Abstract
Listeria monocytogenes utilizes internalin A (InlA; encoded by inlA) to cross the intestinal barrier to establish a systemic infection. Multiple naturally occurring mutations leading to a premature stop codon (PMSC) in inlA have been reported worldwide, and these mutations are causally associated with attenuated virulence. Five inlA PMSC mutations recently discovered among isolates from France and the United States were included as additional markers in our previously described inlA single-nucleotide polymorphism (SNP) genotyping assay. This assay was used to screen >1,000 L. monocytogenes isolates from ready-to-eat (RTE) foods (n = 502) and human listeriosis cases (n = 507) for 18 inlA PMSC mutations. A significantly (P < 0.0001) greater proportion of RTE food isolates (45.0%) carried a PMSC mutation in inlA compared to human clinical isolates (5.1%). The proportion of L. monocytogenes with or without PMSC mutations in inlA was similar among isolates from different RTE food categories except for deli meats, which included a marginally higher proportion (P = 0.12) of isolates carrying a PMSC in inlA. We also analyzed the distribution of epidemic clone (EC) strains, which have been linked to the majority of listeriosis outbreaks worldwide and are overrepresented among sporadic cases in the United States. We observed a significant (P < 0.05) overrepresentation of EC strains in deli and seafood salads and a significant (P < 0.05) underrepresentation of EC strains in smoked seafood. These results provide important data to predict the human health risk of exposure to L. monocytogenes strains that differ in pathogenic potential through consumption of contaminated RTE foods.
Listeria monocytogenes is the etiological agent of listeriosis, a potentially life-threatening food-borne disease that primarily affects individuals with underlying immune-compromising circumstances. Listeriosis is associated with an exceptionally high hospitalization rate of 85 to 90% and 20 to 30% of cases are fatal. L. monocytogenes infections account for nearly 30% of all fatalities attributed to known food-borne pathogens each year in the United States (18). Although L. monocytogenes is readily inactivated by cooking and pasteurization, subsequent cross-contamination of ready-to-eat (RTE) food products exposed to the food processing plant environment following a lethality treatment represents the route through which RTE foods become contaminated (31). L. monocytogenes is a hardy pathogen; it is relatively resistant to acid, is able to grow in high salt concentrations, and is capable of growing at refrigeration temperatures (2, 4). Because L. monocytogenes tolerates intrinsic and extrinsic properties of food typically used to control the growth of pathogens, consumption of RTE foods contaminated by L. monocytogenes represents a significant health risk for immunocompromised individuals. The combined food-borne route of L. monocytogenes transmission and severity of listeriosis prompted establishment of strict regulations regarding the presence of L. monocytogenes in finished RTE foods in many countries, including a “zero tolerance” policy in the United States (30).
Combined epidemiology and subtyping studies support that L. monocytogenes strains differ in their relative likelihood and ability to cause human listeriosis. More than 90% of listeriosis cases have been attributed to three (i.e., 1/2a, 1/2b, and 4b) of the 13 serotypes that L. monocytogenes isolates can be grouped into (17). DNA band-based and sequence-based molecular subtyping approaches showed that L. monocytogenes isolates can be classified into two major genetic lineages (termed lineages I and II), which include the majority of L. monocytogenes isolates, along with two minor lineages (termed lineages III and IV) that correlate with serotype classifications (19, 21, 26, 27, 37, 36, 38). L. monocytogenes isolates from human listeriosis cases are overrepresented among genetic lineage I, despite an apparent increased exposure to lineage II isolates via contaminated foods (9). In addition, a few highly clonal lineage I serotype 4b strains, termed epidemic clones (ECs) I, Ia, and II, have been linked to the majority of listeriosis outbreaks worldwide and are overrepresented among sporadic listeriosis cases in the United States (9, 14).
Although genetic markers associated with enhanced virulence have not been identified, previous studies identified multiple distinct mutations leading to a premature stop codon (PMSC) in the key L. monocytogenes virulence gene, inlA (5, 10, 11, 12, 22-25, 27, 29, 34, 35). The virulence factor internalin A (InlA; encoded by inlA) facilitates the uptake of L. monocytogenes by nonprofessional phagocytic cells that express the human isoform of E-cadherin, and it plays a critical role in crossing the intestinal barrier during the initial stages of an infection (16). L. monocytogenes isolates that carry a PMSC in inlA produce a truncated form of InlA that is secreted rather than anchored to the bacterial cell wall and demonstrate attenuated invasion of Caco-2 human intestinal epithelial cells (5, 22-25, 29, 34). In a recent study, our group characterized a set of paired isogenic mutants with or without a PMSC in inlA using an intragastric guinea pig infection model to show that these mutations appear to be fully responsible for attenuated mammalian virulence (20).
While previous studies investigated the distribution of selected inlA PMSC mutations among specific molecular subtypes known to be associated with those mutations (3, 22, 34, 36), there is a clear need to determine the prevalence of all known PMSC mutations in inlA among a large representative collection of L. monocytogenes isolates from RTE foods and human clinical cases. The objectives of the present study include (i) modifying our existing inlA SNP genotyping assay (34) to incorporate five recently described SNPs leading to a PMSC inlA; (ii) employing this assay to screen a large representative collection of >1,000 L. monocytogenes isolates from RTE foods and human clinical isolates for 18 inlA PMSC mutations; and (iii) analyzing the distribution of strains carrying an inlA PMSC and EC strains among isolate sources, molecular subtypes, and different RTE food product categories.
MATERIALS AND METHODS
Bacterial isolates for SNP genotyping experiments.
