Abstract
In many microorganisms, the key enzyme responsible for catalyzing the last step in triacylglycerol (TAG) and wax ester (WE) biosynthesis is an unspecific acyltransferase which is also referred to as wax ester synthase/acyl coenzyme A (acyl-CoA):diacylglycerol acyltransferase (WS/DGAT; AtfA). The importance and function of two AtfA homologues (AtfA1 and AtfA2) in the biosynthesis of TAGs and WEs in the hydrocarbon-degrading marine bacterium Alcanivorax borkumensis SK2 have been described recently. However, after the disruption of both the AtfA1 and AtfA2 genes, reduced but substantial accumulation of TAGs was still observed, indicating the existence of an alternative TAG biosynthesis pathway. In this study, transposon-induced mutagenesis was applied to an atfA1 atfA2 double mutant to screen for A. borkumensis mutants totally defective in biosynthesis of neutral lipids in order to identify additional enzymes involved in the biosynthesis of these lipids. At the same time, we have searched for a totally TAG-negative mutant in order to study the function of TAGs in A. borkumensis. Thirteen fluorescence-negative mutants were identified on Nile red ONR7a agar plates and analyzed for their abilities to synthesize lipids. Among these, mutant 2 M131 was no longer able to synthesize and accumulate TAGs if pyruvate was used as the sole carbon source. The transposon insertion was localized in a gene encoding a putative cytochrome c family protein (ABO_1185). Growth and TAG accumulation experiments showed that the disruption of this gene resulted in the absence of TAGs in 2 M131 but that growth was not affected. In cells of A. borkumensis SK2 grown on pyruvate as the sole carbon source, TAGs represented about 11% of the dry weight of the cells, while in the mutant 2 M131, TAGs were not detected by thin-layer and gas chromatography analyses. Starvation and lipid mobilization experiments revealed that the lipids play an important role in the survival of the cells. The function of neutral lipids in A. borkumensis SK2 is discussed.
Many microorganisms are able to produce intracellular storage compounds in response to imbalanced growth conditions (47). Polyhydroxyalkanoates (PHAs; linear polyesters), triacylglycerols (TAGs; trioxoesters of glycerol and long-chain fatty acids), and wax esters (WEs; oxoesters of primary long-chain fatty acids and primary long-chain fatty alcohols) are the major substances accumulated in prokaryotes when the organisms are cultivated under conditions characterized by a limited nitrogen or phosphate supply but an excess carbon source (2, 47). TAG accumulation in species belonging to the genera Mycobacterium, Nocardia, Rhodococcus, Streptomyces, and Alcanivorax has been reported previously (47). Some species of these genera are able to accumulate TAGs to levels up to nearly 80% of the cellular dry matter and are also able to mobilize them under carbon-free conditions (2), thereby demonstrating the importance of TAGs in the life cycles of these bacteria. TAGs are nonpolar and water insoluble, and they constitute convenient storage compounds for carbon and energy because they exhibit a minor grade of oxidation and possess a higher calorific value than proteins or carbohydrates (2). Therefore, TAGs provide an energy yield of about 9 kcal/g in comparison to only about 4 kcal/g for carbohydrates and proteins. Furthermore, in the case of TAG-accumulating bacteria present in soil, storage of evaporation-resistant lipids may be a strategy to maintain a basic water supply (47). Moreover, lipid bodies may also provide a deposit for toxic or useless fatty acids during growth on recalcitrant carbon sources (2, 3).
WEs, like TAGs, are nonpolar lipids and are water insoluble by virtue of their hydrophobic components. In Acinetobacter species, WEs are degraded into water-soluble molecules and CO2 during carbon starvation, thereby demonstrating that they serve as an ATP-generating substrate during starvation (16). Accumulation of WEs in species of the genus Acinetobacter (16) but also in marine bacteria such as Marinobacter (25) and Alcanivorax (10, 28, 31) species has been described previously. Also, marine hydrocarbonoclastic bacteria of the genus Alcanivorax are able to export lipids such as TAGs and WEs (31). Accumulation of at least one type of storage lipid can be found in almost all prokaryotes. The presence of these lipids may be an advantage for survival in natural habitats and/or a strong advantage during evolution (47).
In many microorganisms, the key enzyme catalyzing the last step in the biosynthesis of TAGs and WEs is a promiscuous acyltransferase, the wax ester synthase/acyl coenzyme A (acyl-CoA):diacylglycerol acyltransferase (WS/DGAT). WS/DGAT from Acinetobacter baylyi ADP1 (formerly Acinetobacter calcoaceticus ADP1) was the first such enzyme to be described (27), but WS/DGAT homologues have also been found in other neutral lipid-producing species (48). This enzyme exhibits WE synthase (WS) activity, as well as acyl-CoA:diacylglycerol acyltransferase (DGAT) activity. The identification of WS/DGAT homologues and their roles in lipid biosynthesis in Rhodococcus opacus PD630 (1), Streptomyces coelicolor (4), and the oil-degrading marine bacterium Alcanivorax borkumensis SK2 (28) has been reported recently. It was shown that in these bacteria the terminal reaction in TAG and WE biosynthesis is catalyzed by more than one enzyme (AtfA1 and AtfA2 in the case of A. borkumensis SK2). In contrast, biosynthesis of TAGs and WEs in A. baylyi ADP1 is catalyzed by a single WS/DGAT (27). The presence of a reduced but still substantial level of TAG accumulation in a double mutant of A. borkumensis SK2 defective in both acyltransferases encoded in the SK2 genome suggests the existence of an alternative route of TAG synthesis in this bacterium (28). During the screening for additional enzymes involved in lipid accumulation in A. borkumensis, a mutant that was no longer able to synthesize TAGs when pyruvate was the sole carbon source was isolated. In this study, we report analyses of lipid accumulation and mobilization in A. borkumensis and we propose that the intracellularly accumulated lipids play an important role in cellular viability.
MATERIALS AND METHODS
Bacterial strains, media, and cultivation conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. A. borkumensis strains were cultivated aerobically at 30°C and 150 rpm for 72 h in ONR7a medium (15) containing 1% (wt/vol) sodium pyruvate as the sole carbon source. Escherichia coli strains were cultivated in lysogeny broth (LB) medium at 37°C (37). cLB mating medium used for the biparental filter mating technique is a modified LB containing the following per liter: 10 g tryptone, 5 g yeast extract, 0.45 g Na2HPO4·H2O, 2.5 g NaNO3, 16.5 g NaCl, 0.38 g KCl, 0.7 g CaCl2·H2O, and 2% (wt/vol) sodium pyruvate as the carbon source. Cultures were inoculated with a 1% (vol/vol) suspension of precultured cells in the exponential growth phase. Cell growth was determined by measuring the optical density with a Klett-Summerson photoelectric colorimeter (Manostat, New York, NY) and was related to the biomass (the dry weight of cells [CDW]) by establishing standard curves for each strain. When required, the media were supplemented with ampicillin (Ap; 75 mg/liter), kanamycin (Km; 50 mg/liter), chloramphenicol (Cm; 34 mg/liter), nalidixic acid (NDx; 10 mg/liter), and/or tetracycline (Tc; 12.5 mg/liter).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source and/or reference |
|---|---|---|
| A. borkumensis strains | ||
| SK2 (DSM 11573) | Type strain; wild type; TAG and WE producer | 49; DSMZ |
| atfA1ΩKm mutant | atfA1 disruption mutant; Kmr | 28 |
| atfA2ΩSm mutant | atfA2 disruption mutant; Smr | 28 |
| atfA1ΩKm atfA2ΩSm double mutant | atfA1 atfA2 double disruption mutant; Kmr Smr | 28 |
| ABO_1185ΩSm mutant | ABO_1185 disruption mutant; Smr; derivative of SK2 | This study |
| E. coli strains | ||
| SM10 (λpir) | Apr Cmr Kmr pUTmini-Tn5Cm | 13 |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) rpcL nupG endA1 deoR φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK | Invitrogen |
| S17-1 | recA1 thi-1 hsdR17(rK− mK+) proA tra-RP4 | 40 |
| Plasmids | ||
| pBluescript SK(−) | Plasmid used for subcloning of DNA; AprlacPOZ′ | Stratagene |
| pUTmini-Tn5Cm | Suicide vector for mutagenesis with mini-Tn5; Cmr | 13 |
| pCR2.1-TOPO | Plasmid used for subcloning of DNA; Apr | Invitrogen |
| pCR2.1-TOPO::ABO_1185 | Derivative of pCR2.1-TOPO containing 1,908-bp ABO_1185 gene | This study |
| pCR2.1-TOPO::ABO_1185ΩSm | ΩSm cassette cloned into NcoI site of pCR2.1-TOPO::ABO_1185; Apr Smr | This study |
| pSUP202 | Apr Cmr Tcr; carries ColE1 origin and mob site; unable to replicate in A. borkumensis | 40 |
| pSUP202::ABO_1185ΩSm | Fusion of pSUP202 and ABO_1185ΩSm fragment of pCR2.1-TOPO via BamHI/HindIII sites; carries mob site; Cmr Apr Smr | This study |
For abbreviations for genotypes of E. coli, see reference 7. Kmr, kanamycin resistant; Smr, streptomycin resistant; Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Tcr, tetracycline resistant.
