Abstract
Seven different actA subtypes forming two phylogenetic lineages could be distinguished by sequencing the actA gene of Listeria seeligeri isolates from different habitats. Isolates of the two lineages differ in hemolytic as well as phospholipase activities and in the arrangement of the virulence gene cluster. The presence of a serine protease gene resembling orf2110 of L. monocytogenes in some isolates further supports the hypothesis that L. seeligeri is subject to ongoing adaptation to changing environments.
The genus Listeria comprises the species L. monocytogenes, L. ivanovii, L. seeligeri, L. innocua, L. welshimeri, L. grayi, and L. marthii (4, 7, 17). Of these only, L. monocytogenes (15) and L. ivanovii (1, 18) are considered as pathogens. The pathogenicity is closely associated with a virulence gene cluster, although other genes like those coding for internalines are implicated in pathogenesis too (16). Like L. monocytogenes and L. ivanovii, L. seeligeri also carries a virulence gene cluster. Although genes of the virulence cluster are presumably not expressed in a correct or functional fashion (3, 20), L. seeligeri has been associated with human infection in one case so far (14). Recent studies reveal exceptions from the traditional classification into pathogenic and nonpathogenic Listeria species. L. innocua strains carrying a virulence gene cluster similar to that of L. monocytogenes have been isolated (6, 22). Moreover, nonhemolytic L. seeligeri isolates lacking the major part of the virulence gene cluster have been described (21).
L. seeligeri variants.
In this study, a total of 2,492 samples from different habitats of the food chain were investigated with respect to the occurrence of members of the genus Listeria. Of 715 isolates obtained from animals (organs and feces), butcher shops (drains and intermediate products), or humans (feces), a total of 44 could be affiliated with L. seeligeri (Table 1). The L. seeligeri isolates were characterized by sequencing the actA gene, which had been used previously for subtyping of L. monocytogenes (2, 10, 23, 24). Sequencing of 253 bp of the L. seeligeri actA gene with the primers actaf and actar of Volokhov et al. (21) disclosed seven different ActA types (Fig. 1) grouped in two phylogenetic lineages (Fig. 2). While two independent isolates originating from locations not interconnected and 50 km apart are grouped in lineage I, the other 42 L. seeligeri isolates from this study are grouped in lineage II. The two lineages can be distinguished also in the case of other genes of the virulence gene cluster (e.g., the seeligeriolysin gene hly) and genes not affiliated with the virulence gene cluster (e.g., the 16S rRNA coding genes, the iap gene, and the putative protease gene orf2110). Isolates of lineage I exhibit higher hemolytic and phospholipase activities on selective agars than those of lineage II (Fig. 3). ActA sequences of lineage II exhibit a high variability on the protein level (from 79.0% to 99.5% identity), resulting in six different actA subtypes (Fig. 1). Notable differences are an insert of seven additional amino acids in subtypes 1 and 2, a leucine-proline repeat in subtypes 5 to 7, and a stop codon in the actA gene in subtype 4.
TABLE 1.
Occurrence of Listeria spp. in organs of animals, feces of humans and animals, and butcher shopsa in Upper Franconia, Germany
| Origin (n) | % of samples positive for: |
||||
|---|---|---|---|---|---|
| Listeria spp. | L. monocytogenes | L. innocua | L. welshimeri | L. seeligeri | |
| Animal organs (63) | 11.1 | 6.3 | 1.6 | 0.0 | 3.2 |
| Animal feces (31) | 9.7 | 9.7 | 0.0 | 0.0 | 0.0 |
| Drain (522) | 41.8 | 27.2 | 14.0 | 2.5 | 3.4 |
| Sausage meat (789) | 59.1 | 30.4 | 23.8 | 10.4 | 3.0 |
| Human feces (1,087) | 1.9 | 1.1 | 0.7 | 0.4 | 0.0 |
Drains and sausage meat.
FIG. 1.
Hypothetical protein sequences of the seven identified ActA subtypes of L. seeligeri (Ls). The protein sequences correspond to the hypothetical translation products of the 253-bp actA gene fragments (Fig. 2). For every subtype, an example is given. Alignments were done with the program BioEdit. The asterisk within the type 4 sequence denominates a stop codon. Gaps are indicated by the “∼” symbol.
FIG. 2.
