Abstract
We analyzed cryptophyte nucleomorph 18S rRNA gene sequences retained in natural Myrionecta rubra cells and plastid 16S rRNA gene and psbA sequences retained in natural cells of several Dinophysis species collected from Japanese coastal waters. A total of 715 nucleomorph sequences obtained from 134 M. rubra cells and 564 plastid 16S rRNA gene and 355 psbA sequences from 71 Dinophysis cells were determined. Almost all sequences in M. rubra and Dinophysis spp. were identical to those of Teleaulax amphioxeia, suggesting that M. rubra in Japanese coastal waters preferentially ingest T. amphioxeia. The remaining sequences were closely related to those of Geminigera cryophila and Teleaulax acuta. Interestingly, 37 plastid 16S rRNA gene sequences, which were different from T. amphioxeia and amplified from Dinophysis acuminata and Dinophysis norvegica cells, were identical to the sequence of a D. acuminata cell found in the Greenland Sea, suggesting that a widely distributed and unknown cryptophyte species is also preyed upon by M. rubra and subsequently sequestered by Dinophysis. To confirm the reliability of molecular identification of the cryptophyte Teleaulax species detected from M. rubra and Dinophysis cells, the nucleomorph and plastid genes of Teleaulax species isolated from seawaters were also analyzed. Of 19 isolates, 16 and 3 clonal strains were identified as T. amphioxeia and T. acuta, respectively, and no sequence variation was confirmed within species. T. amphioxeia is probably the primary source of prey for M. rubra in Japanese coastal waters. An unknown cryptophyte may serve as an additional source, depending on localities and seasons.
The marine dinoflagellate genus Dinophysis comprises photosynthetic and nonphotosynthetic members and is globally distributed in coastal and oceanic waters (12, 29, 30). Several members of the genus Dinophysis produce potent polyether toxins that can accumulate in filter-feeding bivalves, leading to a syndrome known as diarrhetic shellfish poisoning (DSP) in humans who consume tainted shellfish. These toxic algal species are important not only for their potential impact on public health but also from an ecological point of view because of their dual role as primary and secondary producers in complex microbial food webs. Despite extensive studies over the last 2 decades, little is known about the ecophysiology, toxicology, and bloom mechanisms of DSP-causing species of Dinophysis, primarily due to an inability to culture them. Since the first successful cultivation of Dinophysis acuminata by Park et al. (42), the understanding of Dinophysis biology and ecology has progressed considerably. Three other species (Dinophysis caudata, Dinophysis fortii, and Dinophysis infundibulus) are now available in culture, and it has become clear that these different Dinophysis species require the presence of both cryptophytes and the marine ciliate Myrionecta rubra to grow and proliferate (36, 37, 38). When presented with the marine ciliate M. rubra as prey, the four Dinophysis species mentioned above exhibited phagotrophic behaviors, sequestering the cytoplasm and plastids from within the ciliate through a feeding tube called a peduncle. M. rubra individuals prey upon cryptophytes, harboring their plastids, nuclei, and nucleomorphs; they also utilize the cryptophyte plastids for their own photosynthesis (21, 22, 23, 54). Hence, plastids from the cryptophyte are first transferred to the ciliate M. rubra and then transferred again to the dinoflagellate Dinophysis while their photosynthetic machinery remains functional as kleptoplastids (34). However, whether the plastids of Dinophysis are temporarily sequestered (kleptoplastid) or permanently established is still controversial (9, 34, 35, 41, 43, 50), as is the case of plastids in M. rubra (13, 23). Minnhagen et al. (34) reported that there was no significant difference in plastid DNA content between the G1 and G2 phases in natural Dinophysis norvegica cells, suggesting that this is consistent with kleptoplastidy. On the other hand, Garcia-Cuetos et al. (9) found that several plastid features separated D. acuminata from both the cryptophyte Teleaulax amphioxeia and the ciliate M. rubra based on observations by transmission electron microscopy (TEM) in laboratory culture experiments. Their interpretation of the data was that D. acuminata harbors permanent plastids of cryptophyte origin, not kleptoplastids.
In recent years, it has become clear that plastid sequences of the 16S rRNA gene and psbA (encoding the photosystem II [PSII] reaction center protein D1) in natural Dinophysis cells (except Dinophysis mitra) (27) originate from the cryptophyte genus complex Teleaulax/Geminigera/Plagioselmis (19, 35, 49, 50). Minnhagen and Janson (35) have also reported an exception to this finding in that natural D. acuminata cells isolated from the Greenland Sea contained a plastid 16S rRNA gene sequence that was more closely related to Geminigera cryophila. These characteristics are probably caused by the cryptophyte prey preference of M. rubra in natural environments. Under laboratory conditions, M. rubra can be cultivated by providing the cryptophyte T. amphioxeia (36, 37, 38), Teleaulax acuta (22), or G. cryophila (23), but its prey preference in natural environments has never been investigated.
Therefore, in this study, we investigated the prey organisms of M. rubra and the plastid identities of several Dinophysis species in Japanese coastal waters to reveal their ecological relationships in natural environments. The prey organisms of M. rubra were determined by sequencing the cryptophyte nucleomorph 18S rRNA gene in natural M. rubra cells; this genetic region has high substitution rates and facilitates species identification (16). The origins of plastids in Dinophysis spp. were revealed by sequencing the plastid 16S rRNA gene and psbA in natural Dinophysis cells. We also isolated several Teleaulax-like cryptophytes from Japanese coastal waters and sequenced their nuclear 18S rRNA genes, nucleomorph 18S rRNA genes, plastid 16S rRNA genes, and psbA in order to compare these genes with those in naturally occurring M. rubra and Dinophysis cells.
MATERIALS AND METHODS
Cryptophyte isolation, cultivation, and identification.
Morphologically Teleaulax-like cryptophytes, which have a tail-like protrusion, were isolated from Japanese coastal waters (Fig. 1 and Table 1) and cultured according to the method of Nishitani et al. (38). Nuclear 18S rRNA gene sequences of each cryptophyte were determined for species identification, and the nucleomorph 18S rRNA gene, plastid psbA, and plastid 16S rRNA gene sequences were also determined to compare with those of M. rubra and Dinophysis spp. All sequences of cryptophytes were determined using DNA extracted from cultured cells.
