Abstract
Assessment of the risks posed by severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) on surfaces requires data on survival of this virus on environmental surfaces and on how survival is affected by environmental variables, such as air temperature (AT) and relative humidity (RH). The use of surrogate viruses has the potential to overcome the challenges of working with SARS-CoV and to increase the available data on coronavirus survival on surfaces. Two potential surrogates were evaluated in this study; transmissible gastroenteritis virus (TGEV) and mouse hepatitis virus (MHV) were used to determine effects of AT and RH on the survival of coronaviruses on stainless steel. At 4°C, infectious virus persisted for as long as 28 days, and the lowest level of inactivation occurred at 20% RH. Inactivation was more rapid at 20°C than at 4°C at all humidity levels; the viruses persisted for 5 to 28 days, and the slowest inactivation occurred at low RH. Both viruses were inactivated more rapidly at 40°C than at 20°C. The relationship between inactivation and RH was not monotonic, and there was greater survival or a greater protective effect at low RH (20%) and high RH (80%) than at moderate RH (50%). There was also evidence of an interaction between AT and RH. The results show that when high numbers of viruses are deposited, TGEV and MHV may survive for days on surfaces at ATs and RHs typical of indoor environments. TGEV and MHV could serve as conservative surrogates for modeling exposure, the risk of transmission, and control measures for pathogenic enveloped viruses, such as SARS-CoV and influenza virus, on health care surfaces.
Environmental surfaces have been recognized as likely contributors to the transmission of nosocomial viral infections (25). The question of whether hospital surfaces play a role in the spread of nosocomial viral infection took on particular urgency during the worldwide outbreak of severe acute respiratory syndrome (SARS). SARS was a novel coronavirus infection, and local and institutional outbreaks were driven in part by nosocomial spread; cases of SARS were documented in health care workers, patients, and visitors in health care facilities (20). During outbreaks in health care facilities, surface sampling for SARS coronavirus (SARS-CoV) revealed SARS-CoV nucleic acids on surfaces and inanimate objects (6, 10). This suggests that surfaces could be sources of virus transmission. Assessment of the risk posed by SARS-CoV on surfaces requires data on the survival of the virus on environmental surfaces and data on how this survival is affected by environmental variables, such as air temperature (AT) and relative humidity (RH).
Because working with SARS-CoV requires specially trained personnel working under biosafety level 3 (BSL-3) laboratory containment conditions, there are significant challenges in studying this virus, and only limited data on its survival and response to environmental stressors are available. The use of surrogate coronaviruses has the potential to overcome these challenges and expand the available data on coronavirus survival on surfaces.
In addition to SARS-CoV, there are two pathogenic human coronaviruses that are adapted to propagation and assay in cell culture, 229E and OC43, which could serve as surrogates for SARS-CoV in survival studies. However, previous studies suggested that the survival of 229E and OC43 on surfaces may be shorter than that of SARS-CoV (10, 35). To evaluate surrogates that might serve as more conservative models of SARS-CoV on surfaces, animal coronaviruses were chosen as surrogates for this study. Because SARS-CoV does not fall clearly into either of the two groups of mammalian coronaviruses, the following two potential surrogates representing the two groups were evaluated: transmissible gastroenteritis virus (TGEV), a diarrheal pathogen of swine and a member of coronavirus group 1, and mouse hepatitis virus (MHV), a respiratory and enteric pathogen of laboratory mice and a member of coronavirus group 2 (16). The advantages of using these two viruses as surrogates are the fact that they can be readily propagated and assayed in cell culture systems and the fact that there is no human infection risk. There has been some study of TGEV survival in aerosols (17), but the data on the environmental survival of this potential coronavirus surrogate for SARS-CoV are limited. The use of surrogates for studying the environmental survival of SARS-CoV can increase our understanding of the survival and persistence of this virus on environmental surfaces, the possible role of such surfaces in the transmission of SARS-CoV and other coronaviruses, and the risk posed by contaminated surfaces in outbreak settings. Therefore, this work was undertaken to determine the effect of AT and RH on the survival of the surrogate coronaviruses TGEV and MHV on hard nonporous surfaces.
MATERIALS AND METHODS
Preparation of viral stocks.