Construction of a mini-Tn5 transposon library in A. borkumensis atfA1ΩKm atfA2ΩSm.
To find additional enzymes involved in lipid accumulation in A. borkumensis, transposon mutagenesis was performed using A. borkumensis atfA1ΩKm atfA2ΩSm (28) as the acceptor strain and E. coli SM10 (λpir) harboring pUTmini-Tn5Cm (13) as the donor strain. Transposon mutagenesis was carried out by employing the same biparental filter mating technique described previously (28) for the construction of A. borkumensis atfA1ΩKm atfA2ΩSm. A. borkumensis atfA1ΩKm atfA2ΩSm was grown in ONR7a medium at 30°C and harvested in the stationary growth phase by centrifugation at 4,000 rpm for 50 min at 4°C. The donor strain E. coli SM10 (λpir)(pUTmini-Tn5Cm) was grown overnight at 37°C in LB medium with Cm. Cells were harvested, washed once with cLB mating medium, and concentrated 10-fold in the same medium. The pellets of A. borkumensis atfA1ΩKm atfA2ΩSm and E. coli SM10 (λpir)(pUTmini-Tn5Cm) cells were mixed at a ratio of 4:1 (vol/vol), and correspondent dilutions of this cell mixture were spotted onto a nitrocellulose membrane filter (diameter, 45 mm; pore size, 0.45 μm [Millipore]), which was placed onto a cLB mating agar plate. The plate was incubated at 30°C for 24 h, and the cells were then washed from the filter with 10 mM MgSO4 and resuspended in the same solution. Mutants were selected on ONR7a selection plates containing appropriate antibiotics (NDx and Cm). Chloramphenicol-resistant mutants were transferred with sterile toothpicks onto ONR7a plates containing Nile red (50 μg/ml) to screen for mutants defective in lipid production as described previously (42). n-Hexadecane applied on a filter paper in the lid of each plate was used as a carbon and energy source. After 5 days of incubation, the plates were analyzed under UV light and mutants with decreased or no fluorescence were investigated for their capacity to produce lipids.
Genotypic characterization of mini-Tn5-induced mutants of A. borkumensis atfA1ΩKm atfA2ΩSm.
To identify the mini-Tn5 insertion sites in mutants of A. borkumensis atfA1ΩKm atfA2ΩSm, gene libraries were constructed. Genomic DNA of mini-Tn5 mutants was digested with PstI, and the resulting fragments were ligated into plasmid pBluescript SK(−) (Stratagene). Recombinant E. coli TOP10 clones were selected on LB agar plates containing Cm. The hybrid plasmids of the resulting clones harbored a PstI fragment consisting of mini-Tn5 and genomic DNA adjacent to the mini-Tn5 insertion locus. The recombinant plasmids from these libraries were sequenced using the oligonucleotides M13-forward and M13-reverse (Table 2), which hybridize specifically to pBluescript SK(−). In addition, the mini-Tn5 insertion sites of selected mutants were determined by inverse PCR (IPCR) as described previously (34). For this analysis, the total DNA of the mutant was isolated (32) and digested with PstI. The resulting DNA fragments were circularized with T4 DNA ligase under conditions promoting intramolecular circularization (26). The regions flanking the inserted mini-Tn5 were amplified with two primers which hybridized specifically to both repetitive ends (the I and O ends) of the mini-Tn5 transposon (Table 2). The conditions for the PCR were 30 cycles of 94°C for 1.5 min, 48°C for 1 min, and 72°C for 4 min and a final elongation step at 72°C for 5 min. The PCR products were gel purified and used for sequencing as described below.
TABLE 2.
Oligonucleotides used in this study as primers for PCR analyses and sequencing of recombinant plasmids
| Primer | Sequencea |
|---|---|
| I_end | GGCCGCACTTGTGTA TAAGAGTCAG |
| O_end | GCGGCCAGATCTGATCAAGAGACAG |
| M13-forward | GTAAAACGACGACGGCCGT |
| M13-reverse | CAGGAAACAGCTATGAC |
| 5′_ABO_1185 | TTTTTCTAGAATGCCGCGTACCTTC |
| 3′_ABO_1185 | TTTTGGATCCCTACTGAGCGTTACG |
| 5′_SmR | AAAAAGTCGACCTCACGCCCGGAGCGTAGCGACC |
| 3′_SmR | AAAAAGTCGACAACGACCCTGCCCTGAACCGACG |
Restriction sites used for cloning purposes are underlined.
Cloning of a gene encoding a cytochrome c family protein from A. borkumensis SK2 and disruption of the gene by a biparental filter mating technique.
The gene encoding a cytochrome c family protein (ABO_1185) was amplified from total genomic DNA of A. borkumensis SK2 by tailored PCR using the oligonucleotides 5′_ABO_1185 and 3′_ABO_1185 as primers (Table 2). The resulting PCR product was cloned into the pCR2.1-TOPO vector (Invitrogen), yielding pCR2.1-TOPO::ABO_1185, and E. coli TOP10 was transformed with this plasmid. For inactivation of the ABO_1185 gene, a streptomycin resistance cassette (ΩSm) was amplified by PCR from the vector pCDFDuet-1 by employing the primers 5′_ΩSmR and 3′_ΩSmR (Table 2), digested with NcoI, and cloned into the singular site of ABO_1185, yielding plasmid pCR2.1-TOPO::ABO_1185ΩSm. The plasmid was then digested with BamHI and HindIII and fused to the BamHI-HindIII-restricted mobilizable suicide plasmid pSUP202, yielding plasmid pSUP202::ABO_1185ΩSm. E. coli S17-1 was then transformed with the resulting plasmid for further conjugational transfer of the plasmid into A. borkumensis SK2.
Inactivation of the cytochrome c family protein in A. borkumensis was achieved by conjugational transfer of the suicide plasmid pSUP202::ABO_1185ΩSm from the donor E. coli S17-1 to the recipient A. borkumensis SK2 by a biparental filter mating technique essentially as described above for transposon mutagenesis. Mutants were obtained on ONR7a selection plates containing appropriate antibiotics for selection of mutants and for inhibition of E. coli (NDx and Sm). Hexadecane applied on a filter paper in the lid was used as a carbon and energy source.
DNA manipulation and molecular genetic methods.
DNA manipulations and other standard molecular biology techniques were performed as described previously (37). Plasmid DNA was isolated using the method described by Birnboim and Doly (9). Chromosomal DNA from Tn5-induced mutants and A. borkumensis strains was isolated as described previously (32). Restriction enzymes and ligases were used according to manufacturer instructions.
DNA sequencing and sequence analysis.
Sequencing of the recombinant plasmids was performed by the chain termination method of Sanger et al. (38) by using an ABI Prism 3730 capillary sequencer at the Universitätsklinikum Münster (UKM).
Analysis of lipids by TLC.
The analysis of intra- and extracellular lipids was done by thin-layer chromatography (TLC) as described previously (27). After 72 h of cultivation, cells of Alcanivorax were harvested for 30 min at 7,000 rpm and 4°C and were thereby separated into pellet and supernatant fractions. The cells were washed with 10 mM MgSO4, centrifuged again, and lyophilized. Intracellular lipids were extracted from 5 mg dry cell material by subjecting the material to a vortex in the presence of a chloroform-methanol (2:1 [vol/vol]) mixture and centrifuging to separate the cell debris. The chloroform phase was recovered and evaporated to dryness. Finally, intracellular extracts were resuspended in chloroform-methanol (2:1 [vol/vol]), and aliquots were applied to the TLC plates. Extracellular lipids were extracted directly from the supernatant fraction (corresponding to the amount of dry cell material to analyze), which was mixed with the same volume of chloroform-methanol (2:1 [vol/vol]) and subjected to a vortex. After separation of the two phases, the organic phase was removed and analyzed for extracted lipids. TLC analysis of lipid extracts was done using the solvent system hexane-diethyl ether-acetic acid (80:20:1 or 90:7.5:1 [vol/vol/vol]). Lipids on the plates were visualized by staining with iodine vapor. Triolein, oleic acid, and oleyl oleate were used as reference substances for TAGs, fatty acids, and WEs, respectively.
Quantitative analysis of intracellular lipids by preparative TLC and GC.
For quantification of intracellular lipids, preparative TLC with subsequent fatty acid analysis by gas chromatography (GC) was performed. For this purpose, the lipid extracts of cells were resolved by TLC and spots corresponding to TAGs were scraped from the plates and subjected to sulfuric acid-catalyzed methanolysis (46). For quantitative analysis, tridecanoic acid was used as an internal standard. The TAG content was defined as the ratio of TAGs to the CDW, expressed as a percentage. Fatty acid methyl esters were analyzed using a model 6850 GC (Agilent Technologies, Waldbronn, Germany) equipped with a BP21 capillary column (50 by 0.22 mm; film thickness, 250 nm [SGE, Darmstadt, Germany]) and a flame ionization detector (Agilent Technologies, Waldbronn, Germany). A 2-μl portion of the organic phase was analyzed after split injection (1:5); hydrogen (constant flow rate, 0.6 ml min−1) was used as the carrier gas. The temperatures of the injector and detector were 250 and 275°C, respectively. The following temperature program was applied: 120°C for 5 min, an increase of 3°C min−1 to 180°C, an increase of 10°C min−1 to 220°C, and 220°C for 31 min. Substances were identified by comparison of their retention times with those for authentic standard fatty acid methyl esters (46).
