, III > IV |
| F68 | 0.59 | 0.01 | 240 (100) | 43.7 ± 6.6 | 25.8 ± 5.1 | 33.4 ± 7.0 | 27.3 ± 7.0 | I > II, I > III, I > IV, III > II, III > IV |
| F80 | −0.07 | −0.03 | 231 (96.3) | 2.1 ± 1.5 | 5.0 ± 2.3 | 3.6 ± 2.5 | 4.0 ± 2.5 | II > I, II > III, II > IV, III > I, IV > I |
| F90 | −0.04 | 0.16 | 199 (82.9) | 1.8 ± 2.2 | 3.0 ± 2.7 | 3.4 ± 3.8 | 5.3 ± 3.8 | IV > I, IV > II, IV > III, III > I |
| HEX profile | ||||||||
| R10 | 0.09 | 0.03 | 234 (97.5) | 5.8 ± 3.5 | 2.3 ± 1.3 | 4.4 ± 2.1 | 3.4 ± 2.1 | I > II, I > III, I > IV, III > II, IV > II |
| R12 | −0.06 | 0.12 | 212 (88.3) | 1.3 ± 1.5 | 3.1 ± 2.3 | 3.1 ± 3.2 | 4.7 ± 3.2 | IV > I, IV > II, IV > III, II > I, III > I, |
| R13 | −0.04 | −0.09 | 238 (99.2) | 2.2 ± 1.8 | 4.1 ± 2.2 | 3.1 ± 1.7 | 2.6 ± 1.7 | II > I, II > III, II > IV |
| R20 | −0.05 | −0.10 | 212 (88.3) | 2.3 ± 2.2 | 3.5 ± 2.9 | 1.9 ± 1.5 | 1.3 ± 1.5 | II > I, II > III, II > IV |
| R29 | −0.15 | −0.47 | 238 (99.2) | 9.2 ± 5.2 | 15.3 ± 4.7 | 9.8 ± 3.7 | 7.5 ± 3.7 | II > I, II > III, II > IV |
| R30 | −0.26 | 0.44 | 240 (100) | 10.4 ± 4.1 | 16.6 ± 5.0 | 15.9 ± 5.0 | 25.3 ± 5.0 | IV > I, IV > II, IV > III, II > I, III > I |
| R39 | −0.06 | 0.06 | 233 (97.1) | 1.9 ± 1.4 | 3.8 ± 1.8 | 3.4 ± 2.2 | 4.4 ± 2.2 | IV > I, IV > III, II > I, III > I |
| R43 | 0.68 | 0.03 | 240 (100) | 53.7 ± 8.1 | 33.3 ± 6.0 | 42.7 ± 6.5 | 34.4 ± 6.5 | I > II, I > III, I > IV, III > II, III > IV |
Significant difference between clusters were evaluated by Tukey's multiple comparison (P < 0.05).
Peak area proportions that are significantly higher than those of the other three clusters are shown in boldface.
Five (F42, F57, F68, R10, and R43) of the 18 TRFs had large loadings in the direction in which cluster I was localized (Fig. 2, right area of the diagram). The T-RFLP profiles of cluster I were implicated as bacterial populations, with larger proportions of the bacterial species corresponding to these TRFs, whereas those of clusters II and IV localized in the negative direction were relatively less dominated by these species. The combinations of TRF putatively corresponded to Streptococcus, Granulicatella, Rothia, and Treponema (Table 5) based on the size of TRFs predicted from the 16S rRNA sequences in our oral bacteria database (33). Six TRFs (F32, F53, F80, R13, R20, and R29) had large loadings in the direction in which cluster II was localized (Fig. 2, lower left area of the diagram). The TRF combinations corresponded putatively to Prevotella and Veillonella (Table 5). Seven TRFs (F29, F36, F56, F90, R12, R30, and R39) had large loadings in the direction in which cluster IV was localized (Fig. 2, upper-left area of the diagram), and the TRF combinations putatively corresponded to Neisseria or Haemophilus or Aggregatibacter, Lautropia, Fusobacterium, Porphyromonas, and Parvimonas (Table 5). The T-RFLP profiles of cluster III localized at the center of the diagram were implicated as the intermediate pattern of the other three clusters.
TABLE 5.