Viruses and cell lines were kindly provided by Ralph Baric, University of North Carolina, Chapel Hill, NC. TGEV was grown in swine testicular (ST) cell cultures, and MHV was grown in delayed brain tumor (DBT) cell cultures. Viral stocks were propagated by infecting confluent layers of host cell cultures in flasks, harvesting cell lysates, clarifying the preparations by centrifugation (3,000 × g, 30 min, 4°C), and storing the resulting supernatants as virus stocks at −80°C. Viral titers were determined by using quantal assays for cytopathic effects (CPE) and are expressed below as most probable numbers (MPN). Assays were performed with confluent host cell layers in 24-well plates containing maintenance medium consisting of Eagle's minimum essential medium (MEM), 10% bovine serum replacement (fetal clone II; HyClone, Logan, UT), 10% lactalbumin hydrolysate (Becton Dickinson Co. Sparks, MD), and gentamicin (0.1 mg/ml)-kanamycin (0.05 mg/ml).
Controlled environments.
Nine sets of environmental conditions were tested. The ATs used were 4°C, 20°C, and 40°C. At each AT, RH of 20% ± 3%, 50% ± 3%, and 80% ± 3% were used. Controlled RH environments were created in sealed containers. Environments with 20% RH were created using calcium sulfate granules (Drierite Co., Xenia, OH). Saturated solutions of magnesium nitrate were used for 50% RH environments. For 80% RH environments, saturated solutions of sodium chloride (4°C), ammonium chloride (20°C), and sodium bromide (40°C) were used. AT and RH were monitored using digital monitors (Fisher Scientific, Waltham, MA).
Survival experiments.
SARS-CoV is excreted in body fluids, such as respiratory secretions and feces, that have high levels of protein and other biological organic matter. It is likely that virus deposited on health care surfaces is embedded in such proteinaceous, biological organic matter matrices. To simulate deposition and survival in such matrices, viral inocula were suspended in cell culture medium (which contained proteins, other organic biomolecules, physiological salts, and other constituents and resembled human secretions) and placed on test surfaces. The test surfaces were carriers that were 1-cm2 thin stainless steel coupons with a no. 4 polish. The carriers were prepared by washing them in 0.01% Tween 80, followed by one rinse with 70% ethanol, one rinse with distilled water, and autoclaving. For survival tests, 10 μl containing 104 to 105 MPN of test viruses was inoculated onto three replicate carriers per time point and placed in a controlled RH environment. Control carriers (time zero) were kept at each AT and RH until they were dry and were sampled immediately after drying. The exception was the carrier incubated at 40°C and 80% RH. Under these conditions, virus inactivation took place within a few hours. Consequently, the carriers were sampled beginning at 2 h after inoculation, when the virus inoculum was still wet.
Sampling intervals were chosen after preliminary experiments to assess the maximum length of virus survival were performed. At 4°C and all RH values and at 20°C and 20% RH, carriers were sampled at 7-day intervals for up to 28 days. At 20°C and 50% RH or 80% RH, carriers were sampled at 24-h intervals until virus was no longer detectable. At 40°C, carriers were sampled at 2-h intervals until virus was not detectable. Based on virus recovery experiments, 1.5% beef extract (pH 7.5) was used to elute viruses from carriers (data not shown). At desired time points, carriers were removed, placed in a 24-well plate, and covered with 1 ml beef extract. Viruses were eluted by agitation on a shaking platform (60 rpm) for 20 min at room temperature. Eluted samples were diluted in cell culture medium, and virus infectivity was assayed using the appropriate host cell line. Three replicate carriers were assayed per time point. The virus survival at each time point was expressed as log10(Nt/N0), where Nt is the virus concentration (in MPN/ml) at time t and N0 is the initial virus concentration (in MPN/ml) in the control sample at time zero.
Statistical analysis.