Starvation experiments and mobilization of lipids in ammonium-rich, carbon-free medium.
Cells of A. borkumensis were cultivated in ONR7a medium containing 1% (wt/vol) sodium pyruvate as the sole carbon source at 30°C for 72 h to allow for accumulation of storage lipids. Growth and the total fatty acid content were evaluated at different cultivation times. Cells were harvested, washed twice with ONR7a medium, resuspended in the same medium (containing 1 g/liter ammonium sulfate) without any carbon source, and then incubated at 30°C. At various time points, serial dilutions of culture aliquots were plated onto ONR7a solid medium containing 1% (wt/vol) sodium pyruvate for enumeration of viable cells. Additionally, lipids were extracted from cells at the same time points and subjected to TLC and GC analyses as described above.
RESULTS
Identification and characterization of a mutant of A. borkumensis defective in TAG biosynthesis.
During screening of a mini-Tn5 transposon library in A. borkumensis atfA1ΩKm atfA2ΩSm to identify additional enzymes involved in lipid accumulation in A. borkumensis SK2, mutants defective in neutral lipid production were isolated. About 9,000 mutants were generated by transposon-induced mutagenesis and screened for reduced fluorescence compared to that of the parental strain on ONR7a selection plates containing Nile red under UV light, as done successfully in the past for other lipophilic substances in other organisms (14, 19, 42). Mutants that showed reduced fluorescence under UV light or that were in other aspects phenotypically different from the parental strain were analyzed for growth and the capacity to synthesize neutral lipids. The final biomasses (expressed as grams [CDW] per liter) of 13 transposon-induced mutants and those of the wild type A. borkumensis SK2 and the double mutant A. borkumensis atfA1ΩKm atfA2ΩSm and the levels of neutral lipid production by the mutant, double mutant, and wild-type strains were compared in shake flask experiments. The cells of these strains were cultivated aerobically for 72 h in ONR7a medium with 1% (wt/vol) sodium pyruvate as the sole carbon source. Under the conditions tested, cell densities ranged from 0.79 g/liter (for mutant 2 M93) to about 2.7 g/liter (for the wild type and mutant 2 M231). Growth profiles of the mutants could be divided into two categories based on the cell densities at the end of cultivation: poor growth (for cell densities between 0.79 and 1.5 g/liter) and good growth (for cell densities higher than 1.5 g/liter).
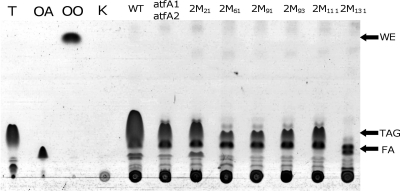
Mutants were also analyzed for their lipid biosynthesis abilities as described in Materials and Methods. Results for a typical TLC plate to assay the accumulation of intracellular lipids in the generated mutants are shown in Fig. 1. Alcanivorax strains produced large amounts of TAGs when the cells were cultivated with pyruvate as the sole carbon source. No presence of WEs was observed, suggesting that Alcanivorax is not able to synthesize this class of neutral lipids de novo. Other lipophilic substances could be observed by TLC analysis, but their amounts were minimal in comparison to TAGs, and they were not analyzed in this study. After separation of extracted lipids from lyophilized cells by TLC, TAGs were scraped from the plate and subjected to GC for quantification using tridecanoic acid as an internal standard. TAGs amounted to an intracellular fatty acid content of about 13% of the CDW of A. borkumensis SK2 during growth on pyruvate (Table 3). All mutants (except 2 M191) showed diminished capacities to synthesize TAGs in comparison to the wild type, as revealed by TLC and GC analyses (Fig. 1 and Table 3). Mutants that yielded only low cell densities (poor-growth strains, e.g., 2 M21, 2 M93, and 2 M193) showed, however, relatively high TAG contents. In contrast, with the exception of mutant 2 M191, which produced TAGs accounting for about 14% of the CDW under the tested conditions, all mutants yielding cell densities higher than 1.5 g/liter showed diminished abilities to accumulate TAGs. Therefore, TAG accumulation in good-growth strains, expressed as a percentage of the CDW, was similar to that in the double mutant A. borkumensis atfA1ΩKm atfA2ΩSm (3% of the CDW).
FIG. 1.
TLC analysis of lipids accumulated in cells of A. borkumensis SK2 and mutant strains. Cells were cultivated aerobically in ONR7a medium containing 1% (wt/vol) sodium pyruvate at 30°C and 150 rpm. After 72 h of cultivation, cells were harvested, washed, and lyophilized and the lipids were extracted with chloroform-methanol (2:1, vol/vol). Lipid extracts obtained from 5 mg lyophilized cells were applied in each lane. The solvent system used for elution was hexane-diethyl ether-acetic acid (90:7.5:1, vol/vol/vol), and lipids were visualized by staining with iodine vapors. Lanes: T, triolein; OA, oleic acid; OO, oleyl oleate; WT, A. borkumensis SK2; atfA1 atfA2, A. borkumensis atfA1ΩKm atfA2ΩSm; K, control (“culture” without cells); and 2 M21 to 2 M131, different mini-Tn5 transposon-induced mutants. FA, fatty acids.
TABLE 3.
Fatty acid contents and compositions of fatty acids of A. borkumensis SK2 and transposon-induced mutantsa
| Strain | Median biomass (g/liter) ± SD | Total TAG content (% CDW) | Amt (mol%) of TAG constituent: |
||||
|---|---|---|---|---|---|---|---|
| C14:0 | C16:0 | C16:1 | C18:0 | C18:1 | |||
| SK2 | 2.7 ± 0.42 | 13 | 4 | 41 | 17 | 3 | 35 |
| atfA1 atfA2 mutant | 2.55 ± 0.07 | 3 | 0 | 45 | 16 | 0 | 39 |
| 2 M21 | 1.02 ± 0.28 | 8 | 2 | 42 | 19 | 0 | 36 |
| 2 M61 | 1.34 ± 0.25 | 6 | 3 | 42 | 20 | 0 | 35 |
| 2 M91 | 2.51 ± 0.55 | 3 | 0 | 42 | 19 | 4 | 36 |
| 2 M93 | 0.79 ± 0.38 | 7 | 0 | 41 | 22 | 0 | 37 |
| 2 M111 | 1.31 ± 0.35 | 4 | 0 | 42 | 21 | 0 | 37 |
| 2 M131 | 2.05 ± 0.21 | ND | ND | ND | ND | ND | ND |
| 2 M141 | 1.07 ± 0.13 | 6 | 3 | 40 | 19 | 2 | 37 |
| 2 M191 | 2.44 ± 0.06 | 14 | 4 | 34 | 8 | 41 | 12 |
| 2 M192 | 2.24 ± 0.06 | 4 | 0 | 34 | 17 | 18 | 31 |
| 2 M193 | 1.08 ± 0.06 | 10 | 2 | 37 | 19 | 3 | 38 |
| 2 M231 | 2.7 ± 0.17 | 4 | 0 | 43 | 21 | 0 | 37 |
| 2 M232 | 1.91 ± 0.16 | 5 | 0 | 41 | 18 | 3 | 39 |
| 2 M233 | 2.56 ± 0.17 | 3 | 0 | 42 | 20 | 0 | 38 |
Cells were cultivated in ONR7a medium containing 1% (wt/vol) sodium pyruvate for 72 h at 30°C. Lipid extracts were obtained from 5-mg samples of lyophilized cells (see the text for details). TAGs were purified by preparative TLC before GC analysis. Tridecanoic acid was used as an internal standard for quantification and was added to the samples before the start of acid-catalyzed methanolysis. For the molar compositions of fatty acids, no trace amounts (<1 mol%) were considered. The values are the averages of two independent determinations. SK2, A. borkumensis wild type; atfA1 atfA2 mutant, A. borkumensis atfA1ΩKm atfA2ΩSm; ND, not determined. 2 M designations denote transposon-induced mutants.
Though a clear tendency was not observed, TAGs accumulated by pyruvate-grown cells consisted mainly of hexadecanoic (C16:0) and octadecenoic (C18:1) acids as principal constituents. The hexadecanoic acid content ranged from 34 to 45 mol%, while the level of octadecenoic acid varied between 12 and 39 mol%. Interestingly, the TAGs produced by mutant 2 M191, which showed the smallest octadecenoic acid fraction (12 mol%), also comprised an unusually high percentage of octadecanoic acid (C18:0). Besides hexadecanoic and octadecenoic acids, hexadecenoic acid (C16:1) was also present as a constituent of TAGs, while only in some strains was the presence of tetradecanoic acid (C14:0) observed (Table 3). Only in the case of mutant 2 M131 could the presence of TAGs not be detected. For that reason, this mutant was investigated in more detail.
In mutant 2 M131, a gene encoding a putative cytochrome c is disrupted.