Bacterial candidates corresponding to the TRF combinations
| TRF combination | Genus | Species |
|---|---|---|
| Cluster I | ||
| F42-R10 | Rothia | R. mucilaginosa and 1 phylotype |
| F42-R43 | Treponema | 4 phylotypes |
| F57-R43 | Streptococcus | S. anginosus, S. downei, S. pyogenes, and 1 phylotype |
| Granulicatella | G. adiacens | |
| F68-R10 | Streptococcus | S. mitis and 1 phylotype |
| F68-R43 | Streptococcus | S. australis, S. cristatus, S. mitis, S. oligofermentans, S. oralis, S. parasanguinis, S. peroris, S. pneumoniae, S. salivarius, S. sanguinis, S. suis, and 31 phylotypes |
| Cluster II | ||
| F32-R20 | Veillonella | V. dispar, V. parvula, and 3 phylotypes |
| F53-R13 | Prevotella | P. dentalis, P. denticola, P. loeschei, P. oulorum, and 16 phylotypes |
| F53-R29 | Prevotella | P. melaninogenica, P. tannerae, and 8 phylotypes |
| F80-R13 | Prevotella | P. veroralis |
| F80-R29 | Prevotella | 1 phylotype |
| Cluster IV | ||
| F29-R30 | Neisseria | N. cinerea, N. elongata, N. subflava, N. weaveri, and 6 phylotypes |
| Haemophilus | H. influenza, H. parainfluenza, H. pittmaniae, H. quentini, and 4 phylotypes | |
| Aggregatibacter | A. actinomycetemcomitans and A. segnis | |
| F36-R30 | Lautropia | L. mirabilis |
| F56-R12 | Parvimonas | P. micra and 5 phylotypes |
| F56-R39 | Fusobacterium | F. nucleatum, F. periodonticum, and 6 phylotypes |
| F90-R12 | Porphyromonas | 6 phylotypes |
The peak area proportions of the characteristic TRFs of each cluster, except for F57 and R39, were significantly greater than those of the other clusters (Table 4). The mean peak area proportion of F57 in cluster I and that of R39 in cluster IV also were greater than those of the other three clusters, although statistical significance was not observed in the difference found in one of the three clusters. The distribution of the peak area proportions of these TRFs strongly influenced the cluster classification of this study.
Other characteristics of the bacterial population structure of each cluster.
Although cluster analysis in this study was influenced mainly by the relative abundance of the dominant TRFs mentioned above, other characteristics of bacterial population structure also were explored from the obtained T-RFLP data. The diversity of the bacterial community, which was determined by the total number of TRFs and the Shannon diversity index of the T-RFLP profile, were significantly lower in cluster I of both the 6-FAM and HEX profiles (Table 6). Cluster II exhibited the highest values among the four clusters on both parameters, although statistical significance was not observed between clusters II and IV with the total number of TRFs in the HEX profiles (Table 6).
TABLE 6.
Diversity of the T-RFLP profiles in each cluster
| Profile and category | No. with T-RFLP profile (means ± SD) in cluster: |
Significant differences between clustersa | |||
|---|---|---|---|---|---|
| I (n = 43) | II (n = 90) | III (n = 62) | IV (n = 45) | ||
| Total no. of TRFs | |||||
| 6-FAM | 17.9 ± 3.8 | 23.7 ± 4.0 | 21.6 ± 3.7 | 21.9 ± 3.2 | II > I, III > I, IV > I, II > III, II > IV |
| HEX | 12.1 ± 2.6 | 15.9 ± 2.5 | 14.3 ± 1.8 | 15.1 ± 1.9 | II > I, III > I, IV > I, II > III |
| Shannon diversity index | |||||
| 6-FAM | 1.9 ± 0.2 | 2.4 ± 0.2 | 2.2 ± 0.1 | 2.3 ± 0.2 | II > I, III > I, IV > I, II > III, II > IV |
| HEX | 1.5 ± 0.2 | 2.0 ± 0.1 | 1.8 ± 0.1 | 1.9 ± 0.1 | II > I, III > I, IV > I, II > III, II > IV |
Significant difference between clusters were evaluated by Tukey's multiple comparison (P < 0.05).
