Abstract
We recently showed that Lactobacillus sakei, a natural meat-borne lactic acid bacterium, can colonize the gastrointestinal tracts (GIT) of axenic mice but that this colonization in the intestinal environment selects L. sakei mutants showing modified colony morphology (small and rough) and cell shape, most probably resulting from the accumulation of various mutations that confer a selective advantage for persistence in the GIT. In the present study, we analyzed such clones, issued from three different L. sakei strains, in order to determine which functions were modified in the mutants. In the elongated filamentous cells of the rough clones, transmission electron microscopy (TEM) analysis showed a septation defect and dotted and slanted black bands, suggesting the presence of a helical structure around the cells. Comparison of the cytoplasmic and cell wall/membrane proteomes of the meat isolate L. sakei 23K and of one of its rough derivatives revealed a modified expression for 38 spots. The expression of six oxidoreductases, several stress proteins, and four ABC transporters was strongly reduced in the GIT-adapted strain, while the actin-like MreB protein responsible for cell shaping was upregulated. In addition, the expression of several enzymes involved in carbohydrate metabolism was modified, which may correlate with the observation of modified growth of mutants on various carbon sources. These results suggest that the modifications leading to a better adaptation to the GIT are pleiotropic and are characterized in a rough mutant by a different stress status, a cell wall modification, and modified use of energy sources, leading to an improved fitness for the colonization of the GIT.
Lactobacilli are Gram-positive bacteria known mostly for their importance in the food industry or their potential beneficial effects on consumers. Their presence in the human gastrointestinal tract (GIT) has frequently been reported, and, although in small amounts (31), 16 different Lactobacillus species have been isolated at least once from human feces (35). Most of those species are probably not autochthonous but rather persisting species introduced in the GIT after food ingestion.
Lactobacillus sakei is known as a food-borne bacterium, frequently isolated from fresh or fermented meat but also found in fish or vegetal fermented products (6, 30). The importance of L. sakei in the meat industry significantly increased in recent years because of its technological properties that are optimal for sausage fermentation (5, 9) and its potential use as a bioprotective culture for the biopreservation of various meat products (3, 5, 36). L. sakei has been detected in human feces (20, 37), and some human strains have been isolated (7, 10). The estimated amount of this species in the fecal samples (106 CFU/g of feces) does not permit a conclusion about whether its presence is strictly food related or not. In addition, although the physiological functions involved in the adaptation to and growth of L. sakei in its preferred environment (meat essentially) are well documented (5, 21), those required for L. sakei transit through and/or colonization of the GIT are yet unknown.
We recently reported that three L. sakei strains from different origins (L. sakei 23K, a meat-borne strain, and L. sakei LTH5590 and FLEC01, both isolated from human feces) can survive the transit through conventional mice and durably colonize the GIT of axenic mice (7). However, we observed that the transit through the GIT of axenic mice led to the appearance of L. sakei subpopulations, characterized by atypical colony morphologies and cell shapes and probably resulting from the accumulation of mutations allowing a better adaptation to the GIT environment. The clones forming small colonies on MRS agar medium (12) were named S and the clones forming rough colonies R. In the present study, we aimed to determine which functions could be involved in this adaptation by comparing S and R clones to their parent strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. sakei 23K is a plasmid-free strain issued from a natural isolate originating from fermented sausage (2). L. sakei LTH5590 (10) and L. sakei FLEC01 (7) were isolated from feces of healthy humans. L. sakei 23K-J10SN2, L. sakei 23K-J30RO2, L. sakei FLEC01-J6SN2, L. sakei FLEC01-J15RN2, L. sakei LTH5590-J21SO2, and L. sakei LTH5590-J40RN2 are clones which have acquired a different colony morphotype on MRS medium after passage through the GIT of axenic mice. They were named from a nomenclature indicating the parent strain they were derived from, the time after transit they were collected (J10, J21, for instance), their colony morphotype (R or S), and the incubation conditions used for their isolation (O2 or N2 for aerobic or anaerobic conditions, respectively), as previously described (7).
Bacteria were routinely grown at 30°C with stirring at 70 rpm either in MRS medium (12) or, for physiological studies, in the chemically defined medium MCD (26) supplemented with 0.5% (wt/vol) various carbon sources. To measure growth rate for various carbon sources, bacteria were first grown overnight in MCD medium containing 0.5% (wt/vol) glucose. Aliquots were then inoculated at an initial optical density at 600 nm (OD600) of 0.05 in fresh MCD medium supplemented with 0.5% (wt/vol) d-glucose (Merck), N-acetyl-d-glucosamine (Sigma), d-mannose (Sigma), d-fructose (Merck), or d-galactose (Sigma) as the sole carbon source. The OD600 was determined every 90 min until stationary phase. Doubling times reached during the exponential phase of growth were calculated. Experiments were repeated at least twice.
Stress response measurement.
To evaluate response to acidic pH, hydrogen peroxide (H2O2), and bile salt stresses, bacteria were collected from 10-ml overnight cultures in MRS medium and resuspended into fresh MRS medium, to which various components were added. For H2O2 stress, bacteria were inoculated at 10% (vol/vol) in fresh MRS medium, to which H2O2 was added at a concentration of 4 or 10 mM. Bacteria were then incubated for 1 h at 30°C, H2O2 stress was stopped by catalase addition, and survival was finally measured by plating bacterial dilutions on MRS plates and CFU counting. For bile salt stress, a sodium deoxycholate solution at 1% was used. Sodium deoxycholate was added at a final concentration of 0.1 or 0.3%, and bacteria were incubated for 3 h at 30°C and treated as described above. For acid stress, drops of concentrated HCl were added in order to obtain pH values ranging from 2.5 to 6.0 every half pH unit (initial MRS pH was around 6.5). Bacteria were inoculated at 1%, incubated for 1 h, and treated as described above. Stress response was measured as the log CFU difference between control (no stress component added) and stress conditions. Experiments were performed at least twice.
Transmission electron microscopy (TEM).