We focused on characterizing the large representative set of human clinical and RTE food L. monocytogenes isolates described by Gray et al. (9). We queried the PathogenTracker database to identify isolates included in this previous study and to obtain molecular epidemiology data (i.e., source, EcoRI ribotype, and RTE food category) for these isolates. The PathogenTracker query returned 1,009 L. monocytogenes isolates, including 502 isolates from RTE foods and 507 isolates from human clinical cases (see supplemental Table 1 [http://ansci.colostate.edu/content/view/88/56/]). The 502 food isolates were obtained through a survey of more than 30,000 samples from selected RTE food product categories (i.e., bagged salads, fresh soft cheeses, soft ripened cheeses, smoked seafood, seafood, and deli salads and meats) collected in Maryland and California during 2000 and 2001 (8). The collection of 507 human clinical isolates included 42 isolates from listeriosis cases reported in Maryland and California in 2000 and 2001, as well as an additional 465 isolates from listeriosis cases in the United States and North America from 1997 to 2001.
TABLE 1.
Description of extension primers incorporated into the SNP genotyping assay developed to differentiate L. monocytogenes isolates with or without a mutation leading to a premature stop codon in inlA
| Primer | Primer concn (μM) | Targeted annealing strand | Multiplex reaction | PMSC mutation type | Size (bp)a | Mutant allelic typeb | Wild-type allelic type | SNP genotyping extension primer sequence (5′-3′)c |
|---|---|---|---|---|---|---|---|---|
| US-A | 0.4 | Antisense | 1 | 3 | 31-32 | G (blue) | C (black) | GCA AAT GAY ATT ACG CTG TA |
| US-B | 0.2 | Antisense | 1 | 4 | 34-35 | C (black) | A (green) | [GCC] GGA GTG TAT ATA GTG AGA ARA AA |
| US-C | 0.2 | Antisense | 1 | 1 | 39-40 | A (green) | T (red) | [CTG CCT G]GG TTA TAC TTT CAA AGG CTG GTA |
| US-D | 0.2 | Antisense | 1 | 2 | 44-45 | T (red) | C (black) | [GAC CGA CCA AGC TCG] CAA CGA CTC AAG CAG TAG ACT AT |
| US-E | 0.2 | Antisense | 1 | 5 | 49-51 | T (red) | C (black) | [GAG ATC GGC AGA GCG GCG AGT A]GC TTT CAG GTT TAA CTA GTC TA |
| US-F | 2.0 | Antisense | 1 | 6 | 54-55 | T (red) | C (black) | [CGT ACA GAC TGG CGA TCG GCA TGC TAC T]AA CAT TTA GYG GAA CYG TGA CR |
| US-G | 0.2 | Sense | 1 | 7 | 62-63 | A (green) | G (blue) | [GGG GGG GTC GTC CTG GCT CAA TCT CGG CCT GGC TAG CT]C TGT GTA GCT GTT AAT ACT AAA TT |
| FR-A | 0.2 | Antisense | 2 | 8 | 30-31 | A (green) | G (blue) | ACA CAG AGC CTG ATA TAA CAT G |
| FR-B | 0.2 | Antisense | 2 | 9 | 33-34 | A (green) | G (blue) | [TTG AT]G CAA AGA AAC AAC CAA AGA AGT G |
| FR-C | 0.2 | Antisense | 2 | 10 | 39-40 | T (red) | A (green) | [AGC AGC AGC TAC G]AA WCA ACG ACT CAA RCA GTA G |
| FR-D | 0.2 | Antisense | 2 | 11 | 44-45 | A (green) | G (blue) | [CCA GCG AAT CAT GCC A]AC GAA AAA ACA GAT GGG AAA AAA T |
| FR-E | 1.0 | Antisense | 2 | 12 | 50-51 | T (red) | A (green) | [CTG CGC ACG ATC GAT GGC TCG ATC T]AA ACM GGY GGA ACT AAM TGG R |
| JP-A | 0.2 | Antisense | 2 | 13 | 57-58 | T (red) | A (green) | [GCA TCC GTC CAC TTC GAT CAA TCG CGC CGC TC]C TGA ACC AGC TAA GCC YGT A |
| FR-F | 0.2 | Antisense | 2 | 14 | 60-61 | T (red) | A (green) or C (black) | [ACT GCA ACT TGC CTA TTA CCC CGT CCA ACT GTC TCC] TTR TTG GKT GGT TTG ATG CC |
| USDA-A | 0.3 | Antisense | 3 | 15 | 32-34 | T (red) | C (black) | GTC TCR CAA ACW GAT YTA GAC |
| USDA-B | 0.3 | Antisense | 3 | 16 | 37-39 | T (red) | G (blue) | [GGG CCG G]AA YCT RAC AAA TTT AAA TCG GCT A |
| USDA-C | 0.3 | Antisense | 3 | 17 | 41-43 | A (green) | T (red) | [GTG CGC CC]C YCC ACT TGG RAT TTT AAC AAA TT |
| USDA-D | 0.3 | Antisense | 3 | 18 | 44-46 | C (black) | T (red) | [ATT GAT TCC TT]G TTY TTA ACM AAT ATT AAT TGG CTT |
The size of the electrophoresed extension product will differ from the expected size based on primer length due to the influence of the weight of each particular fluorescent dye on the mobility shift of the DNA fragments.
The colors (indicated in parentheses) represent which fluorescent dye is incorporated into each dideoxynucleotide triphosphate.
A noncomplementary tail (in brackets and boldface type) was added to the 5′ ends of extension primers to adjust the length to differentiate the sizes of each primer by four to six nucleotides to facilitate the detection of multiple SNPs in a single reaction. Degenerate nucleotides were incorporated into some extension primer sequences to accommodate polymorphic sites in targeted annealing regions, where K = G + C, M = A + C, R = A + G, W = A + T, and Y = C + T.
Modification of a previously developed SNP genotyping assay to detect recently identified PMSC mutations in inlA.