The detection frequencies of each TRF with significantly more or fewer subjects are listed in Table 7. Thirteen TRFs were detected less frequently in cluster I, whereas five and two TRFs in clusters II and IV were detected from more subjects, respectively. The well-known malodor producers, such as Porphyromonas gingivalis (F97 and R65), Porphyromonas endodontalis (F97, F98, and R65), Parvimonas micra (F56 and R12), Prevotella intermedia (F19), and Fusobacterium nucleatum (F56), were included as candidates corresponding to the TRFs less frequently detected in cluster I.
TABLE 7.
Detection frequencies of the TRFs with significantly more or fewer subjects in one cluster than in the other clusters
| TRF | Detection frequency in clustera (%): |
Bacterial candidates corresponding to each TRF | |||
|---|---|---|---|---|---|
| I | II | III | IV | ||
| F19 | 41.9 | 90.0 | 82.3 | 75.6 | Prevotella intermedia, P. nigrescens, and P. pallens |
| F34 | 23.3 | 55.6 | 25.8 | 6.7 | Dialister pneumosintes, 3 phylotypes of Megasphaera, and 1 phylotype of Prevotella |
| F38 | 25.6 | 50.0 | 51.6 | 73.3 | Neisseria polysaccharea and 2 phylotypes, Bergeriella denitrificans, Simonsiella muelleri, Vitreoscilla stercoraria, and 3 phylotypes of Treponema |
| F44 | 30.2 | 71.1 | 54.8 | 48.9 | Actinomyces naeslundii and 4 phylotypes, Eubacterium sulci, E. infirmum, E. saphenum and 6 phylotypes |
| Mogibacterium timidum and M. pumilum, Guggenheimella bovis, and 1 phylotype of Peptococcus | |||||
| F48 | 9.3 | 45.6 | 40.3 | 44.4 | Leptotrichia hofstadii and 8 phylotypes, Kingella denitrificans, Cardiobacterium hominis, Acinetobacter baumannii, and Mycoplasma pneumoniae |
| F52 | 46.5 | 55.6 | 66.1 | 86.7 | Five phylotypes of Prevotella, 2 phylotypes of Treponema, 1 phylotype of Bacteroidales, and 1 phylotype of Ruminococcaceae |
| F56 | 69.8 | 94.4 | 93.5 | 97.8 | Parvimonas micra and 5 phylotypes, Fusobacterium nucleatum, F. periodonticum and 6 phylotypes, and 2 phylotypes of Shuttleworthia |
| F59 | 81.4 | 88.9 | 82.3 | 57.8 | Lactobacillus salivarius |
| F71 | 2.3 | 53.3 | 24.2 | 11.1 | 2 phylotypes of Lachnospiraceae |
| F80 | 86.0 | 97.8 | 100.0 | 97.8 | Prevotella veroralis and 1 phylotype |
| F90 | 65.1 | 84.4 | 87.1 | 91.1 | Six phylotypes of Porphyromonas |
| F97 | 20.9 | 58.9 | 51.6 | 64.4 | Porphyromonas gingivalis, P. endodontalis and 5 phylotypes |
| F98 | 14.0 | 45.6 | 21.0 | 17.8 | Porphyromonas endodontalis and 5 phylotypes |
| R12 | 67.4 | 92.2 | 93.5 | 93.3 | Parvimonas micra and 5 phylotypes, six phylotypes of Porphyromonas, Leptotrichia hofstadii and 4 phylotypes, and 2 phylotypes of Pedobacter |
| R13 | 83.7 | 100.0 | 98.4 | 97.8 | Prevotella dentalis, P. denticola, P. loescheii, P. oulorum, P. oris, P. veroralis and 24 phylotypes |
| R27 | 41.9 | 81.1 | 79.0 | 86.7 | 4 phylotypes of Prevotella |
| R41 | 11.6 | 50.0 | 45.2 | 68.9 | Mogibacterium timidum and M. pumilum, Flexibacter litoralis, 2 phylotypes of Anaerovorax, Guggenheimella bovis, 3 phylotypes of Parvimonas, and 1 phylotype of Bacteroidales |
| R46 | 69.8 | 96.7 | 82.3 | 80.0 | Dialister pneumosintes and Selenomonas ruminantium |
| R51 | 20.9 | 51.1 | 21.0 | 15.6 | Prevotella tannerae and 1 phylotype, and 2 phylotypes of Bacteroidales |
| R65 | 25.6 | 74.4 | 77.4 | 77.8 | Porphyromonas gingivalis, P. endodontalis and 5 phylotypes |
The values significantly higher or lower than those of the other clusters by Tukey's multiple comparison (P < 0.05) are shown in boldface.
