Abstract
We examined the chemical composition of extracellular polymeric substances (EPS) extracted from two natural microbial pellicle biofilms growing on acid mine drainage (AMD) solutions. The EPS obtained from a mid-developmental-stage biofilm (DS1) and a mature biofilm (DS2) were qualitatively and quantitatively compared. More than twice as much EPS was derived from DS2 as from DS1 (approximately 340 and 150 mg of EPS per g [dry weight] for DS2 and DS1, respectively). Composition analyses indicated the presence of carbohydrates, metals, proteins, and minor quantities of DNA and lipids, although the relative concentrations of these components were different for the two EPS samples. EPS from DS2 contained higher concentrations of metals and carbohydrates than EPS from DS1. Fe was the most abundant metal in both samples, accounting for about 73% of the total metal content, followed by Al, Mg, and Zn. The relative concentration profile for these metals resembled that for the AMD solution in which the biofilms grew, except for Si, Mn, and Co. Glycosyl composition analysis indicated that both EPS samples were composed primarily of galactose, glucose, heptose, rhamnose, and mannose, while the relative amounts of individual sugars were substantially different in DS1 and DS2. Additionally, carbohydrate linkage analysis revealed multiply linked heptose, galactose, glucose, mannose, and rhamnose, with some of the glucose in a 4-linked form. These results indicate that the biochemical composition of the EPS from these acidic biofilms is dependent on maturity and is controlled by the microbial communities, as well as the local geochemical environment.
In aqueous environments, microbial cells grow in association with surfaces, leading to the formation of biofilms, which are complex assemblages of microorganisms that are embedded in a matrix of extracellular polymeric substances (EPS) (13). Biofilms are often considered problematic from the human perspective owing to the damage associated with their presence (rusting, dental plaque formation, and the disease cystic fibrosis) (8). The focus of the present study is on microbial biofilms that are instrumental in the generation of acid mine drainage (AMD) (1, 10, 16, 20, 32). The deleterious environmental effects of AMD necessitate research to characterize biofilm structure in such systems, which could provide insight into treatment approaches and environmental control.
The biofilm matrix facilitates structural organization and protects the microbial community (13), and it plays a critical role in metal adsorption and immobilization (1, 4). Given the great diversity of microbial communities that form biofilms, as well as the different environments that they inhabit, it is difficult to make generalizations about biofilm EPS structure and physiological function. Metal chelation by EPS is thought to be an important mechanism in the natural detoxification of heavy metal (e.g., As)-contaminated sites and hence is fundamentally important for their bioremediation (14, 15). In order to better understand the nature and role of EPS in relation to metal binding, it is important to have a clear understanding of the actual sites, capacities, and mechanisms of metal adsorption. These factors are particularly relevant for the EPS in the AMD environment, where various metals are highly abundant.
Detailed composition analysis of an EPS is difficult, as EPS is often a complex mixture of proteins, carbohydrates, lipids, DNA, and humic acid substances (13). Furthermore, even though carbohydrates have been identified as one of the major components of EPS, the biochemical properties of these compounds remain elusive due to their complex structures and unique monomer linkages (3, 17, 21, 24, 27, 36, 38). The nature of carbohydrates within EPS is a dynamic function of the microbial community composition; this is reflected by the complexity of carbohydrates containing diverse sugar residues extracted from biofilm matrices (24, 38, 41). Moreover, the role of polysaccharides in the development and function of biofilm matrices remains speculative.
The present study focused on the composition of EPS in environmental biofilms that grow at the solution-air interface in acid mine drainage (AMD) effluent in the Richmond Mine at Iron Mountain (Redding, CA). The geochemistry and microbial species composition of this site have been well described, which makes it an ideal natural model study system (12, 29, 37, 40). The AMD at this site consists of sulfuric acid-rich solutions (pH ∼1) containing submolar concentrations of Fe(II) and millimolar concentrations of various heavy metals, such as Zn, Cu, and As. Genomic analysis of the biofilms revealed that Leptospirillum group II dominates the community and that smaller amounts of Leptospirillum group III and several archaea are also present (23, 37). The relative proportions of these microbes differ depending on the physical location and developmental stage of the biofilm (24, 38, 41).
Despite extensive efforts to examine the biodiversity of these communities, little is known about their ecophysiology, and understanding the ecophysiology is essential for understanding how microbial consortia assemble and function in their natural habitats. One step toward this goal was a study in which molecular probes were used to examine the biofilm structure and microbial composition during development and organization (40). In parallel, in this study we performed qualitative and comparative chemical analyses of the extracellular matrix in two biofilms at different developmental stages. Importantly, we highlight here how the EPS may affect the ecophysiology of the AMD community.
MATERIALS AND METHODS
Biofilm sampling.
Biofilm samples were collected from the surface of AMD solutions in the Richmond Mine at Iron Mountain in northern California. These samples were immediately frozen in dry ice and then stored at −80°C until they were processed. Biofilms used in this study were obtained from the same site (“AB Muck”) in May and August 2007 (29).
EPS extraction.
The protocol used for EPS preparation was modified from a previously described method (21). Biofilm samples (∼15 ml) were thawed on ice and centrifuged at 15,000 × g for 20 min. Each supernatant, which was the residual AMD solution, was saved. The biofilm pellets were resuspended in ∼30 ml of a cold sulfuric acid solution (0.2 M sulfuric acid, pH 1.1), and the biofilm matrix was broken using a glass hand homogenizer tube and pestle (Wheaton Science Products, Millville, NJ). The cell suspension was stirred at 4°C for 3 h before centrifugation at 15,000 × g for 20 min. The resulting supernatant, referred to below as the “EPS solution,” which contained the total EPS of both the colloidal and capsular fractions, was saved and used for further analysis. The EPS dry weight was calculated by subtracting the cell pellet (after the first centrifugation) dry weight from the biofilm dry weight.
Chemical composition analysis.