Bacterial pellets recovered from MRS cultures (OD600 of 0.5) were fixed with 2% glutaraldehyde in 0.1 M Na cacodylate buffer, pH 7.2, for 3 h at room temperature. Samples were then postfixed with 1% osmium tetroxide containing 1.5% potassium cyanoferrate, gradually dehydrated in ethanol (30% to 100%), and embedded in Epon. Thin sections (70 nm) were collected onto 200 mesh copper grids and counterstained with lead citrate before examination with a Zeiss EM 902 transmission electron microscope at 80 kV (MIMA2; Plateau de Microscopie Electronique, Unité GPL, Jouy-en-Josas, France). Microphotographs were acquired using a MegaView III charge-coupled-device (CCD) camera and analyzed with ITEM software (Eloïse Sarl, Roissy CDG, France).
Preparation of protein samples and labeling of cell wall-associated proteins.
L. sakei 23K and L. sakei 23K-J30RO2 were grown in 250 ml MCD medium supplemented with 0.5% (wt/vol) glucose until an OD600 of 0.8 and centrifuged 30 min at 6,000 × g. Supernatant was discarded, and the bacterial pellet was washed three times with Tris-HCl, 0.1 M, pH 7.5, and stored at −80°C until use.
In order to stain surface-exposed proteins, cells were resuspended in 1 ml Tris-HCl, pH 8.8, and mixed with 400 pmol of CyDye DIGE Fluor Cy5 (Amersham GE Healthcare) diluted in dimethylformamide (DFM) (Sigma) according to the manufacturer's instructions. Samples were incubated at 4°C for 20 min in the dark, then mixed with 1 μl of a 10 mM l-lysine (Sigma) solution, incubated 10 min at 4°C, and washed three times in Tris-HCl, 0.1 M, pH 7.5. Cells were broken by a single pass through a cell disrupter (Basic Z; Constant Systems Ltd., Daventry, United Kingdom) at 2.5 × 105 Pa. Unbroken cells and cell debris were removed by centrifugation at 4,500 × g for 15 min. To ensure that bacteria were not permeable to Cy5, bacteria were collected prior to cell disruption and were observed on an Axio Observer Z1 microscope used in combination with the Apotome system and the Colibri illumination system (all from Carl Zeiss, Goettingen, Germany). Apotome generates deblurred optical sections of fluorescent samples, and the Colibri module is constituted of several light-emitting diodes (LED). For Cy5 excitation, an LED module at 625 nm was used. Images were acquired, stored, and processed using Axio Vision software facilities (Carl Zeiss). No intracellular signal was observed.
Cytoplasmic proteins were obtained by recovering the supernatant after ultracentrifugation at 50,000 × g for 30 min at 4°C and treating the suspension as previously described (32). Membrane vesicles from the pellet obtained after ultracentrifugation were resuspended in 1 ml 10% (wt/vol) sodium dodecyl sulfate (SDS), incubated at 100°C for 15 min, and centrifuged for 10 min at 3,500 × g; supernatant was filtered using 0.22-μm sterile filters. The protein suspension was then treated for 30 min at 37°C with 1 μl benzonase (Merck) in the presence of 10 mM MgSO4 to remove contaminating nucleic acids. Proteins were precipitated by adding trichloroacetic acid to the final concentrations of 1.2 M (Merck) and 0.2 mg ml−1 sodium deoxycholate (Merck). After incubation for 1 h at 4°C in the dark, samples were centrifuged at 16,000 × g for 30 min, and the pellet was washed three times in cold acetone and air dried. Proteins were solubilized as described previously (32) and used for two-dimensional (2-DE) gel electrophoresis.
2-DE.
The protocol for 2-DE gel electrophoresis was performed as previously described (32). Briefly, 350 μg of proteins was focalized in 17-cm IPG strips (pH 4 to 7) for the analysis of the cytoplasmic proteome and in 17-cm strips (pH 3 to 10) for surface proteins (Bio-Rad, Hercules, CA). Strips were rehydrated for 12 h at 50 V using Protean IsoElectric Focusing Cell II (Bio-Rad) and then focused at 300 V for 1 h (exponential), 1,500 V for 1 h (exponential), 10,000 V for 6 h (linear), and 10,000 V for 2.5 h (exponential). The second dimension was performed in 12.5% (wt/vol) polyacrylamide gels (20 by 20 by 0.1 cm) run overnight at 26 mA per plate at 4°C. Gels were stained with BioSafe colloidal Coomassie blue (Bio-Rad) and scanned with an ImageScanner (Amersham Biosciences, Piscataway, NJ) for the analysis of cytoplasmic proteins. To discriminate surface-exposed proteins stained with Cy5 dye in the cell wall/membrane fraction, gels were first scanned with a Typhoon 9410 (Amersham Pharmacia Biotech) at 670 nm to detect Cy5 and then stained with Coomassie blue and scanned with an ImageScanner as described above. Comparative image analysis was performed with Progenesis SameSpots (Nonlinear Dynamics) software by analyzing gels from at least three independent experiments. The relative volume of each spot (percent volume) was obtained from its spot intensity normalized to the sum of the intensities of all spots of the gel. Statistical analysis of the spot expression variation was performed by analysis of variance (ANOVA) provided by the software. Only proteins displaying significantly up- or downregulation in n − 1 gels and with a mean of at least 1.5-fold volume variations were considered and selected for identification through peptide mass fingerprinting.
Peptide mass fingerprinting and protein identification.
Spots were analyzed as described in reference 19. Briefly, target spots were excised and in-gel digested overnight with 250 ng of trypsin (Promega, Madison, WI). The mass of the peptides was determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) on a Voyager DE STR instrument (Applied Biosystems) at the PAPSSO platform of the INRA Center in Jouy-en-Josas. Internal calibration was automatically performed with autolytical fragments of trypsin. L. sakei peptides were searched against the L. sakei database of the annotated genome (5) using Mascot and MS-Fit software. Sequence coverage (above 40%) scores of the search and agreement between theoretical and experimental molecular weights and isoelectric points were taken into account for considering identifications positive.
DNA manipulations, PCR, and sequencing.