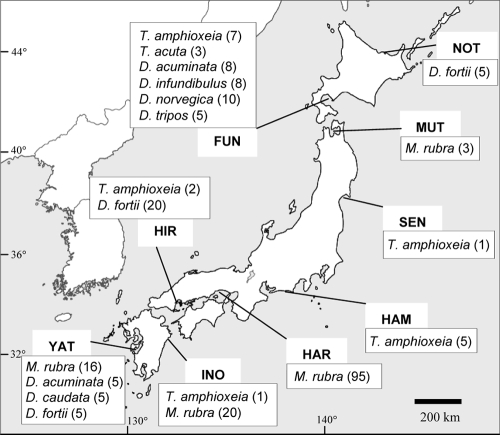
FIG. 1.
Sampling locations of M. rubra, Dinophysis spp., and Teleaulax spp. Numbers in parentheses refer to the number of natural M. rubra or Dinophysis cells isolated and the number of Teleaulax strains in culture (background map courtesy of the Itsuki Corporation; reproduced with permission).
TABLE 1.
The sampling sites and analyzed organisms
| Sampling site | Abbreviation | Position | Sample date | Organism |
|---|---|---|---|---|
| Notoro saline lake | NOT | 44.061°, 144.148° | 7 July 2007 | D. fortii |
| Funka Bay | FUN | 42.553°, 140.680° | 27 September 2008 | T. amphioxeia |
| Funka Bay | FUN | 42.553°, 140.680° | 27 September 2008 | T. acuta |
| Funka Bay | FUN | 42.553°, 140.680° | 8 June 2008 | D. acuminata |
| Funka Bay | FUN | 42.553°, 140.680° | 8 June 2008 | D. infundibulus |
| Funka Bay | FUN | 42.553°, 140.680° | 8 June 2008 | D. norvegica |
| Funka Bay | FUN | 42.553°, 140.680° | 20 August 2007 | D. tripos |
| Mutsu Bay | MUT | 40.945°, 140.863° | 19 August 2008 | M. rubra |
| Sendai Bay | SEN | 38.372°, 141.086° | 10 June 2006 | T. amphioxeia |
| Hamana saline lake | HAM | 34.721°, 137.563° | 15 April 2007 | T. amphioxeia |
| Harima-nada | HAR | 34.760°, 134.591° | 1 November 2007 | M. rubra |
| Hiroshima Bay | HIR | 34.275°, 132.267° | 7 February 2009 | T. amphioxeia |
| Hiroshima Bay | HIR | 34.275°, 132.267° | 8 June 2007 | D. fortii |
| Inokushi Bay | INO | 32.803°, 131.892° | 25 February 2007 | T. amphioxeia |
| Inokushi Bay | INO | 32.803°, 131.892° | 28 January 2008 | M. rubra |
| Yatsushiro Sea | YAT | 32.171°, 130.317° | 19 October 2007 | M. rubra |
| Yatsushiro Sea | YAT | 32.171°, 130.317° | 12 July 2007 | D. acuminata |
| Yatsushiro Sea | YAT | 32.171°, 130.317° | 12 July 2007 | D. caudata |
| Yatsushiro Sea | YAT | 32.171°, 130.317° | 12 July 2007 | D. fortii |
Sample collection of natural M. rubra and Dinophysis cells.
Sampling locations for M. rubra and Dinophysis spp. used in this study are shown in Fig. 1 and Table 1. M. rubra and Dinophysis cells were not collected all together at the same sites except for the Yatsushiro Sea because of the different peaks of blooms between the two species and the limited bloom seasons in Japan. In addition, living Myrionecta cells were very fragile when subjected to oscillation during transportation from the sampling sites to our laboratory. Seawater samples were collected at the surface layer. Single living cells were isolated, washed in several steps in drops of sterile seawater, and transferred into PCR tubes containing 10 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) by micropipetting under an inverted microscope. Species identifications were based on morphological characteristics observed by light microscopy.
Primer design.
For the amplification of nucleomorph 18S rRNA and plastid 16S rRNA genes, specific primers were designed to amplify the target regions in almost all cryptophytes with sequences registered in GenBank (Table 2). For the amplification of nuclear the 18S rRNA gene and plastid psbA, the primers were used as reported by Nishitani et al. (38) and Hackett et al. (11), respectively (Table 2).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) | Annealing site basesf | Reference or source |
|---|---|---|---|
| S1F | AACCTGGTTGATCCTGCCAG | 1-20a | 38 |
| S1R | CTACGAGCTTTTTAACTGCAACAA | 571-594b | 38 |
| S2F | CTGAGAAACGGCTACCACATC | 359-380b | 38 |
| S2R | TGGTAAGTTTTCCCGTGTTGAGTC | 1146-1169b | 38 |
| S3F | AGCTTGCGGCTTAATTTGACTC | 1129-1150b | 38 |
| S3R | CTACGGAAACCTTGTTACGAC | 1707-1727b | 38 |
| Nmorph-F | CATAGGAAGGATTGACAGATTGAG | 1231-1254c | This study |
| Nmorph-R | AAGGCATTCCTCGTTCAAGATG | 1578-1599c | This study |
| pSSU-F | TCGCGTCTGATTAGCTAGTTG | 100-120e | This study |
| pSSU-R | ATGCACCACCTGTGTTCAC | 882-900e | This study |
| psbAFdino | AGCACTGACAACCGTTTATAC | 1-21d | 11 |
| psbAR2 | TCATGCATWACTTCCATACCT | 912-928d | 11 |
DNA extraction, PCR amplification, cloning, and sequencing.