Dendrogram showing phylogenetic relationship among L. seeligeri (Ls) isolates based on partial actA sequences (253 bp) from L. seeligeri isolates obtained in Upper Franconia, Germany, and reference strains L. seeligeri ATCC 35967 (AY878348) and L. seeligeri NRRL (AY510074). The tree was constructed by the minimum evolution method in the MEGA version 4 package. The bootstrap values presented at corresponding branches were evaluated from 1,000 replications. The phylogenetic lineages of the strains are symbolized with Roman numerals and the corresponding actA subtypes with Arabic numerals. The bar indicates 1.0% sequence divergence. Strains marked with an asterisk carried a gene similar to the orf2110 gene of L. monocytogenes F2365 (AE017262).
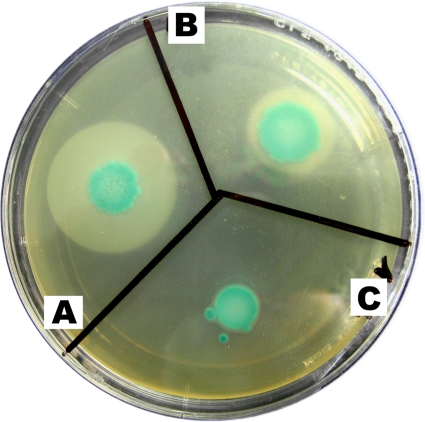
FIG. 3.
Phosphatidylinositol-specific phospholipase activity on ALOA agar plates (Merck, Darmstadt, Germany). For analysis of the phospholipase activity, 10 μl of a overnight culture in brain heart infusion (BHI) liquid medium of L. monocytogenes 1 (A), L. seeligeri 90 (B), and L. seeligeri 187 (C) was dropped onto ALOA agar plates and incubated at 37°C for 48 h.
Virulence clusters of L. seeligeri lineages I and II.
Isolates L. seeligeri 90 and L. seeligeri 187, representing lineage I and lineage II, respectively, exhibit an identical arrangement of the virulence gene cluster with respect to gene organization. Both lineages also show high homology in the housekeeping genes prs and ldh (100% at the protein level), while other virulence gene products vary with respect to their identity from 81.3% to 99.1% (Table 2). The most significant differences between the lineages are observed with the dplcB and orfY genes. While the dplcB gene of lineage II represents a truncated form of the plcB gene, as described previously (3), the dplcB gene of lineage I includes 534 additional nucleotides, resembling the full-length adjacent plcB gene. The hypothetical translation product, a preproenzyme, shows an identity of only 48.5% to the PlcB phospholipase of the same strain. It differs, however, in five out of nine zinc ion coordinating amino acids from PlcB. This would result in a nonfunctional enzyme, as an exchange of Trp1 to Leu1 would impair the binding of a zinc ion at site 1.
TABLE 2.
Similarities and identities between gene and protein sequences of L. seeligeri 90 and 187 and L. monocytogenes F2365
| Lineages compared and comparison parameter | % similarity for comparisona |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| prsb | orfC | orfD | prfA | orfE | plcA | hly | orfK | mpl | actA | dplcB | plcB | orfY | orfX | orfI | orfP | orfB | orfA | ldhb | iapb,c | |
| L. seeligeri 90 and 187 | ||||||||||||||||||||
| Similarity (nt) | 95.9 | 95.1 | 93.7 | 97.8 | 96.1 | 96.3 | 97.5 | 93.3 | 94.2 | 87.8 | 34.9 | 95.1 | 82.1 | 98.2 | 96.8 | 95.2 | 100.0 | 92.7 | 97.0 | 95.6 |
| Identity (aa) | 100.0 | 95.0 | 92.7 | 99.1 | 95.5 | 96.2 | 98.3 | 89.2 | 92.5 | 81.3 | ND | 93.5 | 80.3 | 98.2 | 95.3 | 96.6 | 100.0 | 93.2 | 100.0 | 95.3 |
| L. seeligeri 187 and L. monocytogenes F2365 | ||||||||||||||||||||
| Similarity (nt) | 85.4 | 75.9 | 65.9 | 75.7 | 68.5 | 47.0 | 68.3 | 85.7 | 39.6 | |||||||||||
| Identity (aa) | 95.0 | 73.8 | 63.4 | 81.1 | 64.3 | 21.4 | 62.3 | 97.0 | 39.8 | |||||||||||
| L. seeligeri 90 and L. monocytogenes F2365 | ||||||||||||||||||||
| Similarity (nt) | 87.9 | 76.4 | 65.3 | 75.7 | 69.7 | 46.4 | 68.3 | 85.7 | 39.4 | |||||||||||
| Identity (aa) | 95.0 | 73.8 | 63.7 | 81.5 | 65.2 | 20.8 | 61.6 | 97.0 | 40.2 | |||||||||||
Nucleotide (nt) similarities and amino acid (aa) identities between gene and protein sequences of the two lineages of L. seeligeri isolates 90 (lineage I; GQ862951) and 187 (lineage II; GQ862952) and L. monocytogenes F2365 (AE017262). Alignments were done with the program BioEdit. ND, not determined.