Fourier transform infrared (FTIR) spectroscopy was performed with ethanol-precipitated and dried EPS samples. Initially, EPS was precipitated by adding 3 volumes of 100% cold ethanol and incubated on ice for 2 h. The precipitates were then centrifuged at 17,500 × g for 20 min at 4°C and dried in an oven at 50°C overnight. FTIR spectroscopy was performed with a PE Spectrum GX FTIR system in attenuated total reflection (ATR) mode using a Split Pea (Harrick Scientific Corporation, United States). A pressure applicator was used to hold the samples firmly against the ATR crystal during the scans.
To measure the total protein content, trichloroacetic acid (TCA) (final concentration, 12%) was added to the EPS solution, and the mixture was incubated on ice for 30 min before centrifugation at 15,000 × g for 20 min. The TCA precipitates were washed twice with 10 ml acetone and resuspended in 2 ml of 2-N-morpholinoethanesulfonic acid (MES) buffer (pH 5.0). The protein content was measured using the Bradford assay (Bio-Rad, Hercules, CA) with bovine serum albumin (BSA) as the calibration standard.
The total DNA content in an EPS solution was measured after extraction with 3 volumes of 100% cold ethanol. The mixture was then incubated on ice for 2 h before DNA was recovered by centrifugation at 17,500 × g for 20 min at 4°C. The DNA concentration was measured by fluorimetry using the fluorescent dye 4,6-diamidino-2-phenylindole (DAPI) (5).
The total carbohydrate content was measured using a modified phenol-sulfuric acid method with glucose standards (11). Briefly, 50 μl of an EPS solution was mixed with 125 μl of concentrated sulfuric acid. Then 25 μl of 10% phenol was mixed in, and the mixture was incubated in a 95°C water bath for 5 min. The mixture was cooled and transferred into a 96-well plate (BD Biosciences, San Jose, CA). The absorbance at 490 nm was read with a spectrophotometric plate reader (Synergy HT; BioTek, Winooski, VT).
The metal, cation, and the silica contents of the residual AMD solution and the EPS solution were analyzed by inductively coupled plasma atomic emission spectroscopy (ICP-AES). All standards, blanks, and samples were prepared in ultrapure 2% distilled HNO3. Internal standards were run each time before the samples were run to correct for machine drift.
Solid-state NMR analysis.
Solid-state 13C nuclear magnetic resonance (NMR) spectra were acquired with a Varian-Chemagnetics Infinity 300 nuclear magnetic resonance spectrometer. The static field of the superconducting magnet was ∼7.05 T, and the resonance frequencies of 1H and 13C were ∼300 and 75 mHz, respectively. Cross polarization (CP) (1H-13C) employed an amplitude ramp in the 13C channel, and magic angle sample spinning (MAS) was performed to optimize the signal. The specific experimental parameters included a proton excitation pulse width of 4 μs, a 1H spin lock pulse and decoupling power (w1H/2π) of 62.5 kHz, a contact time of 4 ms, a recycle delay between acquisitions of 1 s, and a MAS frequency (wr/2π) of 12 kHz. The number of acquisitions was 100,000 per sample, with tetramethylsilane as an internal standard.
Glycosyl composition and linkage analyses.
Glycosyl composition and linkage analyses were performed by the Complex Carbohydrate Research Center at the University of Georgia (Athens, GA). For glycosyl composition analysis, methyl glycosides were first prepared from a dry sample obtained by methanolysis in 1 M HCl in methanol at 80°C (18 to 22 h), followed by re-N-acetylation with pyridine and acetic anhydride in methanol (for detection of amino sugars) (26, 42). As an internal standard, 20 μg of inositol was added to the samples before derivatization. The samples were then per-O-trimethylsilylated (TMS) by treatment with Tri-Sil reagent (Pierce) at 80°C for 30 min. Gas chromatography-mass spectrometry (GC-MS) analysis of the TMS methyl glycosides was performed with a Hewlett Packard 6890 GC interfaced with a 5975b mass selective detector (MSD), using an All Tech EC-1 fused silica capillary column (30 m by 0.25 mm [inside diameter]). The monosaccharides were identified by their retention times relative to those of standards, and the carbohydrate character of the monosaccharides was authenticated by their mass spectra.
For glycosyl linkage analysis, the EPS samples were permethylated, depolymerized, reduced, and acetylated; the resultant partially methylated alditol acetates (PMAAs) were analyzed by GC-MS as described previously (7). Briefly, aliquots of samples after dialysis were suspended in about 200 μl of dimethyl sulfoxide (DMSO) and placed on a magnetic stirrer for 5 days. The samples were then permethylated by treatment with sodium hydroxide and methyl iodide in dry DMSO (7). Each sample was subjected to the base NaOH for 10 min, and then methyl iodide was added and the preparation was left for 10 min. Additional methyl iodide was then added, and the preparation was incubated for 40 min. The base was then added, and the preparation was incubated 10 min; finally, more methyl iodide was added, and the preparation was incubated for 40 min. The addition of more methyl iodide and the addition of the NaOH base were necessary to ensure complete methylation of the polymer. Following sample workup, the permethylated material was hydrolyzed using 2 M trifluoroacetic acid in a sealed tube at 121°C for 2 h, reduced with NaBD4, and acetylated using acetic anhydride-trifluoroacetic acid. The resulting PMAAs were analyzed with a Hewlett Packard 5890 GC interfaced with a 5970 MSD in electron impact ionization mode, and separation was performed using a 30-m Supelco 2330 bonded phase fused silica capillary column.
RESULTS
Description of the microbial community and source of the biofilms.
The biofilms used in this study were obtained from the surface of AMD pools at the AB Muck location (29) in May and August 2007. The pH of the AMD solution at the sampling site was between 0.83 and 1.0, and the temperature was between 37 and 41°C. Visual examination and fluorescence in situ hybridization (FISH) analysis indicated that the sample harvested in August represented an early-developmental-stage thin biofilm (developmental stage 1 biofilm [DS1]); this biofilm was dominated by Leptospirillum group II, which accounted for 90% of the population, and there were minor amounts of Leptospirillum group III (3%) and archaea (7%). The sample harvested in May represented a later-growth-stage thicker biofilm (developmental stage 2 biofilm [DS2]) with a more diverse microbial community, including Leptospirillum group II (43%), Leptospirillum group III (28%), and archaea (29%) (9). FISH analyses have shown that the biofilm developmental stages have consistent microbial diversity.