PCR tubes, each containing 10 μl of TE buffer and single cells of Myrionecta or Dinophysis, were boiled at 99°C for 10 min to extract the DNA. In the extraction of cryptophyte DNA, 100 to 1,000 cells/strain collected from the culture were boiled as above. Boiled samples (1.0 μl) were used as templates to amplify the target regions. All PCRs were performed on a PCR thermal cycler (PC-816; Astec, Fukuoka, Japan) in a reaction mixture (20 μl) containing 1.0 μl of template DNA, a 0.2 mM concentration of each deoxynucleoside triphosphate (dNTP), 1× PCR buffer (10 mM Tris-HCl, 50 mM KCl [pH 8.3]; Applied Biosystems, Foster City, CA), 2.5 mM Mg2+, 0.25 U of AmpliTaq Gold (Applied Biosystems), and a 0.2 μM concentration of each primer. For amplifying the nuclear 18S rRNA gene in cryptophytes, three primer pairs (S1F/S1R, S2F/S2R, and S3F/S3R) (Table 2) were used, and the PCR cycling conditions were as follows: 5 min at 94°C, followed by 38 cycles of 30 s each at 94°C, 30 s at 56°C, 1 min at 72°C, with a final elongation for 5 min at 72°C. For amplification of the cryptophyte nucleomorph 18S rRNA gene, the Nmorph-F and Nmorph-R primer pair (Table 2) was used, and the PCR cycling conditions were the same as those mentioned earlier for the nuclear 18S rRNA gene. For amplifications of the plastid 16S rRNA gene, a specifically designed primer pair (pSSU-F and pSSU-R) (Table 2) was used, and the PCR cycling conditions were the same as those of the nuclear 18S rRNA gene, as mentioned above. For amplification of the plastid psbA, the primer pair psbAFdino and psbAR2 (11) was used, and the PCR cycling conditions were the same as those used for the nuclear 18S rRNA gene, except that the annealing temperature was 50°C. Results of the PCRs were checked on ethidium-stained 1.5% agarose gels, and PCR products were then transformed into ECOS Competent Escherichia coli DH5α cells (Nippon Gene, Tokyo, Japan) after ligation into the pGEM T-Easy Vector (Promega, Madison, WI). DNA sequences were determined using a Dynamic ET Terminator Cycle Sequencing Kit (GE Healthcare, Little Chalfont, United Kingdom) with a U19 forward primer (5′-GGTTTTCCCAGTCACGACG-3′) or pUC/M13 reverse primer (5′-TCACACAGGAAACAGCTATGAC-3′) and analyzed on a DNA sequencer (ABI3100; Applied Biosystems). The forward and reverse sequences were aligned in GENETYX software (Software Development, Tokyo, Japan), and the sequences were checked against GenBank using BLASTN (1).
Checking the effectiveness of primers and PCR bias.
To confirm the effectiveness of all the primers (Table 2), we performed PCRs using the following eight extracted DNAs: Chroomonas mesostigmatica (CCMP269), G. cryophila (CCMP2564), Guillardia theta (CCMP327), Hanusia phi (CCMP325), Hemiselmis rufescens (CCMP644), Proteomonas sulcata (CCMP1175), Rhodomonas salina (CCMP1170), and T. amphioxeia (Japanese strain). Successful amplification was confirmed with the eight cryptophyte templates.
To check any bias during PCR and subcloning steps, we conducted a nucleomorph PCR and subcloning experiment using combined DNAs extracted from two cryptophyte species of G. cryophila and T. amphioxeia as a template. The ratio of DNA concentrations was 1:1 in the first test and 1:9 in the second. Sequences were determined under the same conditions as mentioned earlier, and the sequence ratios between the species were roughly reproduced in a total of 35 nucleomorph sequences.
Phylogenetic analysis.
The partial sequences of the nuclear 18S rRNA gene, nucleomorph 18S rRNA gene, plastid 16S rRNA gene, and plastid psbA were aligned using the Clustal W algorithm (52) in MEGA software, version 4.0 (51). Maximum-likelihood analyses were performed with PhyML (10) using an input tree generated by BIONJ with a general time-reversal model (44) that incorporated invariable sites and a discrete gamma distribution (eight categories) (GTR+I+Γ). PhyML bootstrap trees (1,000 replicates) were constructed using the same parameters as the individual maximum-likelihood trees. Posterior probabilities of Bayesian trees were also estimated using MrBayes, version 3.1.1 (17, 46), under the GTR+I+Γ model. One cold and three heated Markov chain Monte Carlo (MCMC) chains were run for 500,000 generations to sample log likelihoods and trees at 100-generation intervals (5,000 trees were saved during MCMC with a burn-in of 1,250 trees).
RESULTS AND DISCUSSION
Genetic analyses in established clonal cultures of cryptophytes.
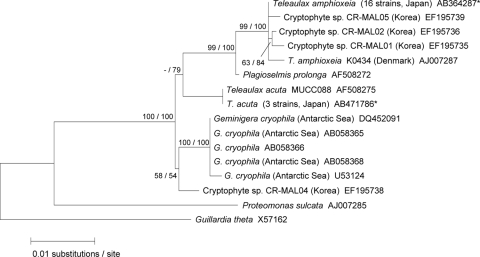
In total, we established 19 clonal strains of Teleaulax-like species from Japanese coastal waters (Fig. 1) and identified 16 strains as T. amphioxeia and three as T. acuta, respectively. These two species were identified based on morphological characteristics (14, 15) and on their nuclear and plastid sequences. The partial nuclear 18S rRNA gene sequences determined in this study were 1,705 bp in T. amphioxeia and 1,706 bp in T. acuta. Sequences of T. amphioxeia in the 16 Japanese strains were identical to each other and showed 0.2% divergence from the sequence of T. amphioxeia (SCCAP K0434) found in Denmark, as reported by Marin et al. (31). These two sequences shared high sequence similarity with those of the cryptophyte species (CR-MAL01, CR-MAL02, and CR-MAL05) reported by Park et al. (40) and formed a clade with five operational taxonomic units (OTUs) (Fig. 2). All nuclear 18S rRNA gene sequences in the three strains of T. acuta were identical to the sequence of the T. acuta MUCC088 strain reported by Deane et al. (5) and formed a sister clade with T. amphioxeia and Plagioselmis prolonga. In the partial nucleomorph 18S rRNA gene (369 bp) sequences of 16 strains of T. amphioxeia and 3 strains of T. acuta isolated from Japanese coastal waters, no intraspecific sequence variations were detected in the two species (Fig. 3). Although we detected several sequence variations (4 to 10 bp) of the nuclear 18S rRNA gene among the Japanese T. amphioxeia strain, the Danish T. amphioxeia strain (K0434), and the Korean cryptophyte species (CR-MAL01), the strains had identical sequences in the plastid 16S rRNA gene (801 bp) and plastid psbA (890 bp) (Fig. 2, 4, and 5), thereby suggesting that these three isolates were the same species and that the sequence variations in the nuclear 18S rRNA genes were likely to be intraspecific variations.