DNA was extracted from L. sakei cultures using the High Pure PCR template preparation kit (Roche) according to the manufacturer's instructions. DNA fragments encompassing the LSA1317 and LSA0104 genes and the LSA0162 to LSA0170, LSA0462-LSA0463, and LSA0551-LSA0552 operons as well as their promoter regions, were amplified by PCR with primers listed in Table S1 in the supplemental material. PCR amplification was performed in a PCT-200 thermocycler (MJ Research). The 100-μl PCR mixtures contained PCR buffer, 1.5 nmol MgCl2 liter−1, 0.2 nmol liter−1 of each dNTP, 0.5 μmol liter−1 of each primer, 1 μg ml−1 of chromosomal DNA, and 2 U of Taq polymerase (Fermentas). The program used was as follows: 95°C for 4 min; 25 cycles of 95°C for 30 s, 55°C for 10 s, and 72°C for 1 min; and a final elongation step at 72°C for 5 min. For sequencing, PCR fragments were treated with 0.1 U shrimp alkaline phosphatase (USB) and 1 U exonuclease I (Biolabs) in 20 mM Tris-HCl, pH 8.0, and 10 mM MgCl2 buffer for 1 h at 37°C and then sent for sequencing to Eurofins MWG Operon (Germany). Primers used for sequencing are listed in Table S2 in the supplemental material.
RESULTS
Morphological changes acquired during transit of L. sakei in the GIT are associated with septation defect or cell wall modification.
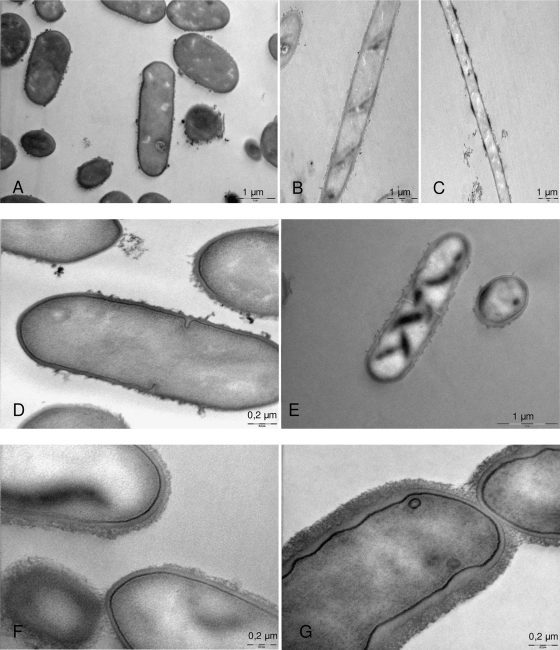
TEM observations showed that long filaments issued from R clone mutants FLEC01-J15RN2 and 23K-J30RO2 were formed by cells lacking internal septation (Fig. 1B, C, and E) compared to their parent strains (Fig. 1A and D). Interestingly, we also noticed the presence of dark transversal bands (absent from wild-type [WT] strains) on the surface of R clones FLEC01-J15RN2 and 23K-J30RO2 (Fig. 1B, C, and E), suggesting a helical structure. In addition, images of the L. sakei LTH5590-J21SO2 S clone (Fig. 1G) revealed an abnormally deformed cell membrane and an apparently thicker cell wall than that of the wild-type parent, LTH5590 (Fig. 1F), which were not observed with the R strains. This confirmed previous scanning electron microscopy observations that showed atypical cell shape and cell surface structures of the bacteria issued from S and R clones (7).
FIG. 1.
Morphology of cells issued from S and R clones of the L. sakei 23K, FLEC01, or LTH5590 strain observed by transmission electron microscopy. L. sakei FLEC01 wild type (A) and its rough clone derivative FLEC01-J15RN2 (B and C); L. sakei 23K wild type (D) and its rough clone derivative L. sakei 23K-J30RO2 (E); L. sakei LTH5590 wild type (F) and it small clone derivative L. sakei LTH5590-J21SO2 (G) grown in MRS liquid medium.
Modification of stress responses in L. sakei R and S clones.
Responses to bile salt, H2O2, or acidic pH stress were evaluated as the difference of survival rates after the stress for the S and R clones and the three parent strains 23K, FLEC01, and LTH5590 from which they were derived. The S clones had, in general, a phenotype close to that of their WT or intermediary between those of their WT and the R clones (Table 1). We noticed that the behaviors of three WT strains regarding bile salt response were slightly different: at 0.3% bile salt, 23K was less sensitive than LTH5590 and FLEC01 (Table 1). However, the R clone 23K-J30RO2 became more sensitive than its WT parent, whereas the two R mutants FLEC01-J15RN2 and LTH5590-J40RN2 were more resistant than their parent strain (Table 1). After 10 mM H2O2 stress, the R clone LTH5590-J40RN2 was the most resistant strain, whereas all other strains showed a high sensitivity (a loss of viability of >5 log CFU). At 4 mM H2O2, all strains showed a loss of viability of ∼3 to 5 log CFU (Table 1). However, LTH5590-J40RN2 was clearly more resistant than its parent, and 23K-J30RO2 was slightly more sensitive (5.0 ± 0.2 versus 3.8 ± 0.1 log CFU) than its parent, L. sakei 23K. Concerning acidic stress, all strains were highly resistant to low pH (loss of viability did not exceed 0.4 log CFU, even at pH 3.0). The mutant 23K-J30RO2 showed a viability decrease but only at pH 3.0 (Table 1).
TABLE 1.
Stress responses of WT and R clonesa
| Strain | Stress response to: |
|||
|---|---|---|---|---|
| Bile salt (0.3%) | H2O2 (4 mM) | H2O2 (10 mM) | pHb | |
| 23K | 0.90 ± 0.10 | 3.20 ± 0.30 | >5 | <0.4 |
| 23K-J10SN2 | 2.10 ± 0.90 | 3.80 ± 0.70 | >5 | ND |
| 23K-J30RO2 | 3.90 ± 0.70 | 5.00 ± 0.20 | >5 | 2.4 ± 0.20c |
| FLEC01 | 5.20 ± 0.00 | 4.05 ± 0.15 | >5 | <0.4 |
| FLEC01-J6SN2 | 4.10 ± 0.10 | 3.40 ± 0.20 | >5 | ND |
| FLEC01-J15RN2 | 3.95 ± 0.15 | 3.40 ± 0.10 | >5 | <0.4 |
| LTH5590 | 5.35 ± 0.05 | >5 | >5 | <0.4 |
| LTH5590-J21SO2 | 3.50 ± 0.10 | 3.80 ± 0.50 | 4.10 ± 0.20 | ND |
| LTH5590-J40RN2 | 1.75 ± 0.15 | 0.60 ± 0.10 | 2.27 ± 1.0 | <0.4 |
Stress response is expressed as the loss of cell viability (Δlog CFU/ml) after the various stress conditions tested, as described in Materials and Methods.