The statistical analysis was done using SAS (SAS Corp., Cary, NC) and GraphPad Prism 5 (GraphPad, San Diego, CA). The parameter log10(Nt/N0) versus time was used to perform a regression analysis for each virus and AT-RH condition. At the level of an individual measurement of log10 inactivation for each day, linear regression was conducted to determine the slope of the inactivation line. Slopes were also determined by linear regression using the mean log10 reduction value for each time point (3 replicates per point). Analysis of covariance (ANCOVA) was used to assess the effects of AT, RH, time, and virus type on viral inactivation at the level of each individual measurement. The ANCOVA model is Yijk = μ0 + (μ1 + αi + βj + γk)t + ɛijk., where Yijk is log10 inactivation and the variables in the model are as follows: αi is the virus (TGEV or MHV; i = 1 or 2), βj is temperature (4°, 20°, and 40°C; j = 1, 2, or 3), γk is RH (20%, 50%, and 80% RH; k = 1, 2, or 3), and t is the number of days exposed to each condition. ANCOVA was also used to determine if there was an interaction between AT and RH as a predictors of virus survival. This adds an interaction term, δjk, to the model described above for effects on log10 inactivation. Using this term allowed evaluation of whether the coefficient days depends on the interaction between AT and RH, as well as each of the AT-RH conditions.
RESULTS
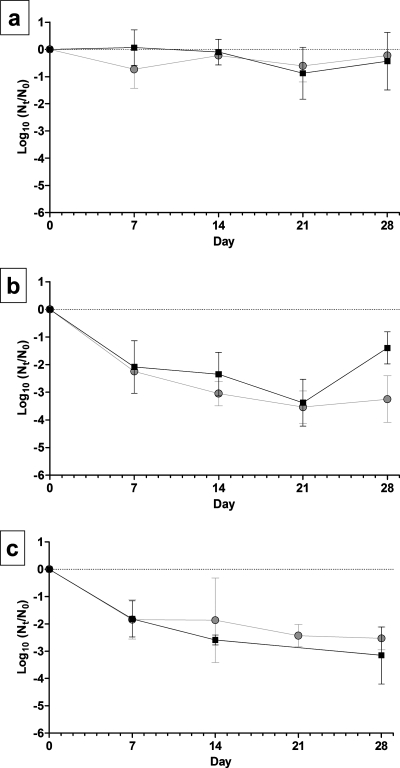
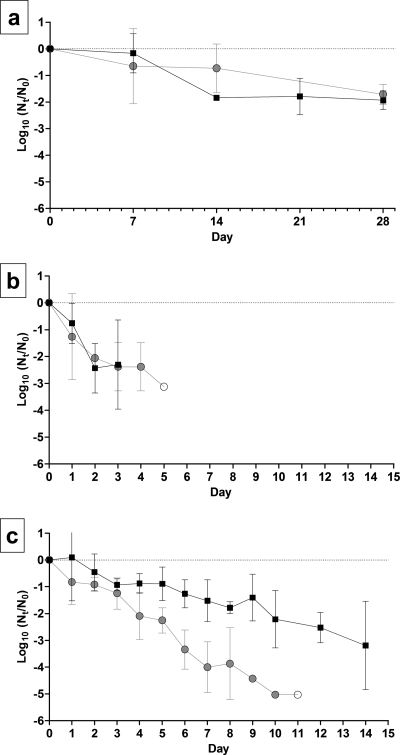
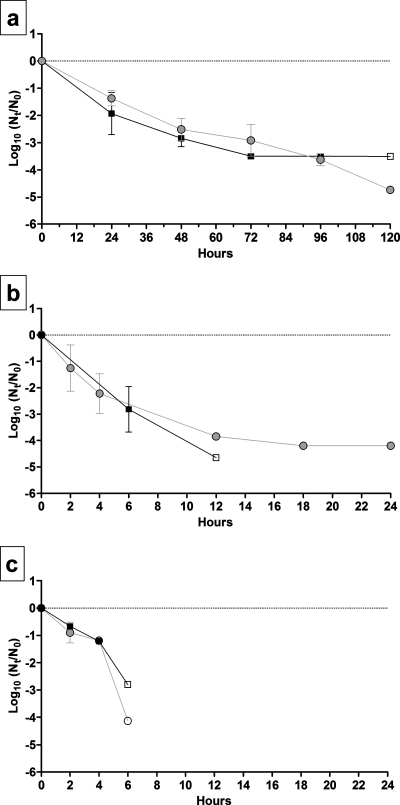
Inactivation of TGEV and MHV over time was measured for nine combinations of AT and RH. The survival of TGEV and MHV at 4°C and three RH levels is shown in Fig. 1. At 4°C, infectious virus deposited on stainless steel surfaces at initial levels of 4 to 5 log10 MPN persisted for as long as 28 days, and the lowest level of inactivation over the 28-day experiment took place at 20% RH. There was a decrease in the observed log10 inactivation rate at 20% and 50% RH from day 21 to day 28. To better simulate the physical state in which viruses in patient secretions are deposited onto surfaces, viruses were not dispersed before inoculation. Therefore, possible clumping and aggregation effects may have resulted in variation in the physical state of the viral inocula on individual carriers, possibly contributing to the variations in the rates of reduction observed. With the exception of the 20% RH model, every linear regression model was statistically significant (P < 0.05) (the coefficient of the linear model was not equal to zero). The slopes of regression lines for TGEV and MHV inactivation at 4°C and each RH are shown in Table 1. The levels of both viruses declined by <0.5 log10 over 28 days at 20% RH. Greater reduction took place at 50% RH, at which the levels of both viruses declined by ∼3.5 log10 after 21 days. At 80% RH, the TGEV level declined by 3.2 log10 over 28 days, and the MHV level declined by 2.5 log10.