FIG. 2.
A maximum-likelihood tree inferred from the cryptophyte nuclear 18S rRNA gene. Sequences obtained in this study are indicated by asterisks. The values at left are in the form bootstrap support values/posterior probabilities. −, a value less than 50.
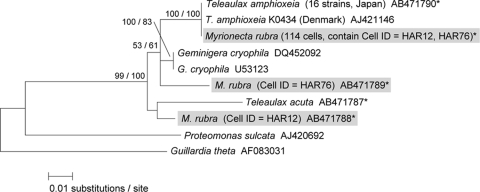
FIG. 3.
A maximum-likelihood tree inferred from the cryptophyte nucleomorph 18S rRNA gene. Sequences obtained in this study are indicated by asterisks, and sequences detected from natural M. rubra cells are further indicated by gray boxes. The values at left are in the form bootstrap support values/posterior probabilities.
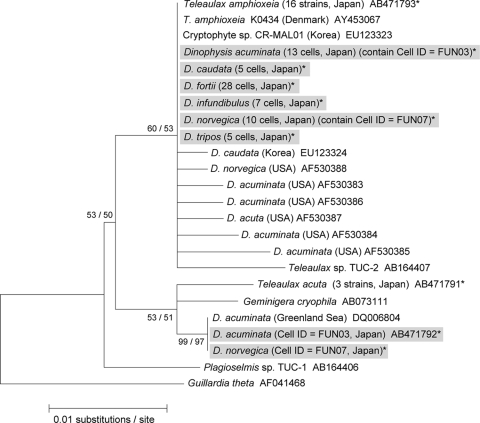
FIG. 4.
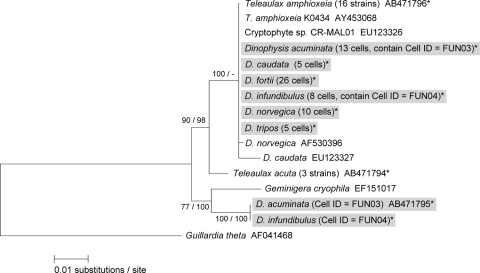
A maximum-likelihood tree inferred from the plastid 16S rRNA gene. Sequences obtained in this study are indicated by asterisks, and sequences detected from natural Dinophysis cells are further indicated by gray boxes. The values at left are in the form bootstrap support values/posterior probabilities.
FIG. 5.
A maximum-likelihood tree inferred from the plastid psbA. Sequences obtained in this study are indicated by asterisks, and sequences detected from natural Dinophysis cells are further indicated by gray boxes. The values at left are in the form bootstrap support values/posterior probabilities.
Genetic analyses in natural M. rubra cells.
In consideration of the possibility that different cryptophyte species coexist in a single M. rubra cell, we checked for a bias in PCR amplification efficiency by using a mixture of nucleomorph primers for DNAs from two species, G. cryophila and T. amphioxeia, at different ratios. When the ratio for G. cryophila and T. amphioxeia DNAs was 1:1, 18 and 17 sequences out of a total of 35 were detected, respectively; i.e., the sequence ratio was 1.0:1.1. Similarly, when the ratio of DNAs was 1:9, 4 and 31 sequences out of 35 were obtained, respectively; i.e., the sequence ratio was 1.0:7.8, which clearly showed that our PCR and cloning procedures provided highly reliable information on the detection of cryptophyte species.
The partial nucleomorph 18S rRNA gene sequences determined in this study comprised 369 bp and showed 8-bp substitutions between T. amphioxeia and G. cryophila. The percentages of cells of those isolated from natural M. rubra that were successfully PCR amplified varied and depended mostly on the sampling locations, ranging from 33% in Mutsu Bay to 100% in Harima-Nada (Table 3). A possible explanation for this might be that there were some artificial errors in the DNA extraction step and/or the PCR amplification. We determined a total of 715 nucleomorph sequences from 114 PCR products of 134 M. rubra cells. Of these, 713 sequences were identical to the sequence of T. amphioxeia (99.7%), while the other 2 sequences (AB471788 and AB471789, representing 0.3%), which were detected from the Harima-Nada station M. rubra cells (cell identifier [ID] HAR12 and HAR76), were closely related to those of G. cryophila and T. acuta (Table 3 and Fig. 3). These two M. rubra cells contained two different cryptophyte sequences in each cell; in each of these cells the sequences detected occurred in the proportion of seven sequences of T. amphioxeia to one other sequence. Knowledge of the availability of suitable prey for M. rubra is very limited, but up to now, M. rubra has been successfully cultured using T. amphioxeia (36, 37, 38), T. acuta (22), and G. cryophila (23) as prey. In addition, Park et al. (40) have isolated several T. amphioxeia-like cryptophyte species inferred from the nuclear 18S rRNA gene phylogeny and have cultured M. rubra by addition of those cryptophytes as prey. Considering these facts, the genera Teleaulax and Geminigera seem to be preferred prey for M. rubra. In this study, however, the nucleomorph sequences of T. acuta and G. cryophila were never detected from natural cells of M. rubra, suggesting that the two species are unlikely to become the prey of M. rubra, at least in Japanese coastal waters. Accordingly, this study found that T. amphioxeia is probably the most important species for M. rubra as a prey source. On the other hand, the nucleomorph analysis of M. rubra in this study also detected two sequences other than T. amphioxeia that were closely related to the sequences of G. cryophila and T. acuta (Fig. 3). The fact suggests that unknown cryptophytes exist and that they serve as an additional prey source for M. rubra. In fact, the possibility exists that unknown cryptophytes serve as a primary prey source for M. rubra, depending on localities and seasons in which T. amphioxeia is less abundant. Thus, it is important to isolate the unknown cryptophytes and conduct further culture experiments with M. rubra. In a recent examination of M. rubra based on ultrastructure by TEM and on the dynamics of photosynthesis, ingestion, and growth rates under laboratory conditions, Hansen and Fenchel (13) presented evidence showing that M. rubra harbors a permanent cryptophyte endosymbiont that has different and larger chloroplasts than the Teleaulax added as prey. Hence, it is possible that the different sequence may have originated from a permanent cryptophyte endosymbiont.