Values were similar for pH values ranging from 3 to 6.0. ND, not determined.
Value is given for pH 3.
L. sakei mutants 23K-J30RO2 and 23K-J10SN2 have improved growth on some carbohydrates.
Ability to grow on various carbon sources of the S and R clones 23K-J10SN2 and 23K-J30RO2 was tested and compared to that of their parent strain, L. sakei 23K (Table 2). No significant differences were observed between the S and R mutants and their parent strain on glucose or mannose. In contrast, L. sakei 23K-J30RO2 exhibited slightly faster growth on N-acetylglucosamine (P < 0.1) and significantly faster growth on fructose, ribose, and galactose. 23K-J10SN2 had a similar but less pronounced phenotype.
TABLE 2.
Growth characteristics of L. sakei 23K, L. sakei 23K-J10SN2, and L. sakei 23K-J30RO2 in MCD medium supplemented with various carbon sourcesa
| L. sakei strain | Doubling timeb |
|||||
|---|---|---|---|---|---|---|
| Glucose | Mannose | Ribose | NAG | Fructose | Galactose | |
| 23K | 86 ± 4 | 98 ± 9 | 117 ± 10 | 102 ± 4 | 113 ± 2 | 117 ± 3 |
| 23K-J10SN2 | 85 ± 7 | 91 ± 8 | 96 ± 6* | 88 ± 8** | 93 ± 8* | 95 ± 5* |
| 23K-J30RO2 | 89 ± 6 | 93 ± 9 | 93 ± 8* | 90 ± 6** | 91 ± 6* | 98 ± 3* |
Carbon sources were added in the medium at the concentration of 0.5% (wt/vol).
Doubling times are expressed in minutes. Results are the mean of three independent repeats. NAG, N-acetylglucosamine; *, values were significantly different from the WT (P < 0.05); **, values were significantly different from the WT (P < 0.1).
Proteomic analysis of a clone forming rough colonies reveals multiple changes.
In order to detect which proteins may be involved in the phenotype of the mutants, we chose to compare the proteomes of a WT strain and of one of its derived mutant. For that purpose, we chose L. sakei 23K as the WT parent, since its genome sequence is known (5), and the R clone L. sakei 23K-J30RO2. Indeed, since this mutant did not show any growth defect in MCD medium supplemented with glucose, no differences due to growth rate were expected from the comparison with the WT. In addition, its R phenotype was shown to be stable in MCD medium (7), avoiding the apparition of revertants and thus avoiding a mixed population that may have led to erroneous proteomic data. As TEM analysis suggested, cell wall modifications in the mutant, both cytosolic and cell wall/membrane compartments, were analyzed.
Twenty-four proteins were downregulated in L. sakei 23K-J30RO2 (Table 3): eight were cytoplasmic, 12 were cell wall or membrane associated, and one (LSA0494), the 30S ribosomal interface protein S30EA, was observed in both compartments. Five out of the 13 proteins of the cell wall/membrane fraction were surface exposed, as revealed by Cy5 labeling. In addition, for three proteins (LSA1366, LSA1188, and LSA1032), products of genes encoding, respectively, the ATP binding subunit of an ABC transporter, the pyruvate oxidase Pox1, and the pyruvate kinase, the differences between the parent and R-derived clone were observed only with the Cy5-labeled fraction and not with the membrane fraction stained with Coomassie blue. This suggests that the relative amounts of these three proteins did not vary, but rather their expositions to the surface of the cell. Among the less expressed spots in the R clone, those identified as proteins LSA0169, LSA0170, LSA1317, LSA0552, and LSA0104 were abundant in the proteome of the L. sakei 23K WT strain but were almost undetectable in the R clone. LSA0169 and LSA0170 are putative general stress proteins of the Asp family and are encoded by the same operon as LSA0165, also downregulated and identified as a putative oxidoreductase. LSA1317 is a putative chromate reductase. LSA0552 (OhrA, a hydroperoxide resistance protein) and LSA0104 (a thiol peroxidase) are both involved in the response to oxidative stresses. Five proteins (LSA0705, LSA0706, LSA0938, LSA0616, and LSA1366) are components of ABC transporters, two proteins (LSA0038 and LSA0836) are involved in general stress response, and three, LSA1188 and LSA1032, mentioned above, and the beta subunit of the pyruvate dehydrogenase complex (LSA1084), are involved in the metabolism of pyruvate. The remaining proteins are involved in amino acid metabolism and protein modification (LSA0463, LSA1134, LSA1426, and LSA1779) or oxidoreduction (LSA0030 and LSA1712). It should be noted that the downregulation of spots LSA0170, LSA0522, and LSA0616 per se is not sufficient to explain the phenotype observed with the R mutant since mutants of the genes encoding these proteins, available in the laboratory (29; M. Zagorec, unpublished results), do not show the same phenotype as that of L. sakei 23K-J30RO2 (M. Champomier-Vergès and M. Zagorec, personal communication).
TABLE 3.