Different constituents of EPS.
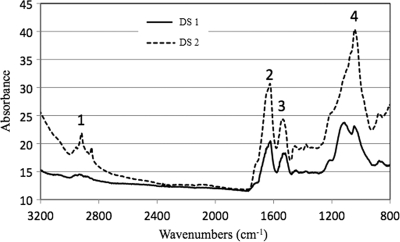
The EPS fractions extracted from the two biofilms differed quantitatively and qualitatively. DS1 contained about 150 mg of EPS per g (dry weight) of biofilm, compared with 340 mg of EPS per g (dry weight) for DS2. This indicated that the ratio of EPS mass to cell mass increases as a biofilm matures from DS1 to DS2 and that EPS accounts for a larger proportion of the mass in the later growth stages. We speculate that the high level of EPS in the later developmental stages provides an important substrate for mixotrophic (archaea) and heterotrophic (fungi) organisms that are more abundant in biofilms in later developmental stages (40). FTIR spectra of EPS extracted from DS1 and DS2 indicated the presence of polysaccharides and nucleic acids (1,300 to 900 cm−1), as well as proteins (1,700 to 1,500 cm−1) (Fig. 1). The spectra had differences both in shape and in absorbance intensity, indicating that there was variation in the composition and quantity of each individual component. The peaks for both protein and carbohydrate were substantially higher for DS2. Additionally, analysis of DS2 revealed a low level of lipids (2,930 to 2,860 cm−1), whereas the lipid peaks were much smaller for DS1.
FIG. 1.
FTIR spectra of EPS from DS1 and DS2 biofilms. Peak 1, —CH— vibrations in lipids; peak 2, amide I in proteins; peak 3, amide II in proteins; peak 4, —COC— group vibrations in carbohydrates, DNA, and RNA.
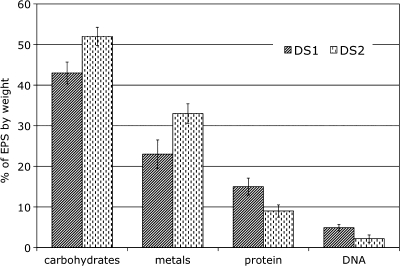
Quantitative measurement of individual components of EPS from DS1 and DS2 samples using biochemical methods (see Materials and Methods) indicated that both samples were comprised of carbohydrates, metals, proteins, and small quantities of DNA (Fig. 2). Metals, the second major constituent of the EPS in these biofilms, showed a pattern similar to that of carbohydrates. Small quantities of protein were detected in both EPS samples, and the values for DNA were the lowest.
FIG. 2.
Comparison of the biochemical compositions of EPS from DS1 and DS2 biofilms. The error bars indicate standard deviations (n = 3).
Solid-state NMR analysis.
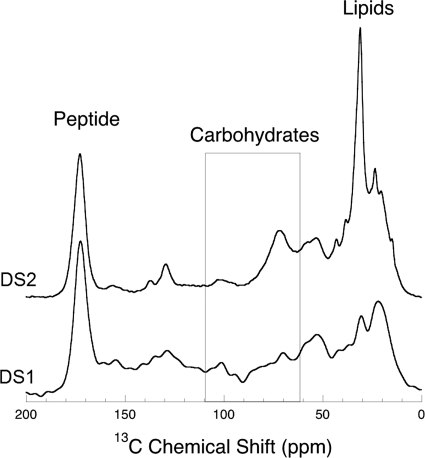
Solid-state 13C NMR spectroscopy also provides a detailed description of the types and abundance of various functional groups associated with the presence of different biomacromolecules. Figure 3 presents 1H-13C CP-MAS spectra of dried DS1 and DS2 biofilms. Due to variations in local electronic structure, 13C in different functional groups resonates at different frequencies (reported as a chemical shift in ppm). Lipids typically resonate in the frequency range from ∼0 to ∼40 ppm. Secondary alcohols of carbohydrates resonate in a range of frequencies from ∼60 to 90 ppm, whereas the glycosidic carbon of polysaccharides resonates at frequencies between 95 and 106 ppm. In the latter case, the resonance frequency of the glycosidic carbon is indicative of the nature of the linkage, i.e., whether the linkage is a linkage through an alpha or beta oxygen. Linkages to beta oxygen resonate in the range from 103 to 106 ppm, whereas linkages to alpha oxygen resonate in the range from ∼95 to 103 ppm. The presence of peptide in biological samples is prominently indicated by the sharp peak at 173 ppm corresponding to the amidyl carbonyl (Fig. 3).
FIG. 3.
Solid-state 13C NMR spectra of EPS from DS1 and DS2 biofilms, revealing the distribution of bonding environments. The spectral regions with relevance to the present study include the regions for peptide-bonded carbon (peptide), polysaccharide glycosidic carbon and secondary alcohols (area in the rectangle), and saturated hydrocarbon (lipids). Differences in the spectral features in the carbohydrate region indicate substantial differences in the compositions of the polysaccharides in the two biofilms.
The solid-state 13C NMR spectra presented in Fig. 3 for both DS1 and DS2 samples reveal differences in the relative abundance and composition of biomacromolecules present in the biofilms. The most obvious difference between the two samples is the difference in their relative lipid levels and lipid structures; e.g., the prominent lipid peak at ∼30 ppm in the DS2 spectrum likely indicates a predominance of long-chain hydrocarbons. The fact that these NMR data indicate that DS2 has proportionally 54% more lipid than DS1 is consistent with what is observed in the high-frequency C-H region of the FTIR spectra for these biofilms (Fig. 1).