TABLE 3.
Summary of the sequence analyses of the cryptophyte nucleomorph gene in natural M. rubra cells and of plastid genes in natural Dinophysis cells detected by single-cell PCR
| Species | Sitec | No. of cells isolatedb | Sequence analysis (no. of cells [no. of sequences analyzed])a |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleomorph |
Plastid 16S rRNA |
psbA |
|||||||||
| PCR | T. amphioxeia | Other | PCR | T. amphioxeia | Other | PCR | T. amphioxeia | Other | |||
| M. rubra | MUT | 3 | 1 | 1 (45) | 0 | ||||||
| M. rubra | HAR | 95 | 95 | 95 (604) | 2 (2) | ||||||
| M. rubra | INO | 20 | 11 | 11 (37) | 0 | ||||||
| M. rubra | YAT | 16 | 7 | 7 (27) | 0 | ||||||
| D. fortii | NOT | 5 | 5 | 5 (75) | 0 | 5 | 5 (88) | 0 | |||
| D. acuminata | FUN | 8 | 8 | 8 (64) | 1 (31) | 8 | 8 (38) | 1 (1) | |||
| D. infundibulus | FUN | 8 | 7 | 7 (43) | 0 | 8 | 8 (45) | 1 (2) | |||
| D. norvegica | FUN | 10 | 10 | 10 (75) | 1 (6) | 10 | 10 (36) | 0 | |||
| D. tripos | FUN | 5 | 5 | 5 (26) | 0 | 5 | 5 (18) | 0 | |||
| D. fortii | HIR | 20 | 19 | 19 (135) | 0 | 17 | 17 (53) | 0 | |||
| D. acuminata | YAT | 5 | 5 | 5 (39) | 0 | 5 | 5 (25) | 0 | |||
| D. caudata | YAT | 5 | 5 | 5 (38) | 0 | 5 | 5 (26) | 0 | |||
| D. fortii | YAT | 5 | 4 | 4 (32) | 0 | 4 | 4 (23) | 0 | |||
| Total | 114 | 114 (713) | 2 (2) | 68 | 68 (527) | 2 (37) | 67 | 67 (352) | 2 (3) | ||
Values are from successful PCR amplifications. Sequences not identified as belonging to T. amphioxeia are grouped as other.
A total of 134 and 71 cells were isolated from M. rubra and Dinophysis spp., respectively.
See Table 1 for identification of the sampling sites.
Currently, 17 genera of cryptophytes are recognized (4). The group contains about 100 freshwater species and a similar number of marine representatives. However, the total number of species has been estimated at about 1,200, which is six times more than are currently known (2), implying that cryptophyte diversity is far from being fully described. Therefore, it is always possible that environmental samples will contain previously undescribed species. Recently, several surveys of environmental sequence diversity using universal primers uncovered the hidden diversity of cryptophytes (8, 32, 33, 45, 48). McDonald et al. (33) detected several novel 16S plastid rRNA gene sequences from environmental clone libraries from the Gulf of Naples that were closely related to G. cryophila (AB073111) or Plagioselmis sp. (AB164406) and represent unknown species. Thus, diverse cryptophyte species have been reported in natural environments. However, in our study we found that only a few cryptophyte species (i.e., some members of the genera Teleaulax and Geminigera) were ingested by M. rubra, suggesting their clear prey selectivity. Studies of natural distribution and abundances of cryptophytes have been undertaken throughout the year using fluorescence in situ hybridization (FISH) probes that specifically bind to Dinophysis plastids (26). Their results have shown that abundances of Teleaulax and Geminigera were much lower than the total abundance of cryptophytes in Iwate Prefecture in northern Japan. In view of the low abundance of Teleaulax and Geminigera in natural environments, why does M. rubra prefer to ingest these cryptophyte species? How does M. rubra they distinguish Teleaulax/Geminigera from other cryptophytes? Food selectivity based on prey size, surface properties, and chemosensory factors has been reported for some herbivorous ciliates (3, 7, 24, 25, 53). Any of these factors may greatly influence the prey selectivity of M. rubra. There may also be some other unknown factors associated with the easier acquirement of Teleaulax/Geminigera plastids as kleptoplastids by M. rubra. Kleptoplastidy of T. amphioxeia by other dinoflagellates, such as Amylax buxus and Amylax triacantha, has also been reported although the vestigial plastid, nucleomorph, and mitochondria were still kept (28). These findings suggest that the process of plastid sequestration by kleptoplastidy is still insufficiently understood.
Plastid genes in natural Dinophysis cells.