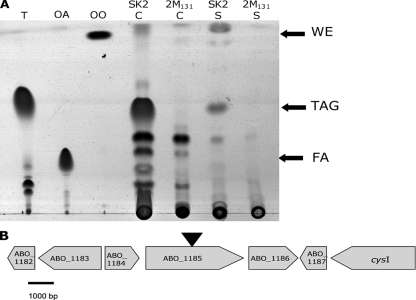
Because mutant 2 M131 was unable to accumulate any TAGs during cultivation in the presence of pyruvate (Fig. 2A), the insertion site of the mini-Tn5 transposon in the genome of this mutant was mapped by IPCR analysis and construction of gene libraries as described in Materials and Methods. These analyses revealed disruption of a gene which encoded a putative cytochrome c family protein (ABO_1185) in the genome of A. borkumensis (39). No other genes known to be involved in the biosynthesis of neutral lipids in bacteria were found adjacent to the insertion site (Fig. 2B). Moreover, since cytochrome c proteins participate in many bacterial processes, no direct correlation between the disruption of the ABO_1185 gene and the biosynthesis of TAGs in A. borkumensis SK2 could be established. Interestingly, when the accumulation of neutral lipids in mutant 2 M131 during cultivation of the cells in the presence of hexadecane, octadecane, or acetate as the sole carbon source was investigated, no significant differences in the pattern or levels of neutral lipid production between this mutant and the wild type were observed (data not shown).
FIG. 2.
Characterization of mutant 2 M131 with regard to lipid formation. (A) TLC analysis of storage lipid accumulation in cells of A. borkumensis SK2 and of mutant 2 M131. Cells were cultivated for 72 h in ONR7a medium containing 1% (wt/vol) sodium pyruvate, and lipid extracts were analyzed by TLC. Lipid extracts were obtained from 3 mg freeze-dried cells and the corresponding amount of the supernatant. Triolein (T), oleic acid (OA), and oleyl oleate (OO) were used as standards. Lanes: SK2 C, extract from cells of A. borkumensis SK2; 2 M131 C, extract from cells of the mutant 2 M131; SK2 S, extract from a supernatant sample from the A. borkumensis SK2 culture; and 2 M131 S, extract from a supernatant sample from the mutant 2 M131 culture. FA, fatty acids. (B) Organization of the mini-Tn5 transposon insertion site and the adjacent region in the genome of mutant 2 M131. The insertion site was mapped to a gene coding for a cytochrome c family protein (ABO_1185) (39). ABO_1182, gene for hypothetical protein; ABO_1183, gene for putative aminotransferase; ABO_1184, gene for hypothetical protein; ABO_1185, gene for cytochrome c family protein; ABO_1186, gene for CorA-like Mg2+ transporter protein; ABO_1187, gene for hypothetical protein; cysI, sulfite reductase (NADPH) gene. The insertion site is shown by an inverted triangle.
Confirmation of the role of ABO_1185 in TAG biosynthesis in A. borkumensis by directed gene insertion mutagenesis.
To confirm that ABO_1185 plays a role in the accumulation of TAGs in A. borkumensis, an isogenic mutant was generated. For this purpose, the gene coding region was disrupted by insertion of a streptomycin resistance gene cassette. By employing a conjugative biparental filter mating technique, a homozygous gene disruption mutant (the ABO_1185ΩSm strain) was generated by homologous recombination via a double-crossover event in A. borkumensis. Diagnostic PCR and DNA sequencing were employed to confirm the correct gene replacement in this mutant.
Biosynthesis and accumulation of TAGs by the wild type, the transposon-induced mutant 2 M131, and the constructed ABO_1185ΩSm mutant of A. borkumensis were analyzed during cultivation of the cells in the presence of pyruvate as the sole carbon source (Fig. 3). Although trace amounts of produced TAGs were still observed in cells of the constructed A. borkumensis ABO_1185ΩSm mutant by TLC analysis, GC analysis showed that the TAG content in this mutant constituted less than 1% of the CDW. In contrast, TAGs produced by A. borkumensis SK2 constituted about 13% of the CDW. In the same way, single mutants defective in the acyltransferase AtfA1 or AtfA2 and the AtfA1 AtfA2 double mutant (28) were still able to synthesize TAGs to yield about 3% of the CDW (Table 4).
FIG. 3.
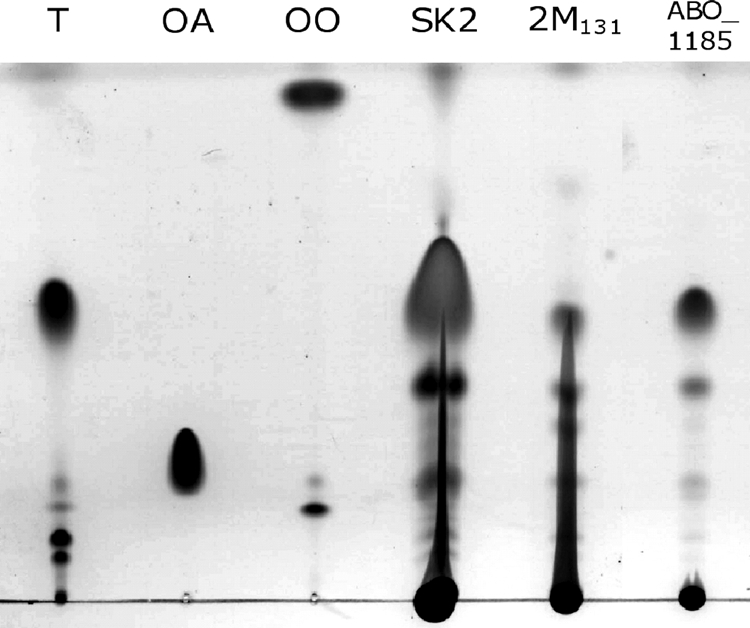
TLC analysis of lipid biosynthesis by A. borkumensis SK2, mutant 2 M131, and the constructed A. borkumensis ABO_1185ΩSm mutant. Cells were cultivated aerobically in ONR7a medium containing 1% (wt/vol) sodium pyruvate at 30°C and 150 rpm. After 72 h of cultivation, cells were harvested, washed, and lyophilized and the lipids were extracted with chloroform-methanol (2:1, vol/vol). Lipid extracts obtained from 5 mg lyophilized cells were applied in each lane. The solvent system used for elution was hexane-diethyl ether-acetic acid (80:20:1, vol/vol/vol), and lipids were visualized by staining with iodine vapor. Lanes: T, triolein; OA, oleic acid; OO, oleyl oleate; SK2, A. borkumensis wild type; 2 M131, transposon-induced mutant; ABO_1185, A. borkumensis ABO_1185ΩSm.
TABLE 4.
Fatty acid contents and compositions of fatty acids of A. borkumensis strainsa
| Strain or relevant genotype | Cultivation time (h) | Total TAG content (% CDW) | Amt (mol%) of TAG constituent: |
||||
|---|---|---|---|---|---|---|---|
| C14:0 | C16:0 | C16:1 | C18:0 | C18:1 | |||
| SK2 | 12 | 0.4 | 0 | 86 | 14 | 0 | 0 |
| 24 | 1.07 | 0 | 52 | 19 | 0 | 28 | |
| 36 | 2.12 | 9 | 46 | 13 | 0 | 33 | |
| 48 | 2.63 | 8 | 47 | 11 | 0 | 34 | |
| 60 | 5.98 | 6 | 39 | 15 | 3 | 37 | |
| 72 | 11.10 | 5 | 45 | 13 | 3 | 33 | |
| atfA1ΩKm | 12 | 0.14 | 0 | 100 | 0 | 0 | 0 |
| 24 | 0.19 | 0 | 100 | 0 | 0 | 0 | |
| 36 | 0.79 | 0 | 53 | 17 | 0 | 30 | |
| 48 | 0.62 | 0 | 55 | 21 | 0 | 24 | |
| 60 | 1.13 | 0 | 52 | 13 | 0 | 35 | |
| 72 | 3.44 | 0 | 49 | 14 | 0 | 38 | |
| atfA2ΩSm | 12 | 0.53 | 0 | 71 | 29 | 0 | 0 |
| 24 | 1.61 | 6 | 49 | 15 | 0 | 30 | |
| 36 | 2.25 | 12 | 44 | 12 | 0 | 32 | |
| 48 | 2.16 | 12 | 45 | 11 | 0 | 32 | |
| 60 | 2.26 | 10 | 45 | 12 | 0 | 33 | |
| 72 | 8.69 | 6 | 49 | 14 | 0 | 31 | |
| atfA1ΩKm | 12 | 0.15 | 0 | 100 | 0 | 0 | 0 |
| atfA2ΩSm | 24 | 0.45 | 0 | 61 | 21 | 0 | 18 |
| 36 | 0.90 | 0 | 51 | 14 | 0 | 35 | |
| 48 | 0.90 | 0 | 52 | 13 | 0 | 35 | |
| 60 | 1.28 | 0 | 53 | 14 | 0 | 33 | |
| 72 | 3.33 | 0 | 50 | 14 | 0 | 36 | |
| 2 M131 | 12 | ND | ND | ND | ND | ND | ND |
| 24 | ND | ND | ND | ND | ND | ND | |
| 36 | ND | ND | ND | ND | ND | ND | |
| 48 | ND | ND | ND | ND | ND | ND | |
| 60 | ND | ND | ND | ND | ND | ND | |
| 72 | ND | ND | ND | ND | ND | ND | |
Cells were cultivated in ONR7a medium containing 1% (wt/vol) sodium pyruvate for 72 h at 30°C. Samples were taken every 12 h and analyzed for growth and lipid content. TAGs were purified by preparative TLC before GC analysis. Tridecanoic acid was used as an internal standard for quantification and was added to the samples before the start of acid-catalyzed methanolysis. For the molar compositions of fatty acids, no trace amounts (<1 mol%) were considered. The values are the averages of two independent determinations. SK2, A. borkumensis wild type; ND, not determined.