Notwithstanding the difference in lipid composition, there is also difference in the secondary alcohol frequency region (65 to 90 ppm) of the carbohydrates (Fig. 3). The fact that the spectral features are so different can be explained only by a significant difference in the distribution of sugar monomers within the polysaccharides (Table 1). The obvious increase in secondary alcohol intensity relative to the glycosidic carbon intensity in DS2 is consistent with a change in carbohydrate composition toward larger sugars. Finally, there are also some differences in glycosidic resonance (95 to 106 ppm) between DS1 and DS2. In both cases the presence of glycosidic intensity spanning the full range of frequency indicates the presence of both α and β oxygen monomer linkages.
TABLE 1.
Sugar substituent compositions of EPS from DS1 and DS2 biofilms
| Sugar or fatty acid | Amt (mol%) ina: |
|
|---|---|---|
| DS1 | DS2 | |
| Galactose | 51.9 ± 5.6 | 18.9 ± 1.9 |
| Glucose | 21.2 ± 2.4 | 16.7 ± 1.8 |
| Heptose | 12.2 ± 1.6 | 33.0 ± 2.2 |
| Rhamnose | 11.1 ± 1.1 | 13.2 ± 1.7 |
| Mannose | 3.7 ± 0.5 | 8.0 ± 1.3 |
| Xylose | ND | 2.6 ± 0.6 |
| 3-Deoxy-2-manno-2-octulosonic acid | ND | 7.8 ± 1.2 |
| 3OH C16 fatty acid | ND | Tr |
| N-Acetylglucosamine | ND | Tr |
The data are averages ± standard deviations for two samples. Tr, trace (<0.5%); ND, not detected (<0.1%).
These solid-state NMR data can also provide an estimate of the maximum amount of 4-linked glucopyranosyl segments in the carbohydrate region of the NMR spectra. First, it was noted that there is no evidence in either spectrum for crystalline cellulose, which characteristically exhibits very sharp resonances for the glycosidic carbon and secondary alcohols. Small segments of 4-linked glucopyranosyl units could be present. In order to place an upper limit on the amount of carbohydrate that is in the 4-linked glucopyranosyl segments, we sought to identify how much Gaussian broadened cellulose could fit within the total carbohydrate regions of the NMR spectra. We used 400-Hz Gaussian broadening to simulate the expected nonhomogeneous broadening that noncrystalline 4-linked glucopyranosyl segments would exhibit in a noncrystalline (i.e., amorphous) solid state. This analysis suggested that up to 33 and 36% of 4-linked glucopyranosyl segments could be accommodated in the total (100%) carbohydrate spectral regions of DS1 and DS2, respectively. These estimates provide only an upper limit, and it is likely the percentages of 4-linked glucopyranosyl are less than these values.
Glycosyl composition and carbohydrate linkage analysis.
To further characterize the carbohydrate composition of the EPS samples, a glycosyl composition analysis was performed after acid hydrolysis. A summary of the glycosyl composition of EPS from the DS1 and DS2 biofilms is given in Table 1. For DS1, the molar percentage of galactose was the highest percentage (∼52%) and was more than twice that of glucose (∼21%). For DS2, heptose was most abundant carbohydrate (∼33%), followed by galactose (∼19%). The galactose, heptose, and mannose contents were significantly different for the DS1 and DS2 samples. Interestingly, xylose was detected only in EPS from DS2 and not from DS1, indicating that xylose production is a function of biofilm maturation. It is not clear which organism(s) produces xylose in the DS2 biofilm. Xylose could be secreted by Leptospirillum group II at later growth stage, or its presence could be due to the increase in microbial abundance of Leptospirillum group III or archaeal species in DS2. The increase in xylose production during biofilm development is reminiscent of the previous observation that xylose is produced only in the stationary phase of a Bordetella culture (17), which may favor the former hypothesis.
Besides the simple sugars detected in the glycosyl analysis, several hallmarks of lipopolysaccharide (LPS), including 3-deoxy-2-manno-2-octulosonic acid (KDO), 3-hydroxylated hexadecanoic acid, and N-acetylglucosamine were detected in EPS from DS2 but not from DS1. The detection of LPS in DS2 is consistent with the results of FTIR and NMR spectrum analyses, in which greater quantities of lipids were observed in EPS from DS2 biofilms (Fig. 1 and 3). In addition, since the amount of KDO detected in DS2 (7.8 mol%) is much greater than the amounts of other components of LPS, it is likely that much of the KDO is associated with the polysaccharides rather than with LPS per se.
Because of the unique environmental conditions of the AMD biofilms, unique features of the structure of carbohydrates in our EPS samples were of immediate interest. Linkage analysis was performed with the EPS from the DS1 and DS2 biofilms (Table 2). Consistent with the results of the glycosyl composition analyses, carbohydrate linkage analysis revealed multiple linked glycosyl sugars, and the majority of them were in the pyranosyl form. Furanosyl hexose was also detected; however, the hexose sugars were not specifically identified in our analysis. Based on a comparison with the glycosyl composition analysis results, we estimated that much of the unidentified furanosyl hexose could be galactose. Consistent with the NMR analysis, the linkage analysis showed that more than 50% of the glucose detected was in the 4-linked form, suggesting the presence of cellulose (β-1,4 glucan). The detection of arabinose in the linkage analysis but not in the glycosyl composition analysis may have been due to the different pretreatments for these two analyses that resulted in different degrees of hydrolysis for arabinose.
TABLE 2.