In our previous study (34), we developed a SNP genotyping assay using the SNaPshot multiplex kit (Applied Biosystems [ABI], Foster City, CA). Since the publication of our previous study, five additional PMSC mutations in inlA have been identified. Specifically, a research group from France recently described a novel mutation leading to a PMSC in a single L. monocytogenes isolate (27), and this SNP was incorporated as the seventh marker into reaction 2 of our existing assay. In addition, a third multiplex reaction (reaction 3) was developed in the present study to incorporate four additional inlA PMSC mutations observed among L. monocytogenes isolates from the United States (35). Extension primers used to interrogate the allelic type for these five additional inlA SNPs were designed as detailed previously (34). DNA samples for isolates newly identified to carry each of novel inlA PMSC mutation were obtained to confirm the presence of these mutations and the ability of the newly designed extension primers to differentiate wild-type and mutant inlA allelic types. Primer concentrations were adjusted to optimize multiplex detection of SNPs from each single base extension in reactions 2 and 3 (Table 1) .
inlA SNP genotyping.
SNP genotyping by multiplex reactions 1, 2, and 3 was performed for the set of L. monocytogenes isolates described above, with the exception of some isolates belonging to ribotype DUP-1062A, since we previously showed that all isolates belonging to ribotype DUP-1062A isolates carry inlA PMSC type 3 (22). The full-length inlA was amplified by PCR using primers and conditions described in supplemental Table S2 (http://ansci.colostate.edu/content/view/88/56/). A 96-well plate format and colorless GoTaq PCR master mix (Promega, Madison, WI) were used to facilitate high-throughput capability. Resultant inlA amplicons were purified by treatment with shrimp alkaline phosphatase (Fermentas, Glen Burnie, MD) and exonuclease I (Fermentas), and purified products were used as a template for single-base-pair extension reactions. Single-base-pair extension PCRs were performed by using the SNaPshot multiplex kit according to the manufacturer's instructions (Applied Biosystems). Single-base-pair extension products were purified and prepared for capillary electrophoresis analyses on an ABI Prism 3100 genetic analyzer as detailed previously (34). The allelic type of each targeted SNP was determined by using GeneScan v.3.7 software (ABI), where specific bins and panels were constructed to distinguish wild-type and mutant allelic types for each marker in all three multiplex reactions.
inlA sequencing.
Targeted inlA sequencing was performed to confirm inlA allelic types for (i) L. monocytogenes isolates that showed one or more ambiguous peaks from SNP typing and (ii) isolates belonging to a molecular subtype (EcoRI ribotype) newly associated with a PMSC in inlA by SNP genotyping. PCR primers and reaction conditions used to amplify the full-length inlA for SNP typing described above were used to amplify inlA for DNA sequencing. PCR products were purified by using a PCR purification kit (Qiagen, Valencia, CA), and DNA concentrations of the purified PCR product were determined by using a spectrophotometer (ND-1000; NanoDrop Technologies). Primers used to amplify inlA along with four internal sequencing primers (supplemental Table S2 [http://ansci.colostate.edu/content/view/88/56/]) were used to sequence targeted regions of inlA or the full-length inlA as necessary. DNA sequencing was performed by Eton Bioscience, Inc. (San Diego, CA). Nucleotide sequences were assembled and aligned with Seqman and Megalign software (DNAStar; Lasergene, Madison, WI), respectively. Sequences are available through the PathogenTracker database.
Statistical analysis.
Categorical data analyses were performed to describe the distribution of L. monocytogenes isolates carrying a PMSC in inlA among (i) RTE foods compared to human clinical cases overall; (ii) RTE foods and human clinical cases belonging to common ribotypes containing at least 10 isolates; and (iii) RTE food categories, including bagged salads, cheeses (the two cheese categories reported in Gray et al. [9] were collapsed into a single category due to small sample sizes), smoked seafood, seafood and deli salads, and deli meats. In addition, categorical data analyses were performed to describe the distribution of L. monocytogenes isolates belonging to an EC strain among different RTE food product categories. Isolates were identified as belonging to an EC strain category based on ribotype data. The EC category included isolates belonging to ribotypes DUP-1038B (EC I), DUP-1042B (EC Ia), DUP-1044A (EC II), and DUP-1053A (EC III) (6, 14). Comparisons were made by using either a chi-square test of independence or the Fisher exact test (as appropriate) by running the frequency procedure as implemented using Statistical Analyses Software (SAS, Cary, NC). One comparison where P = 0.12 was considered to be marginally significant, and all other comparisons where P < 0.05 were considered to be statistically significant.
RESULTS AND DISCUSSION
A modified version of our previously described inlA SNP genotyping assay (34) was used to screen a large representative set of >1,000 L. monocytogenes isolates from RTE foods and human clinical cases for 18 inlA PMSC mutations identified worldwide to date. Categorical data analyses were performed to compare the frequency of inlA PMSC mutations and EC strains among different sources of isolation (RTE food versus human clinical and across different categories of RTE foods) and molecular subtypes. The results from the present study support that (i) L. monocytogenes isolates from RTE foods and human listeriosis cases in the United States carry multiple distinct mutations leading to a PMSC in inlA, and some of these mutations appear to have been accumulated at the population level; (ii) a significant proportion of L. monocytogenes isolated from RTE foods carry a PMSC mutation in inlA, and isolates carrying these mutations only cause human disease on rare occasions; and (iii) the heterogeneous distribution of L. monocytogenes isolates carrying a PMSC mutation in inlA and EC strains among different RTE food categories may play a role in the food attribution of listeriosis.
Multiple distinct mutations have been accumulated in inlA, including a few common mutations and several other rare mutations.