Biosynthesis and accumulation of TAGs by different Alcanivorax strains.
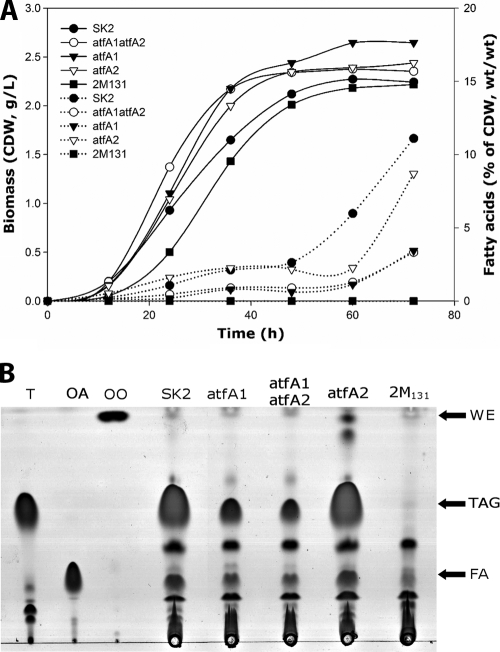
As mentioned above, A. borkumensis SK2 is able to produce and accumulate TAGs as the main storage compound when it is cultivated in the presence of pyruvate as the sole carbon source. Although the export of TAGs has been reported recently (31), in this study we analyzed only the intracellular accumulation of TAGs in order to establish the function of these compounds in A. borkumensis SK2. Recently, it was demonstrated that the amount of accumulated TAGs in this strain diminishes if at least one of the two WS/DGAT homologue genes (atfA1 or atfA2) is disrupted by directed gene insertion mutagenesis (28). However, whether the disruption of the genes had an effect on the growth of A. borkumensis and/or the composition of TAGs was not investigated. Therefore, cell growth and the accumulation and composition of TAGs in cells of A. borkumensis at different cultivation times were analyzed in shake flask experiments. For this analysis, the wild type SK2 and the atfA1 (atfA1ΩKm), atfA2 (atfA2ΩSm), atfA1 atfA2 (atfA1ΩKm atfA2ΩSm), and 2 M131 mutants were cultivated aerobically for 72 h in ONR7a medium containing 1% (wt/vol) sodium pyruvate as the sole carbon source. During the time course of the experiment, samples were withdrawn and analyzed for growth and TAG content (Fig. 4A). For the latter, the total lipid extract was purified by preparative TLC, the spot corresponding to TAGs was scraped, and the fatty acids were quantified by GC using tridecanoic acid as an internal standard. During these cultivation experiments, a lag phase of up to about 10 h of cultivation before the onset of typical exponential growth was observed for all strains (Fig. 4A). After 48 h of cultivation, the cells entered the stationary growth phase. Mutant 2 M131 showed slightly slower growth than the other strains, but the culture reached cell densities similar to those of the other cultures at the end of cultivation. Otherwise, under the conditions tested, no significant differences in the growth characteristics of the Alcanivorax strains were observed. This finding suggested that the disruption of atfA1, atfA2, or both does not have any influence on the growth of A. borkumensis.
FIG. 4.
Profiles of biomass production and fatty acid content and TLC analyses for the different A. borkumensis strains during cultivation with pyruvate as the sole carbon source. Cells were cultivated aerobically in ONR7a medium containing 1% (wt/vol) sodium pyruvate at 30°C and 150 rpm. (A) Samples were taken every 12 h and assayed for biomass and TAG content as described in Materials and Methods. Solid lines represent the growth curves, and dotted lines represent the fatty acid contents. SK2, A. borkumensis wild type; atfA1 atfA2, A. borkumensis atfA1ΩKm atfA2ΩSm; atfA1, A. borkumensis atfA1ΩKm; atfA2, A. borkumensis atfA2ΩSm; 2 M131, transposon-induced mutant. (B) After 72 h of cultivation, cells were harvested, washed, and lyophilized and the lipids were extracted with chloroform-methanol (2:1, vol/vol). Lipid extracts obtained from 5 mg lyophilized cells were applied in each lane. The solvent system used for elution was hexane-diethyl ether-acetic acid (80:20:1, vol/vol/vol), and lipids were visualized by staining with iodine vapor. Lanes: T, triolein; OA, oleic acid; OO, oleyl oleate; SK2, A. borkumensis wild type; atfA1, A. borkumensis atfA1ΩKm; atfA2, A. borkumensis atfA2ΩSm; atfA1 atfA2, A. borkumensis atfA1ΩKm atfA2ΩSm; and 2 M131, transposon-induced mutant. FA, fatty acids.
In contrast, the amount of TAGs accumulated as well as their compositions were influenced, particularly if atfA1 was disrupted. Although accumulation of some TAGs was observed already during the exponential growth phase, most TAGs were synthesized and accumulated after 48 h of incubation, once the cells had entered the stationary growth phase. Therefore, during the next 24 h of incubation, the TAG contents increased from about 2.6 to 11.1% and from 2.2 to 8.7% of the CDW in the wild type SK2 and the atfA2 mutant, respectively. The same tendency was observed for the atfA1 and atfA1 atfA2 mutants; their TAG contents increased from 0.6 to 3.4% and from 0.9 to 3.3% of the CDW, respectively. In contrast, TAG accumulation in cells of 2 M131 could not be detected by GC analysis with the methodology described, which was in concordance with the results obtained by TLC (Fig. 4B).
Hexadecanoic acid (C16:0) was the predominant fatty acid in the early phase of TAG accumulation, accounting for 100 mol% of the fatty acids in the TAGs synthesized by the atfA1 and atfA1 atfA2 mutants and for 86 and 71 mol% of those synthesized by SK2 and the atfA2 mutant, respectively. In SK2 and the atfA2 mutant, the presence of hexadecenoic acid (C16:1) in TAGs was observed already after 12 h of cultivation, whereas in the atfA1 and atfA1 atfA2 mutants, this fatty acid appeared only during the exponential growth phase. In all strains, the molar fraction of C16:1 remained roughly constant until the end of cultivation. Whereas the molar fraction of hexadecanoic acid decreased continuously during the time course of the experiment to about 50% at the end of cultivation, the fraction of octadecenoic acid (C18:1) increased continuously from 0 mol% at the beginning up to about 36 mol% at the end of cultivation. In TAGs produced by A. borkumensis SK2, saturated fatty acids like tetradecanoic (C14:0) and octadecanoic (C18:0) acids were present only at the end of the exponential growth phase or in the stationary growth phase. Neither C14:0 nor C18:0 could be detected in TAGs produced by the atfA1 and atfA1 atfA2 mutants. In TAGs produced by the atfA2 mutant, only C14:0 was detected (Table 4). In this way, it was observed that the amounts of TAGs accumulated by Alcanivorax strains varied to a minor extent during the exponential growth phase but that the molar compositions of the TAGs changed significantly. In contrast, in the stationary growth phase, the compositions of TAGs remained almost constant but their amounts increased significantly (see above).
Mobilization studies of storage lipids in Alcanivorax strains.
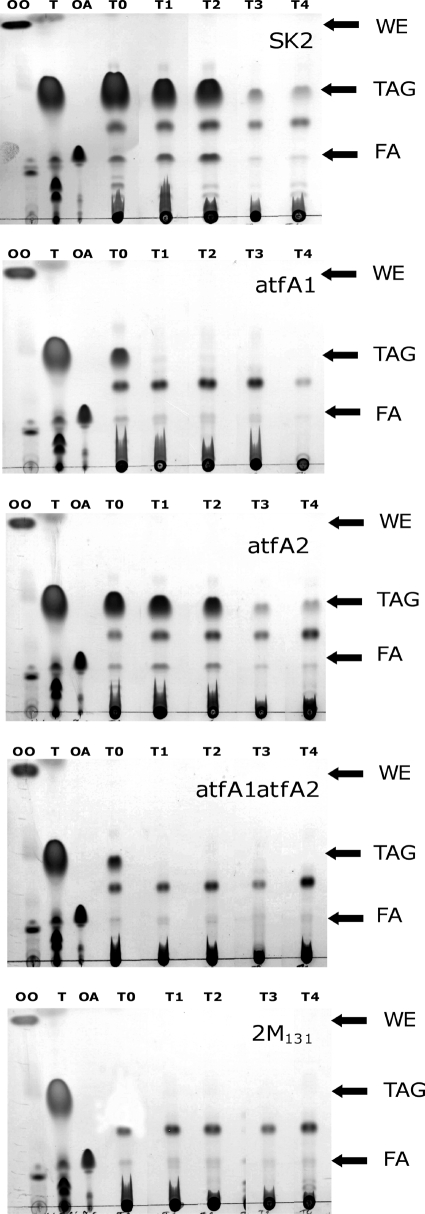
The fact that A. borkumensis accumulates TAGs as intracytoplasmic inclusions does not necessarily imply that these compounds are utilized as endogenous carbon and energy sources. To investigate whether intracellular TAGs are utilized as carbon and energy sources, we conducted TAG mobilization experiments. For this purpose, cells of the wild type SK2 and of the atfA1, atfA2, atfA1 atfA2, and 2 M131 mutants were cultivated for 72 h in ONR7a medium containing pyruvate as the sole carbon source to allow the accumulation of TAGs. Results from TLC analyses of total lipid extracts from these strains after 72 h of cultivation are shown in Fig. 4B. Cells were then harvested, washed twice with 10 mM MgSO4 solution, and resuspended in an ammonium-rich, carbon-free ONR7a medium. This medium did not contain any available carbon source and should favor the mobilization of the stored TAGs. Over the time course of cultivation, samples were taken and analyzed for TAG content.