Linkage analysis of EPS from DS1 and DS2 biofilmsa
| Monosaccharide | Deduced linkage | Amt (mol%) in: |
|
|---|---|---|---|
| DS1 | DS2 | ||
| Glucose (pyranose) | Total | 20.1 | 17.2 |
| Terminally linked glucopyranosyl residue (t-Glc) | 5.7 | 7.2 | |
| 3-Linked glucopyranosyl residue (3-Glc) | 0.6 | 1.1 | |
| 4-Linked glucopyranosyl residue (4-Glc) | 13.1 | 8.2 | |
| 4,6-Linked glucopyranosyl residue (4,6-Glc) | 1.7 | 0.7 | |
| Galactose (pyranose) | Total | 12.1 | 5.6 |
| Terminally linked galactopyranosyl residue (t-Gal) | 1.4 | 0.9 | |
| 4-Linked galactopyranosyl residue (4-Gal) | 2.1 | 1.0 | |
| 3,4-Linked galactopyranosyl residue (3,4-Gal) | 8.6 | 3.7 | |
| Rhamnose (pyranose) | Total | 13.0 | 17.7 |
| Terminally linked rhamnopyranosyl residue (t-Rha) | 12.2 | 15.5 | |
| 2-Linked rhamnopyranosyl residue (2-Rha) | 0.8 | 2.2 | |
| Mannose (pyranose) | Total | 11.8 | 21.9 |
| Terminally linked mannopyranosyl residue (t-Man) | 7.2 | 12.3 | |
| 2-Linked mannopyranosyl residue (2-Man) | 4.6 | 9.6 | |
| Hexose (furanose) | Total | 25.5 | 12.0 |
| Terminally linked hexofuranosyl residue (t-Hexf) | 21.2 | 10.9 | |
| 3-Linked hexofuranosyl residue (3-Hexf) | 1.8 | 1.1 | |
| 6-Linked hexofuranosyl residue (6-Hexf) | 2.5 | 0 | |
| Heptose (pyranose) | Total | 28.4 | 19.5 |
| Terminally linked heptopyranosyl residue (t-Hep pyranose) | 9.9 | 10.6 | |
| 3-Linked heptopyranosyl residue (3-Hep pyranose) | 1.2 | 2.5 | |
| 2-Linked heptopyranosyl residue (2-Hep pyranose) | 0.2 | 1.0 | |
| 3,4-Linked heptopyranosyl residue (3,4-Hep pyranose) | 1.1 | 2.5 | |
| 2,3-Linked heptopyranosyl residue (2,3-Hep pyranose) | 0.5 | 1.9 | |
| 2,6-Linked heptofuranosyl residue (2,6-Hep furanose) | 2.6 | 1.0 | |
| Arabinose (pyranose) | Terminally linked arabinopyranosyl residue (t-Ara pyranose) | 1.0 | 4.2 |
| Xylose (pyranose) | 4-Linked xylopyranosyl residue(4-Xyl pyranose) | 0 | 1.2 |
Hexose and heptose have not been specifically identified.
Metal composition analysis.
Metals extracted from EPS were analyzed, and the relative EPS metal concentrations were compared to the concentrations in the acid AMD solution from which the biofilms were collected (Table 3). In general, the relative concentrations of the metals extracted from EPS were similar to those in the corresponding AMD solution. Fe was the most abundant metal, followed by Al, Mg, and Zn.
TABLE 3.
Metal distribution in the EPS of DS1 and DS2 biofilms and in the AMD solution at the site where the biofilms were collected
| Metal | % of metals ina: |
|||
|---|---|---|---|---|
| DS1 AMD solution | DS1 EPS | DS2 AMD solution | DS2 EPS | |
| Fe | 68.2 ± 3.6 | 71.5 ± 4.3 | 70.8 ± 3.1 | 72.8 ± 2.9 |
| Al | 10.2 ± 2.4 | 13.6 ± 2.1 | 8.4 ± 1.8 | 10.3 ± 2.5 |
| Mg | 7.6 ± 1.2 | 5.6 ± 1.1 | 6.5 ± 1.5 | 3.5 ± 1.3 |
| Zn | 6.8 ± 1.7 | 5.4 ± 1.5 | 5.8 ± 1.3 | 5.2 ± 1.6 |
| Ca | 2.7 ± 0.9 | 4.0 ± 0.7 | 3.7 ± 0.6 | 3.9 ± 0.8 |
| Na | 2.0 ± 0.2 | 1.6 ± 0.3 | 1.9 ± 0.2 | 1.5 ± 0.3 |
| Cu | 1.1 ± 0.3 | 0.7 ± 0.2 | 1.4 ± 0.3 | 0.9 ± 0.2 |
| Si | 1.1 ± 0.3 | 2.0 ± 0.3 | 1.1 ± 0.2 | 2.4 ± 0.3 |
| Mn | 0.17 ± 0.02 | 0.09 ± 0.02 | 0.14 ± 0.02 | 0.07 ± 0.02 |
| Co | 0.0047 ± 0.0003 | 0.0004 ± 0.0001 | 0.0046 ± 0.0002 | 0.0002 ± 0.0001 |
The data are the averages ± standard deviations for two samples.
For the metals analyzed, we found that the concentrations of Si, Mn, and Co varied greatly in the EPS and the AMD solution. The most notable difference was the difference for Si; its concentration in the EPS (∼2%) was twice that in the AMD solution (∼1%) for both DS1 and DS2. We postulate that this was due to the relatively low solubility of Si in the sulfuric acid-based AMD solution compared to the solubilities of the other metals analyzed and that Si was therefore more likely to be trapped in the EPS matrix. In contrast, Mn and Co were much more concentrated in the AMD solution than in the EPS for both biofilms. For Mn, the concentration in the AMD solution (∼0.17%) was twice that in the EPS (∼0.08%) for both DS1 and DS2; the concentration of Co in the AMD solution was 10 to 20 times that in the EPS. We postulate that the strong depletion of Co in the EPS could be due to active utilization of Co by microorganisms in biofilms. Consistent with this hypothesis, a previous proteomic study of proteins extracted from these biofilms indicated that many proteins involved in cobalamin biosynthesis were highly expressed (29). Additionally, cobalamin biosynthesis proteins may also play a role in iron-related metabolism (19), a critical function of the microbial community in the acid mine drainage environment.