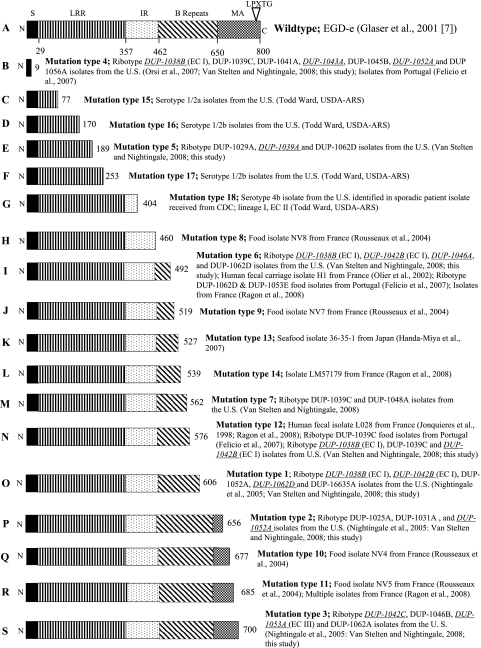
We previously showed that PMSC mutations in inlA are causally associated with attenuated mammalian virulence (20) and developed a multiplex SNP genotyping assay to detect 13 PMSC mutations in inlA (PMSC types 1 to 13) (34). Five additional mutations leading to a PMSC in inlA have been identified since the publication of our previous study (34) and these mutations (termed inlA PMSC types 14 to 18) were incorporated as markers in our inlA SNP genotyping assay. More specifically, PMSC type 14 was recently identified as a novel mutation leading to a PMSC in inlA in a single L. monocytogenes isolate by a research group from France (27; Fig. 1). The mutation leading to inlA PMSC mutation type 14 was incorporated as the seventh inlA PMSC marker into reaction 2 (Table 1). A recent study by Ward et al. (35) described three additional inlA PMSC mutations among L. monocytogenes isolates from the United States (PMSC types 15 to 17; Fig. 1). Described for the first time in the present study, a frameshift mutation where a thymidine was deleted at nucleotide 1165 resulted in a PMSC downstream at InlA codon 404 (PMSC type 18; Fig. 1). As a result, a third multiplex SNP genotyping reaction (reaction 3) was developed to include these four additional markers (inlA PMSC mutation types 15 to 18; Table 1).
FIG. 1.
Full-length InlA (A) and location of premature stop codons in InlA previously identified by research groups in France (H, I, J, L, N, Q, and R), Japan (K), Portugal (B, I, and N), and the United States (B, C, D, E, F, G, I, M, N, O, P, and S). Full-length InlA (A) represents the sequence for L. monocytogenes strain EGD-e. Abbreviations: N, N-terminal end; S, signal sequence; LRR, leucine-rich repeat; IR, intergenic repeat; MA, membrane anchor; C, C-terminal end. Numbers below the EGD-e full-length InlA represent amino acid positions for the beginning of each functional domain. Numbers at the right end for lines B to N represent the amino acid position of each respective premature stop codon in InlA. EcoRI ribotypes newly associated with each InlA PMSC mutation are denoted by italicized and underlined type.
Only a few PMSC mutations in inlA (i.e., PMSC mutation types 1, 3, and 4) appear to have been accumulated at the population level among L. monocytogenes isolates from the United States. Specifically, SNP genotyping revealed that inlA PMSC mutation types 1, 3, and 4 accounted for >90% of the inlA PMSC mutations observed among the isolate set characterized in the present study (Table 2). The mutation leading to PMSC type 3 was the most common inlA PMSC mutation among the isolate set characterized in the present study (Table 2). The 5′ frameshift mutation in a homopolymeric tract of adenine residues (PMSC type 4) was the second most common mutation leading to a PMSC in inlA (Table 2). This homopolymeric tract may serve as a mutational hotspot, since DNA polymerase slippage is more likely to occur in these regions (25). Both inlA PMSC mutation types 3 and 4 were previously described among L. monocytogenes isolates belonging to ribotypes within genetic lineage II (22, 34), providing a possible explanation for the underrepresentation of lineage II isolates among human listeriosis cases. Interestingly, inlA PMSC type 1, which occurs in L. monocytogenes isolates belonging to genetic lineage I (22), also appears to have been accumulated at the population level, suggesting that some lineage I strains may be adapted to survive in the environment.
TABLE 2.
Association of inlA PMSC mutation type with L. monocytogenes isolate origin (human or food)
| inlA PMSC mutation type | No. of isolates with inlA PMSC mutations (% prevalence) among isolates obtained from: |
Total no. of isolates with inlA PMSC mutation types | |
|---|---|---|---|
| Humans | Food | ||
| 1 | 7 (19.4) | 28 (80.0) | 35 |
| 2 | 0 (0) | 3 (100) | 3 |
| 3 | 11 (6.8) | 151 (93.2) | 162 |
| 4 | 4 (10.8) | 33 (89.2) | 37 |
| 5 | 1 (33.3) | 2 (66.6) | 3 |
| 6 | 1 (12.5) | 7 (87.5) | 8 |
| 7 | 0 (0) | 0 (0) | 0 |
| 8 | 0 (0) | 0 (0) | 0 |
| 9 | 0 (0) | 0 (0) | 0 |
| 10 | 0 (0) | 0 (0) | 0 |
| 11 | 0 (0) | 0 (0) | 0 |
| 12 | 2 (50) | 2 (50) | 4 |
| 13 | 0 (0) | 0 (0) | 0 |
| 14 | 0 (0) | 0 (0) | 0 |
| 15 | 0 (0) | 0 (0) | 0 |
| 16 | 0 (0) | 0 (0) | 0 |
| 17 | 0 (0) | 0 (0) | 0 |
| 18 | 0 (0) | 0 (0) | 0 |
| Total | 26 (5.1) | 226 (45.0) | 252 |
On the other hand, the majority of the 18 inlA PMSC mutations, including some inlA PMSC mutations previously identified in isolates from the United States, were observed in a single or few (<10) isolates characterized in the present study (Table 2). It should be noted, however, that previous studies showed that for the most part inlA PMSC mutations occur almost exclusively in isolates from each respective country where those mutations were originally described, with the exception of PMSC types 6 and 12 (5, 10-12, 22-25, 27, 29, 34-36). In fact, a previous study by a French research group showed that a few specific inlA PMSC mutation types were common among L. monocytogenes isolates from France. This 2004 study completed by Jacquet et al. (11) showed 13.8% (62 of 448) of isolates from human listeriosis cases and foods carried three PMSC mutation types (designated InlA2, InlA3, and InlA4 by Jacquet et al. [11]), where InlA2 and InlA3 were accumulated at the population level within France but are rare or have never been detected among L. monocytogenes isolates from the United States (22, 34). In combination with previous studies, our findings contribute to an emerging body of evidence that PMSC mutations in inlA appear to have arisen independently and that some mutations have become accumulated at the population level (3, 5, 10-12, 22-25, 27, 29, 34-36).