Cytosolic and cell wall/membrane-associated proteins downregulated in R clone L. sakei 23K-J30RO2a
| Protein type | Functional category | Function | Locus tag | ANOVAa | Fold vol change |
|---|---|---|---|---|---|
| Cytoplasm | Adaptation to atypical conditions | Putative general stress protein | LSA0169 | 0.003 | 34 |
| Putative general stress protein | LSA0170 | 0.017 | 6.2 | ||
| Universal stress protein (Usp5) | LSA0038 | 0.030 | 4.3 | ||
| Oxidoreduction | Putative chromate reductase (flavoprotein) | LSA1317 | 0.002 | 8.5 | |
| Organic hydroperoxide resistance protein (OhrA) | LSA0552 | 0.030 | 6.5 | ||
| Putative oxidoreductase, short-chain dehydrogenase/reductase family | LSA0165 | 0.005 | 2.7 | ||
| Carbohydrate metabolism | Pyruvate dehydrogenase complex, E1 component, beta subunit (PdhB) | LSA1084 | 0.041 | 3.2 | |
| Amino acid metabolism | Putative 2-hydroxyacid dehydrogenase | LSA0463 | 0.003 | 3.2 | |
| Transcription-translation | 30S ribosomal interface protein S30EA | LSA0494 | 0.007 | 4.0 | |
| Cell wall/membrane | Adaptation to atypical conditions | Universal stress protein (Usp6) | LSA0836 | 0.004 | 2.1 |
| Transport and binding | Oligopeptide ABC transporter, ATP-binding subunit (OppD) | LSA0705 | 0.022 | 3.5 | |
| Oligopeptide ABC transporter, ATP-binding subunit (OppF) | LSA0706 | 0.007 | 3.4 | ||
| Putative drug ABC exporter, ATP-binding subunit | LSA0938 | 0.026 | 3.3 | ||
| Putative glycine/betaine/carnitine/choline ABC transporter, ATP-binding subunit | LSA0616 | 0.023 | 3.1 | ||
| Oxidoreduction | Thiol peroxidase (Tpx) | LSA0104 | 0.003 | 5.3 | |
| Putative aldo/keto reductase (oxidoreductase) | LSA0030 | 0.001 | 2.6 | ||
| Putative nitroreductase (oxidoreductase) | LSA1712 | 0.001 | 2.5 | ||
| Protein modification | ATPase/chaperone (ClpC) | LSA1779 | 0.023 | 2.6 | |
| Amino acid metabolism | Glycine/serine hydroxymethyltransferase (GlyA) | LSA1134 | 0.037 | 2.7 | |
| Putative aminopeptidase | LSA1426 | 0.002 | 3.1 | ||
| Transcription-translation | 30S ribosomal interface protein S30EA | LSA0494 | 0.004 | 3.7 | |
| RNA restriction modification | Putative RNA methyltransferase | LSA1544 | 0.012 | 1.9 | |
| Surface exposed | Adaptation to atypical conditions | Universal stress protein (Usp6) | LSA0836 | 0.004 | 2.1 |
| Transport and binding | Putative glycine/betaine/carnitine/choline ABC transporter, ATP-binding subunit | LSA0616 | 0.0006 | 3.9 | |
| Oligopeptide ABC transporter, ATP-binding subunit (OppD) | LSA0705 | 0.030 | 2.4 | ||
| Oligopeptide ABC transporter, ATP-binding subunit (OppF) | LSA0706 | 0.004 | 2.3 | ||
| Putative ABC exporter, ATP-binding subunit | LSA1366 | 0.046 | 2.1 | ||
| Oxidoreduction | Thiol peroxidase (Tpx) | LSA0104 | 0.037 | 2.4 | |
| Carbohydrate metabolism | Pyruvate oxidase (Pox1) | LSA1188 | 0.012 | 2.3 | |
| Pyruvate kinase (Pyk) | LSA1032 | 0.033 | 1.7 |
Statistical validation was performed by the use of an ANOVA test provided by SameSpots (see Materials and Methods).
Fourteen spots were upregulated in the mutant L. sakei 23K-J30RO2: nine were cytoplasmic, two were found in the cell wall/membrane fraction, and one (LSA1629) was detected at both locations (Table 4). Three spots were detected as surface exposed (i.e., in the Cy5-labeled fraction); among the three spots, LSA0853 was already found in the cell wall/membrane fraction. Four upregulated proteins, GroEL (LSA0359), DnaK (LSA1236), the copper homeostasis protein LSA1440, and the surface-exposed GTP-binding protein LSA1079, are involved in stress response, two are ribosomal proteins with regulatory functions (LSA0007 and LSA1667), one is an ATP-dependent H+ transporter (LSA1127), and one is a cysteinyl tRNA synthase (LSA1681). Interestingly, among the three proteins that were cell wall/membrane-associated proteins and surface exposed, the amount of which was increased in the R clone, we noticed the presence of the actin-like cell shape determining protein MreB (LSA0853). Three spots were identified as enzymes involved in carbohydrate metabolism: 6-phosphofructokinase (LSA1033), l-lactate dehydrogenase (LSA1606), and fructose-biphosphate aldolase (LSA1527). Finally, two proteins, CTP synthase (LSA1629) and xanthine phosphoribosyltransferase (LSA1557), are involved in nucleotide metabolism. Surprisingly, some proteins usually described as cytoplasmic were found in the cell wall/membrane-associated fraction. Such results have already been observed for so-called “moonlight” proteins and particularly with L. sakei (P. Anglade, unpublished results).
TABLE 4.
Cytosolic and cell wall/membrane-associated proteins upregulated in the R clone L. sakei 23K-J30RO2a
| Protein type | Functional category | Function | Locus tag | ANOVAa | Fold vol change |
|---|---|---|---|---|---|
| Cytoplasm | Adaptation to atypical conditions | Chaperonin GroEL | LSA0359 | 0.017 | 2.7 |
| Chaperone protein DnaK | LSA1236 | 0.032 | 2.6 | ||
| Copper homeostasis protein (CutC) | LSA1440 | 0.018 | 1.5 | ||
| Transport and binding | H+-transporting two-sector ATPase (ATP synthase), gamma subunit | LSA1127 | 0.038 | 3.7 | |
| Carbohydrate metabolism | 6-Phosphofructokinase (Pfk) | LSA1033 | 0.044 | 2.5 | |
| Transcription-translation | 30S ribosomal protein S6 (RpsF) | LSA0007 | 0.025 | 2.1 | |
| 50S ribosomal protein L10 (RplJ) | LSA1667 | 0.046 | 2.0 | ||
| Translation | Cysteinyl tRNA synthase (CysS) | LSA1681 | 0.034 | 2.2 | |
| Nucleotide metabolism | CTP synthase (PyrG) | LSA1629 | 0.041 | 3.2 | |
| Xanthine phosphoribosyltransferase (Xpt) | LSA1557 | 0.021 | 1.6 | ||
| Cell wall/membrane | Carbohydrate metabolization | l-Lactate dehydrogenase (l-Ldh) | LSA1606 | 0.007 | 1.5 |
| Nucleotide metabolism | CTP synthase (PyrG) | LSA1629 | 0.003 | 2.5 | |
| Cell division | Cell shape-determining protein (MreB) | LSA0853 | 0.022 | 1.7 | |
| Surface exposed | Adaptation to atypical conditions | GTP-binding protein TypA | LSA1079 | 0.016 | 1.6 |
| Carbohydrate metabolism | Fructose-bisphosphate aldolase (Fba) | LSA1527 | 0.020 | 1.6 | |
| Cell division | Cell shape-determining protein (MreB) | LSA0853 | 0.039 | 2.2 |
See Table 3, footnote a.