Cells of the wild type SK2 mobilized approximately 87% (wt/wt) of the stored lipids after 240 h of cultivation, while cells of the atfA1, atfA2, and atfA1 atfA2 mutants mobilized about 93, 90, and 97% of their stored TAGs, respectively (Table 5). The major portions of accumulated TAGs were mobilized within 24 h by all strains, as revealed by both TLC (Fig. 5) and GC (Table 5) analyses. Similar to TAG biosynthesis by Alcanivorax, fatty acid dissimilation showed a defined pattern in the mobilization experiments. The fraction of saturated fatty acids (represented by C14:0 and C18:0 in the case of A. borkumensis SK2 and C14:0 in the case of the atfA2 mutant) was mobilized earlier than the fraction of unsaturated fatty acids (e.g., C16:1 and C18:1). In general, at the end of the experiment, C16:0 predominated, constituting 100 mol% in the atfA1, atfA2, and atfA1 atfA2 mutants while constituting only 75 mol% in the wild type SK2. Under the conditions tested, it seems that the mobilization of TAGs by A. borkumensis follows a defined pattern, as indicated by the changes in the molar composition of these compounds.
TABLE 5.
Mobilization of stored TAGs by A. borkumensis strainsa
| Strain or relevant genotype | Time (h) | Total fatty acid content (% CDW) | Amt (mol%) of fatty acid constituent: |
||||
|---|---|---|---|---|---|---|---|
| C14:0 | C16:0 | C16:1 | C18:0 | C18:1 | |||
| SK2 | 0 | 11.88 | 7 | 44 | 10 | 6 | 34 |
| 24 | 4.80 | 0 | 49 | 16 | 0 | 35 | |
| 48 | 2.09 | 0 | 50 | 17 | 0 | 33 | |
| 120 | 1.61 | 0 | 78 | 22 | 0 | 0 | |
| 240 | 1.57 | 0 | 75 | 25 | 0 | 0 | |
| atfA1ΩKm | 0 | 3.35 | 0 | 48 | 18 | 0 | 34 |
| 24 | 0.47 | 0 | 100 | 0 | 0 | 0 | |
| 48 | 0.46 | 0 | 100 | 0 | 0 | 0 | |
| 120 | 0.33 | 0 | 100 | 0 | 0 | 0 | |
| 240 | 0.24 | 0 | 100 | 0 | 0 | 0 | |
| atfA2ΩSm | 0 | 8.18 | 6 | 52 | 13 | 0 | 29 |
| 24 | 4.93 | 0 | 68 | 10 | 0 | 21 | |
| 48 | 1.79 | 0 | 58 | 16 | 0 | 27 | |
| 120 | 0.39 | 0 | 100 | 0 | 0 | 0 | |
| 240 | 0.82 | 0 | 100 | 0 | 0 | 0 | |
| atfA1ΩKm | 0 | 2.70 | 0 | 44 | 18 | 0 | 38 |
| atfA2ΩSm | 24 | 0.27 | 0 | 100 | 0 | 0 | 0 |
| 48 | 0.23 | 0 | 100 | 0 | 0 | 0 | |
| 120 | 0.09 | 0 | 100 | 0 | 0 | 0 | |
| 240 | 0.08 | 0 | 100 | 0 | 0 | 0 | |
| 2 M131 | 0 | ND | ND | ND | ND | ND | ND |
| 24 | ND | ND | ND | ND | ND | ND | |
| 48 | ND | ND | ND | ND | ND | ND | |
| 120 | ND | ND | ND | ND | ND | ND | |
| 240 | ND | ND | ND | ND | ND | ND | |
Cells were cultivated in ONR7a medium containing 1% (wt/vol) sodium pyruvate for 72 h at 30°C to allow TAG accumulation. Subsequently, cells were harvested, washed twice with ONR7a medium, and cultivated again in the same medium without a carbon source. Samples were taken at the indicated times and analyzed for lipid content by GC. For this analysis, TAGs were purified by preparative TLC before GC. Tridecanoic acid was used as an internal standard for quantification and was added before the start of acid-catalyzed methanolysis. ND, not determined.
FIG. 5.
Mobilization of stored TAGs by A. borkumensis strains as revealed by TLC analysis. Cells were cultivated in ONR7a medium containing 1% (wt/vol) sodium pyruvate for 72 h at 30°C to promote accumulation of TAGs. Afterwards, cells were harvested, washed twice, and resuspended in carbon-free ONR7a medium to permit the mobilization of TAGs. Samples were taken at the beginning of cultivation (time zero [T0]) and after 24 h (time point 1 [T1]), 48 h (T2), 120 h (T3), and 240 h (T4). Lipid extracts obtained from 5 mg lyophilized cells were applied in each lane. The solvent system used for elution was hexane-diethyl ether-acetic acid (80:20:1, vol/vol/vol), and lipids were visualized by staining with iodine vapor. T, triolein; OA, oleic acid; OO, oleyl oleate; SK2, A. borkumensis wild type; atfA1, A. borkumensis atfA1ΩKm; atfA2, A. borkumensis atfA2ΩSm; atfA1atfA2, A. borkumensis atfA1ΩKm atfA2ΩSm; 2 M131, transposon-induced mutant; FA, fatty acids.
Role of accumulated TAGs in survival under carbon source-free conditions.
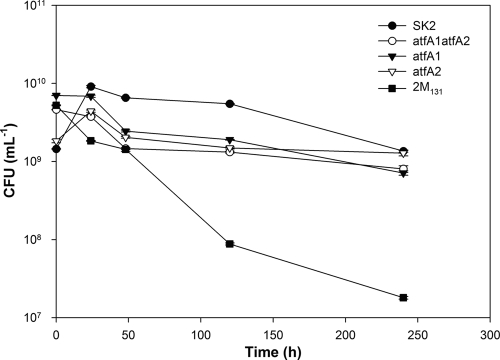
During the mobilization studies, the survival of cells was monitored in parallel by determining numbers of remaining viable cells (measured as the number of CFU per milliliter) in order to establish a correlation between mobilization and cell survival. The wild type SK2 and the atfA2 mutant exhibited increases in viability within the first 24 h of the experiment; thereafter, the viability decreased rapidly in both cases (Fig. 6). This observation paralleled the results of the mobilization experiments (see above), which revealed that SK2 and the atfA2 mutant mobilized the major portion of the accumulated TAGs within the first 24 h of cultivation. The atfA1 and atfA1 atfA2 mutants showed slightly sharper drops in viability than SK2 and the atfA2 mutant during the same period. GC and TLC analyses had also shown for the atfA1 and atfA1 atfA2 mutants that the major portion of accumulated TAGs was assimilated during the first 24 h of cultivation. Apparently, larger amounts of accumulated TAGs provided an advantage to A. borkumensis SK2 and the atfA2 mutant only at the beginning of cultivation. After the TAG contents in A. borkumensis SK2 and atfA2 mutant cells had diminished to only 1.6 and 0.4% of the CDW, respectively, the cells lost their viability very fast. In concordance with these observations, mutant 2 M131, which was completely impaired in producing TAGs, showed a decrease in viability of about 67% already after 24 h of cultivation and this tendency toward decreasing viability remained up to the end of the cultivation experiment.
FIG. 6.
Survival of A. borkumensis strains in carbon-free medium during starvation experiments. Viable cell counts (expressed in CFU) over the time course of carbon starvation were obtained as described in Materials and Methods. Values are means ± errors of results from triplicate determinations.
DISCUSSION
As reported by Kalscheuer et al. (28), inactivation of the two genes coding for WS/DGAT homologues present in the genome of A. borkumensis SK2 did not result in the complete loss of the capability for TAG biosynthesis and accumulation in this bacterium. Accumulation of reduced but still substantial amounts of TAGs in the double mutant A. borkumensis atfA1ΩKm atfA2ΩSm indicated the presence of an alternative non-WS/DGAT-dependent TAG biosynthesis pathway. In this study, we screened a mini-Tn5 transposon library in this double mutant to identify additional enzymes involved in storage lipid accumulation in this hydrocarbonoclastic marine bacterium. Sequence analysis of the A. borkumensis SK2 genome indicated that a third structural homologue of WS/DGAT was unlikely to exist. About 9,000 mutants were analyzed; cells were cultivated on ONR7a plates containing Nile red and screened for fluorescence under UV light. Thirteen mutants which showed no fluorescence or weaker fluorescence than the wild type were analyzed for their abilities to accumulate TAGs. With the exception of mutant 2 M191, all these mutants exhibited only diminished TAG contents. Among these mutants, 2 M131 had completely lost the ability to accumulate TAGs when it was cultivated in the presence of pyruvate as the sole carbon source. The insertion of the transposon in mutant 2 M131 was mapped to a gene coding for a cytochrome c family protein (ABO_1185). The phenotype of the transposon-induced mutant 2 M131 was in principal confirmed by that of the ABO_1185ΩSm disruption mutant. This finding makes it very likely that the cytochrome is somehow related to TAG metabolism.