L. monocytogenes isolates carrying a PMSC in inlA are commonly isolated from RTE foods but only associated with human listeriosis on rare occasions.
Each inlA PMSC mutation type previously identified among L. monocytogenes isolates from the United States (i.e., PMSC types 1 to 7) was detected among the set of isolates characterized in the current study, with the exception of inlA PMSC 7 (Table 2). In our previous study, inlA PMSC mutation type 7 was carried exclusively by L. monocytogenes isolates from raw Norwegian salmon samples obtained from the same seafood processing plant in New York State during 1998 through 2000 (34). Consistent with the findings of previous studies (11, 22, 34), results from inlA SNP genotyping of a large representative set of L. monocytogenes isolates showed that inlA PMSC mutations were significantly (P < 0.0001) more common among L. monocytogenes isolated from RTE foods than among isolates from human listeriosis patients overall (Table 2). More specifically, 26 of 507 (5.1%) human clinical isolates carried a PMSC mutation in inlA, whereas 226 of 502 (45.0%) isolates from RTE foods carried a PMSC mutation in inlA (Table 2). Although humans are frequently exposed to L. monocytogenes isolates carrying virulence-attenuating PMSC mutations in inlA through consumption of contaminated RTE foods, isolates carrying these mutations only cause disease on very rare occasions. Another recent study by our group provided some initial evidence that exposure to a L. monocytogenes strain carrying a PMSC in inlA may confer protection against a subsequent challenge by a fully virulent strain, and additional work is needed to quantify the effect of the presence of these strains in the food supply on maintained population immunity against listeriosis (20). On the other hand, EC strains, which have been associated with the majority of listeriosis outbreaks worldwide and are overrepresented among sporadic listeriosis cases in the United States, were significantly (P < 0.05) underrepresented among L. monocytogenes isolates from RTE foods since these strains only represent 8% of L. monocytogenes isolated from RTE foods, suggesting that these strains have enhanced pathogenic potential (6, 9, 14).
We identified here inlA PMSC mutations that were novel to ribotypes previously shown to include L. monocytogenes isolates with other inlA PMSC mutation types, and we also identified inlA PMSC mutations in ribotypes that had not previously been associated with such a mutation (Fig. 1). To our knowledge, this is the first report wherein inlA PMSC mutations have been described among serotype 4b L. monocytogenes isolates representing EC I (DUP-1038B), EC Ia (DUP-1042B), and EC II (multilocus genotyping [MLGT] value = 1.8; see references 35 and 36) strains, along with serotype 1/2a EC III (DUP-1053A) strains. Full inlA sequencing was performed to confirm the presence of an inlA PMSC among isolates belonging to ribotypes that were not previously associated with an inlA PMSC mutation. Since previous studies showed that inlA evolved through a considerable number of recombination events, the presence of mutations leading to a PMSC in inlA among a small number of isolates representing EC strains is not surprising (22, 25). The frameshift mutation in a 5′ homopolymeric run of adenine residues leading to inlA PMSC mutation type 4 (25) was identified in L. monocytogenes isolates belonging to seven different ribotypes across both major genetic lineages and among isolates from two different countries (Fig. 1).
Chi-square tests of independence indicated that inlA PMSC mutations are not uniformly distributed among molecular subtypes (EcoRI ribotypes), where these mutations were significantly (P < 0.05) more common overall among isolates representing some ribotypes (i.e., DUP-1039C, DUP-1052A, and DUP-1062A) and significantly (P < 0.05) less common among isolates belonging to other ribotypes including, DUP-1038B (EC I), DUP-1039B, DUP-1042B (EC Ia), DUP-1043A, and DUP-1053A (EC III) (Table 3). Our results show that inlA PMSC mutations are significantly underrepresented among L. monocytogenes EC strains, a finding consistent with the common association between these strains and listeriosis epidemics and sporadic cases (6, 9, 14). Categorical data analyses were also performed to describe the distribution of inlA PMSC mutations among different isolate sources (i.e., human clinical and RTE food isolates) for isolates representing each common ribotype. The results showed that inlA PMSC mutations were significantly overrepresented among L. monocytogenes isolates from RTE foods belonging to ribotypes DUP-1038B, DUP-1039C, DUP-1042B, and DUP-1045B (Table 3). In addition, all human clinical and RTE food isolates belonging to ribotype DUP-1062A carried inlA PMSC mutation type 3 (Table 3) (22). Although inlA PMSC mutations are present in a small number of EC strains, EC strains carrying a PMSC in inlA were more commonly isolated from RTE foods and only a single listeriosis case was linked to an EC I strain carrying a PMSC inlA (Table 3). All ribotypes previously identified in the study by Gray et al. (9) to be overrepresented among RTE food isolates compared to human clinical isolates were shown to include at least one isolate carrying a PMSC in inlA in the present study, with the exception of ribotype DUP-1044E (Table 3). Gray et al. (9) described the exclusive isolation of ribotype DUP-1044E isolates from blue-veined and mold-ripened cheese produced by the same facility. This observation is consistent with other previous studies supporting that each food processing plant appears to be characterized by a unique Listeria ecology (13, 15).
TABLE 3.