FIG. 1.
Survival of TGEV and MHV at 4°C and (a) 20% RH, (b) 50% RH, and (c) 80% RH. Squares, TGEV; circles, MHV. The error bars indicate 95% confidence intervals.
TABLE 1.
Slopes of regression lines for virus inactivation at 4°C and 20, 50, and 80% RH
| RH | Replicate | MHV |
TGEV |
||||
|---|---|---|---|---|---|---|---|
| Slope | P | Mean slope | Slope | P | Mean slope | ||
| 20 | 1 | −0.029 | 0.2124 | −0.031 | −0.016 | 0.1082 | −0.021 |
| 2 | −0.036 | 0.0197 | −0.025 | 0.0124 | |||
| 3 | −0.027 | 0.1248 | −0.023 | 0.1792 | |||
| 50 | 1 | −0.161 | 0.0016 | −0.152 | −0.113 | 0.0415 | −0.107 |
| 2 | −0.152 | 0.0042 | −0.111 | 0.0211 | |||
| 3 | −0.143 | 0.0048 | −0.098 | 0.0354 | |||
| 80 | 1 | −0.099 | 0.0067 | −0.106 | −0.139 | 0.0064 | −0.133 |
| 2 | −0.114 | 0.0139 | −0.139 | 0.0052 | |||
| 3 | −0.105 | 0.0171 | −0.122 | 0.0274 | |||
The effect of RH on viral survival at 20°C is shown in Fig. 2. At 20°C, the experiment was terminated at 28 days. At 50% RH, the experiment was terminated at 3 days for TGEV and at 5 days for MHV due to underestimation of the number of sampling points needed. At 80% RH, experiments were terminated when no virus was detectable on the carriers. Inactivation was more rapid at 20°C at all RH levels than at 4°C. Again, there was some variation in the measured viral inocula on individual carriers, which may have contributed to the variations in log10 reductions observed over the course of the experiment. Infectious virus deposited on stainless steel surfaces at titers of 4 to 5 log10 MPN persisted for at least 3 days at 50% RH and for up to 28 days at 20% RH, and the slowest inactivation took place at low RH. The linear regression models for each replicate at each RH were statistically significant (P < 0.05). The slopes of the regression lines for TGEV and MHV inactivation at 20°C and each RH are shown in Table 2. The levels of both viruses declined by 2 log10 over 28 days at 20% RH. The highest rate of inactivation was observed at 50% RH, at which the TGEV level declined by ∼2 log10 by day 3, and the level of MHV declined by ∼3 log10 by day 5. In comparison, at 80% RH, the level of TGEV declined by 1 log10 by day 3 and by 3 log10 over 14 days. The MHV level declined by 2.2 log10 by day 5 and by 5 log10 by day 11 at 80% RH.
FIG. 2.
Survival of TGEV and MHV at 20°C and (a) 20% RH, (b) 50% RH, and (c) 80% RH. Filled squares, TGEV; filled circles, MHV; open circles, value for the sample was below the detection limit of the assay (5 log10 MPN). The error bars indicate 95% confidence intervals.
TABLE 2.