DISCUSSION
In this work, we report on the biochemical composition of the EPS extracted from two biofilms at different developmental stages collected from the same location in the Richmond Mine system. The dynamics of the EPS components that we observed indicate that they are involved in community assembly and suggest that they have particular ecophysiological functions.
The origin of the EPS extracted from DS1 and DS2 was likely to be microbial, based on several observations. First, the layout and geology of the Richmond Mine prohibit influx of excess exogenous organic carbon, and hence, the EPS from DS1 and DS2 biofilms are the result of chemoautolithotrophic primary production (32). Second, because the EPS studied were formed at the solution-air interface, it is less likely that the biofilm matrix contains as much nonbiological materials as biofilms formed in other places in the environment (e.g., in soils). Third, the biofilms look slimy and are soft in texture, resembling microbial pellicles that form in pure cultures. Fourth, our previous FISH analyses (9) have shown that the biofilm developmental stages have consistent diversity, suggesting that the EPS changes that we observed were likely the result of biofilm development instead of the geological environment.
The carbohydrate-to-protein ratios between 3.0 and 6.0 observed for the biofilms in this study were substantially higher than the previously reported ratios between 0.2 and 1.7 observed for various environmental biofilms or biofilms of individual isolates (6, 22, 28, 33). However, our results are consistent with the high ratio (∼2.5) reported for eukaryote-based AMD biofilms (1), suggesting a property that is unique to the AMD communities. The high carbohydrate-to-protein ratios in the AMD biofilms could be caused by a naturally low protein content. Consistently, the protein contents of both the biofilms used in this study (10 to 15%) and the eukaryote-based AMD biofilms (<4%) (1) were substantially lower than those reported for other environmental biofilms, which typically are around 50% (6, 22, 28, 33). The low protein content in the EPS of the AMD biofilms perhaps can be attributed to enzymatic digestion by extracellular proteases and protein degradation under the harsh conditions in AMD. Consistent with these possibilities, proteomic analyses of extracellular proteins have indicated that there are different extents of protein cleavage (18, 34).
Interestingly, LPS components were detected only in EPS from DS2 and not from DS1, suggesting that the association of LPS with the cell membrane becomes looser during biofilm development. This could be due to the separation of cell wall material from the cell surface by shear forces in the natural environment, suggesting that there is detachment of LPS from the outer membrane during biofilm maturation. However, due to higher cell counts in DS2, we cannot rule out the possibility that the LPS in DS2 may be a result of cell lysis. Components of the outer membrane undergo constant turnover and may be excreted and lost from the cell surface as cells age, which could explain the presence of LPS in the EPS preparation of the DS2 biofilm. Consistent with this, Wrangstadh et al. found that marine Pseudomonas sp. strain S9 produced exopolysaccharides that remained cell bound (integral) at the log phase but were peripheral and lost to the external milieu as cells entered the stationary phase (41).
Preliminary inspection of the carbohydrate composition of the EPS from DS1 and DS2 revealed some interesting findings. Despite the extreme conditions of the AMD environment, the carbohydrate composition of the EPS resembles the carbohydrate compositions in other microbial systems (24, 38, 41). However, differences were found when DS1 and DS2 biofilms were compared, which could be related to two factors: (i) the change in the bacterial growth phase for a given species and (ii) the change in the microbial composition for the multispecies biofilms. Consequently, given the diversity in the glycosyl compositions of EPS produced by various microorganisms, it is difficult to predict or justify the specific composition for any given EPS. Similar to the glycosyl composition of EPS produced by microorganisms isolated from paper mill slimes (38, 39), galactose, glucose, rhamnose, and mannose are present in great abundance in the AMD biofilms. Although xylose has been found in some bacterial EPS, such as Paenibacillus and Idiomarina species EPS (2, 25), it is not a common sugar residue associated with EPS. A relatively small quantity of xylose, along with several other components of LPS (such as 3-deoxy-2-manno-2-octulosonic acid, 3-hydroxyl C16 fatty acid, and N-acetylglucosamine), was detected only in DS2, suggesting that xylose may be associated with LPS.
Our use of solid-state NMR and linkage analysis to characterize the polysaccharide composition yielded limited but promising information; a more in-depth analysis of purified EPS fractions is needed to elucidate the structures of the polymers. Also, solid-state NMR unfortunately cannot distinguish between the β-O-4 and β-O-3 linkages of glycosidic carbon atoms. Nevertheless, the substantial intensity in the high-frequency range of glycosidic carbons is consistent with the presence of β linkages, although not conclusively consistent with the presence of β-O-4 linkages associated with cellulose. Furthermore, the presence of 4-linked glucose residues identified by linkage analysis and the observed change in the EPS NMR spectrum before and after cellulase treatment both indicate that cellulose is present in the AMD biofilms (31). Although it is not clear which constituents of EPS are responsible for pellicle formation in AMD biofilms, we speculate that the presence of cellulose facilitates this formation, similar to the cellulose production in Gluconacetobacter pellicle biofilms (30). Floatation is critical for the survival of the AMD biofilm community, as the primary producer in the community, Leptospirillum group II, requires molecular oxygen to live.
Besides carbohydrates and protein, EPS from the AMD biofilms also contains substantial amounts of metals, and the amounts range from 23 to 33% of the total EPS dry mass for DS1 and DS2 biofilms. These values are considerably higher than the reported value (<10%) for previously characterized eukaryote-dominated AMD biofilms (1). The positive correlation of metal content with the amount of carbohydrates in both DS1 and DS2 suggests that the metal absorption capacity is largely due to the presence of carbohydrates in the biofilms. Although the overall metal composition of the EPS closely resembles that of the AMD solution from which the biofilms were collected, it is important to note that the selective binding of certain metals, such as Co and Mn, in the EPS indicates that the EPS matrix plays a role not only in community protection from toxic metals but also in the enrichment of trace elements critical for community survival. The metal-binding capacity of the EPS matrix is likely to be governed by its functional groups, such as carboxyl, phosphate, and sulfate groups, and their ligand-binding preferences for certain metals (35). More detailed characterization of the EPS functional groups is needed to reveal the exact chemical mechanism of metal binding.