Association of inlA PMSC mutations with origin (human or food) and L. monocytogenes ribotype
| Ribotypea | EC type | No. of inlA PMSC mutations/no. of isolates screened (% prevalence) among isolates obtained from: |
||
|---|---|---|---|---|
| Humans | Foodb | Totalc | ||
| DUP-1030A# | 0/8 (0) | 0/8 (0) | 0/16 (0) | |
| DUP-1030B (H)# | 0/9 (0) | 0/0 (0) | 0/9 (0) | |
| DUP-1038B (H) | EC I | 1/63 (1.5) | 5/15 (33.3)*** | 6/78 (7.7)** |
| DUP-1039A (H) | 1/31 (3.2) | 0/12 (0) | 1/43 (2.3)** | |
| DUP-1039B (H)# | 0/43 (0) | 0/18 (0) | 0/61 (0) | |
| DUP-1039C | 5/26 (18.5) | 17/35 (48.6)** | 22/61 (36.1)* | |
| DUP-1042A# | 0/16 (0) | 0/11 (0) | 0/27 (0) | |
| DUP-1042B (H) | EC Ia | 0/72 (0) | 3/18 (16.6)** | 3/90 (3.3)*** |
| DUP-1042C (F)# | 0/0 (0) | 1/14 (7.1) | 1/14 (7.1) | |
| DUP-1043A (F) | 0/12 (0) | 2/30 (6.6) | 2/42 (4.7)** | |
| DUP-1044A (H)# | EC II | 0/28 (0) | 0/11 (0) | 0/39 (0) |
| DUP-1044B (H)# | 0/19 (0) | 0/1 (0) | 0/20 (0) | |
| DUP-1044E (F)# | 0/0 (0) | 0/10 (0) | 0/10 (0) | |
| DUP-1045B | 0/10 (0) | 9/14 (64.3)** | 9/24 (37.5) | |
| DUP-1052A (F) | 6/24 (29.2) | 23/60 (38.3) | 29/84 (34.5)* | |
| DUP-1053A (H) | EC III | 0/41 (0) | 1/24 (4.2) | 1/65 (1.5)*** |
| DUP-1062A (F)# | 11/11 (100) | 149/149 (100) | 160/160 (100)*** | |
| DUP-1062D (F) | 0/1 (0) | 5/28 (17.9) | 5/29 (17.2) | |
| Other (uncommon ribotypes) | 2/93 (2.2) | 11/44 (25)*** | 13/137 (9.5) | |
| Total | 26/507 (5.1) | 226/502 (45.0)*** | 252/1009 (25.0) | |
Letters in parentheses indicate common ribotypes (>10 isolates) characterized by a significant overrepresentation of isolates from human listeriosis patients (H) or RTE foods (F) (9). The χ2 test of independence was not possible for comparisons where 2 or more cells in 2 × 2 table contained a value of 0. Ribotypes that apply to this are indicated by a “#” symbol.
Asterisks indicate significant (***, P < 0.0001; **, P < 0.01) overrepresentation of L. monocytogenes isolates carrying an inlA PMSC in RTE foods compared to isolates from human patients within common ribotypes.
Asterisks indicate significant (***, P < 0.0001; **, P < 0.01; *, P < 0.05) nonuniform distribution (overrepresentation or underrepresentation) of L. monocytogenes isolates carrying an inlA PMSC within a given ribotype.
Distribution of strains carrying a PMSC mutation in inlA and EC strains may play a more important role in the food attribution of listeriosis.
The current L. monocytogenes risk assessment estimated that RTE deli meats are responsible for 89% of listeriosis cases in the United States per annum (33). Declines in the prevalence of L. monocytogenes in RTE deli meats and the incidence of listeriosis, however, have not paralleled each other. Specifically, the prevalence of L. monocytogenes in RTE deli meats declined by almost 7-fold since 1998, while the incidence of listeriosis has yet to decrease by half (1, 32). If contaminated RTE deli meats are in fact responsible for 89% of food-borne listeriosis cases, one would expect to observe a greater decline of listeriosis cases over the last 11 years. We thus hypothesized that a nonuniform distribution of L. monocytogenes isolates carrying a PMSC in inlA or EC strains in deli meats or other RTE food categories may play a more important role in the food attribution of listeriosis than currently acknowledged. To probe this hypothesis, we evaluated the distribution of L. monocytogenes isolates carrying a virulence-attenuating PMSC mutation in inlA and EC strains among different categories of RTE foods, including bag salads, cheese, smoked seafood, deli and seafood salads, and deli meats. The proportion of L. monocytogenes with or without a PMSC mutation in inlA was similar among all RTE food categories except for deli meats, which included a marginally higher proportion (P = 0.12) of isolates carrying a PMSC in inlA (Table 4). On the other hand, we observed a significant (P = 0.006) overrepresentation of EC strains among L. monocytogenes isolated from deli and seafood salads (Table 5). The types of deli salads sampled in the Gombas et al. study included potato salad, tuna salad, pasta salad and coleslaw but not salads containing deli meats (8). A previous study on the isolate set characterized here identified deli and seafood salads as the RTE food product category with the highest levels of L. monocytogenes (8). The combined overrepresentation of EC strains and elevated levels of L. monocytogenes in the deli and seafood RTE food category highlight a critical need to collect additional empirical data and to revise current or develop future risk assessments to predict the human health risk associated with this particular RTE food category. Epidemic clone strains were significantly (P = 0.01) underrepresented among L. monocytogenes from the smoked seafood RTE food product category. Interestingly, there have not been any listeriosis outbreaks linked to smoked seafood, supporting our finding that EC strains, which may have enhanced human pathogenic potential, are not common among this RTE food category. Findings from the present study support that RTE food categories other than deli meats or a nonuniform distribution of EC strains and isolates carrying a virulence-attenuating PMSC mutation in inlA among deli meats, along with other RTE food categories, may play a more important role in the food attribution of listeriosis than currently acknowledged.
TABLE 4.