Partial sequence.
The iap gene encodes the invasion-associated protein P60.
The serine protease gene orf2110.
The orf2110 gene encodes a hypothetical serine protease carrying a V8-like Glu-specific endopeptidase domain (COG3591) (13) as well as three Chw (Clostridium hydrophobic tryptophan) domains (12). The orf2110 gene has been considered specific for L. monocytogenes serotypes 4b, 4d, and 4e (7). In this study, a gene showing high similarity to the orf2110 gene of L. monocytogenes (95.6% to lmof2365_1900 in AAT04669) (11) and to a lesser extent to the analogous gene in L. welshimeri (80.2% similarity; lwe1890 in CAK21308) (5) is described for the first time in L. seeligeri. orf2110 is always located between the genes encoding a hypothetical alkaline protease (lmof2365_1899 in L. monocytogenes F2365 and lwe1889 in L. welshimeri SLCC5334) and a phosphomutase family protein (lmof2365_1901 in L. monocytogenes F2365 and lwe1891 in L. welshimeri SLCC5334). The flanking genes are also present in strains without an orf2110 gene, like in L. innocua (e.g., lin1984 and lin1985 in AL592022 L. innocua strain Clip 11262) (8) and some L. seeligeri strains (Fig. 2). The orf2110 gene is present in the isolates of lineage I as well as in some isolates of lineage II, primarily in ActA types 4 and 5 (Fig. 2). It is noteworthy that the orf2110 genes of L. seeligeri resemble more closely the genes from L. monocytogenes, whereas the 16S and 23S rRNA coding genes as well as the iap gene are phylogenetically related to L. welshimeri (16).
Conclusions.
It has been suggested that L. seeligeri carries an ancestral form of the virulence gene cluster, which has been deleted from the chromosome of L. innocua and L. welshimeri (8) and some L. seeligeri isolates (21). This cluster would have been subsequently optimized by “pathoadaptive mutations” for the infection and survival in mammals during the evolution of L. monocytogenes and L. ivanovii (19). It has been hypothesized that the virulence gene cluster of L. seeligeri and its low expression are adequate for survival in bacteriovorous protozoae like Tetrahymena (9). However, our results suggest that L. seeligeri is subject to ongoing adaptation to changing environments. Accordingly, lineage II appears to be better adapted to natural environments, as indicated for example by the deletion in the dplcB gene, the stop codon within the actA gene, or the loss of the entire virulence gene cluster (21). On the other hand, lineage I might be better adapted to the mammalian hosts, as indicated by higher hydrolytic activities. The concomitant deletion in the orfY gene might also contribute to survival in mammalian hosts as this gene is lacking in L. monocytogenes too.
Acknowledgments
This work was financially supported by the Free State of Bavaria (Bayerisches Staatsministerium für Umwelt und Gesundheit) within the framework of Projekt 82 Informations- und Transferzentrum Lebensmittelsicherheit/-technologie ITL Teilprojekt 82/4: Besondere Aspekte der Lebensmittelsicherheit in kleinen und mittleren Betrieben.
Footnotes
Published ahead of print on 12 March 2010.