DISCUSSION
In the present study, the global composition pattern of bacterial populations in saliva was clearly correlated with the severity of oral malodor. The cluster grouping in this study, which was highly influenced by the peak area proportions of the dominant TRFs frequently detected in the T-RFLP profiles of most subjects (Table 4, Fig. 2), classified the bacterial population structure into four patterns with different proportions of indigenous members. The microbial community compositions belonging to cluster I demonstrated significantly lower levels of malodor productivity than the other patterns of microbiota. The T-RFLP profiles of this cluster were associated with bacterial populations with higher proportions of Streptococcus, Granulicatella, Rothia, and Treponema species than those of the other clusters.
By using the molecular approach of 16S rRNA sequence analysis, differences in bacterial compositions between subjects with high and no or low odor have been comprehensively evaluated in recent studies (9, 11, 30). Although Kazor et al. also mentioned the predominance of Streptococcus salivarius in relation to low malodor, those studies mainly sought to identify bacterial species that were specifically associated with subjects with strong oral malodor. Since most of the bacterial species identified were detected in subjects in both the high and no/low odor groups, Riggio et al. concluded that differences in populations would be quantitative rather than qualitative. Also in the present study, a TRF specifically found in subjects with strong oral malodor was not detected (data not shown). On the other hand, we correlated the relative proportions of the major indigenous bacteria with oral malodor rather than the presence or absence of specific bacterial species. The characteristic bacteria in each cluster were genera commonly found in previous studies regardless of malodor intensity (5, 11, 30).
The bacterial population structures suggested by our results are reasonable and consistent with the preceding suggestion from a metabolic perspective (13). The TRFs with characteristically higher peak area proportions in cluster I correspond to gram-positive saccharolytic species, except for four phylotypes of Treponema. Although Treponema species also were included among the candidates of the corresponding TRFs, it is unlikely that these species were found predominantly in the saliva of many subjects, considering that only a few clones have been obtained from bacterial populations in saliva (12, 25). In both T-RFLP profiles that were strongly associated with oral malodor (clusters II and IV), the TRFs corresponding to gram-positive saccharolytic species also were dominant, but in lower proportions than those of cluster I (Table 4), whereas all of the predicted characteristic bacteria were asaccharolytic or proteolytic gram-negative bacteria. In particular, the Fusobacterium, Porphyromonas, and Parvimonas bacterial species that characterized cluster IV have been found to be active producers of VSCs in vitro (28). The Prevotella and Veillonella species characteristic of cluster II have been indicated to be the most frequently detected H2S-producing bacteria in tongue microbiota and were observed in greater amounts among subjects with oral malodor than in healthy patients (37). However, whether these species are directly involved in the development of oral malodor remains uncertain. Considering that the diversity of the bacterial populations was significantly higher in clusters II and IV than in cluster I (Table 6), the patterns of the bacterial communities may simply reflect the suitability of the environment for the growth of minor but more important species for malodor production. Rather, it is remarkable that cluster I, which was implicated as having higher proportions of gram-positive saccharolytic species, was less diverse than the other clusters (Table 6). In addition, the well-known malodor producers, such as P. gingivalis, were included as candidates corresponding to the TRFs less frequently detected in cluster I (Table 7). Facilitating the growth of these gram-positive saccharolytic species may prevent the growth of minor species and thus may help to reduce the production of oral malodor.