Slopes of regression lines for virus inactivation at 20°C and 20, 50, and 80% RH
| RH | Replicate | MHV |
TGEV |
||||
|---|---|---|---|---|---|---|---|
| Slope | P | Mean slope | Slope | P | Mean slope | ||
| 20 | 1 | −0.052 | 0.0057 | −0.061 | −0.077 | 0.0025 | −0.081 |
| 2 | −0.068 | 0.0005 | −0.079 | 0.0019 | |||
| 3 | −0.063 | 0.0291 | −0.087 | 0.0034 | |||
| 50 | 1 | −0.685 | 0.0004 | −0.685 | −1.083 | 0.0003 | −0.896 |
| 2 | −0.649 | 0.0004 | −0.792 | 0.0029 | |||
| 3 | −0.721 | <0.0001 | −0.814 | 0.0316 | |||
| 80 | 1 | −0.550 | 0.0001 | −0.529 | −0.212 | 0.0001 | −0.212 |
| 2 | −0.531 | 0.0001 | −0.236 | 0.0001 | |||
| 3 | −0.506 | 0.0001 | −0.189 | 0.0001 | |||
Figure 3 shows rates of virus inactivation at 40°C and 20, 50, and 80% RH. Every linear regression model was statistically significant (P < 0.05). The slopes of regression lines for TGEV and MHV inactivation at 40°C and each RH are shown in Table 3. Overall, both viruses were inactivated more rapidly at 40°C than at 20°C. At 20% RH, both viruses survived for up to 5 days. In contrast, the length of survival at 50% RH was 24 h for MHV and <12 h for TGEV, and the length of survival for both viruses at 80% RH was <6 h. At 20% RH, the MHV infectivity declined by 4.7 log10 in 5 days, and the TGEV infectivity declined by 3.5 log10 in 5 days. Unlike the results at 20°C, the loss of infectivity at 40°C was more rapid at 80% RH than at 50% RH. At 40°C and 80% RH, the infectious titers of MHV and TGEV were 4.1 and 2.8 log10 lower at 3 h, respectively.
FIG. 3.
Survival of TGEV and MHV at 40°C and (a) 20% RH, (b) 50% RH, and (c) 80% RH. Filled squares, TGEV; filled circles, MHV; open squares, value for the TGEV sample was below the detection limit of the assay (4 log10 MPN); open circle, value for the MHV sample was below the detection limit of the assay (4 log10 MPN). The error bars indicate 95% confidence intervals.
TABLE 3.
Slopes of regression lines for virus inactivation at 40°C and 20, 50, and 80% RH
| RH | Replicate | MHV |
TGEV |
||||
|---|---|---|---|---|---|---|---|
| Slope | P | Mean slope | Slope | P | Mean slope | ||
| 20 | 1 | −0.984 | 0.0001 | −0.990 | −1.396 | 0.0022 | −1.523 |
| 2 | −0.957 | 0.0003 | −1.586 | 0.0134 | |||
| 3 | −1.028 | 0.0002 | −1.586 | 0.0134 | |||
| 50 | 1 | −9.220 | 0.0015 | −8.426 | −12.440 | <0.0001 | −11.280 |
| 2 | −6.635 | 0.0293 | −11.680 | <0.0001 | |||
| 3 | −9.424 | 0.0034 | −9.720 | <0.0001 | |||
| 80 | 1 | −16.008 | 0.0046 | −15.948 | −10.354 | 0.0006 | −9.857 |
| 2 | −16.092 | 0.0032 | −9.951 | 0.0020 | |||
| 3 | −15.744 | 0.0108 | −9.266 | 0.0089 | |||
ANCOVA modeling appeared to explain some of the variation in virus inactivation (R2 values for replicates 1, 2, and 3, 0.45, 0.52, and 0.41, respectively [P < 0.0001]). A test for equal slopes for a type of virus, performed at the level of individual measurement, indicated that the log10 inactivation did not differ for the two viruses tested (for replicate 1, P = 0.29; for replicate 2, P = 0.20; for replicate 3, P = 0.17). AT and RH are significant predictors of log10 inactivation (P < 0.0001), as is time (for replicate 1, P = 0.03; for replicate 2, P < 0.0001; for replicate 3, P < 0.0001). For all three replicates, the test of equal slopes comparing the RH levels showed that there was a significant difference between 20% RH and 50% RH (for all replicates, P < 0.0001) and between 20% RH and 80% RH (for all replicates, P < 0.0001) but no significant difference between 50% RH and 80% RH (for replicate 1, P = 0.53; for replicate 2, P = 0.77; for replicate 3, P = 0.88).