PCR efficiencies of plastid 16S rRNA genes ranged from 80% to 100% (mean, 96%), depending on the sampling location (Table 3). The partial plastid 16S rRNA gene sequences of Dinophysis spp. determined in this study were 801 bp, and we determined 564 sequences from the successfully amplified PCR products (n = 68). Of these, 527 sequences were identical to the sequence of T. amphioxeia (93.4%). No variation among the sequences was confirmed although Hackett et al. (11) previously detected several variations (less than 16 bp out of ca. 1,200 bp) in plastid 16S rRNA gene sequences in Dinophysis spp. isolated from the East Coast of North America (Fig. 4). The remaining 37 sequences obtained in this study (5.6%), which were all identical and were detected only from the Funka Bay sample (D. acuminata, cell ID FUN03; D. norvegica, cell ID FUN07), showed a close phylogenetic affiliation and a divergence of 0.7% and 0.9% compared with sequences of G. cryophila and T. acuta, respectively (Table 3 and Fig. 4). These Dinophysis cells contained two different cryptophyte sequences in a single cell. Four sequences of T. amphioxeia and 31 other sequences were detected from the D. acuminata cell (cell ID FUN03), and two sequences of T. amphioxeia and six other sequences were detected from the D. norvegica cell (cell ID FUN07). Interestingly, the other sequence (AB471792) was identical to that of a D. acuminata cell isolated from the Greenland Sea (35) although the cryptophyte species was not identified. The plastid 16S rRNA gene sequences of T. acuta and G. cryophila were never detected in natural cells of Dinophysis spp. throughout this study.
The result of the plastid psbA analysis was similar to that of the plastid 16S rRNA gene (Table 3). Depending on the sampling locations, PCR efficiencies ranged from 80% to 100% (mean, 94%). The partial plastid psbA sequences determined in this study were 890 bp, and 355 psbA sequences were determined from the successfully amplified PCR products. Of these, 352 sequences were identical to the sequence of T. amphioxeia (99.4%), and no variation was observed. As in the analysis of the plastid 16S rRNA gene, another three sequences (AB471795), which were all identical and detected only from the Funka Bay sample (D. acuminata, cell ID FUN03; D. infundibulus, cell ID FUN04) (0.6%), showed a close phylogenetic affiliation and a 2.6% divergence from that of G. cryophila (Table 3 and Fig. 5). Six sequences of T. amphioxeia and one other sequence were detected from the D. acuminata cell (cell ID FUN03), and five sequences of T. amphioxeia and two other sequences were detected from the D. infundibulus cell (cell ID FUN04). This unknown cryptophyte may occur in relatively cold waters at high latitudes since this unique sequence was detected from Greenland (35), and the water temperature in Funka Bay when we isolated the Dinophysis cells was ca. 7°C. Because there is an inflow of cold water from around the Sea of Okhotsk into Funka Bay every spring (18, 39, 47), the above unknown cryptophyte may have been transferred from the northern regions into the bay through the coastal Oyashio Current. Thus, it will be necessary to isolate and establish cultures in order to investigate further the role of the unknown cryptophyte as an additional prey species for M. rubra.
The high congruence of M. rubra prey (cryptophyte T. amphioxeia) and Dinophysis plastid identities obtained in this study probably support the kleptoplastidy concept in Dinophysis species. This is primarily because plastid sequences identical with T. amphioxeia sequence were obtained from all Dinophysis cells (D. acuminata, D. caudata, D. fortii, D. infundibulus, D. norvegica, and Dinophysis tripos) collected from Japanese coastal waters, and they are all identical with those collected from the Baltic Sea (20, 35), the North Sea (20, 35), the North Atlantic Ocean (20), a Norwegian fjord (35), and the North American East Coast (11). Second, the other sequences that differ from T. amphioxeia were detected from natural Dinophysis cells in this study (Fig. 4 and 5). If these sequences originated from permanently established plastids in Dinophysis, the sequences should be detected from not only the Funka Bay sample but also the other sampling areas. Although the high congruence can also be interpreted as the result of recent acquisition of plastids (41), this idea is invalidated by the fact that, based on TEM observations, each plastid is lined by only two complete membranes, with remains of a third membrane in D. acuminata (9). Third, we observed the disappearance of relatively large chloroplasts (>5 μm in length) from D. fortii after >4 weeks of incubation without the ciliate prey; only a few small chloroplasts (0.5 to 2 μm in length) remained in the marginal region of the cells, especially in small cells (36). Fourth, Minnhagen et al. (34) reported that there was no significant difference in plastid DNA content between the G1 and G2 phases in natural D. norvegica cells, showing no duplication of plastid genomes coupled with the cell cycle, as expected with kleptoplastidy. In addition, data obtained from monitoring photosynthetic activities by 13C labeling in D. fortii without the ciliate prey clearly indicated that the activities decreased rapidly within 1 week after heavy feeding on M. rubra (S. Nagai et al., unpublished data).
One hypothesis is that D. fortii might ingest M. rubra as merely a heterotrophic nutrient source and utilize the stolen chloroplasts for an ancillary function because growth of D. fortii was observed only during the first 7 days after the removal of prey (36). In observations of Dinophysis cells that were incubated without the ciliate prey for ca. 2 months, cells still retained a few small chloroplasts, implying the possibility of permanent plastids (36, 43). Are they retained kleptoplastids or permanent plastids, or both? To definitively answer these contradictory questions, further study is needed by several approaches such as tracing the cryptophyte plastids in cells of Dinophysis via M. rubra and/or analyzing plastid genomes using more variable regions.
Acknowledgments
This work was supported by a research project grant from the Fisheries Research Agency of Japan, a Grant-in-Aid for Scientific Research (B) from JSPS KAKENHI (grant number 20380116), a Grant-in-Aid for Young Scientists (B) from MEXT KAKENHI (grant number 21780191), and the Sasakawa Grants for Science Fellows.