Most noteworthy in this study were the significant differences in malodor productivity based on the bacterial population structure that were observed following adjustment for potential confounding factors, such as the mean periodontal pocket depth and tongue coating score (Tables 2 and 3). Previous studies indicated that an increase in tongue coating and periodontal disease were two major halitosis-inducing factors (23, 39) that create areas allowing the overgrowth of anaerobic bacteria, many of which have an ability to produce malodorous components. In the present study, differences in bacterial colonization patterns were significantly associated with the intensity of oral malodor independently of these two factors, suggesting that they reflect the oral condition involved in malodor production, which is difficult to evaluate solely by the visual inspection of oral hygiene and periodontal conditions. Considering that the predominant bacterial composition in saliva is relatively stable over time (29, 31), the composition may be implicated in individuals' potential ability to produce oral malodor. On the other hand, the significant relationship between the total bacteria in saliva and oral malodor dissipated after adjustment in the multivariate analysis (Tables 2 and 3). Although it may be controversial to conclude that the total number of bacteria in saliva reflects the amount of bacteria residing in the entire oral cavity, these results indicate that the composition of bacterial populations, rather than the total quantity, is relevant to the development of oral malodor. Current physical or chemical treatments for oral malodor focus on the reduction of the total bacterial count. Although further studies based on the alteration of oral malodor intensity are required, our results suggest the necessity of supplemental treatments to completely cure oral malodor by improving the quality of the indigenous bacterial populations.
In this study, two patterns of bacterial communities (clusters II and IV) were equally implicated as malodor-associated microbiota. Conversely, the total bacterial count was significantly greater in cluster II than in cluster IV (Table 1). The findings suggest that the bacterial populations of clusters II and IV are and are not accompanied by bacterial overgrowth, respectively. The conventional, steadfast mouth-cleaning routine for treating oral malodor may be less effective for subjects possessing bacteria from cluster IV than for those with bacteria from cluster II.
The bacterial prediction made by the present T-RFLP analysis was unable to fully characterize the bacterial community at the species level. Although additional restriction enzyme digestions may reduce the bacterial candidates to those corresponding only to the TRFs, it may interfere with the proportional distribution of bacteria due to the lack of a statistical method capable of dealing with multiple digestion patterns. Cloning and sequencing analyses are more efficient for characterizing bacterial communities at the species level. However, an analysis of 240 subjects remains too laborious and expensive to perform at present, although further insight might be gained by such effort. On the other hand, our main concern in this study was to correlate the patterns of the bacterial community composition with the severity of malodor rather than to identify the causative agents. We consider that the present T-RFLP analysis was sufficient for the purpose intended. Recently, novel high-throughput approaches, such as microbe identification microarrays (27) and bar-coded pyrosequencing (12, 25), have been implemented for the bacterial identification of oral microbial communities. The combinational use of these approaches and innovative cultivation methods may be helpful in identifying the actual malodor producers.
In other organs colonized by commensal microbiota, many studies have reported on the relationship between the global composition of indigenous bacterial populations and human health. For example, the relative proportions of two major bacterial phyla, Firmicutes and Bacteroidetes, in intestinal microbiota are associated with obesity (17), inflammatory bowel disease (6), and experimental type I diabetes (38). Vaginal microbiota shift from the usual lactobacillus-dominated microbiota to diverse communities with anaerobic and facultative bacteria in bacterial vaginosis (7). A variety of bacteria densely colonize the oral cavity as commensal organisms, whereas the relative proportions of indigenous bacteria have not received much attention in relation to oral health. The results of this investigation clearly demonstrate that oral malodor is a symptom based on the characteristic occupation of indigenous oral bacterial populations, rather than solely on bacterial overgrowth due to poor oral hygiene. The observation of oral bacterial populations from a broad ecological view may provide novel insights into human health and other disorders within the oral cavity.
Acknowledgments
This study was supported in part by Grants-in-Aid for Young Scientists 21890187 (T.T.) and 19791645 (N.S.), by Grants-in-Aid for Scientific Research 21592652 (Y.N.), 20592458 (Y.S.), 20592249 (M.Y.), 18592296 (T.H.), 19390541 (Y.Y.), and 21659486 (Y.Y.), and by Grants-in-Aid for Scientific Research (Strategic Research Promotion) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 12 March 2010.