The ANCOVA model with addition of an interaction term explains more of the variation in inactivation than the previous model (R2 values for replicates 1, 2, and 3, 0.75, 0.72, and 0.70, respectively; P < 0.0001). In this model, AT (P < 0.0001), RH (P < 0.0001), and the interaction between AT and RH (P < 0.0001) are all significant predictors of log10 inactivation. As in the first model, virus type was not a significant predictor of inactivation (P = 0.14).
DISCUSSION
This study was the first study to examine the individual and synergistic effects of AT and RH on coronavirus survival on surfaces. The results show that when high numbers of the surrogates TGEV and MHV are deposited, these viruses may survive for days on surfaces at the ambient AT and wide range of RH levels (20 to 60% RH) typical of health care environments. TGEV and MHV may be more resistant to inactivation on surfaces than previously studied human coronaviruses, such as 229E (28). SARS-CoV has been reported to survive for 36 h on stainless steel (35), but the reductions in the levels observed were greater than those seen for either TGEV or MHV at 20°C at any RH in this study. However, the AT and RH conditions for the previous experiment were not reported, making comparisons difficult. Rabenau et al. (23) reported much slower inactivation of SARS-CoV on a polystyrene surface (4 log10 reduction after 9 days; AT and RH conditions not reported), consistent with some observations for TGEV and MHV in the present study. There are some similarities with studies of another enveloped virus, human influenza virus, on surfaces in that at higher RH (50 to 60%), the inactivation kinetics are closer to those of TGEV and MHV (21).
In the experiments in this study, the relationship between inactivation and RH was not monotonic, and survival was greater at low RH, a finding reflected in the results of previous studies of coronaviruses and other enveloped viruses in aerosols. Previous studies of TGEV and human coronavirus 229E in aerosols found that there was greater survival at low RH than at high RH (15, 17). Greater survival of other enveloped viruses, including vaccinia virus, Venezuelan equine encephalitis virus, and influenza virus, at low RH has been observed previously (11, 12, 13, 26, 27).
Overall, virus survival was enhanced by a lower AT. Similar relationships between AT and virus inactivation have been observed for enveloped viruses in liquids (7, 29) and aerosols (11, 15). The coronavirus data obtained in this study suggest that although the rates of viral inactivation are lower at lower ATs, there are still different effects of RH on viral survival at each AT. At 40°C, the same protective effect of low RH was seen at 20% RH compared to that at 50% and 80% RH. Overall, however, inactivation was more rapid at all three RH levels at this high AT. It may be that at 40°C AT effects are the predominant effects that cause viral inactivation and that RH levels play a lesser role than they do at lower ATs. The results of the statistical analysis suggest that RH has a greater effect on viral inactivation than AT, but there are interactions between AT and RH. The relationship between AT, RH, and virus inactivation is still not entirely clear and may vary depending on the virus type (1, 19).
Multiple mechanisms may contribute to viral inactivation on surfaces. Some inactivation may take place when viral capsids accumulate at the air-water interface (AWI) of a solution, causing structural damage (30, 31, 33). Desiccation may also be an important contributor to inactivation on surfaces (1), as loss of water molecules triggers lipid membrane phase changes, cross-linking, Maillard reactions, and peroxide formation (9). Virus inactivation on surfaces may involve both desiccation and interaction at the AWI, with the contribution of each depending on the RH. At a low RH, oxidation and Maillard reactions that occur during rapid desiccation may predominate. Around 80% RH, the rate of loss of water molecules is slowed, the hydrophobicity of the AWI is decreased (19), and the main mechanism may be inactivation at the AWI. Around 50% RH, inactivation at the AWI and desiccation may occur simultaneously; as water molecules are lost, lipid oxidation and Maillard reactions occur (the maximum rates of Maillard reactions occur when the RH is 50 to 80%) (9), possibly providing a partial explanation for why viral inactivation appears to be more rapid at 50% RH than at 20% or 80% RH.