Number of inlA PMSC mutations in various food categories
| Food categorya | No. of isolates (% among total isolates screened for category): |
Total no. of isolates screened | |
|---|---|---|---|
| With inlA PMSC mutations | Without inlA PMSC mutations | ||
| Bag salads | 9 (42.9) | 12 (57.1) | 21 |
| Cheeses | 16 (50) | 16 (50) | 32 |
| Smoked seafood | 41 (40.2) | 61 (59.8) | 102 |
| Deli and seafood salads | 121 (43.8) | 155 (56.2) | 276 |
| Deli meats | 36 (53.7)b | 31 (46.3) | 67 |
| Unknown food source | 3 (75) | 1 (25) | 4 |
| Total | 226 (45.0) | 276 (55.0) | 502 |
Select RTE food categories that represent the 31,707 L. monocytogenes samples collected in the retail food survey. For statistical analysis purposes, the four isolates of unknown origin were removed from the isolate set.
Marginal (P = 0.12) overrespresentation of L. monocytogenes isolates carrying an inlA PMSC within food product categories.
TABLE 5.
Number of EC strains in various food categories
| Food categorya | No. of isolates (% among total isolates screened for category): |
Total no. of isolates screened | |
|---|---|---|---|
| With EC strainsb | Without EC strains | ||
| Bag salads | 2 (9.5) | 19 (90.5) | 21 |
| Cheeses | 2 (6.25) | 30 (93.75) | 32 |
| Smoked seafood | 6 (5.9)* | 96 (94.1) | 102 |
| Deli and seafood salads | 48 (17.4)** | 228 (82.6) | 276 |
| Deli meats | 10 (14.9) | 57 (85.1) | 67 |
| Unknown food source | 0 (0) | 4 (100) | 4 |
| Total | 68 (13.5) | 434 (86.5) | 502 |
Select RTE food categories that represent the 31,707 L. monocytogenes samples collected in the retail food survey. For statistical analysis purposes, the four isolates of unknown origin were left out of the isolate set.
Asterisks indicate significant (*, P < 0.05; **, P < 0.01) nonuniform distribution of EC strains among L. monocytogenes isolates within food product categories.
Conclusions.
Eighteen naturally occurring mutations leading to a PMSC in inlA have been identified worldwide to date (5, 10, 11, 12, 22-25, 27, 29, 34, 35), and PMSC mutations in inlA have been shown to be causally associated with attenuated mammalian virulence (20, 28). Mutations leading to a PMSC in inlA therefore represent a molecular marker for L. monocytogenes virulence attenuation, which can be used to predict the human health risk associated with consumption of foods contaminated by L. monocytogenes. A significantly greater proportion of L. monocytogenes isolates from RTE foods carry a PMSC mutation in inlA compared to isolates from human listeriosis cases. While evaluating the distribution of inlA PMSC mutations among RTE food product categories, we found that the proportion of L. monocytogenes with or without PMSC mutations in inlA was similar among isolates from RTE food categories except for deli meats, which included a marginally higher proportion of virulence-attenuated isolates. On the other hand, we observed that EC strains were overrepresented among L. monocytogenes isolates from deli and seafood salads and underrepresented among isolates from smoked seafood. The results from the present study provide insight into the distribution of L. monocytogenes isolates carrying a PMSC in inlA and EC strains that people are exposed to through consumption of contaminated RTE foods. Furthermore, these data will ultimately facilitate the revision of current and development of future risk assessments that include different exposure frequencies and disease probabilities for isolate populations that carry a virulence-attenuating PMSC mutation in inlA or represent an EC strain.
Acknowledgments
This project was supported by the National Research Initiative of the U.S. Department of Agriculture (USDA) Cooperative State Research, Education and Extension Service (grant 2005-35201-16266).
We thank P. Evans and D. Goldman from the USDA Food Safety Inspection Service for providing DNA samples from isolates carrying inlA PMSC mutation types 15 to 18. We thank S. Brisse, Institut Pasteur, for the generous gift of DNA from an isolate previously reported to carry inlA PMSC type 14. We also thank M. Wiedmann, Cornell University, for generously providing the L. monocytogenes isolate set characterized in the current study and for helpful discussions. In addition, we are indebted to all members of the Colorado State University Food Safety Laboratory for assistance with freezing down isolates and preparation of the bacterial lysates. We are also grateful for technical support and for assistance with the GeneScan software provided by Jason Rivest (Proteomics and Metabolomics Facility, Colorado State University).