REFERENCES
- 1.Barrons, R., and D. Tassone. 2008. Use of Lactobacillus probiotics for bacterial genitourinary infections in women: a review. Clin. Ther. 30:453-468. [DOI] [PubMed] [Google Scholar]
- 2.Burton, J. P., C. N. Chilcott, C. J. Moore, G. Speiser, and J. R. Tagg. 2006. A preliminary study of the effect of probiotic Streptococcus salivarius K12 on oral malodour parameters. J. Appl. Microbiol. 100:754-764. [DOI] [PubMed] [Google Scholar]
- 3.Culligan, E. P., C. Hill, and R. D. Sleator. 2009. Probiotics and gastrointestinal disease: successes, problems and future prospects. Gut Pathog. 1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denepitiya, L., and I. Kleinberg. 1982. A comparison of the microbial compositions of pooled human dental plaque and salivary sediment. Arch. Oral Biol. 27:739-745. [DOI] [PubMed] [Google Scholar]
- 5.Donaldson, A. C., D. McKenzie, M. P. Riggio, P. J. Hodge, H. Rolph, A. Flanagan, and J. Bagg. 2005. Microbiological culture analysis of the tongue anaerobic microflora in subjects with and without halitosis. Oral Dis. 11(Suppl. 1):61-63. [DOI] [PubMed] [Google Scholar]
- 6.Frank, D. N., A. L. St. Amand, R. A. Feldman, E. C. Boedeker, N. Harpaz, and N. R. Pace. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fredricks, D. N., T. L. Fiedler, and J. M. Marrazzo. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353:1899-1911. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg, S., A. Kozlovsky, D. Gordon, I. Gelernter, A. Sintov, and M. Rosenberg. 1994. Cadaverine as a putative component of oral malodor. J. Dent. Res. 73:1168-1172. [DOI] [PubMed] [Google Scholar]
- 9.Haraszthy, V. I., J. J. Zambon, P. K. Sreenivasan, M. M. Zambon, D. Gerber, R. Rego, and C. Parker. 2007. Identification of oral bacterial species associated with halitosis. J. Am. Dent. Assoc. 138:1113-1120. [DOI] [PubMed] [Google Scholar]
- 10.Hartmann, M., and F. Widmer. 2008. Reliability for detecting composition and changes of microbial communities by T-RFLP genetic profiling. FEMS Microbiol. Ecol. 63:249-260. [DOI] [PubMed] [Google Scholar]
- 11.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41:558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keijser, B. J., E. Zaura, S. M. Huse, J. M. van der Vossen, F. H. Schuren, R. C. Montijn, J. M. ten Cate, and W. Crielaard. 2008. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 87:1016-1020. [DOI] [PubMed] [Google Scholar]
- 13.Kleinberg, I., and M. Codipilly. 1995. The biological basis of oral malodor formation, p. 13-40. In M. Rosenberg (ed.), Bad breath: research perspectives. Ramot Publishing, Tel Aviv, Israel.
- 14.Kleinberg, I., and G. Westbay. 1990. Oral malodor. Crit. Rev. Oral. Biol. Med. 1:247-259. [DOI] [PubMed] [Google Scholar]
- 15.Könönen, E., S. Paju, P. J. Pussinen, M. Hyvonen, P. Di Tella, L. Suominen-Taipale, and M. Knuuttila. 2007. Population-based study of salivary carriage of periodontal pathogens in adults. J. Clin. Microbiol. 45:2446-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostelc, J. G., P. R. Zelson, G. Preti, and J. Tonzetich. 1981. Quantitative differences in volatiles from healthy mouths and mouths with periodontitis. Clin. Chem. 27:842-845. [PubMed] [Google Scholar]
- 17.Ley, R. E., P. J. Turnbaugh, S. Klein, and J. I. Gordon. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022-1023. [DOI] [PubMed] [Google Scholar]
- 18.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loesche, W. J., and C. Kazor. 2002. Microbiology and treatment of halitosis. Periodontology 2000 28:256-279. [DOI] [PubMed] [Google Scholar]
- 20.Mager, D. L., A. D. Haffajee, and S. S. Socransky. 2003. Effects of periodontitis and smoking on the microbiota of oral mucous membranes and saliva in systemically healthy subjects. J. Clin. Periodontol. 30:1031-1037. [DOI] [PubMed] [Google Scholar]