At ambient ATs (around 20°C), coronaviruses can survive for 2 days while losing only 1 to 2 log10 infectivity, depending on the RH. Nasopharyngeal aspirates from infected individuals with SARS can have viral loads ranging from 105 to 108 genome templates/ml (8, 14, 22, 34), suggesting that respiratory secretions from SARS patients may contain infectious virus. If deposited on surfaces in these types of secretions, coronaviruses could potentially survive on surfaces in health care environments for days. Evidence of SARS-CoV nucleic acids on surfaces and inanimate objects in hospitals has been reported (6, 10). However, there are no data on the occurrence of infectious SARS-CoV on these surfaces. The dose-response relationship and minimal infectious doses for infection of humans by SARS-CoV and other coronaviruses have also not been defined. Given these gaps in our knowledge, the magnitude of the risk due to virally contaminated surfaces is uncertain and should be examined further.
The survival data for TGEV and MHV suggest that enveloped viruses can remain infectious on surfaces long enough for people to come in contact with them, posing a risk for exposure that leads to infection and possible disease transmission. This risk may also occur for other enveloped viruses, such as influenza virus (3, 4). The potential reemergence of SARS or the emergence of new strains of pandemic influenza virus, including avian and swine influenza viruses, could pose serious risks for nosocomial disease spread via contaminated surfaces. However, this risk is still poorly understood, and more work is needed to quantify the risk of exposure and possible transmission associated with surfaces. Statistical analysis showed that TGEV and MHV do not differ significantly in their inactivation kinetics on surfaces, and both viruses may be suitable models for survival and inactivation of SARS-CoV on surfaces. However, more data on the survival rates and inactivation kinetics of SARS-CoV itself are needed before these relationships with other coronaviruses can be definitively established. However, the findings of this study suggest that TGEV and MHV could serve as conservative surrogates for modeling exposure, transmission risk, and control measures for pathogenic enveloped viruses, such as SARS-CoV and influenza viruses, on health care surfaces.
Acknowledgments
This work was supported by Centers for Disease Control and Prevention grant 1 U01 CI000299-01.
Footnotes
Published ahead of print on 12 March 2010.
REFERENCES
- 1.Abad, F., R. Pinto, and A. Bosch. 1994. Survival of enteric viruses on environmental fomites. Appl. Environ. Microbiol. 60:3704-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Bean, B., B. Moore, B. Sterner, L. Peterson, D. Gerding, and H. Balfour, Jr. 1982. Survival of influenza viruses on environmental surfaces. J. Infect. Dis. 146:47-51. [DOI] [PubMed] [Google Scholar]
- 4.Blachere, F., W. Lindsley, T. Pearce, S. Anderson, M. Fisher, R. Khakoo, B. Meade, O. Lander, S. Davis, and R. Thewlis. 2009. Measurement of airborne influenza virus in a hospital emergency department. Clin. Infect. Dis. 48:438-440. [DOI] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Booth, T., B. Kournikakis, N. Bastien, J. Ho, D. Kobasa, L. Stadnyk, Y. Li, M. Spence, S. Paton, and B. Henry. 2005. Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. J. Infect. Dis. 191:1472-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casanova, L., W. Rutala, D. Weber, and M. Sobsey. 2009. Survival of surrogate coronaviruses in water. Water Res. 43:1893-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu, C., V. Cheng, I. Hung, K. Chan, B. Tang, T. Tsang, K. Chan, and K. Yuen. 2005. Viral load distribution in SARS outbreak. Emerg. Infect. Dis. 11:1882-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, C. 1993. Roles of water molecules in bacteria and viruses. Origins Life Evol. Biosph. 23:29-36. [DOI] [PubMed] [Google Scholar]
- 10.Dowell, S., J. Simmerman, D. Erdman, J. Wu, A. Chaovavanich, M. Javadi, J. Yang, L. Anderson, S. Tong, and M. Ho. 2004. Severe acute respiratory syndrome coronavirus on hospital surfaces. Clin. Infect. Dis. 39:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harper, G. 1961. Airborne micro-organisms: survival tests with four viruses. J. Hyg. 59:479-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harper, G. 1963. The influence of environment on the survival of airborne virus particles in the laboratory. Arch. Virol. 13:64-71. [DOI] [PubMed] [Google Scholar]