Spot disappearance in the R mutant does not result from cis-acting mutations.
Since several spots, some being encoded by the same operon, strongly diminished or even became undetectable in the R clone, one could interpret this as the result of mutations in or upstream from the corresponding genes. Therefore, we amplified and sequenced those genes, with their promoter regions, from both L. sakei 23K and L. sakei 23K-J30RO2 and compared the DNA sequences. We chose operons and genes coding for proteins with the highest volume change according to Progenesys SameSpots image analysis: the operons LSA0162 to LSA0170 (from which the genes encoding spots corresponding to LSA0165, LSA0169, and LSA0170 disappeared in the mutant, with the other genes encoding proteins not being detectable in any strain), LSA0551-LSA0552, and LSA0462-LSA0463 and the two monocistronic genes LSA0104 and LSA1317. However, DNA sequences showed 100% identity between the parent and derivative strains, thus indicating that mutations suppressing the expression of those spots were located neither in their structural genes nor in their promoter regions.
DISCUSSION
We previously showed that colonization of L. sakei in the GIT of axenic mice induced the emergence of subpopulations, having the advantage to colonize the GIT and to partially displace the preexisting bacterial population (7). This confirmed that food is the preferred environment for L. sakei but suggested that mutations can be acquired for a better adaptation to the GIT in axenic mice. Here we show that mutants acquired a modified response to various stresses that can be encountered in the GIT and an increased growth ability on some carbon sources that are present in the GIT.
The appearance of phenotypic heterogeneity within an isogenic microbial population during transit through the GIT of mice has already been described for the Gram-negative E. coli MG1655 strain: within 10 days, small granular colonies reached more than 90% of the total population but could not completely displace the original population (17). This phenotype results from mutations in the E. coli EnvZ/OmpR system, a master regulator controlling more than 100 target genes. The authors proposed that mutations in regulatory genes promote a better adaptation and evolution than some improving a single function. Proteomic analysis performed with an R mutant issued from L. sakei 23K showed some variations, suggesting a modification in sugar catabolism and/or a pyruvate rerouting. Indeed, whereas l-lactate dehydrogenase (LSA1606), 6-phosphofructokinase (LSA1033), and 1,6-fructose-bisphosphate aldolase (LSA1527) are overexpressed, pyruvate oxidase (LSA1188), pyruvate kinase (LSA1032), and a subunit of the pyruvate dehydrogenase complex are underexpressed. Although no growth difference was observed between the parent strain and the R mutant on glucose and mannose, the mutant grew faster on ribose, galactose, fructose, and N-acetylglucosamine. This may lead to a better fitness due to more efficient use of alternative carbon sources that are present in the GIT, an environment poor in energy sources, particularly in glucose. These characteristics are similar to those reported for E. coli MG1655 mutants isolated from feces of streptomycin-treated mice, which showed improved growth on several sugars (27), some shown indeed to be utilized by E. coli in the GIT (1).
Proteomic analysis performed on an R clone suggested a modification of the stress status of the cells, which may correlate with modification of the survival rate after various stresses were applied. Four proteins involved in stress response (LSA0038, LSA0836, LSA0169, and LSA0170) were remarkably underrepresented in the proteome of the R clone; in contrast, the chaperones GroEL (LSA0359) and DnaK (LSA1236), the copper homeostasis protein CutC (LSA1440), and one GTP binding protein, TypA (LSA1079), all involved in stress response, were upregulated. The two paralogs LSA0169 and LSA0170 showed some similarity with the protein Gls24 of Enterococcus faecalis, and a gls24 mutant exhibited phenotypes and modification of protein expression partially similar to those of the L. sakei 23K-J30RO2 R mutant (16). Some proteins involved in transcription and regulation were also differently expressed in the mutant. The wild-type L. sakei 23K expresses the LSA0494 ribosomal protein four times more than the R clone. In contrast, L. sakei 23K-J30RO2 overexpresses RpsF (LSA0007) and RplJ (LSA1667). Those two ribosomal proteins are overproduced in L. sakei, E. faecalis, and Listeria monocytogenes after a high-pressure treatment (23). RpsF is also reported to be upregulated in B. subtilis after cold shock (25), while both RpsF and RplJ are downregulated in stationary phase in L. monocytogenes (15). We also noticed that the L. sakei R clone overproduces an ATP-dependent H+ transporter involved in the maintenance of the internal pH but was, however, less resistant than its parent after an external shock at pH 3.0. Cysteinyl tRNA synthase (LSA1681) is also upregulated in L. sakei 23K-J30RO2. The importance of sulfur amino acids in the mouse GIT colonization and in particular in the resistance to acid stress has already been reported. The luxS gene (absent from the L. sakei 23K genome), coding for the autoinducer-2 and involved in the metabolism of cysteine and methionine, has an essential role in the persistence of Lactobacillus reuteri 100-23 and Lactobacillus rhamnosus GG in the GIT (28, 34). In addition, Bifidobacterium longum 8809dpH, a mutant resistant to low pH, overexpresses proteins involved in the metabolism of sulfur amino acids compared to its parental strain (33). The increased amount of LSA1681 in the R mutant of L. sakei might thus contribute to its improved adaptation to the GIT. Several oxidoreductases (LSA1317, LSA0552, LSA0165, LSA0104, LSA0030, and LSA1712) involved in detoxification or oxidative stress protection were strongly downregulated in the R clone. This may explain the slightly higher sensitivity to H2O2 in the mutant, although observed in laboratory conditions. The modified expression of all these proteins suggests that the R mutant has acquired a different stress status than that of the parent strain.
Recently, the appearance of a heterogeneous population composed of microcolonies characterized by elongated cells and lack of septation after exposure to acid stress was described for Lactobacillus plantarum WCFS1 (22). As for L. plantarum WCFS1, the long cells or filaments of our R and S clones were lacking septa. In addition, we observed a modification of the membrane shape but only in the R mutant derived from LTH5590, suggesting that the factors affected in the various S and R clones are different. This is consistent with our previous observation of many different growth patterns in the collection of S and R clones (7). Interestingly, we noticed in our TEM observations the presence of transversal slanted black bands crossing filamentous cells located apparently at the surface of cells issued from R clones. Our proteomic analysis showed that the actin-like protein MreB, located at the surface of the cells, is upregulated in the R mutant. MreB is a protein involved in cell wall formation, cell elongation, and cell shape determination. MreB polymers can form dots and helical filamentous structures associated with the membranes of the cell (18, 24). Although further experiments would be required to demonstrate it, it is tempting to hypothesize that the helical structures observed with R mutants may be composed of MreB, which is overexpressed, and may consequently “modify” the cell shape but does not affect cell division. That could also explain the atypical morphology of colonies (rough) of the mutant and might possibly modify their adhesion properties, which are important for colonization of the GIT.