- 21.Mager, D. L., L. A. Ximenez-Fyvie, A. D. Haffajee, and S. S. Socransky. 2003. Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30:644-654. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki, H., M. Arao, K. Okamura, K. Kawaguch, A. Toyofuku, K. Hoshi, and K. Yaegaki. 1999. Tentative classification for halitosis patients and its treatment needs. J. Niigata Dent. J. 32:11-15. [Google Scholar]
- 23.Miyazaki, H., S. Sakao, Y. Katoh, and T. Takehara. 1995. Correlation between volatile sulphur compounds and certain oral health measurements in the general population. J. Periodontol. 66:679-684. [DOI] [PubMed] [Google Scholar]
- 24.Nakano, Y., T. Takeshita, N. Kamio, S. Shiota, Y. Shibata, M. Yasui, and Y. Yamashita. 2008. Development and application of a T-RFLP data analysis method using correlation coefficient matrices. J. Microbiol. Methods 75:501-505. [DOI] [PubMed] [Google Scholar]
- 25.Nasidze, I., J. Li, D. Quinque, K. Tang, and M. Stoneking. 2009. Global diversity in the human salivary microbiome. Genome Res. 19:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oho, T., Y. Yoshida, Y. Shimazaki, Y. Yamashita, and T. Koga. 2001. Characteristics of patients complaining of halitosis and the usefulness of gas chromatography for diagnosing halitosis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 91:531-534. [DOI] [PubMed] [Google Scholar]
- 27.Paster, B. J., and F. E. Dewhirst. 2009. Molecular microbial diagnosis. Periodontology 2000 51:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Persson, S., M. B. Edlund, R. Claesson, and J. Carlsson. 1990. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol. Immunol. 5:195-201. [DOI] [PubMed] [Google Scholar]
- 29.Rasiah, I. A., L. Wong, S. A. Anderson, and C. H. Sissons. 2005. Variation in bacterial DGGE patterns from human saliva: over time, between individuals and in corresponding dental plaque microcosms. Arch. Oral Biol. 50:779-787. [DOI] [PubMed] [Google Scholar]
- 30.Riggio, M. P., A. Lennon, H. J. Rolph, P. J. Hodge, A. Donaldson, A. J. Maxwell, and J. Bagg. 2008. Molecular identification of bacteria on the tongue dorsum of subjects with and without halitosis. Oral Dis. 14:251-258. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto, M., Y. Takeuchi, M. Umeda, I. Ishikawa, and Y. Benno. 2003. Application of terminal RFLP analysis to characterize oral bacterial flora in saliva of healthy subjects and patients with periodontitis. J. Med. Microbiol. 52:79-89. [DOI] [PubMed] [Google Scholar]
- 32.Scully, C., S. Porter, and J. Greenman. 1994. What to do about halitosis. BMJ 308:217-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeshita, T., Y. Nakano, T. Kumagai, M. Yasui, N. Kamio, Y. Shibata, S. Shiota, and Y. Yamashita. 2009. The ecological proportion of indigenous bacterial populations in saliva is correlated with oral health status. ISME J. 3:65-78. [DOI] [PubMed] [Google Scholar]
- 34.Takeshita, T., Y. Nakano, and Y. Yamashita. 2007. Improved accuracy in terminal restriction fragment length polymorphism phylogenetic analysis using a novel internal size standard definition. Oral Microbiol. Immunol. 22:419-428. [DOI] [PubMed] [Google Scholar]
- 35.Tonzetich, J. 1971. Direct gas chromatographic analysis of sulphur compounds in mouth air in man. Arch. Oral Biol. 16:587-597. [DOI] [PubMed] [Google Scholar]
- 36.Tonzetich, J., and S. K. Ng. 1976. Reduction of malodor by oral cleansing procedures. Oral Surg. Oral Med. Oral Pathol. 42:172-181. [DOI] [PubMed] [Google Scholar]
- 37.Washio, J., T. Sato, T. Koseki, and N. Takahashi. 2005. Hydrogen sulfide-producing bacteria in tongue biofilm and their relationship with oral malodour. J. Med. Microbiol. 54:889-895. [DOI] [PubMed] [Google Scholar]
- 38.Wen, L., R. E. Ley, P. Y. Volchkov, P. B. Stranges, L. Avanesyan, A. C. Stonebraker, C. Hu, F. S. Wong, G. L. Szot, J. A. Bluestone, J. I. Gordon, and A. V. Chervonsky. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455:1109-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yaegaki, K., and K. Sanada. 1992. Volatile sulfur compounds in mouth air from clinically healthy subjects and patients with periodontal disease. J. Periodont. Res. 27:233-238. [DOI] [PubMed] [Google Scholar]