- 13.Hemmes, J., K. C. Winkler, and S. M. Kool. 1960. Virus survival as a seasonal factor in influenza and poliomyelitis. Nature 188:430-431. [DOI] [PubMed] [Google Scholar]
- 14.Hung, I. F., V. C. Cheng, A. K. Wu, B. S. Tang, K. H. Chan, C. M. Chu, M. M. Wong, W. T. Hui, L. L. Poon, D. M. Tse, K. S. Chan, P. C. Woo, S. K. Lau, J. S. Peiris, and K. Y. Yuen. 2004. Viral loads in clinical specimens and SARS manifestations. Emerg. Infect. Dis. 10:1550-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ijaz, M., A. Brunner, S. Sattar, R. Nair, and C. Johnson-Lussenburg. 1985. Survival characteristics of airborne human coronavirus 229E. J. Gen. Virol. 66:2743-2748. [DOI] [PubMed] [Google Scholar]
- 16.Jackwood, M. W. 2006. The relationship of severe acute respiratory syndrome coronavirus with avian and other coronaviruses. Avian Dis. 50:315-320. [DOI] [PubMed] [Google Scholar]
- 17.Kim, S., M. Ramakrishnan, P. Raynor, and S. Goyal. 2007. Effects of humidity and other factors on the generation and sampling of a coronavirus aerosol. Aerobiologia 23:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Mbithi, J., V. Springthorpe, and S. Sattar. 1991. Effect of relative humidity and air temperature on survival of hepatitis A virus on environmental surfaces. Appl. Environ. Microbiol. 57:1394-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald, L., A. Simor, S. IhJen, S. Maloney, M. Ofner, C. KowTong, J. Lando, A. McGeer, L. MinLing, and D. Jernigan. 2004. SARS in healthcare facilities, Toronto and Taiwan. Emerg. Infect. Dis. 10:777-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noyce, J., H. Michels, and C. Keevil. 2007. Inactivation of influenza A virus on copper versus stainless steel surfaces? Appl. Environ. Microbiol. 73:2748-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peiris, J., S. Lai, L. Poon, Y. Guan, L. Yam, W. Lim, J. Nicholls, W. Yee, W. Yan, and M. Cheung. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabenau, H. F., J. Cinatl, B. Morgenstern, G. Bauer, W. Preiser, and H. W. Doerr. 2005. Stability and inactivation of SARS coronavirus. Med. Microbiol. Immunol. 194:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Sattar, S. 2004. Microbicides and the environmental control of nosocomial viral infections. J. Hosp. Infect. 56(Suppl. 2):S64-S69. [DOI] [PubMed] [Google Scholar]
- 26.Schaffer, F., M. Soergel, and D. Straube. 1976. Survival of airborne influenza virus: effects of propagating host, relative humidity, and composition of spray fluids. Arch. Virol. 51:263-273. [DOI] [PubMed] [Google Scholar]
- 27.Shechmeister, I. 1950. Studies on the experimental epidemiology of respiratory infections. III. Certain aspects of the behavior of type A influenza virus as an air-borne cloud. J. Infect. Dis. 87:128-132. [DOI] [PubMed] [Google Scholar]
- 28.Sizun, J., M. Yu, and P. Talbot. 2000. Survival of human coronaviruses 229E and OC43 in suspension and after drying on surfaces: a possible source of hospital-acquired infections. J. Hosp. Infect. 46:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tennant, B., R. Gaskell, and C. Gaskell. 1994. Studies on the survival of canine coronavirus under different environmental conditions. Vet. Microbiol. 42:255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, S., M. Flury, M. Yates, and W. Jury. 1998. Role of the air-water-solid interface in bacteriophage sorption experiments. Appl. Environ. Microbiol. 64:304-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, S., and M. Yates. 1999. Bacteriophage inactivation at the air-water-solid interface in dynamic batch systems. Appl. Environ. Microbiol. 65:1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reference deleted.
- 33.Trouwborst, T., S. Kuyper, J. C. de Jong, and A. Plantinga. 1974. Inactivation of some bacterial and animal viruses by exposure to liquid-air interfaces. J. Gen. Virol. 24:155-165. [DOI] [PubMed] [Google Scholar]
- 34.Wong, S. C., J. K. Chan, K. C. Lee, E. S. Lo, and D. N. Tsang. 2005. Development of a quantitative assay for SARS coronavirus and correlation of GAPDH mRNA with SARS coronavirus in clinical specimens. J. Clin. Pathol. 58:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization. 2003. First data on stability and resistance of SARS coronavirus compiled by members of WHO laboratory network. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/sars/survival_2003_05_04/en/index.html.