In summary, the results of the composition analysis of EPS from AMD biofilms in this study reinforced the complex biochemical nature of EPS, which can be influenced by microbial composition as a function of biofilm developmental stage. In order to systematically control for the variables, analysis of more biofilms at different growth stages is required. Additionally, more knowledge of the EPS polysaccharide arrangements and linkages should lead to a better understanding of metal-polysaccharide interactions at the molecular level.
Acknowledgments
Funding for this study was provided by U.S. Department of Energy Office of Science, Genome Sciences Program grant DE-FG02-05ER64134. Work at Lawrence Livermore National Laboratory was performed under the auspices of the U.S. Department of Energy under contract DE-AC52-07NA27344. The W. M. Keck Solid State NMR facility at the Geophysical Laboratory, Carnegie Institution of Washington, received support from the W. M. Keck Foundation, the NSF, and the Carnegie Institution of Washington.
We thank Vincent Denef and other members of the Banfield geomicrobiology group for field sample collection and helpful discussions; Mona Hwang for laboratory assistance; and Zhiri Wang of the Complex Carbohydrate Research Center at the University of Georgia for glycosyl linkage analysis. Solid-state 13C NMR spectra were acquired at the W. M. Keck Solid State NMR facility at the Geophysical Laboratory, Carnegie Institution of Washington. We are very grateful to the reviewers for insightful comments which greatly improved the manuscript.
Footnotes
Published ahead of print on 12 March 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Aguilera, A., V. Souza-Egipsy, P. S. Martin-Uriz, and R. Amils. 2008. Extracellular matrix assembly in extreme acidic eukaryotic biofilms and their possible implications in heavy metal adsorption. Aquat. Toxicol. 88:257-266. [DOI] [PubMed] [Google Scholar]
- 2.Aguilera, M., M. Monteoliva-Sanchez, A. Suarez, V. Guerra, C. Lizama, A. Bennasar, and A. Ramos-Cormenzana. 2001. Paenibacillus jamilae sp. nov., an exopolysaccharide-producing bacterium able to grow in olive-mill wastewater. Int. J. Syst. Evol. Microbiol. 51:1687-1692. [DOI] [PubMed] [Google Scholar]
- 3.Beech, I., L. Hanjagsit, M. Kalaji, A. L. Neal, and V. Zinkevich. 1999. Chemical and structural characterization of exopolymers produced by Pseudomonas sp. NCIMB 2021 in continuous culture. Microbiology 145:1491-1497. [DOI] [PubMed] [Google Scholar]
- 4.Beech, I. B., J. A. Sunner, and K. Hiraoka. 2005. Microbe-surface interactions in biofouling and biocorrosion processes. Int. Microbiol. 8:157-168. [PubMed] [Google Scholar]
- 5.Brunk, C. F., K. C. Jones, and T. W. James. 1979. Assay for nanogram quantities of DNA in cellular homogenates. Anal. Biochem. 92:497-500. [DOI] [PubMed] [Google Scholar]
- 6.Bura, R., M. Cheung, B. Liao, J. Finlayson, B. C. Lee, I. G. Droppo, G. G. Lepard, and S. N. Liss. 1998. Composition of extracellular polymeric substances in the activated sludge floc matrix. Water Sci. Technol. 37:325-333. [Google Scholar]
- 7.Ciucanu, I., and F. Kerek. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131:209-217. [Google Scholar]
- 8.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 9.Denef, V. J., L. H. Kalnejais, R. S. Mueller, P. Wilmes, B. J. Baker, B. C. Thomas, N. C. VerBerkmoes, R. L. Hettich, and J. F. Banfield. 2010. Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc. Natl. Acad. Sci. U. S. A. 107:2383-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Druschel, G. K., B. J. Baker, T. H. Gihring, and J. F. Banfield. 2004. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 5:13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois, M., K. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1951. A colorimetric method for the determination of sugars. Nature 168:167. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, K. J., T. M. Gihring, and J. F. Banfield. 1999. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl. Environ. Microbiol. 65:3627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flemming, H. C., T. R. Neu, and D. J. Wozniak. 2007. The EPS matrix: the “house of biofilm cells.” J. Bacteriol. 189:7945-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flemming, H. C., and J. Wingender. 2001. Relevance of microbial extracellular polymeric substances (EPSs). Part I. Structural and ecological aspects. Water Sci. Technol. 43:1-8. [PubMed] [Google Scholar]
- 15.Flemming, H. C., and J. Wingender. 2001. Relevance of microbial extracellular polymeric substances (EPSs). Part II. Technical aspects. Water Sci. Technol. 43:9-16. [PubMed] [Google Scholar]
- 16.Hallberg, K. B., K. Coupland, S. Kimura, and D. B. Johnson. 2006. Macroscopic streamer growths in acidic, metal-rich mine waters in North Wales consist of novel and remarkably simple bacterial communities. Appl. Environ. Microbiol. 72:2022-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irie, Y., A. Preston, and M. H. Yuk. 2006. Expression of the primary carbohydrate component of the Bordetella bronchiseptica biofilm matrix is dependent on growth phase but independent of Bvg regulation. J. Bacteriol. 188:6680-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeans, C., S. W. Singer, C. S. Chan, N. C. Verberkmoes, M. Shah, R. L. Hettich, J. F. Banfield, and M. P. Thelen. 2008. Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J. 2:542-550. [DOI] [PubMed] [Google Scholar]