Footnotes
Published ahead of print on 19 March 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, R. A. 1992. Diversity of eukaryotic algae. Biodivers. Conserv. 1:267-292. [Google Scholar]
- 3.Christaki, U., J. R. Dolan, S. Pelegri, and F. Rassoulzadegan. 1998. Consumption of picoplankton-size particles by marine ciliates: effects of physiological state of the ciliate and particle quality. Limnol. Oceanogr. 43:458-464. [Google Scholar]
- 4.Clay, B. L., P. Kugrens, and R. E. Lee. 1999. A revised classification of Cryptophyta. Bot. J. Linn. Soc. 131:131-151. [Google Scholar]
- 5.Deane, J. A., I. M. Strachan, G. W. Saunders, D. R. A. Hill, and G. I. McFadden. 2002. Cryptomonad evolution: nuclear 18S rDNA phylogeny versus cell morphology and pigmentation. J. Phycol. 38:1236-1244. [Google Scholar]
- 6.Douglas, S. E., C. A. Murphy, D. F. Spencer, and M. W. Gray. 1991. Cryptomonad algae are evolutionary chimaeras of two phylogenetically distinct unicellular eukaryotes. Nature 350:148-151. [DOI] [PubMed] [Google Scholar]
- 7.Fenchel, T., and P. R. Jonsson. 1988. The functional biology of Strombidium sulcatum, a marine oligotrich ciliate (Ciliophora, Oligotrichina). Mar. Ecol. Prog. Ser. 48:1-15. [Google Scholar]
- 8.Fuller, N. J., C. Campbell, D. J. Allen, F. D. Pitt, K. Zwirglmaier, F. L. Gall, D. Vaulot, and D. J. Scanlan. 2006. Analysis of photosynthetic picoeukaryotes diversity at open ocean sites in the Arabian Sea using a PCR biased towards marine algal plastids. Aquat. Microb. Ecol. 43:79-93. [Google Scholar]
- 9.Garcia-Cuetos, L., Ø. Moestrup, P. J. Hansen, and N. Daugbjerg. 2010. The toxic dinoflagellate Dinophysis acuminata harbors permanent chloroplasts of cryptomonad origin, not kleptochloroplasts. Harmful Algae 9:25-38. [Google Scholar]
- 10.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 11.Hackett, J. D., L. Maranda, H. S. Yoon, and D. Bhattacarya. 2003. Phylogenetic evidence for the cryptophyte origin of the plastid of Dinophysis (Dinophysiales, Dinophyceae). J. Phycol. 39:440-448. [Google Scholar]
- 12.Hallegraeff, G. M. 1995. Harmful algal blooms: a global overview, p. 1-22. In G. M. Hallegraeff, D. M. Andersen, and A. D. Cembella (ed.), Manual on harmful marine microalgae. Intergovernmental Oceanographic Commission manuals and guides no. 33. United Nations Educational, Scientific, and Cultural Organization, Paris, France.
- 13.Hansen, P. J., and T. Fenchel. 2006. The bloom-forming ciliate Mesodinium rubrum harbours a single permanent endosymbiont. Mar. Biol. Res. 2:169-177. [Google Scholar]
- 14.Hill, D. R. A. 1992. Teleaulax acuta (Butcher) Hill (Cryptophyceae). Baltic Sea phytoplankton identification sheet no. 12. Ann. Bot. Fennici 29:173-174. [Google Scholar]
- 15.Hill, D. R. A. 1992. Teleaulax amphioxeia (Conrad) Hill, comb. nov. (Cryptophyceae). Baltic Sea phytoplankton identification sheet no. 13. Ann. Bot. Fennici 29:175-176. [Google Scholar]
- 16.Hoef-Emden, K., B. Marin, and M. Melkonian. 2002. Nuclear and nucleomorph SSU rDNA phylogeny in the Cryptophyta and the evolution of cryptophyte diversity. J. Mol. Evol. 55:161-179. [DOI] [PubMed] [Google Scholar]
- 17.Huelsenbeck, J. P., F. Ronquist, R. Nielsen, and J. P. Bollback. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310-2314. [DOI] [PubMed] [Google Scholar]
- 18.Isoda, Y., and M. Kishi. 2003. A summary of Coastal Oyashio symposium. Bull. Coast. Oceanogr. 41:1-3. [Google Scholar]
- 19.Janson, S. 2004. Molecular evidence that plastids in the toxin-producing dinoflagellate genus Dinophysis originate from the free-living cryptophyte Teleaulax amphioxeia. Environ. Microbiol. 6:1102-1106. [DOI] [PubMed] [Google Scholar]
- 20.Janson, S., and E. Granéli. 2003. Genetic analysis of the psbA gene from single cells indicates a cryptomonad origin of the plastid in Dinophysis (Dinophyceae). Phycologia 42:473-477. [Google Scholar]
- 21.Johnson, M. D., D. Oldach, C. F. Delwiche, and D. K. Stoecker. 2007. Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra. Nature 445:426-428. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, M. D., and D. K. Stoecker. 2005. Role of feeding in growth and photophysiology of Myrionecta rubra. Aquat. Microb. Ecol. 39:303-312. [Google Scholar]
- 23.Johnson, M. D., T. Tengs, D. Oldach, and D. K. Stoecker. 2006. Sequestration, performance, and functional control of cryptophyte plastids in the ciliate Myrionecta rubra (Ciliophora). J. Phycol. 42:1235-1246. [Google Scholar]
- 24.Jonsson, P. R. 1986. Particle size selection, feeding rates and growth dynamics of marine planktonic oligotrichous ciliates (Ciliophora: Oligotrichina). Mar. Ecol. Prog. Ser. 33:265-277. [Google Scholar]
- 25.Kamiyama, T., and S. Arima. 2001. Feeding characteristics of two tintinnid ciliate species on phytoplankton including harmful species: effects of prey size on ingestion rates and selectivity. J. Exp. Mar. Bio. Ecol. 257:281-296. [DOI] [PubMed] [Google Scholar]
- 26.Koike, K., A. Nishiyama, K. Takishita, A. Kobiyama, and T. Ogata. 2007. Appearance of Dinophysis fortii following blooms of certain cryptophyte species. Mar. Ecol. Prog. Ser. 337:303-309. [Google Scholar]
- 27.Koike, K., H. Sekiguchi, A. Kobiyama, K. Takishita, M. Kawachi, K. Koike, and T. Ogata. 2005. A novel type of kleptoplastidy in Dinophysis (Dinophyceae): presence of haptophyte-type plastid in Dinophysis mitra. Protist 156:225-237. [DOI] [PubMed] [Google Scholar]