REFERENCES
- 1.Cummins, A. J., A. K. Fielding, and J. McLauchlin. 1994. Listeria ivanovii infection in a patient with AIDS. J. Infect. 28:89-91. [DOI] [PubMed] [Google Scholar]
- 2.den Bakker, H. C., X. Didelot, E. D. Fortes, K. K. Nightingale, and M. Wiedmann. 2008. Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC Evol. Biol. 8:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouin, E., J. Mengaud, and P. Cossart. 1994. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 62:3550-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graves, L. M., L. O. Helsel, A. G. Steigerwalt, R. E. Morey, M. I. Daneshvar, S. E. Roof, R. H. Orsi, E. D. Fortes, S. R. Milillo, H. C. den Bakker, M. Wiedmann, B. Swaminathan, and B. D. Sauders. 2009. Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. doi: 10.1099/ijs.0.014118-0. [DOI] [PubMed]
- 5.Hain, T., C. Steinweg, C. T. Kuenne, A. Billion, R. Ghai, S. S. Chatterjee, E. Domann, U. Kärst, A. Goesmann, T. Bekel, D. Bartels, O. Kaiser, F. Meyer, A. Pühler, B. Weisshaar, J. Wehland, C. Liang, T. Dandekar, R. Lampidis, J. Kreft, W. Goebel, and T. Chakraborty. 2006. Whole-genome sequence of Listeria welshimeri reveals common steps in genome reduction with Listeria innocua as compared to Listeria monocytogenes. J. Bacteriol. 188:7405-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson, J., K. Jinneman, G. Stelma, B. G. Smith, D. Lye, J. Messer, J. Ulaszek, L. Evsen, S. Gendel, R. W. Bennett, B. Swaminathan, J. Pruckler, A. Steigerwalt, S. Kathariou, S. Yildirim, D. Volokhov, A. Rasooly, V. Chizhikov, M. Wiedmann, E. Fortes, R. E. Duvall, and A. D. Hitchins. 2004. Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes. Appl. Environ. Microbiol. 70:4256-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khelef, N., M. Lecuit, C. Buchrieser, D. Cabanes, O. Dussurget, and P. Cossart. 2006. Listeria monocytogenes and the genus Listeria, p. 404-476. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: a handbook on the biology of bacteria. Bacteria: Firmicutes, Cyanobacteria, 3rd ed., vol. 4. Springer, New York, NY. [Google Scholar]
- 8.Kreft, J., J. A. Vázquez-Boland, S. Altrock, G. Domínguez-Bernal, and W. Goebel. 2002. Pathogenicity islands and other virulence elements in Listeria. Curr. Top. Microbiol. Immunol. 264:109-125. [PubMed] [Google Scholar]
- 9.Ly, T. M. C., and H. E. Müller. 1990. Interactions of Listeria monocytogenes, Listeria seeligeri, and Listeria innocua with protozoans. J. Gen. Appl. Microbiol. 36:142-150. [Google Scholar]
- 10.Meinersmann, R. J., R. W. Philips, M. Wiedmann, and M. E. Berrang. 2004. Multilocus sequencing typing of Listeria monocytogenes by use of hypervariable genes reveals clonal and recombination histories of three lineages. Appl. Environ. Microbiol. 70:2193-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparison of serotype 4b and 1/2a strains of the food borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nölling, J., G. Breton, M. V. Omelcheko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qui, J. Hitti, GTC Sequencing Center Production, Finishing, and Bioinformatics Teams, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucalle, M. J. Daly, G. N. Bennett, E. V. Konin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasad, L., Y. Leduc, K. Hayakawa, and L. T. Delbaere. 2004. The structure of a universally employed enzyme: V8 protease from Staphylococcus aureus. Acta Crystallogr. D Biol. Crystallogr. 60:256-259. [DOI] [PubMed] [Google Scholar]
- 14.Rocourt, J., H. Hof, A. Schrettenbrunner, R. Malinverni, and J. Bille. 1986. Acute purulent Listeria seeligeri meningitis in an immunocompetent adult. Schweiz. Med. Wochenschr. 116:248-251. [PubMed] [Google Scholar]
- 15.Schlech, W. F., III. 2000. Foodborne listeriosis. Clin. Infect. Dis. 31:770-775. [DOI] [PubMed] [Google Scholar]
- 16.Schmid, M. W., E. Y. W. Ng, R. Lampidis, M. Emmerth, M. Walcher, J. Kreft, W. Goebel, M. Wagner, and K.-H. Schleifer. 2005. Evolutionary history of the genus Listeria and its virulence genes. Syst. Appl. Microbiol. 28:1-18. [DOI] [PubMed] [Google Scholar]
- 17.Seeliger, H. P. R., and D. Jones. 1986. The genus Listeria, p. 1235-1245. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins, Baltimore, MD. [Google Scholar]
- 18.Snapir, Y. M., E. Vaisbein, and F. Nassar. 2006. Low virulence but potentially fatal outcome—Listeria ivanovii. Eur. J. Intern. Med. 17:286-287. [DOI] [PubMed] [Google Scholar]
- 19.Sokurenko, E. V., D. L. Hasty, and D. E. Dykhuizen. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 20.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. Gonzáluez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volokhov, D., J. George, C. Anderson, R. E. Duvall, and A. D. Hitchins. 2006. Discovery of natural atypical nonhemolytic Listeria seeligeri isolates. Appl. Environ. Microbiol. 72:2439-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volokhov, D. V., S. Duperrier, A. A. Neverov, J. George, C. Buchrieser, and A. D. Hitchins. 2007. The presence of the internalin gene in natural atypically hemolytic Listeria innocua strains suggests descent from L. monocytogenes. Appl. Environ. Microbiol. 73:1928-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiedmann, M., J. L. Bruce, C. Keating, A. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in their pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]