- 19.Jiao, Y., A. Kappler, L. R. Croal, and D. K. Newman. 2005. Isolation and characterization of a genetically tractable photoautotrophic Fe(II)-oxidizing bacterium, Rhodopseudomonas palustris strain TIE-1 Appl. Environ. Microbiol. 71:4487-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lear, G., D. Niyogi, J. Harding, Y. Dong, and G. Lewis. 2009. Biofilm bacterial community structure in streams affected by acid mine drainage. Appl. Environ. Microbiol. 75:3455-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lima, L. F., S. Habu, J. C. Gern, B. M. Nascimento, J. L. Parada, M. D. Noseda, A. G. Goncalves, V. R. Nisha, A. Pandey, V. T. Soccol, and C. R. Soccol. 2008. Production and characterization of the exopolysaccharides produced by Agaricus brasiliensis in submerged fermentation. Appl. Biochem. Biotechnol. 151:283-294. [DOI] [PubMed] [Google Scholar]
- 22.Liu, H., and H. H. Fang. 2002. Extraction of extracellular polymeric substances (EPS) of sludges. J. Biotechnol. 95:249-256. [DOI] [PubMed] [Google Scholar]
- 23.Lo, I., V. J. Denef, N. C. Verberkmoes, M. B. Shah, D. Goltsman, G. DiBartolo, G. W. Tyson, E. E. Allen, R. J. Ram, J. C. Detter, P. Richardson, M. P. Thelen, R. L. Hettich, and J. F. Banfield. 2007. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446:537-541. [DOI] [PubMed] [Google Scholar]
- 24.Manca, M. C., L. Lama, R. Improta, E. Esposito, A. Gambacorta, and B. Nicolaus. 1996. Chemical composition of two exopolysaccharides from Bacillus thermoantarcticus. Appl. Environ. Microbiol. 62:3265-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mata, J. A., V. Bejar, P. Bressollier, R. Tallon, M. C. Urdaci, E. Quesada, and I. Llamas. 2008. Characterization of exopolysaccharides produced by three moderately halophilic bacteria belonging to the family Alteromonadaceae. J. Appl. Microbiol. 105:521-528. [DOI] [PubMed] [Google Scholar]
- 26.Merkle, R. K., and I. Poppe. 1994. Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol. 230:1-15. [DOI] [PubMed] [Google Scholar]
- 27.Morikawa, M., S. Kagihiro, M. Haruki, K. Takano, S. Branda, R. Kolter, and S. Kanaya. 2006. Biofilm formation by a Bacillus subtilis strain that produces gamma-polyglutamate. Microbiology 152:2801-2807. [DOI] [PubMed] [Google Scholar]
- 28.Prolund, B., R. Palmgren, K. Keiding, and P. H. Nielsen. 1996. Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res. 30:1749-1758. [Google Scholar]
- 29.Ram, R. J., N. C. Verberkmoes, M. P. Thelen, G. W. Tyson, B. J. Baker, R. C. Blake II, M. Shah, R. L. Hettich, and J. F. Banfield. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915-1920. [PubMed] [Google Scholar]
- 30.Ross, P., R. Mayer, and M. Benziman. 1991. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55:35-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schrenk, M. O., G. D. Cody, M. Thelen, and J. F. Banfield. 2006. Detection of cellulose and comparative biopolymeric analyses of low pH, sub-aerial biofilms of the Richmond Mine, Iron Mountain, California, poster, abstract B11A-1007. Am. Geophys. Union Fall Meet. 2006, San Francisco, CA.
- 32.Schrenk, M. O., K. J. Edwards, R. M. Goodman, R. J. Hamers, and J. F. Banfield. 1998. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans: implications for generation of acid mine drainage. Science 279:1519-1522. [DOI] [PubMed] [Google Scholar]
- 33.Sheng, G. P., H. Q. Yu, and Z. Yu. 2005. Extraction of extracellular polymeric substances from the photosynthetic bacterium Rhodopseudomonas acidophila. Appl. Microbiol. Biotechnol. 67:125-130. [DOI] [PubMed] [Google Scholar]
- 34.Singer, S. W., C. S. Chan, A. Zemla, N. C. VerBerkmoes, M. Hwang, R. L. Hettich, J. F. Banfield, and M. P. Thelen. 2008. Characterization of cytochrome 579, an unusual cytochrome isolated from an iron-oxidizing microbial community. Appl. Environ. Microbiol. 74:4454-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stone, A. T. 1997. Reactions of extracellular organic ligands with dissolved metal ions and mineral surfaces, p. 309-334. In J. F. Banfield and K. H. Nealson (ed.), Reviews in mineralogy, vol. 35. Mineralogical Society of America, Washington, DC.
- 36.Tallon, R., P. Bressollier, and M. C. Urdaci. 2003. Isolation and characterization of two exopolysaccharides produced by Lactobacillus plantarum EP56. Res. Microbiol. 154:705-712. [DOI] [PubMed] [Google Scholar]
- 37.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 38.Verhoef, R., P. de Waard, H. A. Schols, M. Ratto, M. Siika-aho, and A. G. Voragen. 2002. Structural elucidation of the EPS of slime producing Brevundimonas vesicularis sp. isolated from a paper machine. Carbohydr. Res. 337:1821-1831. [DOI] [PubMed] [Google Scholar]
- 39.Verhoef, R., H. A. Schols, A. Blanco, M. Siika-aho, M. Ratto, J. Buchert, G. Lenon, and A. G. Voragen. 2005. Sugar composition and FT-IR analysis of exopolysaccharides produced by microbial isolates from paper mill slime deposits. Biotechnol. Bioeng. 91:91-105. [DOI] [PubMed] [Google Scholar]
- 40.Wilmes, P., J. P. Remis, M. Hwang, M. Auer, M. P. Thelen, and J. F. Banfield. 2009. Natural acidophilic biofilm communities reflect distinct organismal and functional organization. ISME J. 3:266-270. [DOI] [PubMed] [Google Scholar]
- 41.Wrangstadh, M., U. Szewzyk, J. Ostling, and S. Kjelleberg. 1990. Starvation-specific formation of a peripheral exopolysaccharide by a marine Pseudomonas sp., strain S9. Appl. Environ. Microbiol. 56:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.York, W. S., A. G. Darvill, M. McNeil, T. T. Stevenson, and P. Albersheim. 1986. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118:3-40. [Google Scholar]