In eukaryotic cells, oxidative stress can disrupt the actin cytoskeleton by oxidative modification of the sulfhydryl group of its conserved Cys374 (11). Residues Cys113 and Cys324 of the MreB proteins are highly conserved in prokaryotes, and a role for the MreB protein in the response to oxidative stress was proposed for Vibrio parahaemolyticus by demonstrating how the configuration of the MreB cytoskeleton can change under stressing conditions (8). In particular, it was proposed that the oxidation of this protein could act as a neutralizer for reactive species generated when the cell is stressed. The overexpression of MreB detected in the L. sakei R strain might thus have a secondary effect as an alternative response toward oxidative stresses, compensating for the downregulation of several oxidoreductases.
It is also possible that transport of some molecules could be modified in the R clone, since five spots (LSA0705, LSA0706, LSA0938, LSA0616, and LSA1366) identified as components of ATP-dependent transporters for oligopeptides or osmoprotectants or for drug export were downregulated in L. sakei 23K-J30RO2. These results suggest the improved fitness, involving cell permeability, of the R mutant for transit through the GIT. Similarly, reduced permeability of the membranes has been reported for the E. coli MG1655 ompB mutants, naturally adapted to the GIT environment (17). As well, a decreased expression of genes encoding transporters has been observed during the transit of Lactobacillus johnsonii NCC533 in the jejunum and cecum intestine (13).
Sequencing of five DNA regions coding for proteins, the expression of which was strongly diminished in the R clone, did not reveal any mutation (7). We think that, as described for E. coli GIT-adapted mutants, only one or a few mutations having pleiotropic effects occurred. In addition, the modification of carbohydrate metabolism and the stress status of the cells may have secondary multiple effects, explaining the proteome differences we observed between the R strain and its parent.
Several methods were used to investigate the bacterial functions involved in GIT colonization. A proteomics-based analysis of E. coli cells collected directly from mice feces has been reported (1). Several transcriptomics analyses, aiming at the identification of bacterial genes expressed in the GIT of mice and using microarrays, in vivo expression technology (IVET), or resolvase IVET (R-IVET), have also been reported for various lactobacilli (4, 13, 14, 38, 39). Alternatively, our approach combining proteomic and phenotypic analyses of L. sakei mutants adapted to GIT colonization allowed us to define functions that are important for improving the fitness of this meat-borne bacterium in the GIT.
Supplementary Material
Acknowledgments
Fabrizio Chiaramonte is the recipient of a fellowship within the framework of the EC EST project LabHealth, Marie Curie contract MEST-2-CT-2004-514428.
We gratefully thank Christian Hertel for providing the strain L. sakei LTH5590, Christine Longin and Sophie Chat from the MIMA2 platform (Microscopie et Imagerie des Micro-organismes, Animaux et Aliments), INRA, Jouy en Josas (France), for TEM analysis, and Christian Bordat from Unité Nurélice, UR909, INRA, Jouy en Josas (France), for Cy5 Apotome microscopy analysis.
Footnotes
Published ahead of print on 5 March 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alpert, C., J. Scheel, W. Engst, G. Loh, and M. Blaut. 2009. Adaptation of protein expression by Escherichia coli in the gastrointestinal tract of gnotobiotic mice. Environ. Microbiol. 11:751-761. [DOI] [PubMed] [Google Scholar]
- 2.Berthier, F., M. Zagorec, M. Champomier-Vergès, S. D. Ehrlich, and F. Morel-Deville. 1996. Efficient transformation of Lactobacillus sake by electroporation. Microbiology 142:1273-1279. [DOI] [PubMed] [Google Scholar]
- 3.Bredholt, S., and Nesbakken, T. 1999. Protective cultures inhibit growth of Listeria monocytogenes and Escherichia coli O157.H7 in cooked, vacuum and gas-packaged meat. Int. J. Food Microbiol. 53:43-52. [DOI] [PubMed] [Google Scholar]
- 4.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaillou, S., M. C. Champomier-Vergès, M. Cornet, A. M. Crutz-Le Coq, A. M. Dudez, V. Martin, S. Beaufils, E. Darbon-Rongère, R. Bossy, V. Loux, and M. Zagorec. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23:1527-1533. [DOI] [PubMed] [Google Scholar]
- 6.Champomier, M. C., M. C. Montel, F. Grimont, and P. A. D. Grimont. 1987. Genomic identification of meat lactobacilli as Lactobacillus sake. Ann. Inst. Pasteur Microbiol. 138:751-758. [DOI] [PubMed] [Google Scholar]
- 7.Chiaramonte, F., S. Blugeon, S. Chaillou, P. Langella, and M. Zagorec. 2009. Behavior of the meat-borne bacterium Lactobacillus sakei during its transit through gastrointestinal tract of axenic and conventional mice. Appl. Environ. Microbiol. 75:4498-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu, S.-W., S.-Y. Chen, and H.-C. Wong. 2008. Localization and expression of MreB in Vibrio parahaemolyticus under different stresses. Appl. Environ. Microbiol. 74:7016-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocconcelli, P. S. 2007. Starter cultures: bacteria, p. 137-145. In F. Toldrà (ed.), Handbook of fermented meat and poultry. Blackwell Publishing, Oxford, United Kingdom.