Footnotes
Published ahead of print on 5 March 2010.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2009. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2008. MMWR Morb. Mortal. Wkly. Rep. 58:333-337. [PubMed] [Google Scholar]
- 2.Cole, M. B., M. V. Jones, and C. Holyoak. 1990. The effect of pH, salt concentration, and temperature on the survival and growth of Listeria monocytogenes. J. Appl. Bacteriol. 69:63-72. [DOI] [PubMed] [Google Scholar]
- 3.Ducey, T. F., B. Page, T. Usgaard, M. K. Borucki, K. Pupedis, and T. J. Ward. 2007. A single-nucleotide-polymorphism-based multilocus genotyping assay for subtyping lineage I isolates of Listeria monocytogenes. Appl. Environ. Microbiol. 73:133-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felicio, M. T., T. Hogg, P. Gibbs, P. Teixeira, and M. Wiedmann. 2007. Recurrent and sporadic Listeria monocytogenes contamination in Alheiras represents considerable diversity, including virulence-attenuated isolates. Appl. Environ. Microbiol. 73:3887-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fugett, E., E. Fortes, C. Nnoka, and M. Wiedmann. 2006. International Life Sciences Institute North America Listeria monocytogenes strain collection: development of standard Listeria monocytogenes strain sets for research and validation studies. J. Food Prot. 69:2929-2938. [DOI] [PubMed] [Google Scholar]
- 7.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusnoik, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simones, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 8.Gombas, D. E., Y. Chen, R. S. Clavero, and V. N. Scott. 2003. Survey of Listeria monocytogenes in ready-to-eat foods. J. Food Prot. 66:559-569. [DOI] [PubMed] [Google Scholar]
- 9.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Handa-Miya, S., B. Kimura, H. Takahashi, M. Sato, T. Ishikawa, K. Igarashi, and T. Fugii. 2007. Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int. J. Food Microbiol. 117:312-318. [DOI] [PubMed] [Google Scholar]
- 11.Jacquet, C., M. Doumith, J. I. Gordon, P. M. V. Martin, P. Cossart, and M. Lecuit. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 12.Jonquieres, R., H. Bierne, J. Mengaud, and P. Cossart. 1998. The inlA gene of Listeria monocytogenes L028 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66:3420-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabuki, D. Y., A. Y. Kuaye, M. Wiedmann, and K. J. Boor. 2004. Molecular subtyping and tracking of Listeria monocytogenes in Latin-style fresh-cheese processing plants. J. Dairy Sci. 87:2803-2812. [DOI] [PubMed] [Google Scholar]
- 14.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 11:1811-1829. [DOI] [PubMed] [Google Scholar]
- 15.Lappi, V. R., J. Thimothe, K. K. Nightingale, K. Gall, V. N. Scott, and M. Wiedmann. 2004. Longitudinal studies on Listeria in smoked fish plants: impact of intervention strategies on contamination patterns. J. Food Prot. 67:2500-2514. [DOI] [PubMed] [Google Scholar]
- 16.Lecuit, M., H. Ohayon, L. Braun, J. Mengaud, and P. Cossart. 1997. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 65:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLauchlin, J. 1990. Distribution of serovars of Listeria monocytogenes isolated from different categories of patients with listeriosis. Eur. J. Clin. Microbiol. Infect. Dis. 9:210-213. [DOI] [PubMed] [Google Scholar]
- 18.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCraig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nightingale, K. K., R. A. Ivy, A. J. Ho, E. D. Fortes, B. L. Njaa, R. M. Peters, and M. Wiedmann. 2008. inlA premature stop codons commonly found in Listeria monocytogenes isolated from food are responsible for virulence attenuation and confer protective immunity against infection by fully virulent L. monocytogenes. Appl. Environ. Microbiol. 74:6570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nightingale, K. K., K. Windham, and M. Wiedmann. 2005. Evolution and molecular phylogeny of Listeria monocytogenes from human and animal cases and foods. J. Bacteriol. 187:5537-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nightingale, K. K., K. Windham, K. E. Martin, M. Yeung, and M. Wiedmann. 2005. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA leading to expression of truncated and secreted internalin A and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71:8764-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olier, M., F. Pierre, S. Rousseaux, J. P. Lemaitre, A. Rousset, P Piveteau, and J. Guzzo. 2003. Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 71:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olier, M., F. Pierre, J. P. Lemaitre, C. Divies, A. Rousset, and J. Guzzo. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 25.Orsi, R. H., D. Ripoll, M. Yeung, K. K. Nightingale, and M. Wiedmann. 2007. Recombination and positive selection contribute to evolution of Listeria monocytogenes inlA. Microbiology 153:2666-2678. [DOI] [PubMed] [Google Scholar]
- 26.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. U. S. A. 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ragon, M., T. Wirth, F. Hollandt, R. Lavenir, M. Lecuit, A. Le Monnier, and S. Brisse. 2008. A new perspective on Listeria monocytogenes evolution. PloS Pathog. 4:e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roldgaard, B. B., J. B. Andersen, T. B. Hansen, B. B. Christensen, and T. R. Licht. 2009. Comparison of three Listeria monocytogenes strain in a guinea-pig model simulating food-borne exposure. FEMS Microbiol. Lett. 291:88-94. [DOI] [PubMed] [Google Scholar]
- 29.Rousseaux, S., M. Olier, J. P. Lamaitre, P. Piveteau, and J. Guzzo. 2004. Use of PCR-restriction fragment polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 70:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shank, F. R., E. L. Elliot, I. K. Wachsmuth, and M. E. Losikoff. 1996. US position on Listeria monocytogenes in foods. Food Control 7:229-234. [Google Scholar]
- 31.Tompkin, R. B. 2002. Control of Listeria monocytogenes in the food-processing environment. J. Food Prot. 65:709-725. [DOI] [PubMed] [Google Scholar]
- 32.U.S. Department of Agriculture, Food Safety and Inspection Service. 2008. The FSIS microbiological testing program for ready-to-eat (RTE) meat and poultry products, 1990-2008. U.S. Department of Agriculture, Washington, DC. http://www.fsis.usda.gov/science/micro_testing_RTE/index.asp.
- 33.U.S. Food and Drug Administration, Food Safety and Inspection Service, and Centers for Disease Control and Prevention. 2003. Quantitative assessment of the relative risk to public health from foodborne Listeria monocytogenes among selected categories of ready-to-eat foods. U.S. Food and Drug Administration, Washington, DC. http://www.foodsafety.gov/∼dms/lmr2-toc.html.
- 34.Van Stelten, A., and K. K. Nightingale. 2008. Development and implementation of a multiplex single-nucleotide polymorphism genotyping assay for detection of virulence-attenuating mutations in the Listeria monocytogenes virulence-associated gene inlA. Appl. Environ. Microbiol. 74:7365-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward, T. J., P. Evans, M. Wiedmann, T. Usgaard, S. E. Roof, S. G. Stoika, and K. Hise. Molecular and phenotypic characterization of Listeria monocytogenes from FSIS surveillance of RTE foods and processing facilities. J. Food Prot., in press. [DOI] [PubMed]
- 36.Ward, T. J., T. F. Ducey, T. Usgaard, K. A. Dunn, and J. P. Bielawski. 2008. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 74:7629-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 15:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]