- 28.Koike, K., and K. Takishita. 2008. Anucleated cryptophyte vestiges in the gonyaulacalean dinoflagellates Amylax buxus and Amylax triacantha (Dinophyceae). Phycol. Res. 56:301-311. [Google Scholar]
- 29.Lessard, E. J., and E. Swift. 1986. Dinoflagellates from the North Atlantic classified as phototrophic or heterotrophic by epifluorescence microscopy. J. Plankton Res. 8:1209-1215. [Google Scholar]
- 30.Maestrini, S. Y. 1998. Bloom dynamics and ecophysiology of Dinophysis spp., p. 243-265. In D. M. Anderson, A. D. Cembella, and G. M. Hallegraeff (ed.), Physiological ecology of harmful algal blooms. NATO ASI series, vol. G 41. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 31.Marin, B., M. Klingberg, and M. Melkonian. 1998. Phylogenetic relationships among the Cryptophyta: analyses of nuclear-encoded SSU rRNA sequences support the monophyly of extant plastid-containing lineages. Protist 149:265-276. [DOI] [PubMed] [Google Scholar]
- 32.Massana, R., V. Balagué, L. Guillou, and C. Pedrós-Alió. 2004. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol. Ecol. 50:231-243. [DOI] [PubMed] [Google Scholar]
- 33.McDonald, S. M., D. Sarno, D. J. Scanlan, and A. Zingone. 2007. Genetic diversity of eukaryotic ultraphytoplankton in the Gulf of Naples during an annual cycle. Aquat. Microb. Ecol. 50:75-89. [Google Scholar]
- 34.Minnhagen, S., W. F. Carvalho, P. S. Salomon, and S. Janson. 2008. Chloroplast DNA content in Dinophysis (Dinophyceae) from different cell cycle stages is consistent with kleptoplasty. Environ. Microbiol. 10:2411-2417. [DOI] [PubMed] [Google Scholar]
- 35.Minnhagen, S., and S. Janson. 2006. Genetic analyses of Dinophysis spp. support kleptoplastidy. FEMS Microbiol. Ecol. 57:47-54. [DOI] [PubMed] [Google Scholar]
- 36.Nagai, S., G. Nishitani, Y. Tomaru, S. Sakiyama, and T. Kamiyama. 2008. Predation on the ciliate Myrionecta rubra by the toxic dinoflagellate Dinophysis fortii and observation of sequestration of ciliate chloroplasts. J. Phycol. 44:909-922. [DOI] [PubMed] [Google Scholar]
- 37.Nishitani, G., S. Nagai, S. Sakiyama, and T. Kamiyama. 2008. Successful cultivation of the toxic dinoflagellate Dinophysis caudata (Dinophyceae). Plankton Benthos Res. 3:78-85. [Google Scholar]
- 38.Nishitani, G., S. Nagai, Y. Takano, S. Sakiyama, K. Baba, and T. Kamiyama. 2008. Growth characteristics and phylogenetic analysis of the marine dinoflagellate Dinophysis infundibulus (Dinophyceae). Aquat. Microb. Ecol. 52:209-221. [Google Scholar]
- 39.Ohtani, K., and Y. Akiba. 1970. Studies on the change of hydrographic conditions in the Funka Bay I. The annual change of the water of the bay. Bull. Fac. Fish. Hokkaido Univ. 20:303-312. [Google Scholar]
- 40.Park, J. S., G. Myung, H. S. Kim, B. C. Cho, and W. Yih. 2007. Growth responses of the marine photosynthetic ciliate Myrionecta rubra to different cryptomonad strains. Aquat. Microb. Ecol. 48:83-90. [Google Scholar]
- 41.Park, M. G., M. Kim, S. Kim, and W. Yih. Does Dinophysis caudata (Dinophyceae) have permanent plastids? J. Phycol., in press.
- 42.Park, M. G., S. Kim, H. S. Kim, G. Myung, Y. G. Kang, and W. Yih. 2006. First successful culture of the marine dinoflagellate Dinophysis acuminata. Aquat. Microb. Ecol. 45:101-106. [Google Scholar]
- 43.Park, M. G., J. S. Park, M. Kim, and W. Yih. 2008. Plastid dynamics during survival of Dinophysis caudata without its ciliate prey. J. Phycol. 44:1154-1163. [DOI] [PubMed] [Google Scholar]
- 44.Rodríguez, F., J. L. Oliver, A. Marín, and J. R. Medina. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485-501. [DOI] [PubMed] [Google Scholar]
- 45.Romari, K., and D. Vaulot. 2004. Composition and temporal variability of picoeukaryotes communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol. Oceanogr. 49:784-798. [Google Scholar]
- 46.Ronquist, F., and J. P. Huelsenbeck. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 47.Sakurai, Y. 2007. An overview of the Oyashio ecosystem. Deep Sea Res. II 54:2526-2542. [Google Scholar]
- 48.Shalchian-Tabrizi, K., J. Bråte, R. Logares, D. Klaveness, C. Berney, and K. S. Jakobsen. 2008. Diversification of unicellular eukaryotes: cryptomonad colonizations of marine and fresh waters inferred from revised 18S rRNA phylogeny. Environ. Microbiol. 10:2635-2644. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi, Y., K. Takishita, K. Koike, T. Maruyama, T. Nakayama, A. Kobiyama, and T. Ogata. 2005. Development of molecular probes for Dinophysis (Dinophyceae) plastid: a tool to predict blooming and explore plastid origin. Mar. Biotechnol. 7:95-103. [DOI] [PubMed] [Google Scholar]
- 50.Takishita, K., K. Koike, T. Maruyama, and T. Ogata. 2002. Molecular evidence for plastid robbery (kleptoplastidy) in Dinophysis, a dinoflagellate causing diarrhetic shellfish poisoning. Protist 153:293-302. [DOI] [PubMed] [Google Scholar]
- 51.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verity, P. G. 1988. Chemosensory behavior in marine planktonic ciliates. Bull. Mar. Sci. 43:772-782. [Google Scholar]
- 54.Yih, W., H. S. Kim, H. J. Jeong, G. Myung, and Y. G. Kim. 2004. Ingestion of cryptophyte cells by the marine photosynthetic ciliate Mesodinium rubrum. Aquat. Microb. Ecol. 36:165-170. [Google Scholar]