Cytochromes are electron transfer proteins that carry heme as a prosthetic group. They form a diverse group of proteins having only a few features in common (33). Most bacterial cytochromes function either in photosynthetic electron transport or in aerobic and anaerobic respiration, whereby ATP formation is coupled to the oxidation of reduced substrates such as organic substances, hydrogen, reduced sulfur, and metals (44). In contrast to other heme proteins, heme in cytochrome c is covalently bound to the polypeptide (8). Involvement of cytochrome c-type proteins in processes related to lipid biosynthesis has been described already (24, 45). For example, a mutant of Pseudomonas putida P8 harboring a mini-Tn5 insertion within the cytochrome c operon that provokes the turnoff genes involved in the attachment of heme to cytochrome c-type proteins exhibits remarkable reduction of cis-trans isomerization capability for unsaturated fatty acids (24). Cis-trans isomerization of unsaturated fatty acids may protect bacterial cells from, e.g., exposure to elevated temperatures (30), high levels of harmful compounds (22, 23), or elevated salinity. It has also been reported previously that in Rhodobacter capsulatus, mutations in either olsA or olsB, involved in the synthesis of an ornithine lipid [OL; α-N-(3-acyloxyacyl)-ornithine], greatly decrease the amounts of various c-type cytochromes and other proteins in cell membranes (5, 6). Genetic complementation of appropriate mutants (olsAB negative) with the corresponding genes concomitantly eliminates both the protein and the lipid defects. Thus, two seemingly unrelated aspects, namely, OL biosynthesis and the steady-state amounts of some extracytoplasmic proteins, including cytochromes c, are interconnected under certain physiological conditions (6).
Because mutant A. borkumensis 2 M131 did not show significant differences in the accumulation of TAGs in comparison to the wild type when it was cultivated on alkanes such as hexadecane and octadecane, the results obtained in this study suggest that the transposon insertion in ABO_1185 has an effect on de novo fatty acid biosynthesis. Accumulation of TAGs by this mutant during growth on alkanes may be explained by assimilation through a different route, like β-oxidation, in which there is no need for de novo fatty acid synthesis.
It has been reported repeatedly that the last step of TAG biosynthesis in some bacteria is not catalyzed by a single enzyme (1, 4). In fact, 15 atfA homologues in Mycobacterium tuberculosis H37Rv have been identified and characterized (11, 41). In addition, at least 10 genes encoding putative Atf enzymes occur in R. opacus PD630 (1). Furthermore, multiple TAG biosynthesis routes in S. coelicolor (4) have been described. Genes with high degrees of homology to A. baylyi strain ADP1 atfA were also identified in the genome databases for several Gram-negative strains (48). Small amounts of TAGs in an atfA deletion mutant of A. baylyi strain ADP1 (27) and in the A. borkumensis atfA1 atfA2 double mutant (28), as well as in the atf1 disruption mutant of R. opacus strain PD630 (1), support the hypothesis of the existence of an alternative TAG biosynthesis pathway.
The fatty acid accumulation profiles of the A. borkumensis strains investigated in this study during growth in medium containing pyruvate as the sole carbon source showed the occurrence of hexadecanoic and octadecenoic acids as major components of accumulated TAGs. Hexadecanoic acid is the most universally distributed fatty acid, and it occurs in almost all bacteria, along with minor fractions of other saturated fatty acids (usually tetradecanoic and octadecanoic acids) (17). These fatty acids were probably synthesized de novo from acetyl-CoA. Propionyl-CoA was apparently not formed in appreciable quantities by A. borkumensis SK2 during growth on pyruvate, since fatty acids with odd-numbered chain lengths were absent. In addition, TAGs produced by A. borkumensis SK2 contained tetradecanoic and octadecanoic acids as constituents. Neither fatty acid was observed in TAGs produced by atfA1 and atfA1 atfA2 mutants, suggesting that mainly atfA1 is involved in the synthesis of TAGs containing saturated fatty acids with even-numbered chain lengths. Regarding the biosynthesis of TAGs in A. borkumensis, probably the lack of ammonium at the end of cultivation triggered the biosynthesis of these lipids. The relatively high carbon-to-nitrogen ratio of the ONR7a medium (about 50) may provoke some TAG biosynthesis already during the exponential growth phase, although a remarkable increase in the TAG content of the cells in the stationary phase was subsequently observed.
There are three criteria for a compound to function in energy storage: (i) it is accumulated under conditions in which there is an excess energy supply from exogenous sources; (ii) it is utilized when the supply of energy from exogenous sources is no longer sufficient for optimal maintenance of the cell, either for growth and division or for maintenance of viability and other processes; and (iii) it is degraded to produce energy, which can be utilized by the cells, thereby providing a biological advantage over those cells which do not have such a compound (12). In this study, we investigated the intracellular accumulation and mobilization of TAGs in Alcanivorax. It was clearly demonstrated that TAGs are true storage compounds in A. borkumensis SK2 because (i) they were accumulated under imbalanced growth conditions (C/N ratio, about 50); (ii) they were utilized as a supply of carbon and energy under carbon-free cultivation conditions, as demonstrated by viability studies and GC and TLC analyses; and (iii) accumulation of larger amounts of TAGs in the SK2 and atfA2 strains than in the atfA1, atfA1 atfA2, and 2 M131 mutants provided an advantage for survival of the SK2 and atfA2 strains, at least at the beginning of cultivation when the lipids were mobilized. TAGs were readily mobilized in this phase if the presence of ammonium in the medium allowed proliferation of cells.
A. borkumensis is a slowly growing, alkane-degrading Gram-negative bacterium present only at low titers in pristine waters. However, the population of this species can dramatically increase after an oil spill, and it can become the most abundant microorganism in oil-polluted waters (20, 29, 43). The predominance of A. borkumensis in early stages of petroleum degradation has also been reported in a microcosm study (36) and a field-scale study (35). Alcanivorax spp. seem to be early colonizers after an oil spill event, because after an initial rapid increase, populations of Alcanivorax spp. decline to much lower numbers within a few weeks. This phenomenon is related to the depletion of saturated hydrocarbons, suggesting a specialization of Alcanivorax strains to degrade this class of compounds (21). Moreover, the importance of A. borkumensis SK2 as an essential organism in the first steps of marine hydrocarbon degradation has been shown recently (18). TAGs and WEs but not PHAs are the principal storage lipids present in hydrocarbonoclastic marine bacteria such as A. borkumensis (28). The rapid decline of A. borkumensis populations after depletion of saturated hydrocarbons suggests rapid mobilization of the storage compounds. Storage lipid accumulation seems to provide a growth advantage only during short periods of starvation. Results presented here may provide an explanation for the temporally limited predominance of A. borkumensis after an oil spill.
Acknowledgments
We are very grateful for financial support of this study by the Deutsche Forschungsgemeinschaft (DFG) to A.S. (through grant no. STE.386/76-3). E.M.-P. is indebted to the National Council on Science and Technology (CONACyT, México) and the German Academic Exchange Service (DAAD) for doctoral scholarship awards.
E.M.-P. thanks B. Andreessen and K. Kampmann for critically reading the manuscript and for helpful discussions.
Footnotes
Published ahead of print on 19 March 2010.
REFERENCES
- 1.Alvarez, A. F., H. M. Alvarez, R. Kalscheuer, M. Wältermann, and A. Steinbüchel. 2008. Cloning and characterization of a gene involved in triacylglycerol biosynthesis and identification of additional homologous genes in the oleaginous bacterium Rhodococcus opacus PD630. Microbiology 154:2327-2335. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, H. M., and A. Steinbüchel. 2002. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 60:367-376. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez, H. M., M. F. Souto, A. Viale, and O. H. Pucci. 2001. Biosynthesis of fatty acids and triacylglycerols by 2,6,10,14-tetramethyl pentadecane-grown cells of Nocardia globerula 432. FEMS Microbiol. Lett. 200:195-200. [DOI] [PubMed] [Google Scholar]
- 4.Arabolaza, A., E. Rodriguez, S. Altabe, H. Alvarez, and H. Gramajo. 2008. Multiple pathways for triaclyglycerol biosynthesis in Streptomyces coelicolor. Appl. Environ. Microbiol. 74:2573-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aygun-Sunar, S., R. Bilaloglu, H. Goldfine, and F. Daldal. 2007. Rhodobacter capsulatus OlsA is a bifunctional enzyme active in both ornithine lipid and phosphatidic acid biosynthesis. J. Bacteriol. 189:8564-8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aygun-Sunar, S., S. Mandaci, H. G. Koch, I. V. J. Murray, H. Goldfine, and F. Daldal. 2006. Ornithine lipid is required for optimal steady-state amounts of c-type cytochromes in Rhodobacter capsulatus. Mol. Microbiol. 61:418-435. [DOI] [PubMed] [Google Scholar]
- 7.Bachmann, B. J. 1987. Linkage map of Escherichia coli K-12, edition 7, p. 807-877. In F. C. Neidhardt, J. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, DC.