- 10.Dal Bello, F., J. Walter, W. P. Hammes, and C. Hertel. 2003. Increased complexity of the species composition of lactic acid bacteria in human feces revealed by alternative incubation condition. Microb. Ecol. 45:455-463. [DOI] [PubMed] [Google Scholar]
- 11.Dalle-Donne, I., R. Rossi, A. Milzani, P. Di Simplicio, and R. Colombo. 2001. The actine cytoskeleton response to oxidants: from small heat shock protein phosphorylation to changes in the redox state of actin itself. Free Radic. Biol. Med. 31:1624-1632. [DOI] [PubMed] [Google Scholar]
- 12.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 13.Denou, E., B. Berger, C. Barretto, J. M. Panoff, F. Arrigoni, and H. Brussow. 2007. Gene expression of commensal Lactobacillus johnsonii strain NCC553 during in vitro growth and in the murine gut. J. Bacteriol. 189:8109-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vos, W. M., P. A. Bron, and M. Kleerebezem. 2004. Post-genomics of lactic acid bacteria and other food-grade bacteria to discover gut functionality. Curr. Opin. Biotechnol. 15:86-93. [DOI] [PubMed] [Google Scholar]
- 15.Folio, P., P. Chavant, I. Chafsey, A. Belkorchia, C. Chambon, and M. Hébraud. 2004. Two-dimensional electrophoresis database of Listeria monocytogenes EGDe proteome and proteomic analysis of mid-log and stationary growth phase cells. Proteomics 4:3187-3201. [DOI] [PubMed] [Google Scholar]
- 16.Giard, J. C., A. Rincé, H. Capiaux, Y. Auffray, and A. Hartke. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182:4512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraud, A., S. Arous, V. Gaboriau-Routhiau, M. De Paepe, J. C. Bambou, S. Rakotobe, A. B. Lindner, F. Taddei, and N. Cerf-Bensussan. 2008. Dissecting the genetic components of adaptation of Escherichia coli to the mouse gut. PLoS Genet. 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graumann, P. L. 2007. Cytoskeletal elements in bacteria. Annu. Rev. Microbiol. 61:589-618. [DOI] [PubMed] [Google Scholar]
- 19.Guillot, A., C. Gitton, P. Anglade, and M. Y. Mistou. 2003. Proteomic analysis of Lactococcus lactis, a lactic acid bacterium. Proteomics 3:337-354. [DOI] [PubMed] [Google Scholar]
- 20.Heilig, H. G., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hüfner, E., T. Markieton, S. Chaillou, A.-M. Crutz-Le Coq, M. Zagorec, and C. Hertel. 2007. Identification of Lactobacillus sakei genes induced during meat fermentation and their role in survival and growth. Appl. Environ. Microbiol. 73:2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingham, C. J., M. Beerthuyzen, and J. van Hylckama Vlieg. 2008. Population heterogeneity of Lactobacillus plantarum WCFS1 microcolonies in response to and recovery from acid stress. Appl. Environ. Microbiol. 74:7750-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jofré, A., M. Champomier-Vergès, P. Anglade, F. Baraige, B. Martín, M. Garriga, M. Zagorec, and T. Aymerich. 2007. Protein synthesis in lactic acid and pathogenic bacteria during recovery from a high pressure treatment. Res. Microbiol. 158:512-520. [DOI] [PubMed] [Google Scholar]
- 24.Jones, L. J., R. Carballido-López, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 25.Kaan, T., G. Homuth, U. Mäder, J. Bandow, and T. Schweder. 2002. Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology 148:3441-3455. [DOI] [PubMed] [Google Scholar]
- 26.Lauret, R., F. Morel-Deville, F. Berthier, M. C. Champomier-Vergès, P. Postma, S. D. Ehrlich, and M. Zagorec. 1996. Carbohydrate utilization in Lactobacillus sake. Appl. Environ. Microbiol. 62:1922-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leatham, M. P., S. J. Stevenson, E. J. Gauger, K. A. Krogfelt, J. J. Lins, T. L. Haddock, S. M. Autieri, T. Conway, and P. S. Cohen. 2005. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 73:8039-8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lebeer, S., I. J. J. Claes, T. L. A. Verhoeven, C. Shen, I. Lambrichts, J. L. Ceuppens, J. Vanderleyden, and S. C. J. de Keersmaecker. 2008. Impact of luxS and suppressor mutations on the gastrointestinal transit of Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 74:4711-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marceau, A., M. Zagorec, S. Chaillou, T. Méra, and M. C. Champomier-Vergès. 2004. Evidence for involvement of at least six proteins in adaptation of Lactobacillus sakei to cold temperatures and addition of NaCl. Appl. Environ. Microbiol. 70:7260-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Najjari, A., I. Ouzari, A. Boudabous, and M. Zagorec. 2008. Method for reliable isolation of a collection of Lactobacillus sakei strains originating from Tunisian seafood and meat products. Int. J. Food Microbiol. 121:342-351. [DOI] [PubMed] [Google Scholar]
- 31.Rajilic-Stojanovic, M., H. Smidt, and W. M. de Vos. 2007. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9:2125-2136. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez, B., M. C. Champomier-Vergès, P. Anglade, F. Baraige, C. G. de los Reyes-Gavilan, A. Margolles, and M. Zagorec. 2005. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187:5799-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez, B., M. C. Champomier-Vergès, M. Del Carmen Collado, P. Anglade, F. Barraige, Y. Sanz, C. G. de los Reyes-Gavilan, A. Margolles, and M. Zagorec. 2007. Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl. Environ. Microbiol. 73:6450-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tannock, G. W., S. Ghazally, J. Walter, D. Loach, H. Brooks, G. Cook, M. Surette, C. Simmers, P. Bremer, F. Dal Bello, and C. Hertel. 2005. Ecological behavior of Lactobacillus reuteri 100-23 is affected by mutation of the luxS gene. Appl. Environ. Microbiol. 71:8419-8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaughan, E. E., M. C. de Vries, E. G. Zoetendal, K. Ben-Amor, A. D. Akkermans, and W. M. de Vos. 2002. The intestinal LABs. Antonie Van Leeuwenhoek 82:341-352. [PubMed] [Google Scholar]
- 36.Vermeiren, L., F. Devlieghere, and J. Debevere. 2006. Co-culture experiments demonstrate the usefulness of Lactobacillus sakei 10A to prolong the shelf-life of a model cooked ham. Int. J. Food Microbiol. 108:68-77. [DOI] [PubMed] [Google Scholar]
- 37.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter, J., N. C. K. Heng, W. P. Hammes, D. M. Loach, G. W. Tannock, and C. Hertel. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 69:2044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter, J., P. Chagnaud, G. W. Tannock, D. M. Loach, F. Dal Bello, H. F. Jenkinson, W. P. Hammes, and C. Hertel. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71:979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.