- 8.Bengtsson, J., H. Tjalsma, C. Rivolta, and L. Hederstedt. 1999. Subunit II of Bacillus subtilis cytochrome c oxydase is a lipoprotein. J. Bacteriol. 181:685-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bredemeier, R., R. Hulsch, J. O. Metzger, and L. Berthe-Corti. 2003. Submersed culture production of extracellular wax esters by the marine bacteria Fundibacter jadensis. Mar. Biotechnol. 5:579-583. [DOI] [PubMed] [Google Scholar]
- 11.Daniel, J., C. Deb, V. S. Dubey, T. D. Sirakova, B. Abomoelak, H. R. Morbidoni, and P. E. Kolattukudy. 2004. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 186:5017-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawes, E. A., and P. J. Senior. 1973. The role and regulation of energy reserve polymers in micro-organisms. Adv. Microbiol. Physiol. 10:135-266. [DOI] [PubMed] [Google Scholar]
- 13.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz, G., M. Melis, B. Batetta, F. Angius, and A. M. Falchi. 2008. Hydrophobic characterization of intracellular lipids in situ by Nile Red red/yellow emission ratio. Micron 39:819-824. [DOI] [PubMed] [Google Scholar]
- 15.Dyksterhouse, S. E., J. P. Gray, R. P. Herwig, J. C. Lara, and J. T. Staley. 1995. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 45:116-123. [DOI] [PubMed] [Google Scholar]
- 16.Fixter, L. M., M. N. Nagi, J. G. McCormack, and C. A. Fewson. 1986. Structure, distribution and function of wax esters in Acinetobacter calcoaceticus. J. Gen. Microbiol. 132:3147-3157. [Google Scholar]
- 17.Fulco, A. J. 1983. Fatty acid metabolism in bacteria. Prog. Lipid Res. 22:133-160. [DOI] [PubMed] [Google Scholar]
- 18.Gertler, C., G. Gerdts, K. N. Timmis, M. M. Yakimov, and P. N. Golyshin. 2009. Populations of heavy fuel oil-degrading marine microbial community in presence of oil sorbent materials. J. Appl. Microbiol. 107:590-605. [DOI] [PubMed] [Google Scholar]
- 19.Greenspan, P., E. P. Mayer, and S. D. Fowler. 1985. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100:965-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harayama, S., H. Kishira, Y. Kasai, and K. Shutsubo. 1999. Petroleum biodegradation in marine environments. J. Mol. Microbiol. Biotechnol. 1:63-70. [PubMed] [Google Scholar]
- 21.Head, I. M., D. M. Jones, and F. M. Röling. 2006. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4:173-182. [DOI] [PubMed] [Google Scholar]
- 22.Heipieper, H. J., R. Diefenbach, and H. Keweloh. 1992. Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl. Environ. Microbiol. 58:1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heipieper, H. J., J. Weber, J. Sikkema, H. Keweloh, and J. A. M. de Bont. 1994. Mechanisms of resistance of whole cells to toxic organic solvents. Trends Biotechnol. 12:409-425. [Google Scholar]
- 24.Holtwick, R., H. Keweloh, and F. Meinhardt. 1999. cis/trans isomerase of unsaturated fatty acids of Pseudomonas putida P8: evidence for a heme protein of the cytochrome c type. Appl. Environ. Microbiol. 65:2644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holtzapple, E., and C. Schmidt-Dannert. 2007. Biosynthesis of isoprenoid wax ester in Marinobacter hydrocarbonoclasticus DSM 8798: identification and characterization of isoprenoid coenzyme A synthetase and wax ester synthases. J. Bacteriol. 189:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, G., L. Zhang, and R. G. Birch. 2000. Rapid amplification and cloning of Tn5 flanking fragments by inverse PCR. Lett. Appl. Microbiol. 31:149-153. [DOI] [PubMed] [Google Scholar]
- 27.Kalscheuer, R., and A. Steinbüchel. 2003. A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J. Biol. Chem. 278:8075-8082. [DOI] [PubMed] [Google Scholar]
- 28.Kalscheuer, R., T. Stöveken, U. Malkus, R. Reichelt, P. N. Golyshin, J. S. Sabirova, M. Ferrer, K. N. Timmis, and A. Steinbüchel. 2007. Analysis of storage lipid accumulation in Alcanivorax borkumensis: evidence for alternative triacylglycerol biosynthesis routes in bacteria. J. Bacteriol. 189:918-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasai, Y., H. Kishira, T. Sasaki, K. Syutsubo, K. Watanabe, and S. Harayama. 2002. Predominant growth of Alcanivorax strains in oil-contaminated and nutrient-supplemented sea water. Environ. Microbiol. 4:141-147. [DOI] [PubMed] [Google Scholar]
- 30.Loffeld, B., and H. Keweloh. 1996. cis/trans isomerization of unsaturated fatty acids as possible control mechanism of membrane fluidity in Pseudomonas putida P8. Lipids 31:811-815. [DOI] [PubMed] [Google Scholar]
- 31.Manilla-Pérez, E., C. Reers, M. Baumgart, S. Hetzler, R. Reichelt, U. Malkus, R. Kalscheuer, M. Wältermann, and A. Steinbüchel. 2010. Analysis of lipid export in hydrocarbonoclastic bacteria of the genus Alcanivorax: identification of lipid export-negative mutants of A. borkumensis SK2 and A. jadensis T9. J. Bacteriol. 192:643-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marmur, J. 1961. A procedure for the isolation of desoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 33.Mathews, F. S. 1985. The structure, function and evolution of cytochromes. Prog. Biophys. Mol. Biol. 45:1-56. [DOI] [PubMed] [Google Scholar]
- 34.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Röling, W. F., M. G. Milner, D. M. Jones, F. Fratepietro, R. P. J. Swanell, F. Daniel, and I. M. Head. 2004. Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with buried oil. Appl. Environ. Microbiol. 70:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Röling, W. F., M. G. Milner, D. M. Jones, K. Lee, F. Daniel, R. P. J. Swanell, and I. M. Head. 2002. Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil-spill bioremediation. Appl. Environ. Microbiol. 68:5537-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
- 38.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneiker, S., V. A. P. Martins dos Santos, D. Bartels, T. Bekel, M. Brecht, J. Buhrmester, T. N. Chernikova, R. Denaro, M. Ferrer, C. Gertler, A. Goesmann, O. V. Golyshina, F. Kaminski, A. N. Khachane, S. Lang, B. Linke, A. C. McHardy, F. Meyer, T. Nechitaylo, A. Pühler, D. Regenhardt, O. Rupp, J. S. Sabirova, W. Selbitschka, M. M. Yakimov, K. N. Timmis, F. J. Vorhölter, S. Weidner, O. Kaiser, and P. N. Golyshin. 2006. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat. Biotechnol. 24:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784-791. [Google Scholar]
- 41.Sirakova, T. D., V. S. Dubey, C. Deb, J. Daniel, T. A. Korotkova, B. Abomoelak, and P. E. Kolattukudy. 2006. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology 152:2717-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spiekermann, P., B. H. A. Rehm, R. Kalscheuer, D. Baumeister, and A. Steinbüchel. 1999. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 171:73-80. [DOI] [PubMed] [Google Scholar]
- 43.Syutsubo, K., H. Kishira, and S. Harayama. 2001. Development of specific oligonucleotide probes for the identification and in situ detection of hydrocarbon-degrading Alcanivorax strains. Environ. Microbiol. 3:371-379. [DOI] [PubMed] [Google Scholar]
- 44.Thöny-Meyer, L. 1997. Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 61:337-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Wallbrunn, A., H. H. Richnow, G. Neumann, F. Meinhardt, and H. J. Heipieper. 2003. Mechanism of cis/trans isomerization of unsaturated fatty acids in Pseudomonas putida. J. Bacteriol. 185:1730-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wältermann, M., H. Luftmann, D. Baumeister, R. Kalscheuer, and A. Steinbüchel. 2000. Rhodococcus opacus strain PD630 as a new source of high-value single-cell oil? Isolation and characterization of triacylglycerols and other storage lipids. Microbiology 146:1143-1149. [DOI] [PubMed] [Google Scholar]
- 47.Wältermann, M., and A. Steinbüchel. 2005. Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J. Bacteriol. 187:3607-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wältermann, M., T. Stöveken, and A. Steinbüchel. 2007. Key enzymes for biosynthesis of neutral lipid storage compounds in prokaryotes: properties, function and occurrence of wax ester synthases/acyl-CoA:diacylglycerol acyltransferases. Biochimie 89:230-242. [DOI] [PubMed] [Google Scholar]
- 49.Yakimov, M. M., P. N. Golyshin, S. Lang, E. R. B. Moore, W. R. Abraham, H. Lünsdorf, and K. N. Timmis. 1998. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 48:339-348. [DOI] [PubMed] [Google Scholar]